Abstract
The WWOX gene is a tumour suppressor gene affected in various types of malignancies. Numerous studies showed either loss or reduction of the WWOX expression in variety of tumours, including breast, ovary, liver, stomach and pancreas. Recent study demonstrated that breast cancer patients exhibiting higher WWOX expression showed significantly longer disease-free survival in contrast to the group with lower relative WWOX level. This work was undertaken to show whether similar phenomena take place in colon tumours and cell lines. To assess the correlation of WWOX gene expression with prognosis and cancer recurrence in 99 colorectal cancer patients, we performed qRT-PCR analysis. We also performed analysis of WWOX promoter methylation status using MethylScreen method and analysis of loss of heterozygosity (LOH) status at two WWOX-related loci, previously shown to be frequently deleted in various types of tumours. A significantly better disease-free survival was observed among patients with tumours exhibiting high level of WWOX (hazard ratio = 0.39; p = 0.0452; Mantel–Cox log-rank test), but in multivariate analysis it was not an independent prognostic factor. We also found that although in colorectal cancer WWOX expression varies among patients and correlates with DFS, the exact mode of decrease in this type of tumour was not found. We failed to find the evidence of LOH in WWOX region, or hypermethylation in promoter regions of this gene. Although we provide the evidence for tumour-suppressive role of WWOX gene expression in colon, we were unable to identify the molecular mechanism responsible for this.
Keywords: BCL2, Colorectal cancer, CpG methylation, LOH, Quantitative RT-PCR, WWOX
Introduction
The WWOX (WW domain containing oxidoreductase) gene is located in the chromosome 16 region 16q23.3–24.1, also known as common fragile site FRA16D [1], an area which was found to be frequently affected by allelic losses in breast and other cancers [2]. WWOX expression was reported to be higher in the testis, ovary and prostate, i.e. tissues where its activity is regulated hormonally [1]. On this basis, WWOX was speculated to be involved in regulation of the steroids signalling pathways. Studies on biological role of WWOX in tumourigenesis showed that its function in cellular metabolism is likely to modulate gene expression by interactions with other proteins involved in cell cycle/apoptosis control and transcription factors. Up to now several partner proteins were identified, i.e. p73, AP-2γ[3], ErbB-4 [4], Runx2 [5] and members of Dvl protein family [6]. It was also shown that WWOX protein physically binds to two cytoplasmic regions of ErbB-4, which were previously verified to be responsible for interactions with Yap proteins. This competition for the ErbB-4 binding sites may prevent ErbB-4 transactivation and may lead to dysregulation of cell signalling [4]. Regardless of its function in cell metabolism, WWOX is considered as a tumour suppressor gene in various types of malignancies, including: breast [7], ovarian and lung cancer [2]. The evidence for its tumour suppressor activity was demonstrated for the first time in several cancer cell lines [7]. Since then numerous studies showed either loss or reduction of the WWOX expression in a variety of human tumours of breast, ovary, liver, stomach, pancreas, oesophagus, lung and haematopoietic malignancies [8]. Latest studies showed that WWOX gene is a bona fide tumour suppressor gene (reviewed in [3]), however the most common mechanism of decreasing WWOX expression in cancer cells is through hemizygous deletions (especially in breast cancer), while point mutations are very rare [1]. Recently, a set of complex deletions was found at FRA16D in the HCT116 colon cancer cell line, which was responsible for removing fragments of WWOX gene [9].
Another mechanism of reducing WWOX transcriptional level which was vastly studied is CpG islands hypermethylation of WWOX promoter and coding region. It seems that this mechanism may play some role in downregulation of WWOX expression in several cancer cell lines, for example tumours of pancreas and prostrate [10], breast, lung and bladder [11]; however first reports on methylation at the WWOX promoter region in thirteen breast cancer cell lines revealed that despite dramatic difference in WWOX expression, there was no methylation present at this region in any studied cell line [7]. Płuciennik et al. have shown that breast cancer patients exhibiting higher levels of WWOX expression exhibited significantly longer DFS in contrast to the group with relatively lower WWOX transcript levels [12]. Similarly, Aqeilan et al. showed prognostic relevance of WWOX and ErbB4 proteins in breast cancer [13].
With all results cited above, the lack of studies regarding role of WWOX gene and its protein product in tumourigenesis in colon and especially homozygous deletions in WWOX region found in HCT116 colon cancer cell line, as reported by [9], prompted us to undertake present work. The aim of our research was to evaluate the role of deletions in WWOX gene, its expression and prognostic value in patients with CRC (colorectal cancer). We also evaluated methylation of WWOX gene promoter region and the correlations of WWOX expression level with other well-known cancer/cell cycle-related genes, as: pro-apoptotic BAX, anti-apoptotic BCL2, cell cycle regulators: cyclins D1 (CCND1) and E1 (CCNE1) both regarded as playing an important role in tumourigenesis, tumour suppressor gene TP73 which encodes for the p73 protein, proliferation marker - Ki-67 and one ERBB4 isoform transcript—JM-a/CVT-1.
