Abstract
Although cryo-electron microscopy (cryo-EM) of biological macromolecules has made important advances in the past few years, the level of current technical performance is still well below what the physics of electron scattering would allow. It should be possible, for example, to use cryo-EM to solve protein structures at atomic resolution for particle sizes well below 80 kDa, but currently this has been achieved only for particles at least 10 times larger than that. In this review, we first examine some of the reasons for this large gap in performance. We then give an overview of work that is currently in progress to 1), improve the signal/noise ratio for area detectors; 2), improve the signal transfer between the scattered electrons and the corresponding images; and 3), reduce the extent to which beam-induced movement causes a steep fall-off of signal at high resolution. In each case, there is substantial reason to think that cryo-EM can indeed be made to approach the estimated physical limits.
Introduction
The ability to experimentally determine the structures of individual macromolecules, complex macromolecular machines, and subcellular devices is of great value in biochemistry and cell biology because it allows investigators to test existing hypotheses as well as to generate information that may lead to the formulation of new hypotheses (1). A wide range of biophysical tools are available for characterizing and determining structure, including (but not limited to) spectroscopy, crystallography, and various forms of microscopy. Our goal in this review is to give an update on what currently is being accomplished by cryo-electron microscopy (cryo-EM), in the context of what is achievable in principle and what improvements in technology are still required before cryo-EM can approach the level of performance allowed by physics.
Cryo-EM is generally understood to mean EM of biological specimens in an unstained, frozen-hydrated state. This technique provides structural information with improved validity, and (as is increasingly the case) can even provide sufficient information to build structural models at atomic resolution (Fig. 1). Automated data collection and ever-increasing computational capacity are making it possible to record and process data from unprecedented numbers of images. Applications of this technique include studies on assembly pathways (2) and conformation dynamics (3) for large molecular machines. In addition, cryo-EM tomography of suitably thin specimens provides three-dimensional (3D) images of cellular ultrastructure in a more lifelike state than can be achieved by traditional methods such as plastic sectioning (4).
Figure 1.
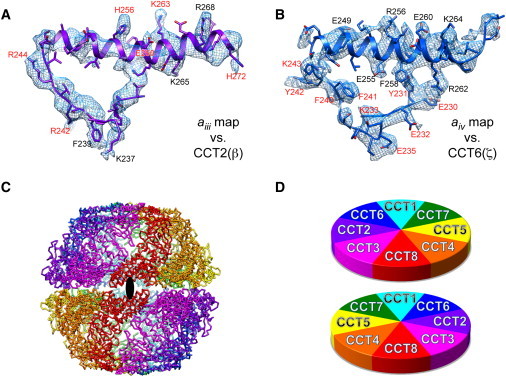
Determination of the atomic-resolution structure of a multiprotein complex by cryo-EM. The eukaryotic chaperonin, TRiC/CCT, is a dimer of hetero-octomeric rings of homologous but not identical subunits. (A and B) Two examples are shown of segmented densities and their corresponding interpretation in terms of fitting the primary structures of the β and ζ chains, respectively. (C) A ribbon diagram of the assembled structure. (D) A schematic diagram of how the two rings stack together. This figure was kindly prepared by Y. Cong from data published by Cong et al. (35).
There are limits, of course, as to how much structural information can be obtained by cryo-EM. The primary limiting factor is radiation damage (5) and, more specifically, the poor signal/noise ratio (SNR) in images recorded with optimal electron exposures (6). It is safe to assume that computational methods will continue to improve, enabling investigators to more fully extract the information that does exist in these noisy images (7). At the same time, experimental methods can also be improved so as to more fully relay the information initially carried in the scattered electron wave onto the recorded image intensity.
In this mini-review, we examine various reasons for the large gap that still exists between the SNR in images and that observed in the scattered electron wave. In addition, we summarize some of the experimental studies that are currently under way with the aim of closing this gap as much as possible.
