Abstract
Microtubule (MT) length and location is tightly controlled in cells. One novel family of MT-associated proteins that regulates MT dynamics is the MT-severing enzymes. In this work, we investigate how katanin (p60), believed to be the first discovered severing enzyme, binds and severs MTs via single molecule total internal reflection fluorescence microscopy. We find that severing activity depends on katanin concentration. We also find that katanin can remove tubulin dimers from the ends of MTs, appearing to depolymerize MTs. Strikingly, katanin localizes and severs at the interface of GMPCPP-tubulin and GDP-tubulin suggesting that it targets to protofilament-shift defects. Finally, we observe that binding duration, mobility, and oligomerization are ATP dependent.
Introduction
Microtubules (MTs) are noncovalent cytoskeletal polymers found in most eukaryotic cells. They form complex and dynamic arrays that are involved in mitosis, cell motility, intracellular transport, secretion, and the maintenance of cell shape and cell polarization (1). MTs rely upon their intrinsic polymerization dynamics to carry out these diverse activities. In vitro, MTs stochastically interconvert between growth and shrinkage, a process termed dynamic instability (2). Comparisons of MT dynamics in cells and in vitro have revealed large discrepancies in growth and shrinkage rates as well as catastrophe (switch from growing to shrinking) and rescue (switch from shrinking to growing) frequencies. These differences are due to MT-associated proteins that regulate dynamics by promoting the assembly, stability, or destabilization of filaments (3). A recent addition to these MT-associated proteins is the class of MT-severing enzymes (4).
MT-severing enzymes use ATP to break the MT lattice in the middle. They are members of the highly diverse AAA+ (ATPases associated with a variety of cellular activities) protein superfamily (5) that often act as hexameric rings (6). There are three families of this class: katanin, spastin, and fidgetin. Katanin was the first discovered as an enzymatic factor in sea urchin egg extracts (7). It is known that katanin has a 60 kD catalytic subunit (p60) and an 80 kDa targeting and regulatory subunit (p80) (5,7).
Homologs of p60 and p80 katanin have been identified in species ranging from plants to humans (4). In plants, katanin is involved in the release of MTs nucleated from branch points (8). In Chlamydomonas, it severs doublet MTs in the flagellum during deflagellation (9). In mitosis of Drosophila S2 cells, katanin localizes to kinetochores and chromosomes and is required for the depolymerization of chromosome-attached MT plus ends during anaphase A (10). During interphase, katanin is localized to the cortex of S2 cells where it regulates MT length and dynamics (11). In differentiated neurons, katanin regulates axonal outgrowth and branching (12,13).
Although katanin was identified 20 years ago, it is notoriously difficult to purify and work with, so biophysical studies have made slow progress. Previous work with katanin has shown that p60 uses the energy of ATP hydrolysis to cause the severing of MTs, and the ATPase activity is stimulated by MTs (5,7). Structural analysis by rotary shadowing-electron microscopy of p60 and p60/p80 katanin (5), gel filtration, and fluorescence resonance energy transfer have shown that katanin forms a transient hexamer in the presence of both ATP and MTs (6). Furthermore, the nucleotide state and presence of MTs is thought to regulate p60 oligomerization (6). Elegant mathematical models have been used to argue that katanin likely severs at lattice defects (14), however, direct evidence that katanin can identify and remove defects from the lattice is lacking.
At present, there are many open questions about how and where katanin performs its function. To determine where and how katanin acts on the MT lattice, we have purified a green-fluorescent protein (GFP) labeled, full-length catalytic subunit (p60) of Drosophila Katanin, GFP-Katanin-60, using the baculovirus expression system. With this protein, we performed the first, to our knowledge, in vitro single-molecule study of MT severing. Our findings reveal multiple GFP-Katanin-60s bind rapidly to and disassociate quickly from severing sites before severing occurs. Interestingly, we find that GFP-Katanin-60 removes tubulin from the ends of taxol-stabilized MTs, which appears as depolymerization. This activity is ATP-dependent and varies with the concentration of GFP-Katanin-60. In addition, we report that defects in the MT lattice such as dislocations and the ends of the lattice appear to enhance katanin activity. Finally, we find that the GFP-Katanin-60 binding, diffusion, and oligomerization state depend on the nucleotide state of the monomer.
Materials and Methods
Reagents
Reagents were purchased from Sigma (St. Louis, MO) unless indicated otherwise.
Protein
Drosophila melanogaster Katanin-60 tagged with 6×His-GFP at the amino-terminus (GFP-Katanin-60) was expressed using the Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA) and purified using metal affinity chromatography to get a clean band of protein of 92 kDa (see Fig. 1 B). Katanin-60 coding sequences were polymerase chain reaction (PCR) amplified and then subcloned into pFasBac HT, which resulted in the fusion of a 6×His Ni2+ binding sequence to the amino terminus of GFP. Recombinant baculovirus was prepared according to manufacturer's protocol.
Figure 1.
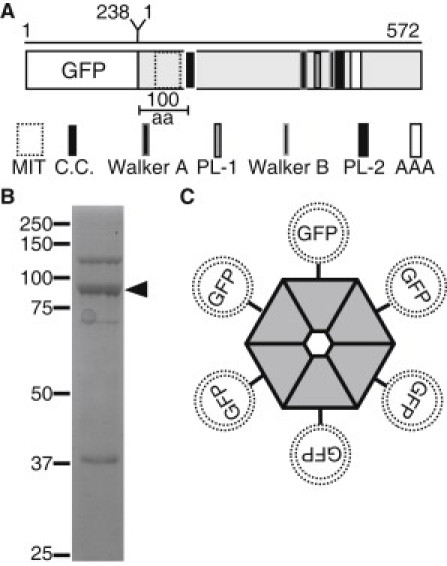
Description of domain architecture and purification of GFP-Katanin-60 from Sf9 insect cells. (A) Schematic depiction, approximately to scale, of domain architecture of GFP-Katanin-60. The amino acid length of GFP and Katanin-60 are indicated at the top of the figure. (aa, amino acids; MIT, microtubule interaction and trafficking domain; C.C., coiled-coil; PL-1, Pore Loop-1; PL-2, Pore Loop-2; AAA, AAA minimum consensus ATPase domain, Walker A, Walker B) (B) A coomassie stained SDS-PAGE gel of purified 6×His-tagged GFP-Katanin-60 after protein purification. Arrow marks the position of GFP-Katanin-60 (92 kD) that is the major band and molecular mass markers in kDa indicated at the left. (C) Cartoon of hexameric GFP-Katanin-60 ring showing the location of GFP at NH-terminal.
