Abstract
Motor proteins of the kinesin family move actively along microtubules to transport cargo within cells. How exactly a single motor proceeds on the 13 narrow lanes or protofilaments of a microtubule has not been visualized directly, and there persists controversy on the relative position of the two kinesin heads in different nucleotide states. We have succeeded in imaging Kinesin-1 dimers immobilized on microtubules with single-head resolution by atomic force microscopy. Moreover, we could catch glimpses of single Kinesin-1 dimers in their motion along microtubules with nanometer resolution. We find in our experiments that frequently both heads of one dimer are microtubule-bound at submicromolar ATP concentrations. Furthermore, we could unambiguously resolve that both heads bind to the same protofilament, instead of straddling two, and remain on this track during processive movement.
Introduction
Kinesin-1 motor proteins are involved in intracellular transport along microtubules (MTs) in most eukaryotic cells and have been widely studied as prototypical mechanoenzymes (1–3). Kinesin-1 motors are homodimers and can move for hundreds of nanometers parallel to the axis of a MT, taking an 8 nm step for each ATP molecule hydrolyzed (4–7). MTs (hollow cylinders of ∼25 nm diameter) in cells consist of 13 protofilaments which, in turn, are made of head-to-tail polymerized heterodimers of α/β tubulin (each ∼4 nm diameter). Each Kinesin-1 monomer consists of a (∼5 nm diameter) head domain, containing the MT binding site and the adenosine 5′-triphosphate (ATP) binding pocket, followed by an extended α-helical stalk enabling dimerization to the functional form (8). Kinesin head domains bind to the tubulin dimers and interact with both subunits (9). It is well established that the heads alternate in binding successive tubulin dimers in an asymmetric hand-over-hand mechanism during processive motion (10–12). Most evidence points to one unbound head when the motor waits for the next ATP molecule in the ATP waiting state (13–15). Experiments in which the position of a single fluorescently labeled head was followed (16,17) indicate, somewhat in contradiction, that both heads are 8 nm apart while waiting for ATP at low ATP concentration. At high ATP, when the ATP waiting state is not rate limiting, evidence quite definitely points to a two-head-bound state (15,17–19).
Models of motor motion have assumed tracking on either a single protofilament or on two neighboring protofilaments (20). Direct visualization of motion using light microscopy has been elusive because of the difficulty to achieve nanometer-spatial resolution for both heads at the same time. It has been suggested that kinesin binds with both heads along one protofilament from helical reconstructions of (static) electron microscopy images of tightly decorated MTs (21) and from fluorescence resonance energy transfer (FRET) experiments (15). Further evidence came from an analysis of the step size distribution in optical tweezers experiments which did not reveal an alternating step size (22). Moving on parallel protofilaments might entail alternating steps, but, given that the hand-over-hand mechanism is asymmetric, the finding does not strictly exclude a dual protofilament trajectory.
A technique with which one can perform, in principle, dynamic imaging at atomic resolution, is atomic force microscopy (AFM). AFM can achieve true atomic resolution on hard surfaces in ultrahigh vacuum (23), and even in liquids (24). The mechanical softness of proteins limits resolution (25) and makes samples susceptible to mechanical damage. Immobilized proteins immersed in solution can, however, still be imaged with nanometer resolution (26). Ando et al. have pioneered high-speed (sub-second) AFM imaging of biological samples in liquid and have visualized the dynamics of myosin motor proteins (27,28). We previously found that protofilaments and individual monomers of tubulin could be resolved by AFM if the tip force was limited to <100 pN (29). To visualize individual kinesin motors on MTs, we have operated the AFM in dynamic mode using small cantilevers with an average scan force of 20 pN while maintaining single-protein resolution at sub-minute frame rates.
Materials and Methods
Sample preparation
MTs, prepared as described in (29), were diluted to 7.5 μg/mL in PEM80 buffer (80 mM Pipes, pH 6.9, 1 mM EGTA, 2 mM MgCl2,) containing 8 μM taxol (all chemicals from Sigma-Aldrich, St. Louis, MO). Buffers were cleaned by ultracentrifugation for 1 h at 120,000 × g. To attach the negatively charged MTs to a substrate, clean glass coverslips were silanized with positively charged trimethoxysilylpropyl-diethylenetriamine (29). A 20 μL drop of MT solution was incubated on a coverslip for 10 min. To prevent nonspecific binding of kinesin to the surface, we washed the sample with 20 μg/mL casein in PEM80 including taxol. Before imaging, another 20 μL PEM80 including taxol were added to prewet the cantilever, resulting in a total sample volume of ∼40 μL.