What Physics Allows
The amount of structural information that can be extracted from cryo-EM images is ultimately limited by the SNR in such images. The cross section for electron scattering sets a limit on how large the numerator can be, and the shot noise associated with optimal electron exposures sets a physical limit on how small the denominator can be.
It is well established that thin cryo-EM specimens are accurately modeled as weak phase objects (8,9). As Zernike (10) pointed out, one can intentionally defocus the image of a phase object to generate visible contrast, and this is still the norm in cryo-EM. A better alternative, however, would be to use a quarter-wave plate to apply a 90° phase shift to the scattered wave relative to the unscattered wave. At the resolution needed to estimate the size and shape of a protein, the phase shift for 200 keV electrons transmitted through water (mass density 1 g/cm3) is ∼36 mradian/nm (see, for example, Fig. 3 A of Wang et al. (11)). The corresponding phase shift through the protein will scale approximately as the mass density, and, in addition, the mass density of amorphous ice is ∼6% less than that of liquid water. The relative phase shift for a protein embedded in vitreous ice should thus be ∼14 mradian/nm. The amount of phase contrast, ΔI/<I>, for cryo-EM images of proteins can thus be ∼0.028 T (i.e., twice the phase modulation in the transmitted wave (9)), where T is the thickness of an object in nanometers. In highly defocused images, on the other hand, the contrast is five or 10 times smaller than this.
The optimal electron exposure that can be used for cryo-EM depends somewhat on the resolution (6,12). For example, it is counterproductive to use exposures (with 300 keV electrons) higher than 2000 electrons/nm2 to image features at high resolution, whereas exposures five times larger than that can be used to image features at very low resolution. Although the shot noise associated with these low-dose limits is easily calculated for an ideal detector, in practice the noise is significantly greater due to the imperfect detective quantum efficiency (DQE) of currently available detectors (13).
The smallest 3D feature that produces a detectable signal in an image, even under ideal conditions, depends on 1), the density of the feature relative to its surround (i.e., the contrast, or signal); and 2), the exposure used to record the image (i.e., the noise). Table 1 specifies how large the density difference must be for the signal to be ≥3σ, assuming a perfect detector. Of interest, the dose-fractionation theorem tells us that the SNR required to detect a feature is the same in a single, two-dimensional (2D) projection as it is in a full, 3D reconstruction, provided that the same total electron exposure is used (14,15). As noted in the table legend, the density resolution indicated in Table 1 is 14 times more sensitive than that reported in a previous study (16) in which it was assumed that the amplitude-contrast model rather than the phase-contrast model was applicable. The main conclusion that can be drawn from this table is that a resolution of ∼2 nm is physically achievable in cryo-EM images of individual particles, but only for features (e.g., small domains) with the highest attainable difference in density (contrast) relative to their surround. Features that have smaller density differences relative to their surround must be correspondingly larger in size to be distinguished from one another.
Table 1.
The density resolution that is achievable for object detection (revised from Saxberg and Saxton (16))
| p (e nm−2) | 0.5 nm | 1 nm | 2 nm | 3 nm | 5 nm |
|---|---|---|---|---|---|
| 30 | 31 | 7.7 | 1.9 | 0.86 | 0.31 |
| 100 | 17 | 4.3 | 1.1 | 0.48 | 0.17 |
| 300 | 10 | 2.5 | 0.62 | 0.27 | 0.10 |
| 103 | 5.4 | 1.4 | 0.34 | 0.15 | 0.06 |
| 3 × 103 | 3.1 | 0.79 | 0.19 | 0.09 | 0.03 |
| 104 | 1.7 | 0.43 | 0.11 | 0.05 | 0.02 |
| 3 × 104 | 1.0 | 0.25 | 0.06 | 0.03 | 0.01 |
| 105 | 0.54 | 0.14 | 0.04 | 0.02 | 0.006 |
The entries in the table represent the minimum difference of specific gravity that can be discriminated from one voxel to the next, on the basis of intensity differences in the pixels that correspond to the 2D projected positions of the respective voxels. The minimum detectable density difference is expressed as a function of voxel size (indicated at the head of columns 2–6) and the total electron exposure value (indicated in column 1), to ensure that ΔI/σ = ≥3. (The less-stringent criterion that the intensity difference should be 3σ, adopted by Saxberg and Saxton (rather than the more commonly used criterion of 5σ), has been retained in this table.) The previously published entries have been divided by a factor of 14 to reflect the increased sensitivity of phase contrast relative to amplitude contrast, which was the model used earlier by Saxberg and Saxton. This factor of 14 emerges from a comparison of the contrast estimated if the phase contrast is ∼0.028 times the thickness in nanometers (see the discussion in the “What Physics Allows” section) to the contrast estimated by Saxberg and Saxton. This factor also agrees well with experimental estimates of the ratio of amplitude and phase contrast (63,64).