Materials and methods
Patients and samples
The CRC samples analysed herein were obtained from 99 cases of primary colorectal tumours treated at the Oncology Clinic, Medical University of Łódź. Only patients without previous familial history of CRC and those who did not receive preoperative radiotherapy were enrolled to this study. From these, only 50 had complete history of disease and reliable DFS observations (thus only these patients could be included to survival analysis). Experiments involving human subjects were conducted according to the Declaration of Helsinki: the study was approved by the Ethics Committee at Medical University of Łódź. The mean age of the patients was 61.3 years (median, 63 years; for women, 60 years; for men, 63 years; range, 30–86 years). Median follow-up period was 42.5 months. More detailed characteristics of the patients are shown in Table 1, together with WWOX mRNA level and results of Mann–Whitney U test. Tumours were classified according to the International Union Against Cancer staging and grading criteria. The tissue samples were examined histologically and stored at −80°C in RNAlater (Ambion, Inc.) until RNA extraction.
Table 1.
Correlations of WWOX expression with clinical characteristics of the patients
| Feature | n | WWOX mRNA median (range) | P (Mann–Whitney U) |
|---|---|---|---|
| Sex | |||
| Women | 51 | 1.49 (1.23–3.744) | 0.3985 |
| Men | 48 | 1.51 (0.57–2.193) | |
| Localisation of the primary tumoura | |||
| Rectum | 34a | 1.69 (0.29–3.74) | 0.8301 |
| Sigmoid colon | 37a | 1.42 (1.23–2.47) | |
| Descending colon | 6a | ||
| Splenic flexure | 3 | ||
| Transverse colon | 4 | ||
| Ascending colon | 3 | ||
| Cecum | 14 | ||
| Lymphocytic infiltration | |||
| Absent | 57 | 1.68 (1.23–3.87) | 0.1908 |
| Present | 41 | 1.35 (0.29–3.46) | |
| Unknown | 1 | ||
| Metastasis to the lymph nodes# | |||
| Absent | 55 | 1.65 (1.24–4.26) | 0.1591 |
| Present | 34 | 1.57 (0.57–3.52) | |
| Unknown | 10 | ||
| Grading (differentiation) | |||
| G1 | 10 | 1.09 (0.01–21.38) | 0.7331 (G1/G2) |
| G2 | 60 | 1.51 (1.24–3.66) | 0.7643 (G2/G3) |
| G3 | 29 | 1.60 (0.57–4.26) | 0.7153 (G1/G3) |
| Dukes' stage | |||
| A | 26 | 1.119 (0.21–3.66) | 0.6489 (A/B) |
| B | 29 | 1.858 (0.32–4.78) | 0.4133 (B/D) |
| C | 26 | 2.475 (1.31–7.35) | 0.8777 (A/C) |
| D | 16 | 1.342 (0.86–4.89) | 0.3166 (A/D) |
| Unknown | 2 | ||
| Relapse during follow-up | |||
| No | 34 | 2.10 (0.97–5.729) | 0.1008 |
| Yes | 27 | 1.32 (0.162–2.06) | |
| Unknown | 38 | ||
| Demise during follow-up | |||
| No | 46 | 2.10 (1.26–5.729) | 0.2370 |
| Yes | 45 | 1.40 (0.57–2.475) | |
| Unknown | 8 | ||
aThe localisation of primary tumours from three patients was ambiguous, thus they were qualified to two groups
Cancer cell lines
We used cell lines derived from tumours of colon (HCT116, SW480, SW620, HT-29) and two breast cancer cell lines (MDA-MB-231, MCF-7), which served as a control of our results, as both were previously studied for WWOX expression [7, 14]. Cell culture was performed according to the vendor's protocol. In brief, HT-29 and HTC116 cell lines were grown in McCoy's 5a medium with addition of 1% L-Glutamine; SW480 and SW620 were cultured in RPMI1640; MCF-7 and MDA-MB-231 cell lines were cultured in DMEM Advanced Medium with 1% L-Glutamine; MCF-7 cells were also supplemented with addition of bovine insulin to the final concentration 0.01 mg/ml. All media were supplemented with 10% foetal bovine serum and 1% PSN antibiotic mixture (penicillin, streptomycin and neomycin; all ingredients Sigma, Germany). Atmosphere consisted of 95% of air and 5% of CO2; incubation temperature was 37°C.
Real-time quantitative RT-PCR analysis
All RNA extractions and cDNA synthesis were performed as described elsewhere [12]. All real-time RT-PCR reactions were performed in duplicate, except the samples in which the analysis outcome was questionable. If this had happened, another two replicates were analysed. Detection of the amplification product was enabled with EvaGreen® dye (Biotium Inc., Hayward CA, USA), according to the manufacturer's recommendations in Corbett Research RG-3000 platform (Corbett Life Science, Sydney, Australia), in total reaction volume of 10 or 25 μl. Expression levels were normalised using the panel of four genes: β2-microglobulin B2M, histone H3F3A, ribosomal proteins RPS17 and RPLP0, which were selected using the geNorm applet [15]. Relative expression was calculated with the mathematical model allowing for correction of reaction efficiency and using the Universal Human Reference RNA (Stratagene, La Jolla, CA, USA) as a reference. Primer sequences used in this study are shown in Table 2; detailed PCR protocols are available upon request from the corresponding author.
Table 2.