Kinesin was then added at a 1:20 to 1:3 molar ratio to tubulin dimers in the presence of ∼15 μM nonhydrolyzable ATP analog adenosine 5′-(β,γ-imido) triphospate (AMP-PNP). For most single-kinesin motility assays the AMP-PNP was washed out by rinsing the sample 5 times with 40 μL PEM80 buffer including taxol and 0.5–2 μM ATP. Some experiments were performed without ever adding AMP-PNP (including Fig. 3, A–D), see also Fig. S3 in the Supporting Material for more details. All experiments were performed at room temperature.
Figure 3.
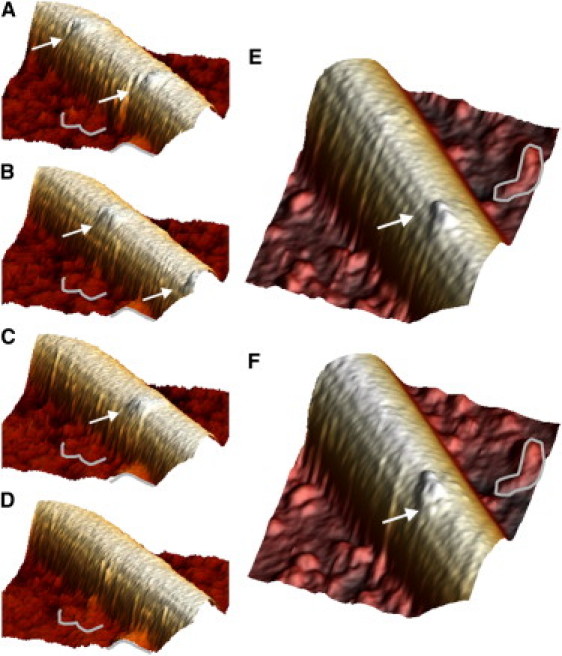
Single kinesin motors moving along a MT. (Movies S1 and S2). (A–D) Four frames (30 s acquisition time per frame) of a movie: This was the only movie in which we observed two kinesin motors moving; see Fig S2 for a top-view rendering. The average speed was 3 nm/s at 0.5 μM ATP. (A) Two kinesin dimers, indicated by arrows. (B) Both motors have proceeded (only the last head is still visible of the leading dimer). (C) The second kinesin has proceeded further. (D) The second kinesin has also disappeared. (E and F) Two frames from another sample show a displacement of 16 nm. The two kinesin heads are clearly visible. Fiduciary marks on the background were used to correct for drift. The position of the arrow is fixed in both frames with respect to the fiduciary marks.
We tested several dimeric kinesin constructs for imaging and found that truncated motors gave the best results, most likely because the long stalks in full-length motors tend to obscure the images of bound heads. Here, we used Neurospora crassa kinesin truncated at amino acid 433 (NcKin433) (a kind gift from Günther Wöhlke, Technical University Munich, Garching, Germany), a member of the Kinesin-1 family (30). This construct has been shown to form dimers and maintain processivity (31). Kinesin was prepared as described in (31). Although NcKin moves about three times faster than human Kinesin-1 at saturating levels of ATP, its Michaelis-Menten constant for ATP hydrolysis is ∼10× higher (Km ∼0.2 mM ATP) than that of human Kinesin-1 (32). This allowed us to run NcKin at low speed for imaging in μM concentrations of ATP.
Tip-sample dilation simulation
The model of the MT in Fig. 1 was created using an extrusion of the axial projection of the electron-density map of MTs (kindly provided by K. Downing, Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, California), (see also (33)). Kinesin heads were modeled as 4 nm spheres placed on top of protofilaments. The tip-sample dilation simulation was performed with a parabolic tip () (34). For the radius r we chose 15 nm as this best reproduced the measured 70 nm average width of the MTs.
Figure 1.
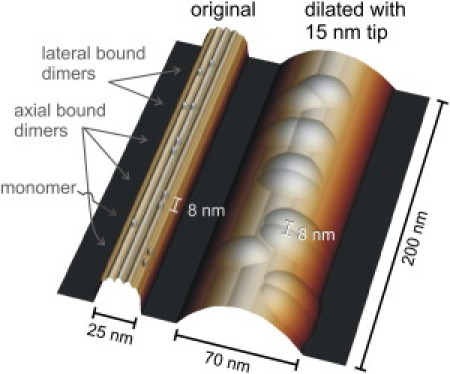
Simulation of the tip-sample dilation occurring in imaging a MT decorated with kinesin. The left image shows a MT with 25 nm diameter and 13 protofilaments. The MT is decorated with kinesin motors in different orientations, with either both heads bound to two parallel protofilaments, along one protofilament, or with just one single head bound. The right image shows the dilation caused by scanning with a 15 nm parabolic tip. The lateral dimensions get exaggerated by the tip-sample dilation, but the heads remain clearly visible and their axial spacing is not affected.