The values attainable for cryo-EM specimens are restricted to the entries in the clear cells of the table. The dark shading indicates that the density values are greater than the density difference between protein and vitreous ice, and thus it is not possible to realize the corresponding resolution, at the corresponding electron exposure, for cryo-EM specimens. The clear background indicates that the density values are in the range that can occur in cryo-EM specimens. Entries are shown in bold font if we believe that the corresponding resolutions should be achievable at the corresponding electron exposures. When the entries are not shown in bold font, it is questionable whether the indicated resolution will be achievable, because of the high level of radiation damage that would occur. In particular, when the entries are shown with double strikeout, we expect that radiation-induced bubbling would have occurred at the corresponding electron exposures, and as a result the corresponding resolution will not be achievable.
However, one can improve the resolution at which images and 3D reconstructions have acceptable SNR indefinitely by merging data from independent images of identical objects. In this case, the only physical limitation is that the SNR in individual images must be high enough for one to correctly align and orient the particles. (In this regard, it is important to note that it is easier to merge data from large particles than from small ones, since large structures provide more signal to use when computing the best alignment.) As originally estimated by Henderson (17), for example, it should be possible to merge data at high resolution from proteins as small as ∼40 kDa, and it should require only ∼12,000 particles to produce a density map that is interpretable at atomic resolution, regardless of the particle size. These estimates are actually rather conservative (18), a point that was subsequently put on a more secure mathematical basis by Rosenthal and Henderson (19).
What is (And is not, But should be) Currently Achieved
In recent years, investigators have made rapid advances in the use of cryo-EM to obtain high-resolution maps of icosahedral virus particles (20–28). For the first time, cryo-EM has begun to produce density maps in which side-chain residues are as easily recognized (and in some cases even more easily recognized) than they are in x-ray crystal structures of the same particle. Structurally stable icosahedral particles thus join well-ordered 2D crystals (29,30) and helical assemblies (31,32) as objects for which cryo-EM is the method of choice for determining structures at atomic resolution. Except for the work on the bacterial flagellum (31), however, the number of particles for which data must be merged has remained well in excess of one million asymmetric units, which is more than 100 times the estimated number that would be needed if the SNR of the data were close to what should be physically achievable (17).
High-resolution structures have also been obtained for chaperonins, including Mm-cpn (33) and GroEL (34). Again, these are relatively large particles with high symmetry (16-fold and 14-fold, respectively), and both of these factors make it easier to achieve high resolution. Therefore, Cong et al.'s (35) use of cryo-EM to propose a chain-trace (atomic) model of TriC/CCT (Fig. 1) may be close to the limit of what is currently achievable, because this chaperonin particle consists of two rings made of eight related but distinct polypeptides and thus has only twofold symmetry. As a result, the interpretation of the density map is not always as convincing as it is for maps that these authors and others have obtained for icosahedral virus particles. Indeed, some consider the interpretation of this density map to be controversial, since the assignment of the relative positions of individual subunits differs from that proposed in another study (36). Even so, the quality of the cryo-EM map achieved by Cong et al. represents a major step forward.