Real-time RT-PCR primers and reaction conditions used for expression analysis of specified genes
| Gene name | Gene primers (For/Rev) | Annealing temperature (°C) | Detection temperature (°C) | PCR product size (bp) |
|---|---|---|---|---|
| (5′ → 3′) | ||||
| BAX | For: AGAGGTCTTTTTCCGAGTGGCAGC | 56 | 81 | 137 |
| Rev: TTCTGATCAGTTCCGGCACCTTG | ||||
| BCL2 | For: TTGGCCCCCGTTGCTTTTCCTC | 56 | 81 | 122 |
| Rev: TCCCACTCGTAGCCCCTCTGCGAC | ||||
| B2M | For: TGAGTGCTGTCTCCATGTTTGA | 50 | 81 | 88 |
| Rev: TCTGCTCCCCACCTCTAAGTTG | ||||
| CCND1 | For: GTCCTACTACCGCCTCACACGCTTCCTCTCCAG | 63 | 86 | 160 |
| Rev: TCCTCTTCCTCCTCCTCGGCGGCCTTG | ||||
| CCNE1 | For: TTCTTGAGCAACACCCTCTTCTGCAGCC | 68 | 68 | 138 |
| Rev: TCGCCATATACCGGTCAAAGAAATCTTGTGCC | ||||
| ERBB2 | For: TGACCTGCTGGAAAAGGGGGAGCG | 63 | 83 | 150 |
| Rev: TCCCTGGCCATGCGGGAGAATTCAG | ||||
| ERBB4 | For: ACACAGCCCTCCTCCTGCCTACAC | 56 | 76 | 95 |
| Rev: AGGGCACAGACACTCCTTGTTCAGC | ||||
| H3F3A | For: AGGACTTTAAAACAGATCTGCGCTTCCAGAG | 65 | 72 | 76 |
| Rev: ACCAGATAGGCCTCACTTGCCTCCTGC | ||||
| Ki-67 | For: TCCTTTGGTGGGCACCTAAGACCTG | 56 | 81 | 156 |
| Rev: TGATGGTTGAGGCTGTTCCTTGATG | ||||
| RPLP0 | For: ACGGATTACACCTTCCCACTTGCTGAAAAGGTC | 65 | 72 | 69 |
| Rev: AGCCACAAAGGCAGATGGATCAGCCAAG | ||||
| RPS17 | For: AAGCGCGTGTGCGAGGAGATCG | 64 | 72 | 87 |
| Rev: TCGCTTCATCAGATGCGTGACATAACCTG | ||||
| TP73 | For: AACCACGAGCTCGGGAGGGACTTCAAC | 63 | 81 | 159 |
| Rev: TTCCGTCCCCACCTGTGGTGGCTC | ||||
| WWOX | For: GAGCTGCACCGTCGCCTCTCCCCAC | 63 | 77 | 150 |
| Rev: TCCCTGTTGCATGGACTTGGTGAAAGGC |
For, forward primer; Rev, reverse primer
LOH analysis
In order to determine the LOH status of the 16q23.3–24.1 region, we used two sequence-tagged site (STS) markers: D16S3096 and D16S518. They are located at: eighth intron and second intron, respectively, of WWOX gene. The D16S518 marker is the most frequently affected with LOH in breast cancer (up to 77% in some populations, as described in [16]). Primer sequences used were according to UniSTS database (http://www.ncbi.nlm.nih.gov/). HRM analysis of ampilification products was performed in a LightCycler 480 (Roche Diagnostics, Poland) with EpiTect HRM PCR Kit (Qiagen, Germany).
Analysis of WWOX methylation status
To assess the methylation status of one 5′-upstream region involved in regulation of WWOX expression (from −508 to −174 bp) and region adjacent to and containing WWOX promoter (from −171 to +239 bp) we used novel bisulfite-free alternative technology MethylScreen, utilising the real-time quantitative PCR assay on templates generated by combined restriction digest using: methylation-sensitive restriction enzymes (MSRE), methylation-dependent restriction enzymes (MDRE), combined double digest (both MSRE and MDRE) and mock digestion [17]. The enzymes used in this study were: HhaI, HpaII (MSRE) and McrBC (MDRE; New England Biolabs, Ipswich, MA, USA); all digestions were performed according to the manufacturer's instructions on 500 ng of patient's DNA. All PCRs were performed in total volume of 50 μl, with 4 μl of respective digested sample DNA, 1 μl of each primer (10 mM). The sequences of the primers used were as follows: −508 bp region; For—5′-ACAGAAGCCCAGGACAACAGCATGG-3′; Rev—5′-ACCACGAAGCTGAAATCCAGTCTCCG-3′; −171-bp region; For—5′-AGACTGGATTTCAGCTTCGTGGTCG-3′; Rev—5′-AAGCTCCTTAACAGTTACTTTCACTTTGCAC-3. Cycle conditions were: 95°C for 5 min followed by 55 cycles of 94°C for 30 s, 55°C/30 s, 72°C/90 s and 77°C/15 s (fragment −508 bp) or 95°C for 5 min followed by 50 cycles of 94°C for 30 s, 50°C/30 s, 72°C/90 s and 80°C/15 s (fragment −171 bp).
Statistical analysis
Spearman's rank correlation test was used to analyse possible linear associations between all the gene expression levels. Disease-free survival was estimated with the Kaplan–Meier method. The significance of differences between survival rates was verified using the log-rank (Mantel–Cox) test. Disease-free survival was calculated according to Kaplan–Meier method. Multivariate survival analysis was performed using Cox's regression model. Values of p < 0.05 (confidence level > 95%) were considered statistically significant.