Sf9 (Spodoptera frugiperda) insect cells were grown in SFM-900 II media (Gibco, catalogue No.10902, Carlsbad, CA) supplemented with 100× antibiotic/antimycotic (Life Technologies, Carlsbad, CA) to a final working concentration of 0.5× using the shaker culture method (15). Expression of GFP-Katanin-60 was performed in 0.5 L Erlenmeyer flasks (Sigma-Aldrich, catalogue No. CLS430422) containing 250 mL of SFM media complemented with 5% HI FBS (Gibco, catalogue No. 16140) serum and using a multiplicity of infection of 1.0 pfu/cell. The cells were harvested at 72 h post infection by 1000 × g centrifugation and resuspended in lysis buffer (50 mM Tris [pH 8.5], 250 mM NaCl, 5 mM MgCl2, 50 μM ATP, 1 mM PMSF, 7 mM 2-mercaptoethanol, 200 mM Imidazole, 10% sucrose). Cells were homogenized by two passes through a Microfluidizer (15,000–20,000 psu Nitrogen) (Avestin, Ottawa, Ontario, Canada). Cell debris was removed by centrifugation (16,000 rpm for 30 min at 4°C). Protein was affinity purified by incubating the supernatant in batch with Ni2+-NTA beads (Qiagen, catalogue No. 30210, Valencia, CA) in agitation for 60 min at 4°C, washed twice with wash buffer (50 mM Tris [pH 8.5], 250 mM NaCl, 5 mM MgCl2, 50 μM ATP, 1 mM PMSF, 7 mM 2-mercaptoethanol, 200 mM Imidazole, 10% sucrose), and eluted with elution buffer (50 mM Tris [pH 8.5], 250 mM NaCl, 5 mM MgCl2, 50 μM ATP, 1 mM PMSF, 7 mM 2-mercaptoethanol, 0.5 M Imidazole, 10% sucrose), followed by buffer exchange in Severing Buffer I (10% sucrose, 20 mM HEPES, 300 mM NaCl, 3 mM MgCl2, 5 mM DTT, 50 μM ATP, pH 7.0), and concentrated with centrifugal filter units (Millipore, catalogue No. UFC801024, Billerica, MA).
Katanin concentrations were estimated by comparison to bovine serum albumin standards using a commercial Bicinchoninic Acid BCA (Thermo Scientific, Rockford, IL). Experiments with GFP-Katanin-60 were performed with freshly purified protein within 2 days of purification. Only ∼1 in 3 preparations functioned properly, as has been reported previously (14).
MTs
Tubulin was purified from pig brains according to previous methods (16). Rhodamine-tubulin and biotin-labeled tubulin were purchased from Cytoskeleton (Denver, CO). Guanalyl-(α,β)-methylene diphosphate (GMPCPP, Jena Biosciences, Jena, Germany)-stabilized MTs were grown at 37°C from a 5:1 mixture of unlabeled and rhodamine tubulin. Taxol-stabilized MTs were grown at 37°C from a 30:1 mixture of unlabeled and rhodamine tubulin in PEM-100 (100 mM Pipes, 2 mM EGTA, 2 mM MgCl2, pH 6.9 with KOH). Polarity-marked MTs were made using bright GMPCPP MTs as seeds to nucleate the growth of MTs in the presence of taxol.
Severing and depolymerization assays
Microscope chambers were constructed with 22 mm × 30 mm coverslips separated by double-sided tape (Scotch 3M, St. Paul, MN) to create channels 0.1 mm deep, 3 mm wide, and 30 mm long. Glass coverslips (22X30-1.5, Fisherbrand, catalogue No. 12-544-A, Pittsburgh, PA) were biologically cleaned before silanization in 0.05% dichlorodimethylsilane in trichloroethylene. To immobilize MTs, channels were incubated with 0.2% antibody antitubulin (clone 2.1) or 15% antibody antibiotin (clone BN-34) in PEM-100 for 5 min, followed with 5% Pluronic F-127 in PEM-100 for 5 min. Rhodamine MTs at 50 μg/mL were added and allowed to bind for 5 min. Channels were rinsed with chamber wash buffer (PEM-100, 20 μM Taxol) before addition of the imaging solution (Severing buffer II (20 mM HEPES, 100 mM NaCl, 3 mM MgCl2, 10% sucrose, (pH 7.0) plus 20 μM Taxol) supplemented with 0.1% Pluronic F-127, 0.5 mg/mL bovine serum albumin, 2 mM ATP or 5 mM AMPPNP or 4 mM Hexokinase (as described in the text), 10 mM DTT, 40 mM glucose, 40 μg/mL glucose oxidase, 16 μg/mL catalase, and GFP-Katanin-60 at different concentrations.
Imaging
Single molecule and epifluorescence images were acquired with either an electron multiplier CCD (EM-CCD) Cascade II camera (Photometrics, Tucson, AZ) or an IXON EM-CCD camera (Andor, South Windsor, CT). Total internal reflection fluorescence (TIRF) microscopy used a home-build laser system around a Nikon Eclipse Ti microscope and a high numerical aperture objective (60×, NA = 1.49) (Nikon, Melville, NY). TIRF excitation was achieved using a 488-nm Argon-ion air laser to visualize GFP. MTs were imaged in epifluorescence using a Xe-Hg lamp. An oxygen scavenging system of glucose oxidase and catalase was employed to reduce photobleaching and photodamage during illumination with the laser. In addition, neutral density filters and shuttering were employed to attenuate laser light to the desired level. The standard exposure time was 500 ms, and the frame interval was 5, 10, or 20 s. To measure binding dwell times and diffusion, we imaged with 100 ms exposures without delay to capture short time binding events. Control experiments without GFP-Katanin-60 or with a dead-mutant E393Q (6) were performed in parallel on the same day to make sure we did not photodamage MTs during regular imaging.