Although the ribosome is a completely asymmetric particle, it was initially a bit surprising, given the large size of this particle, that attempts to obtain a high-resolution density map remained stalled at a resolution of ∼0.6 nm (37–39). However, it has become clear that the existence of multiple, biochemically relevant conformational states represents both a challenge and a special opportunity for structural studies by cryo-EM (40). The challenge lies in the need to collect and process n-fold as many images as would otherwise be the case (where n is the number of conformationally distinct states that are present in the sample). On the other hand, we have the opportunity to visualize the structural changes and mechanistic trajectory of a molecular machine under physiologically relevant buffer conditions, free from the constraints of crystal packing.
Other factors besides structural heterogeneity still limit the scope of what currently can be achieved by cryo-EM. It is still quite challenging to detect particles and merge data from the resultant noisy images for single particles as small as 500 kDa, and 3D reconstructions are rarely attempted for particles half that size. In addition, even when density maps are obtained at atomic resolution, it is usually necessary to merge data from millions rather than from only tens of thousands of particles. As is discussed below, the currently limiting factors include the imperfect performance of detectors used in EM, the poor contrast transfer that is achieved in highly defocused images, and additional loss of signal due to radiation-induced specimen (or image) movement. However, it is likely that robust and nearly perfect experimental performance will eventually be achieved in all three of these areas, in which case cryo-EM could make even more significant contributions to the field of structural biology.
Areas in Which Significant Technological Improvement Seems to be Possible
Noise-free detectors for EM images
Estimates of how many particles are needed to produce a 3D reconstruction at atomic resolution, and the smallest particle size for which this would be possible, assume not only perfect contrast transfer in the EM images but also the use of a noise-free detector. Although photographic film has long been the standard to beat (especially for 300 keV electrons), it leaves much room for improvement in terms of detective quantum efficiency under low-exposure conditions. New types of area detectors that are currently being developed for EM not only improve on the readout speed of CCD cameras but also promise to improve the point-spread function (i.e., resolution) relative to the pixel size of the detector (13,41,42). In principle, such detectors could also be operated in the electron-counting mode, thus approaching the limit of an ideal detector.
Ideal contrast transfer
The potential importance of Zernike phase contrast can be quantified by numerical simulation of cryo-EM images (43). As an example, we show in Fig. 2 that the improved signal at low spatial frequencies results in a significant increase in the height of the cross-correlation peak for particles as small as 100 kDa, which in turn translates into much more accurate alignment of the particles.
Figure 2.
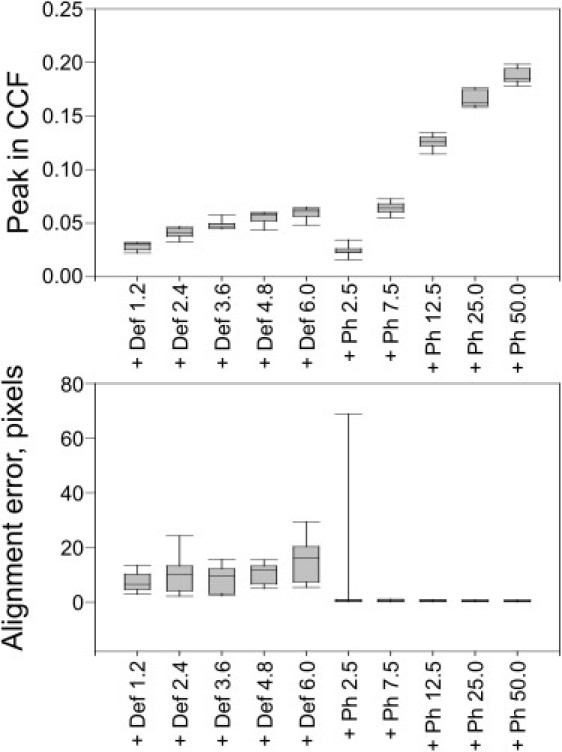
Quantitative estimate of one of the improvements expected from development of in-focus (Zernike) phase contrast for cryo-EM. As part of work that is not yet published, images of a 100 kDa enzyme embedded in vitreous ice were simulated for the case of defocus-based phase contrast, at defocus values (indicated by the abbreviation Def) ranging from 1.2 to 6.0 μm, and for Zernike phase contrast at cut-on frequencies (indicated by the abbreviation Ph) ranging from 1/(2.5 nm) to 1/(50 nm). Box plots indicate the minimum, lower quartile, median, upper quartile, and maximum for the distribution of values obtained for multiple simulated images. (A) Plot showing the distribution of CCF peak heights when the simulated images are cross correlated with a perfect, noise-free image. (B) Plot showing the alignment error, i.e., the distance of the CCF peak from the position corresponding to correct alignment.