Results
Correlation of WWOX expression with clinical parameters
Relative WWOX expression in CRC tissues ranged from 0 to 123.18 (median 7.66). Results of statistical analysis of WWOX expression in groups of patients divided according to the classical clinical markers are presented in Table 1. We did not find any statistically significant relations between groups of patients stratified according to their basic clinicopathological features; however, we found a tendency for higher relative WWOX expression in samples from patients without relapse during the follow-up period (2.10 versus 1.32 units; p = 0.1008). This finding was then confirmed by analysis of DFS based on relatively high and low WWOX expression, which showed significant differences. The cut-off point for discrimination between ‘high’ and ‘low’ expression of WWOX was 2.70 (units of relative expression). This point was selected with the X-tile software [18]. We found that this cut-off value applied to the standard Kaplan–Meier DFS analysis yielded a significantly better DFS observed among patients with tumours in which the level of WWOX mRNA was classified as high (hazard ratio; HR = 0.39; p = 0.0452; Cox–Mantel log-rank test; Fig. 1), however in multivariate analysis it was not an independent prognostic factor (p = 0.8027). We also conducted survival analysis of patients stratified according to the localisation of primary tumour. Although we found disparity between DFS in patients with primary tumour localised in rectum versus all other localisations, this was not statistically significant (HR = 0.48; p = 0.1566).
Fig. 1.
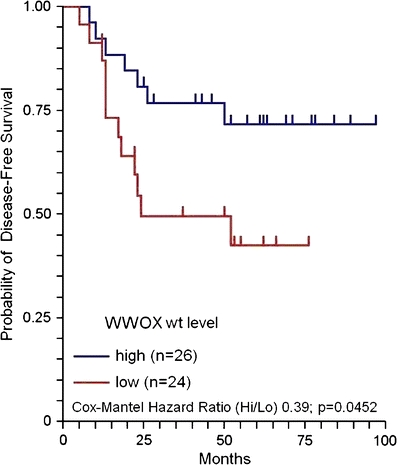
Results of DFS analysis in patients stratified according to the WWOX mRNA level (Kaplan–Meier test)
Analysis of WWOX expression in cell lines
HCT116 colon cancer cell line, although previously found to harbour homozygous deletion in WWOX gene, surprisingly showed 70% level of WWOX expression, in comparison with MCF-7 cells (all results in Table 3). Reason for this results is suggested by the work of Alsop et al. [9]; authors imply that HCT116 homozygous deletions are within WWOX intron, so they should not affect its expression (apart for the supposed role of WWOX Δ6–8 transcript in decreasing WWOXwt transcription, which nowadays is not supported by solid evidence). Indeed, they previously found two WWOX transcripts: variant 1 (WWOXwt) and variant 4 (WWOX Δ6–8) in this cell line [2], although Northern blots presented showed that WWOXwt appears in low abundance. This discrepancy could be in part linked to different techniques used, as suggested by Ding et al.: the correlation between the Northern and qRT/PCR results for 24 genes studied was r = 0.39; after excluding outlier genes the correlation coefficient rised to 0.72, still far from ideal [19]. We also found that HT-29 cell line, originating from a colorectal adenocarcinoma, showed very low level of WWOX expression. The difference in WWOX expression between SW480 and SW620 cell lines suggest that there is a room for stating that WWOX has some role (not fully identified yet) in the progression of CRC tumours: SW480 cells, originating from primary tumour showed 0.230 of WWOX relative expression; whereas in SW620 cells, from metastatic tumour of the same patient, relative WWOX expression was even lower, 0.175. This result is similar to the difference in WWOX relative expression among patients with lymph node metastases and patients in which there was no nodal metastasis present (1.65 vs. 1.57), however that relationship was not statistically significant (p = 0.1591). MCF7 cells showed the highest expression of the studied cell lines; accordingly, aggressive and highly metastatic breast cancer cell line MDA-MB-231 had the lowest WWOX expression (60-fold lower than MCF-7), which is in accordance with previous reports [2, 3].
Table 3.
WWOX expression in the studied cell lines
| Cell lines | Average WWOX relative expression |
|---|---|
| HCT116 | 1.075 |
| HT29 | 0.080 |
| SW480 | 0.230 |
| SW620 | 0.175 |
| MCF-7 | 1.497 |
| MDA-MB-231 | 0.025 |
Analysis of methylation and LOH status of WWOX gene locus
In the studied population of patients, we did not find any significant hemizygosity suggesting LOH at the investigated loci in CRC tumours samples. Cell lines exhibited differences in surveyed markers (Table 4), however this had no connection to the expression level of WWOX. For instance SW620 cells, which showed retention of both alleles as the only one cell line studied, exhibited almost tenfold lower WWOX expression than MCF-7 cells, which expression was the highest observed here.
Table 4.
LOH analysis in two WWOX gene regions in human cancer cell lines
| STS marker | Cell lines | |||||
|---|---|---|---|---|---|---|
| HCT116 | HT-29 | SW-480 | SW-620 | MCF-7 | MDA-MB-231 | |
| D16S3096 | R | LOH | R | R | LOH | LOH |
| D16S518 | LOH | R | LOH | R | LOH | LOH |
LOH, loss of one allele; R, retention of both alleles; NI, non-informative result
There was no significant methylation of WWOX promoter in patients' samples: we found that only eight (8.1%) of patients had low methylation and two (2%) had moderate methylation at 5′-upstream region (−508 to −174 bp), whereas only seven (7.1%) of patients exhibited low methylation at region adjacent to and containing WWOX promoter (from −171 to +239 bp). Of the patients, 12.1% had non-informative results of analysis at 5′ upstream region, while at the promoter region the number of non-informative cases was 8.1% (all results in Table 5). None of the cell lines used in this study showed methylation of WWOX promoter region (data not shown). Statistical analysis of correlations between methylation level or LOH at studied loci and WWOX relative expression did not prove that there was any relationship between those parameters in our study.