Photobleaching
Photobleaching of GFP-Katanin-60 complexes was performed in the same chambers described previously except anti-GFP antibodies at 0.01 mg/mL were used to specifically bind GFP-Katanin-60 complexes either in the presence of 2 mM ATP or 2 mM AMPPNP. Movies were taken without delay with 100 ms intervals and the laser illumination was constant to induce photobleaching. Photobleaching steps were counted manually. The photobleaching data were analyzed by measuring the intensity of small bright spots over time using ImageJ. The intensity was plotted over time, and for data with clear steps, the number of steps was determined manually. For data with many bleach steps that occur simultaneously, we measured the intensity of the last few bleaches and measured the intensity of the complex at the initial frames. Using this data we determined the number of GFPs in the complex by dividing the intensity of the complex by the intensity of a single GFP, as described previously (17,18).
Data analysis
The data were recorded as 16-bit images using NIS Elements AR software (Nikon). The data were exported at 16-bit tiff files for each color. All movies were aligned using the ImageJ plug-in stackreg in translation mode (19). The motion of single GFP-Katanin-60 molecules was analyzed using kymographs generated with the Multiple Kymograph plug-in for ImageJ (J. Rietdorf and A. Seitz, European Molecular Biology Laboratory, Heidelberg, Germany). The rates of depolymerization were calculated by measuring the average angle of loss of signal at the end of MTs (see Fig. 4 B). One over the tangent of the angle gave the rate of depolymerization in distance pixel per time pixel. The time interval and distance scale were used to convert the rate data. The frequency of severing was obtained by counting the number of severs in every MT by hand. The number of severing events in a single movie was divided by the total length of the MT at the start of the movie and divided by the total time of the movie. The frequency of binding was determined by counting the number of binding events manually. The duration of association was measured from kymographs. The frequency of severing at interfaces between GMPCPP-tubulin and GDP-tubulin regions was obtained by counting the number of severs in every interface of MTs, and then the number of severing events at interfaces was divided by the total number of interfaces in the MT. The frequency of severing at interfaces was compared with the frequency of severing at 1 μm far away from the interface.
Figure 4.
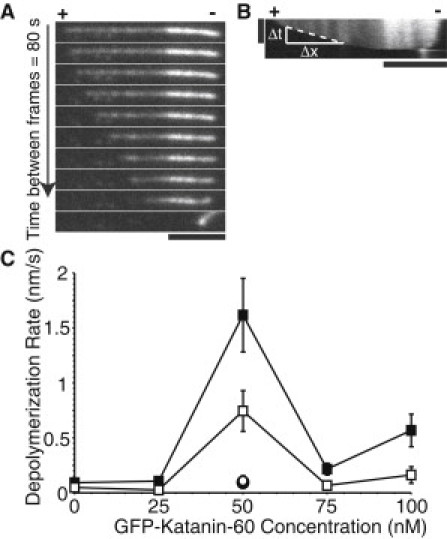
GFP-Katanin-60 depolymerizes MTs in an ATP and concentration dependent manner. (A) Time series of rhodamine polarity-marked MTs with 50 nM GFP-Katanin-60. The (+) indicates the plus end and the (−) indicates the minus end. Time between frames is 80 s; scale bar, 5 μm. (B) Kymograph from MT showing depolymerization is faster at plus end (+). Kymographs were used to measure depolymerization rates by drawing a line and measuring the change in distance over the change in time. Vertical scale bar, 10 min; horizontal scale bar, 5 μm. (C) Quantitative measurement of GFP-Katanin-60 MT depolymerization expressed as rate of depolymerization of MTs at plus end [0 nM (N = 19), 25 nM (N = 39), 50 nM (N = 77), 75 nM (N = 53), 100 nM (N = 76), black squares], minus end [0 nM (N = 19), 25 nM (N = 39), 50 nM (N = 164), 75 nM (N = 49), 100 nM (N = 66), white squares] in the presence of 2 mM ATP, with AMPPNP at plus end (N = 12, white circle), and hexokinase at plus end (N = 16, black circle). N-values represent the number of MTs analyzed, the points on the plot represent the mean value, and the error bars represent the mean ± SE.
Results
Baculovirus expression and domain organization architecture of GFP-Katanin-60
We expressed and purified full-length GFP-labeled Drosophila Katanin-60 to test its activity on MTs using single molecule assays. The 572 amino acids of the catalytic subunit, Katanin-60 contains the most important features of the MT-severing AAA ATPases, an amino MT-interacting and trafficking domain, and the highly conserved AAA ATPase domain at the C-terminus. This domain is longer than 200 amino acids and has conserved domains and motifs responsible for the ATPase and severing activity (Fig. 1 A). We obtained soluble GFP-Katanin-60 as a band of 92 kDa (Fig. 1 B). The N-terminal position of GFP does not interfere with the residues involved in the oligomerization of Katanin-60 in the presence of ATP (Fig. 1 C). We also expressed and purified an inactive mutant Katanin-60 E393Q. In this mutant, the Walker B motif's highly conserved glutamic acid (E) residue 393 is replaced with glutamine (Q) to create a substrate trap (Fig. S1 in the Supporting Material) (6).
Katanin severs MTs in vitro and severing is concentration dependent
It has been previously shown that katanin severs MTs and requires ATP for this activity (7), however, imaging of GFP-Katanin-60 during severing in vitro has not, to our knowledge, been previously shown. We attached microtubules to coverslips using surface-absorbed antitubulin or antibiotin antibodies and then added polarity-marked MTs, as described in the Materials and Methods. After a 3 min initial imaging phase performed in the absence of Katanin, we added various concentrations of purified GFP-Katanin-60 with ATP and the reaction was imaged over time using TIRF and epifluorescence. Images were recorded at 20 s intervals to reduce photobleaching, and an oxygen scavenging system was employed. GFP-Katanin-60 appeared as distinct bright dots distributed along the MTs (Fig. 2). The GFP signal of complexes fluctuated during imaging, indicative of different oligomerization states of the katanin or varying numbers of katanin complexes (Fig. 2).
Figure 2.