AFM
The AFM (Dulcinea, Nanotec, Madrid, Spain) was operated in buffer at room temperature with BL150 cantilevers (30 × 60 μm, 0.03 N/m, Olympus, Tokyo, Japan). The cantilevers were oscillated with an amplitude of ∼5 nm at ∼7 kHz. Fast scans were run only in one direction (trace). To minimize the tip-sample interaction, the tip was elevated from the surface during the retrace. The maximal scan speed was 4 μm/s. This allowed us to decrease the acquisition time to 3 s for a 156 nm × 156 nm area scanned with 64 scan lines. Most images were recorded at lower scan rates and with 128 scan lines to obtain higher resolution.
Within a recorded movie the slow scan direction was always in the same direction. The waiting time between sequential images was less then 0.5 s. The frame acquisition time for the movies we recorded in the presence of ATP varied between 10 and 100 s, with 77% between 10 and 33 s. The movies shown in the Supporting Material were recorded on 8 different samples, each time using fresh buffers and a new cantilever. On average one movie consisted of 19 frames (Table 1).
Table 1.
Motor statistics in AFM movies performed in the presence of ATP
| MTs imaged | 13 |
| Movies recorded | 23 |
| Frames recorded | 441 |
| Kinesin motors imaged | 1061 |
| Average number of motors per frame | 2.41 |
| Number of motors appearing | 63 |
| Average rate of motors appearing, per frame (λapp) | 0.168 |
| Number of motors disappearing | 103 |
| Average rate of motors disappearing, per frame (λdis) | 0.102 |
From the deflection versus time curves, we estimated that the tip was in contact with the sample for ∼10% of the cycle (35). At an estimated maximum force of 50 pN, this gives an average loading rate >1 μN/s. From the average deflection of the cantilever during scanning, we estimated an average scan force of 20 pN. To estimate the total contact time per frame between the tip and the kinesin dimer, we measured that a kinesin dimer occupied ∼400 nm2 in a scan (1.6% of a 156 nm × 156 nm = 24,000 nm2 scan). After multiplying with the 10% contact time per cycle, we concluded that the tip is in actual contact with the kinesin dimer during < 0.2% of the acquisition time per frame (i.e., ∼20 ms for a 10 s scan).
The AFM method we applied proved gentle enough to be able to repeatedly scan the same section of a MT 50–100 times before damage occurred. Lateral drift was typically <5 nm/min during scanning. Background features were used as fiduciary marks to correct for lateral drift in the recorded images by maximizing the cross correlation between frames. All image processing was performed with WSxM software (Nanotec) (36).
Results and Discussion
AFM can resolve single kinesin motors bound to the MT
Two main concerns arise when AFM is used to map the topography of fragile biological molecules. First, because the radius of the tip (∼15 nm) is typically comparable to the dimensions of the molecule, the image will be affected by the tip shape (34). Second, the tip mechanically touches the sample during scanning, which could result in deformation, displacement, or even destruction of the scanned object. To predict the effects of the tip-sample dilation, we created a 3D structure of a MT with kinesins bound in the different possible orientations (Fig. 1, left). We then simulated the scanning of the MT with a 15 nm radius parabolic tip. Fig. 1, right shows the resulting dilated image. The lateral dimensions are exaggerated by the tip. The apparent width of the microtubule of 70 nm is comparable to the widths we found in the experiments. Height and spacing of the two kinesin heads, in contrast, are faithfully reproduced because the z-direction experiences no dilation effect and the top of the MT is flat. This allows us to distinguish clearly the different binding scenarios for a double-headed kinesin molecule as described below. Because the AFM tip exerts force on the MT-motor complexes, it is crucial to explore how much this affects the sample. The effect is expected to depend on direction and magnitude as well as duration and rate of force application. From previous work we know that MTs get irreversibly deformed at forces exceeding 0.3 nN (33). In optical trapping experiments, forces around 10 pN, applied parallel to the MT axis, were necessary to pull kinesins off the MT at loading rates of 2–18 pN/s (37). We believe that this does not occur in our experiments. Although the forces applied in AFM are higher than 10 pN, they are directed toward the MT. Furthermore, we used three orders of magnitude higher loading rates (on the order of μN/s, see Materials and Methods). The probability of release is inversely proportional to the loading rate (38), and we therefore expect a substantially higher unbinding force for the kinesin-MT complex at the loading rates we used.