Table 5.
Results of the WWOX promoter regions methylation analysis in CRC patients
| Methylation status | Number of cases (%) | |
|---|---|---|
| 5′-Upstream region | WWOX promoter region | |
| (−508 to −174 bp) | (−171 to +239 bp) | |
| ‘0’ | 77 (77.8%) | 84 (84.8%) |
| ‘1’ | 8 (8.1%) | 7 (7.1%) |
| ‘2’ | 2 (2.0%) | 0 (0%) |
| NI | 12 (12.1%) | 8 (8.1%) |
| Sum | 99 | 99 |
‘0’, no methylation found (difference between the Mock and the MDRE less than one cycle); ‘1’, low methylation (difference between the Mock and the MDRE 1 ≥ 1.49 cycle); ‘2’, intermediate methylation (difference between the Mock and the MDRE >1.5 cycle); NI, non-informative result
Correlation of WWOX transcript level with expression of other genes
We found that WWOX wt expression is correlated (with statistical significance; Spearman rank correlation test used) with number of surveyed genes (presented in order of lowering probability): significant negative correlation with CCNE1 expression (−0.3579; p = 0.0005), which in general is regarded as a marker of bad prognosis and is directly associated with tumourigenesis. We also found a significant positive correlation with BCL2/BAX ratio (0.3480; p = 0.0006). Positive correlations were found between WWOX expression and ERBB4 and BCL2 (all results in Table 6).
Table 6.
Correlations between WWOX wt expression and other genes (Spearman test)
| Gene name | Spearman rank correlation coefficient | p Value |
|---|---|---|
| CCNE1 | −0.3579 | 0.0005 |
| BCL2/BAX ratio | 0.3480 | 0.0006 |
| BAX/BCL2 ratio | −0.3308 | 0.0012 |
| Ki67 | −0.2913 | 0.0046 |
| ERBB4 | 0.2473 | 0.0242 |
| BCL2 | 0.2066 | 0.0372 |
| ERBB2 | −0.1957 | 0.0709 |
| BAX | −0.1684 | 0.0906 |
Discussion
In the presented study, we analysed the expression of WWOX gene in 99 tumours from patients with colorectal cancer. In several reports it was shown that WWOX expression is lowered in various types of tumours (mentioned above). Moreover, many authors have shown that suppressed transcription of WWOX is associated with more aggressive phenotype of breast cancer [12], non-small cell lung cancer [20] and ovarian cancer [21]. Here, we show that relatively high WWOX expression corresponds with better disease-free survival of CRC patients hazard ratio (HR = 0.39; p = 0.0452; Mantel–Cox log-rank Test, Fig. 1) in comparison with those with lowered WWOX transcription. This supports the view that loss of WWOX expression is associated with tumourigenesis in different types of cancers. Such an idea was additionally proven by in vitro and in vivo studies which showed that elevated WWOX expression suppresses tumourigenicity of different cancer cell lines: breast [7], lung [22] and prostate [23]. However, WWOX expression cannot be used as an independent prognostic marker in CRC, since results of multivariate analysis excluded this marker from analysis on early stages (results not shown). Despite the frequent suppression of WWOX expression in many cancers, complete gene inactivation by deletion of one allele and second mutation or homozygous deletion is very rare [9]. Based on the observations, it was postulated that WWOX inactivation is driven by hemizygous deletions, which was recently proven with mouse model using targeted deletion of WWOX gene [24]. In our analysis of 16q23.3–24.1 region we did not find any evidence for LOH in the two studied WWOX-associated loci in CRC. We used two STS (sequence-tagged site) markers (D16S3096 and D16S518) which are most often afflicted by hemizygous deletions in all kinds of cancers, for instance: breast ductal carcinoma in situ lesions [16], breast cancer metastases [25], hepatocellular carcinoma [26], non-small cell lung cancer [27], oesophageal squamous cell carcinoma [28], gastric carcinoma [29], but none of the STS markers displayed LOH in our set of colorectal cancer samples.