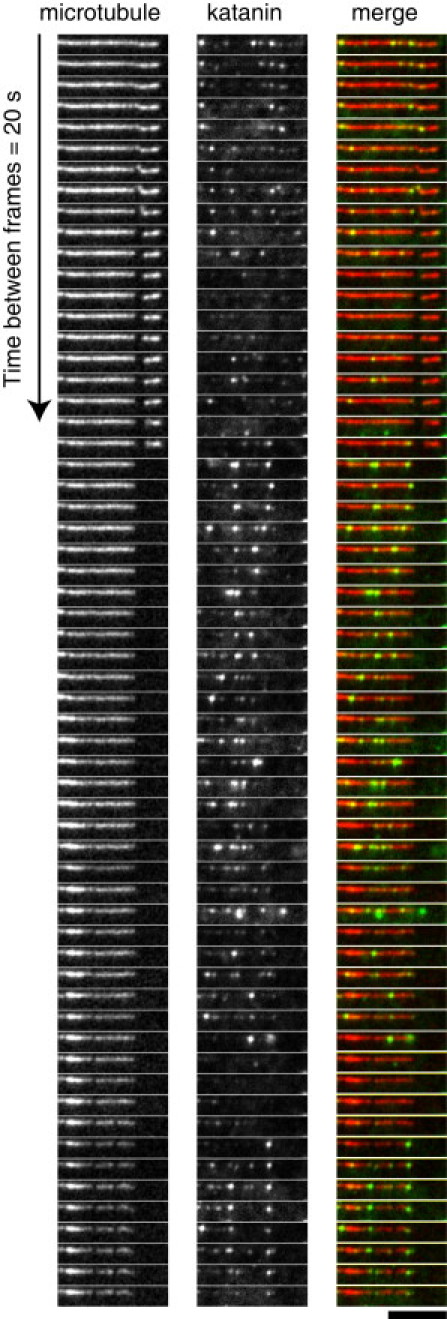
Localization of GFP-Katanin-60 during activity on MTs. Time series of MT-severing assays using rhodamine polarity-marked MTs with 200 nM GFP-Katanin-60 at 20 s intervals. From left to right, MTs (red in merge), GFP-Katanin-60 (green in merge), and merge. Scale bar, 5 μm.
When MTs were incubated with 100 nM GFP-Katanin-60, the whole MT lattice was disassembled before the end of 20 min recording. In the presence of 50 nM GFP-Katanin-60, breaks or gaps appeared within the lattice of taxol-stabilized MTs within 1 min (Fig. 2). Strikingly, the single molecule imaging showed that GFP-Katanin-60 molecules were localized at zones of severing and they remained bound to freshly severed ends (Fig. 2).
To measure the effect of katanin concentration on severing, we performed imaging assays with increased concentrations of GFP-Katanin-60 (0 nM, 25 nM, 50 nM, 75 nM, 100 nM, and 200 nM) for three active preparations (Fig. 3). The frequency of severing in each condition was determined as described in the Materials and Methods. In the absence of GFP-Katanin-60 (0 nM), no severing was observed. The frequency of severing at 50 nM GFP-Katanin-60 was 9.9 (± 2.2) × 10−5 severs/μm∗s and it reached a peak of 19.6 (± 3.6) × 10−5 severs/μm∗s at 100 nM. The highest concentrations of GFP-Katanin-60 (200 nM) showed a reduction in the frequency of severing.
Figure 3.
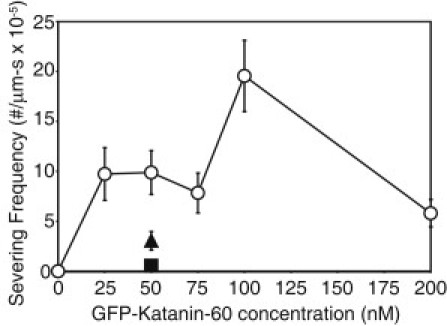
Quantitative measurement of GFP-Katanin-60 MT severing expressed as severing frequency of MTs in the presence of 2 mM ATP [0 nM (N = 18); 25 nM (N = 39); 50 nM (N = 80); 75 nM (N = 48); 100 nM (N = 34); 200 nM (N = 77), circles], in the presence of AMPPNP (N = 11, triangle) in the presence of hexokinase to use up residual ATP (N = 10, square). N-values represent the number of MTs analyzed, the points on the plot represent the mean value, and the error bars represent the mean ± SE.
We performed several control experiments to check that the severing was caused by katanin and not due to photodamage. First, the absence of GFP-Katanin-60 (Fig. 3) or the addition of the mutant GFP-Katanin-60 E393Q (Fig. S1 A) did not induce severing. This indicates that MT severing is due to the presence and activity of GFP-Katanin-60 and not due to photodamage or another enzyme that copurified. Depletion of ATP with hexokinase (apo state) lowered the severing frequency more than fivefold. The nonhydrolyzable ATP analog, AMPPNP, abated the severing frequency more than two and a half-fold (Fig. 3). Combined, these results indicate GFP-Katanin-60 is actively severing MTs, and our preparations that sever have repeatable activity levels. It has been reported previously that katanin is very unstable. We did find this as well, but the benefit of single-molecule assays is that it is easy and rapid to determine functioning protein from dysfunctioning protein. No data were taken with preparations that did not quickly sever MTs.
Katanin removes tubulin from MT plus ends in a concentration dependent manner
While examining the effect of GFP-Katanin-60 concentration on MT severing, depolymerization from the MT ends was observed. To determine if the MT plus or minus end was depolymerizing faster, we used polarity marked MTs. We observed that severing was faster from the plus end compared to the minus end (Fig. 4, A and B). Although Fig. 4 A shows an example of a MT with very little minus end growth, even MTs with longer minus end segments showed slower depolymerization from the minus end. Furthermore, MTs without a GMPCPP seed had a slower depolymerization rate from one end. Thus, we do not believe the slower minus end rate is a consequence of katanin's inability to depolymerize GMPCPP MTs.
To characterize the effect of katanin concentration on MT depolymerization, polarity-marked MTs were incubated with increasing concentrations of GFP-Katanin-60 (0 nM, 25 nM, 50 nM, 75 nM, and 100 nM,). The rate of depolymerization at plus and minus ends was determined as described in the Materials and Methods. Without GFP-Katanin-60, the plus end depolymerized at a basal rate of 0.092 ± 0.022 nm/s, whereas the minus end depolymerized at 0.047 ± 0.012 nm/s. In the presence of 50 nM GFP-Katanin-60, the rate of depolymerization at plus ends reached a peak to 1.62 ± 0.34 nm/s, corresponding to 0.2 tubulin dimers/s (Fig. 4 C). In addition, the rate of depolymerization at minus ends was always lower than plus ends, around 0.74 ± 0.18 nm/s. At concentrations higher than 50 nM, the rate of depolymerization decreased. Interestingly, the maximal depolymerization rate occurs at a lower concentration (50 nM) compared to the maximal severing rate (100 nM), perhaps implying that the ends are an early target for the enzyme.