Although it was recognized more than 60 years ago that miniature devices could be designed to provide in-focus (Zernike) phase contrast in the electron microscope (44), the first major breakthrough occurred when Nagayama (45) used a focused ion beam to drill holes as small as 0.3 μm into a thin carbon-film phase plate. In addition, Nagayama and Danev (46) used both a final coating of evaporated carbon and continuous heating of the phase plate to minimize electrostatic charging. Although the useful lifetime of such phase plates seems to be restricted by an unknown aging mechanism (47), tremendous improvements in cryo-EM image contrast have been demonstrated for a wide range of specimens (48–50). Earlier work with such phase plates (51) also stimulated interest in the development of microfabricated electrostatic phase plates (52–54), and new schemes have also been proposed to combine phase plates with aberration correctors, or to use a focused laser as a phase plate (55). It thus seems certain that further developments in phase-plate technology will be achieved.
Minimizing beam-induced movement
Finally, we turn to the point that the signal in cryo-EM images shows a much steeper falloff at high resolution than is observed in the electron diffraction pattern of the same specimen (56). In addition, the amount of falloff varies from one image to the next, and even from one area to another within a single image. Following up on the observation that the power spectra of individual particle images in a data set can be classified according to the amount and direction of movement (57), we have noted that particles in different classes tend to be clustered together in different areas of a given micrograph (Fig. 3). One model of such effects proposes that radiation damage produces stochastically distributed stress within the sample, which in turn produces random shifts (strain) of different areas by as much as a few angstroms or more. Another model is that random fluctuations in specimen charging result in spatially varying movement of different parts of the image. In any event, beam-induced movement that occurs while the image is recorded is responsible for the steep falloff of signal.
Figure 3.
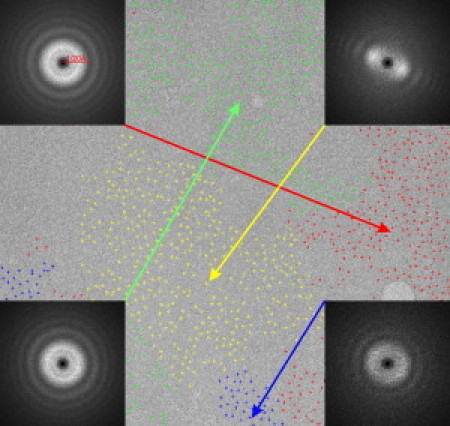
Example showing variation in the amount of beam-induced movement in a cryo-EM image of 70S ribosomes supported on a continuous carbon film. (We thank Dr. Joachim Frank for providing a large set of micrographs that were analyzed in this way.) The noisy power spectra of individual ribosome particles were classified by multivariate statistical analysis followed by hierarchical ascendant classification. The positions of individual members in each class are shown with different colors, and the average power spectra for each class are shown as insets in the corners of the figure. The first zero in the contrast transfer function is at a resolution of 20 Å, as indicated in the top-left inset. The results of this experiment show that members of each class are clustered into separate areas of the micrograph, implying that entire regions of the specimen move as a coherent block. Not shown here is the additional fact that most micrographs showed comparable results, but the patterns of movement varied substantially from one micrograph to the next.