To test if we could actually image kinesin at sufficient resolution without removing it from the MT, we immobilized MTs on a silanized glass surface (29) and blocked the nonspecific binding of kinesin to the surface with casein proteins. We then added truncated N. crassa kinesin-1 (NcKin433), at a ratio 1 motor per 3–20 tubulin dimers in the presence of AMP-PNP. We tried two different scan modes, jumping and dynamic mode. In jumping mode, where for each pixel a high-speed force-distance curve is performed at a rate of ∼250 Hz and the cantilever deflection is used as a feedback signal (29,39), we could resolve individual motors on top of the MTs. In dynamic mode, where the tip is oscillated around 7 kHz and the amplitude is used as a feedback signal, we obtained comparable resolution, and we could scan motor-decorated MTs up to 10 times without removing or displacing any kinesins. This indicates that the tip force was not sufficient to tear off AMP-PNP-bound kinesins. We chose to use dynamic mode for the following experiments because this allowed us higher frame rates than jumping mode. Fig. S1 shows three MTs, each with a height of 25 nm. Protofilaments are well resolved as lines parallel to the MT axis. The kinesins are visible as blobs bound to the MTs, although the orientation of the heads cannot be resolved at this resolution.
Kinesin binds with both heads on a single protofilament in the presence of AMP-PNP
To determine the binding mode of individual kinesin motors on the MT, we increased the resolution of our scans by imaging only small portions (150 nm × 150 nm with ∼1 pixel/nm) of several MTs decorated sparsely with kinesin motors in the AMP-PNP state such that isolated kinesin dimers could be identified (Fig. 2 A). We imaged 60 isolated motors with both heads bound with 8 nm distance from head to head along single protofilaments. We also observed four single heads, but we did not see, in any image, a pair of heads that were bound on two neighboring protofilaments. Fig. 2 B shows an individual motor, with both heads bound to one protofilament. Binding with both heads in the presence of AMP-PNP is in agreement with the finding that the unbinding force of a kinesin dimer is doubled in the presence of AMP-PNP, compared to the nucleotide-free or the AMP-PNP + ADP state (37). The average height of individual motors, such as shown in Fig. 2, was 3.0 nm (n = 17), slightly less than the 4.5 nm we have measured in an earlier study on fully decorated MTs (40). The small discrepancy might be due to the absence of a lattice of neighboring motors, which is likely to screen individual motors from lateral forces applied by the AFM tip. This explanation is also supported by the reduced apparent width of the motors as compared to the simulation in Fig. 1. This effect is likely to occur when a slightly flexible object gets pushed aside when the tip impacts off center. In three images of two different pairs of heads we could even resolve ∼10 nm long extensions extending laterally from the heads that might represent the short stalk of ∼1 nm diameter expected for NcKin433 (Fig. 2 B, inset). The images show that single dimers of kinesin, bound in the AMP-PNP state, can be imaged with good contrast with our AFM method. We conclude that in this state both heads bind 8 nm apart to the same protofilament.
Figure 2.