We performed several control experiments. In the absence of GFP-Katanin-60 and in the presence of 50 nM of Katanin-60 mutant E393Q, we observed basal rates of depolymerization (Fig. S1 B). We repeated assays in the presence of 50 nM GFP-Katanin-60 with AMPPNP or hexokinase to deplete residual ATP. We found that depolymerization activity depended on ATP because both ATP depleted or AMPPNP decreased the rate of depolymerization back to basal levels (0.094 ± 0.038 and 0.108 ± 0.048 nm/s, respectively, Fig. 4 C).
Katanin targets to MT defects in vitro
To further determine where GFP-Katanin-60 targets on the MTs, we created MTs with lattice defects at observable locations. We chose to examine polarity-marked MTs with bright GMPCPP seeds, and dim elongation segments polymerized in the presence of taxol. Previous studies have shown that 96% of GMPCPP MTs have 14 protofilaments when polymerized in vitro (20). Furthermore, polymerization of MTs in the presence of taxol creates 12 to 13 protofilaments MTs (21). Thus, our MTs should have a lattice shift in protofilament number at the seed elongation junction (within the resolution of our light microscope).
We observed that GFP-Katanin-60 molecules arrived and remained attached just at the border between GMPCPP seeds and the taxol-stabilized elongation segment of MTs that we called interfaces (Fig. 5 A). Then, GFP-Katanin-60 molecules remained attached on interfaces until they severed them. We saw severing at interfaces over 50–200 nM GFP-Katanin-60 concentrations (Fig. 5 A), and we quantified the frequency of severing at these interfaces and compared them with the frequency of severing that occurred at a location 1 μm away from the interfaces (Fig. 5 B). The frequency of severing at interfaces was always ∼50% more likely compared to 1 μm away. For instance, at 75 nM GFP-Katanin-60, there were 0.60 severs/min at the interface, whereas there were only 0.39 severs/min 1 μm away.
Figure 5.
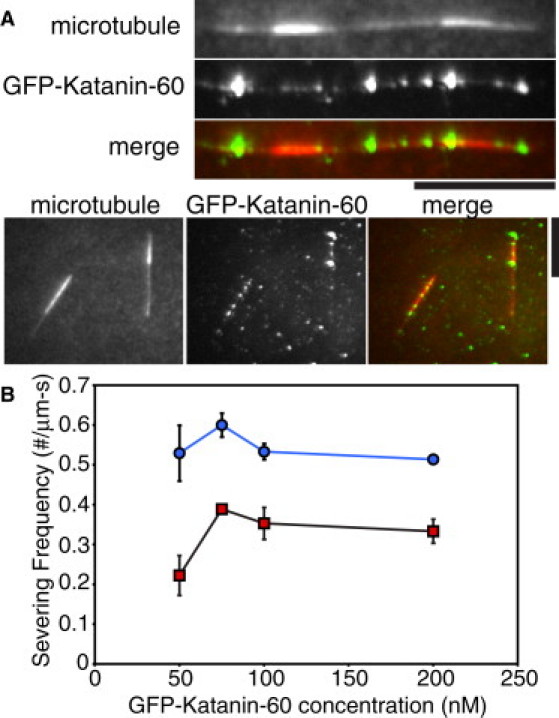
GFP-Katanin-p60 severs at interfaces between GMPCPP and GDP-taxol MT segments. (A) The MT frames depict the MTs as they existed at the beginning of the movie before severing (red in merge). The GFP-Katanin-60 frame is a z-projection of the SD of a time series of GFP-Katanin-60 binding (green in merge). Scale bars, 5 μm. (B) Measurement of frequency of severing at 50 nM (N = 9), 75 nM (N = 18), 100 nM (N = 17), and 200 nM (N = 21) GFP-Katanin-60 at interfaces (blue circles) and 1 μm away from interfaces (red squares). N-values represent the number of MTs analyzed, the points on the plot represent the mean value, and the error bars represent the mean ± SE.
Katanin binding, mobility, and hexamerization is nucleotide dependent
It has been shown that the binding and hydrolysis of ATP is required for hexamerization and severing activity of katanin (6). To examine binding, diffusion, and hexamerization we imaged GFP-Katanin-60 in different nucleotide states using ATP, AMPPNP, and hexokinase to deplete residual ATP. We also used photobleaching in the presence of ATP and AMPPNP to measure the hexamerization state of the complexes.
For lattice binding and diffusion, we created kymographs of our movies to determine the location and duration of GFP-Katanin-60 binding. In the presence of ATP, GFP-Katanin-60 bound and detached rapidly from MTs (Fig. 6 A). To determine the duration and diffusion coefficient, we imaged with 100 ms intervals in the GFP channel (Fig. 6 B). Very few molecules bound and diffused for more than a few seconds (Table 1). A histogram of the rate of diffusion was an exponential decay with a decay length, or average diffusion coefficient of 0.003 ± 0.0005 μm2/s (error is the fit error to the exponential decay, Fig. S2).
Figure 6.
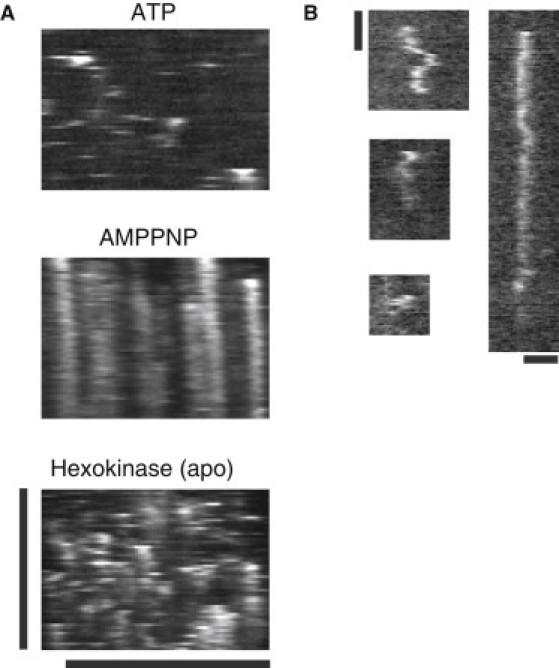
GFP-Katanin-60 dynamics depends on the nucleotide state. (A) Kymographs of 50 nM GFP-Katanin-60 in the presence of 2 mM ATP, 5 mM AMPPNP, and 5 mM hexokinase. Vertical scale bar, 20 min; horizontal scale bar, 5 μm for all images. (B) Examples of kymographs of GFP-Katanin-60 diffusing along taxol-stabilized MTs in the presence of 2 mM ATP. Vertical scale bar, 1 min; horizontal scale bar, 1 μm.