We also tested the status of methylation in the promoter region of WWOX gene, presumably resulting in lowered WWOX expression, which was shown in several studies [11, 22]. Nevertheless, there are data showing that the methylation status of WWOX promoter region does not contribute to the decrease of WWOX expression in breast cancer cell lines and prostate tumours [7, 30] which is also in the case of CRC patients studied herein. To our knowledge, this is the first report on methylation status of WWOX gene in CRC patients or CRC cell lines. Nevertheless, results of our MethylScreen analysis were very similar to the previously cited work by Bastian et al. [30], who analysed CpG island hypermethylation in a set of 13 gene loci (including WWOX) in 78 prostate carcinomas, 32 benign prostate hyperplasias and four prostate cell lines (LNCaP, DU145, PC3, BPH-1) using MethyLight PCR. They found only one case showing WWOX promoter region methylation; none of the benign samples were methylated in WWOX locus [30]. Moreover, none of the cell lines surveyed (LNCaP, DU145, PC3, BPH-1) exhibited methylation of WWOX [30]. Interestingly, previous studies showed loss of WWOX expression in as much as 84% (37 of 44 tumour samples) [23] and involvement of promoter methylation in decreasing of WWOX expression in prostate cancer cell lines LNCaP, DU145 and PC-3 [23]. We hypothesise that this striking discrepancy between the two abovementioned papers could arise because of the two different strategies of study: Bastian et al showed the exact methylation status of WWOX by using MethyLight PCR, whereas Qin et al. used methylation-specific PCR (MSP). One should remember that MSP is gel-based technique and provides rather qualitative results, whereas PCR-based techniques are able to discriminate between different levels of methylation. Qin et al. also assumed that increased WWOX mRNA and protein expression in prostate cancer-derived cells after treatment with 5-aza-2′-deoxycytidine (AZA; a DNA methyltransferase inhibitor) and trichostatin A (a histone deacetylase inhibitor), is a result of demethylation of only WWOX promoter region. However, one should be aware of the fact that these agents are not specific and they change the global methylation/acetylation status of the cell, including all hypothetical and/or unknown regulators of WWOX expression.
Recently, a paper by Kosla et al. showed that both methylation of WWOX promoter region and LOH at D16S518, D16S3096 and D16S504 have influence on WWOX expression in glioblastoma multiforme tumours [31]. In this work, we did not find any evidence for such a relationship, which may suggest that these mechanisms are tissue specific.
We found that in population of Polish patients studied herein WWOX expression correlated with several genes involved in cell cycle/apoptosis or interacting with WWOX. The strongest correlation found was negative association of WWOX expression level with that of CCNE1 (−0.3579; p = 0.0005). Cyclin E1 is thought to be a potential predictor of systemic therapy, because of the cell cycle alterations induced by its overexpression: decreased length of the G1 phase, faster transition from G1 to S phase and increased genomic instability [32]. Moreover, overexpression of CCNE1 and amplification in breast cancer human breast epithelial cells results in chromosomal instability and worse prognosis [32]. In colorectal cancer cells it was found that combined treatment of these cells with various cytotoxic drugs (e.g. c-myc antisense phosphorothioate oligonucleotides, taxol, 5-fluorouracil (5-FU), doxorubicin and vinblastine) resulted in growth arrest of these cells in the G2/M and S phases, noticeable apoptotic effect and the reduction of mRNA levels of BCL2, BCLxL, CDK2, cyclin E1, CDK1 and cyclin B1, while increasing the mRNA levels of p21, p27, BAX and caspase-3 [33].
We also found correlation of WWOX transcription level with the BCL2/BAX expression ratio (0.3480 p = 0.0006). This relationship would mean that in CRC patients with higher WWOX expression, the tumours/its cells are less prone to apoptosis. This seemingly paradoxical finding has been also recently reported by Reeve's group in CRC patients. The impact of tumour proliferation on the grade of malignancy in CRC is not clear, especially when markers well established for breast cancer are used (e.g. Ki-67, PCNA) that is why the group used a self-devised colon-specific gene-proliferation signature (GPS) [34], including 36 genes commonly expressed (upregulated) in an exponentially growing in vitro CRC model and in human colon proliferative crypt compartments. Among the GPS genes, there are 15 cell cycle related, for instance CCNA2 (cyclin A2), CDC2 (cell division cycle 2, G1 to S and G2 to M, transcript variant 1). After stratification of colorectal tumours into high and low GPS groups by K-means clustering method, authors found that reduced GPS expression was associated with shorter DFS in CRC patients [34]. Authors also validated the GPS on public microarray data from two independent breast cancer experiments and found that in breast cancer group with increased GPS had significantly shorter DFS [34]. It is worth mentioning that among the 36 GPS genes there are only two involved in apoptosis—MADL2, which is anti-apoptotic and ITGB3BP (integrin beta 3 binding protein, NRIF3) shown to induce rapid and profound apoptosis in various breast cancer cell lines [35]. In a previous report, it was found that in breast cancer patients, the median expression of WWOX was almost 13-fold lower in tumours exhibiting BCL2/BAX ratio lower than 2 [12]. Similarly, in the presented work: colorectal tumours in which BCL2/BAX ratio was lower than 2, showed WWOX median expression 0.791, whereas in samples with higher BCL2/BAX ratio it was 4.590 (5.81-fold difference; p = 0.0025). We also hypothesise that WWOX expression regulation in CRC, or in colon tissue/cell lines in general, could be similar to the E-cadherin (CDH1) gene/E-Cad protein. This well-known