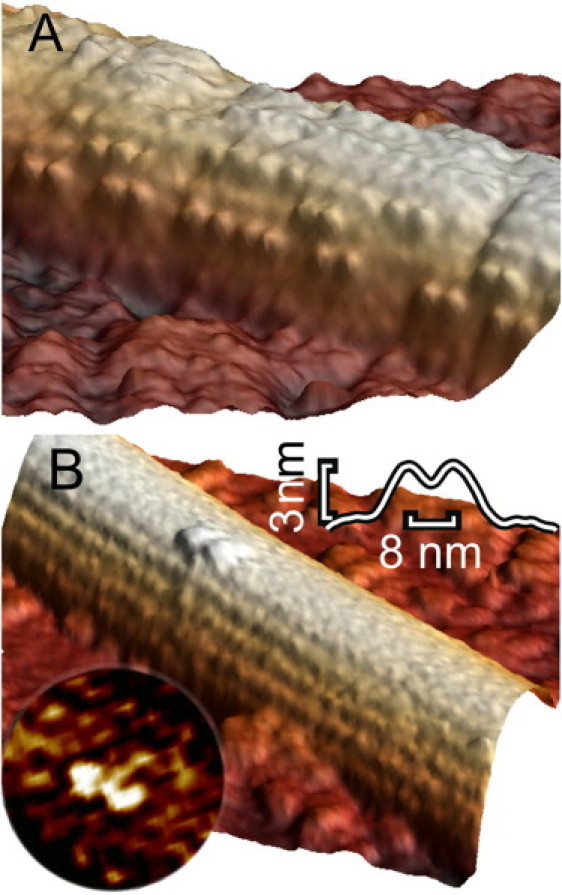
Individual kinesin motors immobilized on MTs in the presence of 15 μM AMP-PNP. Both images are 3D-rendered. (A) Intermediate kinesin concentration (<1 motor/tubulin dimer). Individual motors could be clearly distinguished. Heads always appeared in pairs, aligned parallel to the MT axis. (B) Low kinesin concentration (<1 motor/10 tubulin dimers). Isolated motors could be seen. Both heads were bound to the same protofilament. (Upper inset) Averaged axial profile of 17 kinesin molecules, interhead spacing: 8 nm, height: 3.0 nm. (Lower inset) Laplacian filtered image of another kinesin molecule showing clearly the two heads and a ∼10 nm long structure that might represent the stalk.
Kinesin moves with both heads along a single protofilament in the presence of ATP
The question remains if kinesin dimers also track along a single protofilament while they move. Using the distinctive capability of AFM to image a dynamic system in buffer, we prepared samples like the ones for Fig. 2, but now with 0.5–2 μM of ATP. For most samples (including the one shown in Fig. 3, E and F) we initially added low concentrations of AMP-PNP, which we washed out when adding ATP, to increase the chance to find single motors bound to a MT at the beginning of the experiment. The sequence of frames in Fig. 3 shows the motility of three motors in two samples (see also Movies S1, S2, and S3). In each frame both heads were bound to a single protofilament. In the subsequent frames the motors had moved unidirectionally, but stayed on the same protofilament. In total, we observed 20 isolated kinesin dimers that proceeded along a single protofilament; we found 14 1-step, four 2-step, and two 3-step events. The topographical profiles along the protofilament axis produced for all images bracketing the in total 28 individual steps are shown in Fig. S3. The profiles show that the two heads are in most cases well resolved. For an additional six kinesins we also observed a lateral displacement to a different protofilament.
We conclude that NcKin433 preferably tracks along a single protofilament, although side-stepping did occur with a probability of 6/(28 + 6) ≈ 0.18. Lateral displacements were also observed by 2D tracking of fluorescently labeled motors (41). In this work, Yildiz et al. (41) found, in remarkable agreement, that for human kinesin-1 the probability for side-stepping was 0.13. In the presence of AMP-PNP, we never found a motor bound with its heads on two neighboring protofilaments simultaneously. The speeds we found in the different experiments ranged from 2 to 9 nm/s, and were consistent with the velocity versus ATP-concentration curve of NcKin (32), with the variation explained mainly by the stochastic timing of steps at limiting concentrations of ATP (6).
It appears likely, at the given low motor concentration, that what we observed was in most cases one and the same motor in the successive video frames. Another possibility is, however, that one motor unbound and a new motor happened to land in a neighboring position. We controlled against this possibility by analyzing the statistics of the events. Binding of a second motor on the same protofilament within a few binding sites in a subsequent frame has a probability that we can estimate from the overall number of observed motor appearances and the total number of binding sites we could monitor. Binding of a third, fourth, etc., motor in the following frames, again on the same protofilament and in the same direction, becomes exponentially unlikely. We thus evaluated all 441 frames of the movies we acquired during the experiments in which we found motility. The average number of motors per frame was 2.4 (see Table 1 for details). We counted 63 motors that newly appeared in a frame due to landing or walking into the field of view (rate of appearance: λapp = 0.17 per frame), and 103 motors that disappeared in a frame due to detachment or motility (rate of disappearance λdis = 0.10 per frame per motor). Frames were 156 nm × 156 nm in size and typically showed diagonally oriented microtubules with a length of ∼180 nm. Typically, we could only resolve fine structure and bound motors on the top three protofilaments. With an average motor velocity on the order of several nm/s and frame-acquisition times of around 30 s, motors can perform several 8 nm steps from frame to frame. We thus counted as potential motility events all those events during which a motor was repeatedly imaged on the same protofilament, but away from the first appearance site. For multiple-step events, movement had to be consistently in the same direction. To estimate the expected number of false motility events from random rebinding, given the average rates we determined, we assume that appearance and disappearance of motors are independent of each other and follow Poisson statistics (see Supporting Material for details).