Although several methods for minimizing this movement have been investigated, only partial reduction has been achieved by techniques such as limiting the size of the illuminated area (58,59) or improving the electrical conductivity of the support film (60,61). Other approaches that might have been expected to be effective, such as extreme reduction in the size of the illuminated area or fractionation of the electron exposure into extremely small steps, did not improve the situation (62). In contrast to these limited improvements, images of paraffin crystals grown on thick carbon support films consistently showed the full, theoretically expected contrast (signal) at a resolution of ∼0.4 nm (62), and produced calculated Fourier transforms such as the one shown in Fig. 4. This recent result thus establishes that it is physically possible to overcome beam-induced movement for at least some types of radiation-sensitive specimens. Although this result is encouraging, one can think of many reasons why additional methods may still be needed to achieve comparable improvements for specimens embedded in vitreous ice.
Figure 4.

Example of the calculated Fourier transform of the image of a monolayer crystal of paraffin (tetratetracontane) grown on a 35-nm-thick carbon film. The three sets of quasi-hexagonal reflections, all at a resolution of ∼0.4 nm, have essentially the full, theoretically expected amplitude based on the measured intensities of the electron diffraction pattern of comparable crystals. This is a cropped (lower-resolution) version of the same diffraction pattern published by Glaeser et al. (62), where it is shown (using insets with stretched contrast) that diffraction spots for this image remain detectable out to a resolution of 0.15 nm.
Summary: Goals Still to be Achieved
As useful as cryo-EM already is for structural studies in biochemistry and cell biology, it nevertheless performs at a level well below what it might. In principle, it should be possible to determine structures at atomic resolution for protein complexes that are as much as 20 times smaller than is currently the case. In addition, such structures should require data sets that are 100 times smaller than what is currently needed. If the SNR of images could be made to approach what physics will allow, one could also expect considerable improvement in how well the conformational substates can be distinguished within a heterogeneous population. As indicated above, however, the detectors used to record images must be made virtually noise-free, as would be the case for a pixilated electron counter. The phase-contrast transfer function must also be made uniformly close to 1.0, as it would be for images recorded with a charging-free quarter-wave plate. Finally, the problem of beam-induced movement of specimens embedded in vitreous ice must be overcome, as was recently done for specimens attached firmly to a thick carbon support film. Significant progress on at least some, if not all, of these objectives is certain to be made over the next few years, resulting in greatly expanded opportunities in structural biology.
Acknowledgments
This work was supported as part of ENIGMA, a Scientific Focus Area Program supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics:GTL Foundational Science through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy, and by Grant RPG0039 from the Human Frontiers Science Program.
References
- 1.Glaeser R.M. Cryo-electron microscopy of biological nanostructures. Phys. Today. 2008;61:48–54. [Google Scholar]
- 2.Mulder A.M., Yoshioka C., Williamson J.R. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer N., Konevega A.L., Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 4.Leis A., Rockel B., Baumeister W. Visualizing cells at the nanoscale. Trends Biochem. Sci. 2009;34:60–70. doi: 10.1016/j.tibs.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Glaeser R.M. Retrospective: radiation damage and its associated “information limitations”. J. Struct. Biol. 2008;163:271–276. doi: 10.1016/j.jsb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Baker L.A., Smith E.A., Rubinstein J.L. The resolution dependence of optimal exposures in liquid nitrogen temperature electron cryomicroscopy of catalase crystals. J. Struct. Biol. 2010;169:431–437. doi: 10.1016/j.jsb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Harrison S.C. Virology. Looking inside adenovirus. Science. 2010;329:1026–1027. doi: 10.1126/science.1194922. [DOI] [PubMed] [Google Scholar]