tumour suppressor, which is located in the vicinity of WWOX locus (16q22.1) was reported to have decreased expression in various cancers, including CRC. However, the exact mode of CDH1 expression regulation was largely unknown when studies were performed to identify the ‘classical’ ways of downregulating gene expression. Early works on downregulation of E-Cad expression due to the mutations in CDH1 gene showed that the mutation rate in this gene was low [36]. Also, polymorphisms found in the CDH1 and its promoter region seems to have at least ambiguous significance in regulation of E-Cad expression, because studies on greater number of patients showed no such associations [37]. Epigenetic changes (methylation status) in CDH1 gene region in tumours were also studied, but results of these analyses are also unclear and seem to depend mostly on the technique used in the survey (this situation is very much alike to the one of WWOX methylation). Once again, when methylation was studied using MSP-based methods, it seemed that this kind of regulation has great influence on E-cad protein level [38], whereas study done using the qPCR-based method (MethyLight) showed extremely modest level of CDH1 promoter methylation and there was no correlation between DNA methylation and E-cad protein level (neither in tumour tissues nor the normal mucosae; total 142 pairs of matching tissues) [39]. Also in a paper mentioned earlier, Bastian et al. described differences between the CDH1 promoter methylation status they found in prostate carcinomas and previously published results of such analyses in this kind of tumour [30]. The next step in resolving this complexity was the showing of different repressor proteins that contribute to E-cad transcription regulation. Up to this date many of these transacting factors were discovered, including: Snail, Slug, Twist, SIP1/ZEB2, deltaEF1/ZEB2, as reviewed in [38]. Recently, a paper by Guler et al. described a relationship between the “triple negative” breast tumours phenotype and reduced expression of WWOX with elevated expression of AP-2γ (as shown by using tissue microarrays), although the authors did not find direct correlation between WWOX neither AP-2α nor Ap-2γ expression levels [40]. In summary, in this study we found that WWOX expression varies among patients and correlates with DFS, however we were unable to identify the molecular reason of lowered WWOX transcription. Our data suggest that unlike other tumours, WWOX expression in colorectal cancer is affected by different mechanisms than small deletions or methylation of promoter region. These findings, the ambiguous nature of role of the WWOX promoter methylation in expression regulation and the previous studies showing a wide array of proteins interacting with WWOX (e.g. YAP, ErbB-4, Dvl family) seem to suggest that there is a place to hypothesise that phenomena similar to CDH1 expression regulation may occur in WWOX expression regulation in colon.
Acknowledgements
This study was founded by Polish Ministry of Science and Higher Education grants N N401 233934 and N N402 195635.
We would like to thank Ms. Ewa Latkowska for her excellent technical support, also Mrs, Agnieszka Piastowska-Ciesielska and Mrs Magdalena Nowakowska for the cell lines cultures studied in this paper.
Conflicts of interest
None
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60(8):2140–2145. [PubMed] [Google Scholar]
- 2.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc. Natl Acad. Sci. USA. 2001;98(20):11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6(2):249–259. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, et al. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65(15):6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 5.Aqeilan RI, Hassan MQ, de Bruin A, Hagan JP, Volinia S, Palumbo T, et al. The WWOX tumor suppressor is essential for postnatal survival and normal bone metabolism. J. Biol. Chem. 2008;283(31):21629–21639. doi: 10.1074/jbc.M800855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouteille N, Driouch K, Hage PE, Sin S, Formstecher E, Camonis J, et al. Inhibition of the Wnt/beta-catenin pathway by the WWOX tumor suppressor protein. Oncogene. 2009;28(28):2569–2580. doi: 10.1038/onc.2009.120. [DOI] [PubMed] [Google Scholar]
- 7.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, et al. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61(22):8068–8073. [PubMed] [Google Scholar]
- 8.Aqeilan RI, Croce CM. WWOX in biological control and tumorigenesis. J. Cell. Physiol. 2007;212(2):307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- 9.Alsop AE, Taylor K, Zhang J, Gabra H, Paige AJ, Edwards PA. Homozygous deletions may be markers of nearby heterozygous mutations: the complex deletion at FRA16D in the HCT116 colon cancer cell line removes exons of WWOX. Genes Chromosom. Cancer. 2008;47(5):437–447. doi: 10.1002/gcc.20548. [DOI] [PubMed] [Google Scholar]
- 10.Kuroki T, Yendamuri S, Trapasso F, Matsuyama A, Aqeilan RI, Alder H, et al. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin. Cancer Res. 2004;10(7):2459–2465. doi: 10.1158/1078-0432.CCR-03-0096. [DOI] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, et al. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24(9):1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 12.Pluciennik E, Kusinska R, Potemski P, Kubiak R, Kordek R, Bednarek AK. WWOX–the FRA16D cancer gene: expression correlation with breast cancer progression and prognosis. Eur. J. Surg. Oncol. 2006;32(2):153–157. doi: 10.1016/j.ejso.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Aqeilan RI, Donati V, Gaudio E, Nicoloso MS, Sundvall M, Korhonen A, et al. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67(19):9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- 14.Iliopoulos D, Fabbri M, Druck T, Qin HR, Han SY, Huebner K. Inhibition of breast cancer cell growth in vitro and in vivo: effect of restoration of Wwox expression. Clin. Cancer Res. 2007;13(1):268–274. doi: 10.1158/1078-0432.CCR-06-2038. [DOI] [PubMed] [Google Scholar]