Table 1.
Binding duration and frequency depend on nucleotide
| Nucleotide | Duration of association (s) | Frequency of binding (No. binding/μm-s) |
|---|---|---|
| ATP | 29.8 ± 1.8, N = 275 | 5097.4 ± 606.3 |
| AMPPNP | 734.6 ± 34.1, N = 173 | 2380.7 ± 359.4 |
| Hexokinase (apo) | 357.3 ± 19.1, N = 173 | 3354.6 ± 564.7 |
In the presence of 5 mM AMPPNP, GFP-Katanin-60 displayed strong binding along MTs, but no diffusion was observed (Fig. 6 A). Most GFP-Katanin-60 remained attached for the entire 20 min of imaging (Table 1). However, because AMPPNP was not hydrolyzed, GFP-Katanin-60 did not sever or detach from MTs. When ATP was removed from the chamber by adding 4 mM hexokinase, GFP-Katanin-60 bound and diffused for intermediate times (Table 1, Fig. 6 A).
To examine how nucleotide affected the oligomerization state of the GFP-Katanin-60 complexes, we performed single complex photobleaching. In the presence of 2 mM ATP, bleaching steps are clearly visible (Fig. 7 A). These steps were easily counted manually. We created a histogram of the percentage of complexes displaying each number of steps (see Fig. 7 C). We also performed photobleaching in the presence of a nonhydrolyzable analog of ATP, AMPPNP at 2 mM. For the AMPPNP photobleaching, it is more difficult to discern steps compared to ATP because the AMPPNP complexes have more GFPs (Fig. 7 B). In complexes with more GFPs, there is a higher probability that two GFPs will bleach together (as seen in Fig. 7 B). To analyze the data, we determined the intensity change due to photobleaching the final GFP and the maximum intensity, as previously described (17). We assumed that all GFPs have the same intensity and divided the total intensity by the intensity of one GFP to compute the total number of GFPs (Fig. 7 B). Using this method, we estimated the number of bleach events for AMPPNP data, and created a histogram of the percentage of complexes displaying each number of steps (Fig. 7 C).
Figure 7.
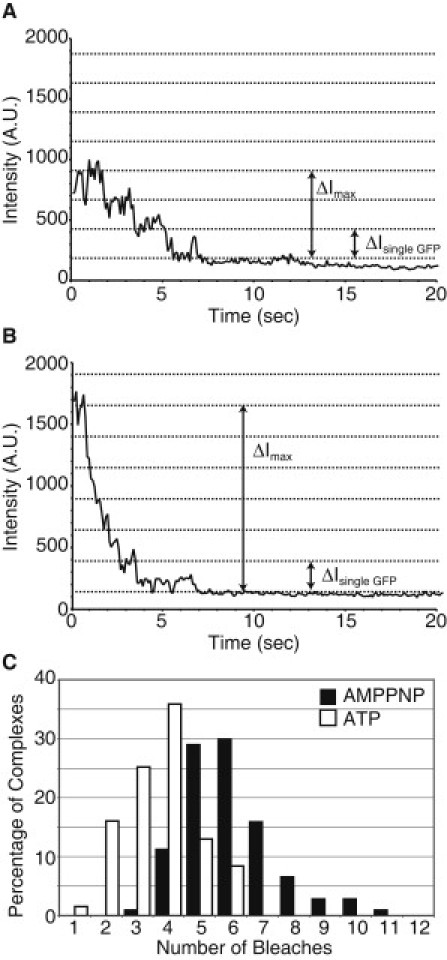
Photobleaching and fluorescence intensity analysis of GFP-Katanin-60. (A) Representative example of three-step photobleaching event in the presence of 2 mM ATP. (B) Example of multiple photobleaching events in the presence of 2 mM AMPPNP. (C) Probability distribution of number of bleach events per complex in the presence of 2 mM ATP (white bars) and 2 mM AMPPNP (black bars). The average number of bleaching events is 4/complex for ATP; whereas the average number of bleaching events is 6/complex for AMPPNP.
In the presence of ATP, the maximum number of bleach events displayed by a GFP-Katanin-60 complex was six, which corresponds to a hexamer. The most likely number of bleach events was four, which is only a tetramer. In the presence of AMPPNP, complexes are most likely to display six bleach events, thus they are hexamers. We also see complexes that contain up to 11 GFPs, which could be oligomerization of hexamers.
Discussion
We have, for the first time, to our knowledge, imaged a MT-severing enzyme during the act of MT severing and depolymerization. The method of single molecule imaging allows us to determine information about binding and mobility as well as severing and depolymerization activity of GFP-Katanin-60. For functional preparations, the amino-terminal GFP label does not appear to affect the activity of our severing protein because the start time of severing and severing activity of our Drosophila GFP-Katanin-60 are similar to those previously reported for p60 from sea urchin (5). On the other hand, the GFP and His tags could enhance aggregation of the protein that we see increasing over time. We believe that the reduced activity of the protein at 200 nM (Fig. 3) is due to aggregation of the protein.
We have found, as expected, that the severing and depolymerization activities of GFP-Katanin-60 depend on the concentration of protein, the activity of the ATPase site, and the presence of nucleotide. Interestingly, we find that activity is inhibited at high concentrations of GFP-Katanin-60 (200 nM). This is likely due to aggregation of the protein. To test if 200 nM or 400 nM GFP-Katanin-60 aggregates during the assay, we performed centrifugation assays in the presence of all the ingredients in the chamber. We did not see GFP-Katanin-60 in the pellets at 200 nM or 400 nM, but we did see some GFP-Katanin-60 in the pellets of our stock of protein (Fig. S3). Thus, our protein could be aggregating in the stock solution. For this reason, we only use fresh protein that is never frozen and thawed.