- 8.Frank J. Oxford University Press; New York: 2006. Three-Dimensional Electron Microscopy of Macromolecular Assemblies—Visualization of Biological Molecules in their Native State. [Google Scholar]
- 9.Glaeser R.M., Downing K., Frank J. Oxford University Press; New York: 2007. Electron Crystallography of Biological Macromolecules. [Google Scholar]
- 10.Zernike F. How I discovered phase contrast. Science. 1955;121:345–349. doi: 10.1126/science.121.3141.345. [DOI] [PubMed] [Google Scholar]
- 11.Wang L.G., Bose P.S., Sigworth F.J. Using cryo-EM to measure the dipole potential of a lipid membrane. Proc. Natl. Acad. Sci. USA. 2006;103:18528–18533. doi: 10.1073/pnas.0608714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bammes B.E., Jakana J., Chiu W. Radiation damage effects at four specimen temperatures from 4 to 100 K. J. Struct. Biol. 2010;169:331–341. doi: 10.1016/j.jsb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMullan G., Chen S., Faruqi A.R. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwen B.F., Downing K.H., Glaeser R.M. The relevance of dose-fractionation in tomography of radiation-sensitive specimens. Ultramicroscopy. 1995;60:357–373. doi: 10.1016/0304-3991(95)00082-8. [DOI] [PubMed] [Google Scholar]
- 15.Hoppe W., Hegerl R. Some remarks concerning the influence of electron noise on 3D reconstruction. Ultramicroscopy. 1981;6:205–206. [Google Scholar]
- 16.Saxberg B.E.H., Saxton W.O. Quantum noise in 2D projections and 3D reconstructions. Ultramicroscopy. 1981;6:85–90. [Google Scholar]
- 17.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 18.Glaeser R.M. Review: electron crystallography: present excitement, a nod to the past, anticipating the future. J. Struct. Biol. 1999;128:3–14. doi: 10.1006/jsbi.1999.4172. [DOI] [PubMed] [Google Scholar]
- 19.Rosenthal P.B., Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Wolf M., Garcea R.L., Harrison S.C. Subunit interactions in bovine papillomavirus. Proc. Natl. Acad. Sci. USA. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J.Z., Settembre E.C., Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc. Natl. Acad. Sci. USA. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W., Baker M.L., Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 23.Yu X.K., Jin L., Zhou Z.H. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., Settembre E., Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Jin L., Zhou Z.H. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z.H. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr. Opin. Struct. Biol. 2008;18:218–228. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H.R., Jin L., Zhou Z.H. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D.-H., Baker M.L., Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Löwe J., Li H., Nogales E. Refined structure of αβ-tubulin at 3.5 A resolution. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 30.Gonen T., Cheng Y.F., Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonekura K., Maki-Yonekura S., Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 32.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J. Mol. Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J.J., Baker M.L., Chiu W. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludtke S.J., Baker M.L., Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Cong Y., Baker M.L., Chiu W. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. USA. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amit M., Weisberg S.J., Horovitz A. Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J. Mol. Biol. 2010;401:532–543. doi: 10.1016/j.jmb.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 37.Schuette J.C., Murphy F.V., 4th, Spahn C.M. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villa E., Sengupta J., Frank J. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. USA. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armache J.P., Jarasch A., Beckmann R. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. USA. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spahn C.M.T., Penczek P.A. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr. Opin. Struct. Biol. 2009;19:623–631. doi: 10.1016/j.sbi.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battaglia M., Contarato D., Radmilovic V. Characterisation of a CMOS active pixel sensor for use in the TEAM microscope. Nucl. Instrum. Methods Phys. Res. A. 2010;622:669–677. [Google Scholar]
- 42.Li S.D., Matis H.S., Kleinfelder S. Modeling and analysis of charged-particle CMOS image sensor arrays. IEEE Trans. Nucl. Sci. 2009;56:1062–1068. [Google Scholar]