- 15.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T, Sahin A, Aldaz CM. Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res. 1996;56(24):5605–5609. [PubMed] [Google Scholar]
- 17.Holemon H, Korshunova Y, Ordway JM, Bedell JA, Citek RW, Lakey N, et al. MethylScreen: DNA methylation density monitoring using quantitative PCR. Biotechniques. 2007;43(5):683–693. doi: 10.2144/000112597. [DOI] [PubMed] [Google Scholar]
- 18.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 19.Ding Y, Xu L, Jovanovic BD, Helenowski IB, Kelly DL, Catalona WJ, et al. The methodology used to measure differential gene expression affects the outcome. J. Biomol. Tech. 2007;18(5):321–330. [PMC free article] [PubMed] [Google Scholar]
- 20.Donati V, Fontanini G, Dell'Omodarme M, Prati MC, Nuti S, Lucchi M, et al. WWOX expression in different histologic types and subtypes of non-small cell lung cancer. Clin. Cancer Res. 2007;13(3):884–891. doi: 10.1158/1078-0432.CCR-06-2016. [DOI] [PubMed] [Google Scholar]
- 21.Nunez MI, Rosen DG, Ludes-Meyers JH, Abba MC, Kil H, Page R, et al. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer. 2005;5(1):64. doi: 10.1186/1471-2407-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantor JP, Iliopoulos D, Rao AS, Druck T, Semba S, Han SY, et al. Epigenetic modulation of endogenous tumor suppressor expression in lung cancer xenografts suppresses tumorigenicity. Int. J. Cancer. 2007;120(1):24–31. doi: 10.1002/ijc.22073. [DOI] [PubMed] [Google Scholar]
- 23.Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S, et al. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66(13):6477–6481. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 24.Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc. Natl Acad. Sci. USA. 2007;104(10):3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Driouch K, Dorion-Bonnet F, Briffod M, Champeme MH, Longy M, Lidereau R. Loss of heterozygosity on chromosome arm 16q in breast cancer metastases. Genes Chromosom. Cancer. 1997;19(3):185–191. doi: 10.1002/(SICI)1098-2264(199707)19:3<185::AID-GCC8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Yakicier MC, Legoix P, Vaury C, Gressin L, Tubacher E, Capron F, et al. Identification of homozygous deletions at chromosome 16q23 in aflatoxin B1 exposed hepatocellular carcinoma. Oncogene. 2001;20(37):5232–5238. doi: 10.1038/sj.onc.1204674. [DOI] [PubMed] [Google Scholar]
- 27.Yendamuri S, Kuroki T, Trapasso F, Henry AC, Dumon KR, Huebner K, et al. WW domain containing oxidoreductase gene expression is altered in non-small cell lung cancer. Cancer Res. 2003;63(4):878–881. [PubMed] [Google Scholar]
- 28.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, et al. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62(8):2258–2260. [PubMed] [Google Scholar]
- 29.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, et al. Loss of WWOX expression in gastric carcinoma. Clin. Cancer Res. 2004;10(9):3053–3058. doi: 10.1158/1078-0432.CCR-03-0594. [DOI] [PubMed] [Google Scholar]
- 30.Bastian PJ, Ellinger J, Heukamp LC, Kahl P, Muller SC, von Rucker A. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur. Urol. 2007;51(3):665–674. doi: 10.1016/j.eururo.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Kosla K, Pluciennik E, Kurzyk A, Jesionek-Kupnicka D, Kordek R, Potemski P, et al. Molecular analysis of WWOX expression correlation with proliferation and apoptosis in glioblastoma multiforme. J Neurooncol. 2011;101(2):207–13. doi: 10.1007/s11060-010-0254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunt KK, Keyomarsi K. Cyclin E as a prognostic and predictive marker in breast cancer. Semin. Cancer Biol. 2005;15(4):319–326. doi: 10.1016/j.semcancer.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Abaza MS, Al Saffar A, Al Sawan S, Al Attiyah R. c-myc antisense oligonucleotides sensitize human colorectal cancer cells to chemotherapeutic drugs. Tumour Biol. 2008;29(5):287–303. doi: 10.1159/000156706. [DOI] [PubMed] [Google Scholar]
- 34.Anjomshoaa A, Lin YH, Black MA, McCall JL, Humar B, Song S, et al. Reduced expression of a gene proliferation signature is associated with enhanced malignancy in colon cancer. Br. J. Cancer. 2008;99(6):966–973. doi: 10.1038/sj.bjc.6604560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Das S, Yamada T, Samuels HH. The NRIF3 family of transcriptional coregulators induces rapid and profound apoptosis in breast cancer cells. Mol. Cell. Biol. 2004;24(9):3838–3848. doi: 10.1128/MCB.24.9.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Efstathiou JA, Liu D, Wheeler JM, Kim HC, Beck NE, Ilyas M, et al. Mutated epithelial cadherin is associated with increased tumorigenicity and loss of adhesion and of responsiveness to the motogenic trefoil factor 2 in colon carcinoma cells. Proc. Natl Acad. Sci. USA. 1999;96(5):2316–2321. doi: 10.1073/pnas.96.5.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenab M, McKay JD, Ferrari P, Biessy C, Laing S, Munar GM, et al. CDH1 gene polymorphisms, smoking, Helicobacter pylori infection and the risk of gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) Eur. J. Cancer. 2008;44(6):774–780. doi: 10.1016/j.ejca.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 38.van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008;65(23):3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Zhao Y, Wu C, Ho KS, Koh PK, Chong SF, et al. Modest promoter methylation of E-cadherin gene in sporadic colorectal cancers: a quantitative analysis. Cancer Biomark. 2008;4(2):111–120. doi: 10.3233/cbm-2008-4207. [DOI] [PubMed] [Google Scholar]
- 40.Guler G, Huebner K, Himmetoglu C, Jimenez RE, Costinean S, Volinia S, et al. Fragile histidine triad protein, WW domain-containing oxidoreductase protein Wwox, and activator protein 2gamma expression levels correlate with basal phenotype in breast cancer. Cancer. 2009;115(4):899–908. doi: 10.1002/cncr.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]