We did not observe the katanin to bind continuously along the MT, even at high concentrations (Fig. 5, A and B). We believe that specific binding sites were saturated such as lattice defects or post-translationally modified dimers. Because we use pig brain tubulin, there is a mix of post-translational modifications. It is possible that post-translational modifications are regulating GFP-Katanin-60 activity in our assays. Indeed, this is supported by cellular studies that reveal katanin has a preference for acetylated MTs (22). Future in vitro work with tubulin of single modification type will be needed to definitively determine if post-translational modifications signal to GFP-Katanin-60.
The depolymerization activity of GFP-Katanin-60 was very interesting because our MTs were taxol-stabilized. Depolymerization was caused by GFP-Katanin-60 ATPase because we showed the lack of ATP or mutated GFP-Katanin-60 stopped depolymerization. We have previously reported that the ends of MTs remain blunt in the presence of Katanin-60 (11). This implies that GFP-Katanin-60 is not removing dimers by peeling back protofilaments, which is the mechanism employed by depolymerizing kinesin enzymes, such as Kinesin-13s. Furthermore, the depolymerization rate of GFP-Katanin-60 is much lower than that of MCAK, the first identified kinesin-13 (23). MCAK depolymerization has a peak rate of 65 nm per second at 10 nM, corresponding to 105 tubulin dimers removed per second per MT end (24). In comparison, our fastest depolymerization rate was 1.6 nm per second or ∼5 dimers per second per end. We believe that GFP-Katanin-60 is performing the same activity on ends during depolymerization as it does on the lattice during severing. Thus, GFP-Katanin-60 selectively removes individual tubulin dimers from the ends. Unlike severing that peaks at 100 nM, depolymerization peaks at 50 nM and decreases at 100 nM. We believe this is because the faster severing activity masks the slower depolymerization activity at high concentrations.
Why would GFP-Katanin-60 target MT ends? The dimers at the end of the filament should be more accessible and have a lower energy barrier for removal from the filament, because they have fewer longitudinal bonds (Fig. 8 A). In addition, katanin may be targeted to the ends, because that location is a defect in the lattice, and previous reports indicate that katanin localizes to lattice defects (14). Using polarity-marked MTs, we found that depolymerization was faster from the plus end than from the minus end. Although we currently do not have a full explanation for why GFP-Katanin-60 prefers the plus end, this end is more labile, and perhaps these dimers have the lowest energy barrier for removal. It is an interesting coincidence that we see enhanced activity at plus ends, and GFP-Katanin-60 localizes to MT plus ends in the cell (10,11) (Fig. 8 B) We are currently examining the other severing proteins to determine if they all have the same depolymerization ability and the same preference for the plus end.
Figure 8.
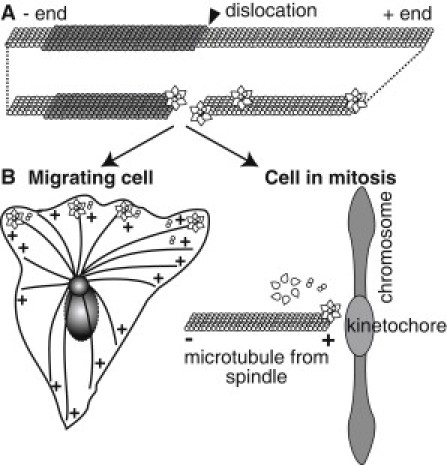
Illustration of proposed GFP-Katanin-60 activities in vitro and in the cellular context. (A) The GFP-Katanin-60 assembles as a hexamer in the presence of ATP, and associates with defects in the lattice, such as protofilament shifts and MT ends. Once GFP-Katanin-60 binds, it rapidly removes dimers from active sites at plus ends and at defects. (B) In the cell, the same katanin could regulate MT plus ends to alter length and dynamics at the leading edge of migrating cells or at the kinetochores in the mitotic spindle.
MTs made in vitro often have dislocations or shifts in protofilament number throughout the lattice (25). We created MTs with a bright GMPCPP-tubulin region and a dim GDP-tubulin region that should have a protofilament shift between the seed and the elongation segment. We found that the interface between these regions was indeed a target for GFP-Katanin-60 binding and severing (Fig. 4, A–C). Although we favor a mechanism of defect localization (Fig. 8 A), it is possible that GFP-Katanin-60 is recognizing the dimer nucleotide state and preferentially removing dimers located where the nucleotide state changes. This would be a novel localization scheme that we cannot rule out with our current data. We are working on future methods of localizing lattice defects using fluorescence microscopy.
Another interesting observation is that GFP-Katanin-60 subunits bind to and diffuse along MTs with a diffusion coefficient of 0.0033 μm2/s. This is a recurring theme in MT-associated proteins—especially enzymes—that appear to localize to their sites of activity or aid in transport and tethering by loosely binding to the MT (24,26,27). The diffusion coefficient for GFP-Katanin-60 is twice that of p150Glued dynactin (27) and approximately the same as another MT-associated AAA enzyme, cytoplasmic dynein (28). This interaction may be caused by the microtubule interacting and traffic domain, and this would be an interesting avenue for future study. We found that the diffusion depended on nucleotide state of the enzyme. For instance, we observe motionless, tight binding of GFP-Katanin-60 in the presence of AMPPNP. This implies that hydrolysis enables dissociation of katanin from MTs. The dependence on nucleotide state implies that some portion of the enzyme is altered or masked during the catalytic cycle. Future work on the binding and mobility of severing proteins should examine how this effect depends on ionic strength and on different portions of the katanin protein.
It has been shown that the nucleotide state can alter the oligomerization of katanin using bulk biochemical assays such as gel filtration and centrifugation (6). Photobleaching of single complexes can give a more accurate picture of the distribution of the number of monomers in the complex (Fig. 7 C). Indeed, we find that ATP induces oligomerization, but these are mostly tetramers, and not the expected hexamers. This is likely because the katanin is hydrolyzing ATP at a slow basal rate, and hydrolysis alters the nucleotide state and the oligomerization state. AMPPNP is a nonhydrolyzable analog of ATP, and we find that it enhances the number of hexamers in the population (Fig. 7 C). In addition, we observe some complexes with up to 11 GFP-Katanin-60s. Some AAA enzymes can form stacked rings to create larger complexes (29). Others are thought to be able to oligomerize into 7-mers or 8-mers and are not restricted to hexamers (29). We do not know if this can happen with GFP-Katanin-60. Coincidentally, complexes that are performing severing often dim and brighten as we watch (Fig. 2, Movie S1). It is possible that this intensity variation may be due to variable oligomerization during activity.