- 43.Chang W.H., Chiu M.T.K., Tu I.P. Zernike phase plate cryoelectron microscopy facilitates single particle analysis of unstained asymmetric protein complexes. Structure. 2010;18:17–27. doi: 10.1016/j.str.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Boersch H. Uber die Kontraste von Atomen im Electronenmikroskop. Z. Naturforschg. 1947;2a:615–633. [Google Scholar]
- 45.Nagayama K. Development of phase plates for electron microscopes and their biological application. Eur. Biophys. J. 2008;37:345–358. doi: 10.1007/s00249-008-0264-5. [DOI] [PubMed] [Google Scholar]
- 46.Nagayama K., Danev R. Phase contrast electron microscopy: development of thin-film phase plates and biological applications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:2153–2162. doi: 10.1098/rstb.2008.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danev R., Glaeser R.M., Nagayama K. Practical factors affecting the performance of a thin-film phase plate for transmission electron microscopy. Ultramicroscopy. 2009;109:312–325. doi: 10.1016/j.ultramic.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murata K., Liu X.A., Chiu W. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–912. doi: 10.1016/j.str.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danev R., Nagayama K. Single particle analysis based on Zernike phase contrast transmission electron microscopy. J. Struct. Biol. 2008;161:211–218. doi: 10.1016/j.jsb.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 50.Danev R., Kanamaru S., Nagayama K. Zernike phase contrast cryo-electron tomography. J. Struct. Biol. 2010;171:174–181. doi: 10.1016/j.jsb.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Danev R., Nagayama K. Transmission electron microscopy with Zernike phase plate. Ultramicroscopy. 2001;88:243–252. doi: 10.1016/s0304-3991(01)00088-2. [DOI] [PubMed] [Google Scholar]
- 52.Schultheiss K., Perez-Willard F., Schroder R.R. Fabrication of a Boersch phase plate for phase contrast imaging in a transmission electron microscope. Rev. Sci. Instrum. 2006;77:033701–033704. [Google Scholar]
- 53.Cambie R., Downing K.H., Jin J. Design of a microfabricated, two-electrode phase-contrast element suitable for electron microscopy. Ultramicroscopy. 2007;107:329–339. doi: 10.1016/j.ultramic.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Majorovits E., Barton B., Schröder R.R. Optimizing phase contrast in transmission electron microscopy with an electrostatic (Boersch) phase plate. Ultramicroscopy. 2007;107:213–226. doi: 10.1016/j.ultramic.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Mueller H., Jin J.A., Glaeser R.M. Design of an electron microscope phase plate using a focused continuous-wave laser. N. J. Phys. 2010;12:073011. doi: 10.1088/1367-2630/12/7/073011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henderson R., Glaeser R.M. Quantitative analysis of image contrast in electron micrographs of beam-sensitive crystals. Ultramicroscopy. 1985;16:139–150. [Google Scholar]
- 57.Sander B., Golas M.M., Stark H. Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. J. Struct. Biol. 2003;142:392–401. doi: 10.1016/s1047-8477(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 58.Downing K.H. Spot-scan imaging in transmission electron microscopy. Science. 1991;251:53–59. doi: 10.1126/science.1846047. [DOI] [PubMed] [Google Scholar]
- 59.Bullough P., Henderson R. Use of spot-scan procedure for recording low-dose micrographs of beam-sensitive specimens. Ultramicroscopy. 1987;21:223–229. [Google Scholar]
- 60.Typke D., Downing K.H., Glaeser R.M. Electron microscopy of biological macromolecules: bridging the gap between what physics allows and what we currently can get. Microsc. Microanal. 2004;10:21–27. doi: 10.1017/S1431927604040164. [DOI] [PubMed] [Google Scholar]
- 61.Yoshioka C., Carragher B., Potter C.S. Cryomesh: a new substrate for cryo-electron microscopy. Microsc. Microanal. 2010;16:43–53. doi: 10.1017/S1431927609991310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glaeser R.M., McMullan G., Henderson R. Images of paraffin monolayer crystals with perfect contrast: minimization of beam-induced specimen motion. Ultramicroscopy. 2011;111:90–100. doi: 10.1016/j.ultramic.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toyoshima C., Unwin N. Contrast transfer for frozen-hydrated specimens: determination from pairs of defocused images. Ultramicroscopy. 1988;25:279–291. doi: 10.1016/0304-3991(88)90003-4. [DOI] [PubMed] [Google Scholar]
- 64.Yonekura K., Braunfeld M.B., Agard D.A. Electron energy filtering significantly improves amplitude contrast of frozen-hydrated protein at 300kV. J. Struct. Biol. 2006;156:524–536. doi: 10.1016/j.jsb.2006.07.016. [DOI] [PubMed] [Google Scholar]