Katanin's severing and depolymerization activities may well both be important in cells. Stabilized MTs in cells can be stable by MT-associated proteins that bind to the sides or by stabilizing end caps. These end caps could be in the form of dynamic end-binding proteins, such as EB-1, or dynamic caps that hold MTs at the kinetochores, such as Ncd-80 (Fig. 8 B). Stable end caps could hold MT ends at nucleating centers, such as γ-tubulin and the recently discovered patronin (30). If severing enzymes can cut loose these tethers and further depolymerize the MT, these activities may be involved in regulation of MT arrays during mitosis (10), interphase (11), and oriented migration (11,31,32). Alternatively, severing enzymes may cut to allow the activity of depolymerizing kinesins in cells.
Conclusions
We have used single molecule imaging to directly observe the MT severing protein katanin as it binds to, diffuses along, and severs the MT. Overall, our data lead us to two main conclusions 1), katanin preferentially targets MT defects within the lattice and at the ends (Fig. 8 A), and 2), nucleotide state affects katanin oligomerization and affinity. Further biophysical and biochemical studies of katanin and the katanin-like proteins may reveal intrinsic differences between these proteins. Along with katanin, there are two additional severing protein types the activities of which have not been fully explored: spastin and fidgetin. These are particularly interesting, because mutations in the genes encoding these proteins are known to result in neuromuscular disorders and birth defects (33,34). Future studies on the biophysical activities of these proteins and their mutant forms will elucidate the molecular mechanisms of these proteins in cells and also how they dysfunction in disease.
Acknowledgments
We thank Lynn Liu and Carey Fagerstrom for help with this project, and Professors Patricia Wadsworth and Jeanne Hardy for the use of cell culture and microfluidizer facilities.
J.D.D-V. and M.M. were supported on a March of Dimes Basil O'Connor Starter Grant 5-FY09-46 to J.L.R. D.Z. was supported on a National Institutes of Health grant to D.S.
Supporting Material
References
- 1.Conde C., Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 2.Desai A., Mitchison T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 3.Valiron O., Caudron N., Job D. Microtubule dynamics. Cell. Mol. Life Sci. 2001;58:2069–2084. doi: 10.1007/PL00000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roll-Mecak A., McNally F.J. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman J.J., Mahr J., McNally F.J. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/s0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 6.Hartman J.J., Vale R.D. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 7.McNally F.J., Vale R.D. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 8.Murata T., Sonobe S., Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 9.Lohret T.A., McNally F.J., Quarmby L.M. A role for katanin-mediated axonemal severing during Chlamydomonas deflagellation. Mol. Biol. Cell. 1998;9:1195–1207. doi: 10.1091/mbc.9.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D., Rogers G.C., Sharp D.J. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D., Grode K.D., Sharp D.J. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 2011;13:361–369. doi: 10.1038/ncb2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad F.J., Yu W., Baas P.W. An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 1999;145:305–315. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu W., Qiang L., Baas P.W. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol. Biol. Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis L.J., Odde D.J., Gross S.P. The importance of lattice defects in katanin-mediated microtubule severing in vitro. Biophys. J. 2002;82:2916–2927. doi: 10.1016/S0006-3495(02)75632-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss S.A., Godwin G.P., Whitford W.G. Insect cell culture in serum-free media. In: Richardson C.D., editor. Baculovirus Expression Protocols. Humana Press; Totowa, NJ: 1995. pp. 79–95. [DOI] [PubMed] [Google Scholar]
- 16.Shelanski M.L., Gaskin F., Cantor C.R. Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. USA. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross J.L., Dixit R. Multiple color single molecule TIRF imaging and tracking of MAPs and motors. Methods Cell Biol. 2010;95:521–542. doi: 10.1016/S0091-679X(10)95026-7. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks A.G., Perlson E., Holzbaur E.L. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 2010;20:697–702. doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thévenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 20.Hyman A.A., Chrétien D., Wade R.H. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J. Cell Biol. 1995;128:117–125. doi: 10.1083/jcb.128.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnal I., Wade R.H. How does taxol stabilize microtubules? Curr. Biol. 1995;5:900–908. doi: 10.1016/s0960-9822(95)00180-1. [DOI] [PubMed] [Google Scholar]
- 22.Sudo H., Baas P.W. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J. Neurosci. 2010;30:7215–7226. doi: 10.1523/JNEUROSCI.0048-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wordeman L., Mitchison T.J. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helenius J., Brouhard G., Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 25.Chrétien D., Metoz F., Wade R.H. Lattice defects in microtubules: protofilament numbers vary within individual microtubules. J. Cell Biol. 1992;117:1031–1040. doi: 10.1083/jcb.117.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross J.L., Wallace K., Holzbaur E.L. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 27.Culver-Hanlon T.L., Lex S.A., King S.J. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat. Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z., Sheetz M.P. One-dimensional diffusion on microtubules of particles coated with cytoplasmic dynein and immunoglobulins. Cell Struct. Funct. 1999;24:373–383. doi: 10.1247/csf.24.373. [DOI] [PubMed] [Google Scholar]
- 29.Hanson P.I., Whiteheart S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 30.Goodwin S.S., Vale R.D. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkelstein E., Chang W., Bulinski J.C. Roles of microtubules, cell polarity and adhesion in electric-field-mediated motility of 3T3 fibroblasts. J. Cell Sci. 2004;117:1533–1545. doi: 10.1242/jcs.00986. [DOI] [PubMed] [Google Scholar]
- 32.Tran A.D., Marmo T.P., Bulinski J.C. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J. Cell Sci. 2007;120:1469–1479. doi: 10.1242/jcs.03431. [DOI] [PubMed] [Google Scholar]
- 33.Hazan J., Fonknechten N., Weissenbach J. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 34.Cox G.A., Mahaffey C.L., Frankel W.N. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat. Genet. 2000;26:198–202. doi: 10.1038/79923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