In Table 2 we compare the numbers of actually observed events with the average numbers expected and give the probability of obtaining at least the observed numbers from random rebinding. The small probabilities listed in Table 2 show that the random rebinding scenario is very unlikely as an explanation for our observations. Additional evidence comes from the fact that on all six MTs on which we observed multiple events, the direction of motion between successive frames was always the same. Thus, it is most likely that our recordings, including Movies S1, S2, and S3, largely show individual moving kinesin motors.
Table 2.
Probability analysis to control against false events
| Steps | Events expected | Events observed | Probability |
|---|---|---|---|
| 1 | 4.9 | 28 | 1∗10−12 |
| 2 | 1.2∗10−2 | 8 | 1∗10−20 |
| 3 | 3.1∗10−5 | 2 | 5∗10−10 |
Expected numbers of 1-, 2-, 3-step false motility events λ1,λ2,λ3, (second column), actually observed events (third column), and the cumulative probabilities to obtain the respective number of observations or more from a Poissonian process of detachment/attachment with the given average rates (fourth column). For the numbers of observed events we also counted each 2-step event as two 1-step events, and each 3-step event as two 2-step and three 1-step events.
Due to the relatively slow rate of recording movie frames, we, of course, may miss steps in the motor motion. Nevertheless, to obtain an idea of the fidelity with which the motors track the protofilaments over longer distances, we have plotted the axial height profiles of all motor dimers observed in a row of consecutive frames (Fig. S3), referenced to the respective first image. The displacements clearly fall on an 8 nm grid (Fig. 4), consistent with the tubulin dimer periodicity (7,42). Motors were thus predominantly captured at resting positions spaced multiples of 8 nm apart on the same protofilament. The maximal run length of 96 nm observed was limited by the scanned area and the frame rate rather than by the intrinsic run length of the motor. As an alternative and more rapid approach to observe motor dynamics, we performed consecutive line scans across the MT. The spatial resolution along one axis is sacrificed for increased temporal resolution. These recordings showed motors crossing the scan line, producing a characteristic transient height increase of ∼3 nm. Individual heads could not be distinguished (Fig. S4).
Figure 4.
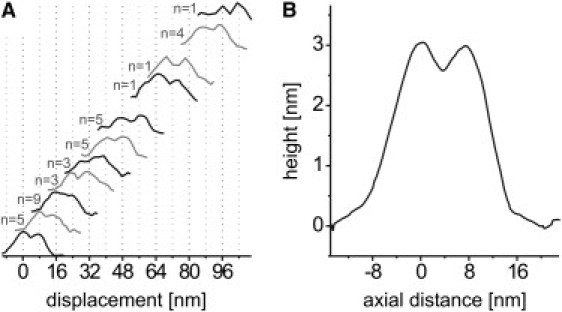
Aligned topographical profiles (parallel to the protofilament axis) of moving kinesin dimers, showing the 8 nm periodicity of the resting positions. (A) Displacements found for the 20 isolated moving kinesin dimers, recorded with different samples. Each event comprised 2 to 4 frames. Each curve represents a profile taken along the protofilament with kinesin bound. The position of the profiles from a single movie was referenced to fiduciary marks in the background to correct for drift. The x axis shows the axial displacement of the respective kinesin compared to the first or any of the previous frames. Most displacement distances were observed more than once, in which case the curves were averaged. Curves are plotted with a vertical offset for clarity. (B) Average of all profiles after subtracting the respective multiple of 8 nm. The heads, both bound to the MT and spaced 8 nm apart, can be clearly distinguished.
In conclusion, we found that NcKin433 kinesin, while moving in the presence of low ATP, could be observed with both heads bound on the same protofilament. This observation suggests that kinesin can spend considerable time with both heads bound to the MT firmly enough to withstand the forces of the AFM tip, presumably while waiting for the next ATP molecule to arrive.
The waiting state of kinesin has received much recent attention. Most studies agree that at high (mM) ATP concentrations, kinesin spends most of its time with both heads bound to the MT (15,17–19). At low (μM) ATP concentrations, the ATP waiting state becomes rate limiting in the mechanochemical cycle of kinesin. Most experiments performed at low ATP point to a one-head-bound ATP waiting state (14,15,43). Single-molecule FRET experiments from Verbrugge et al. (19) with sub-ms temporal resolution suggest that even at high ATP during the ATP waiting state only one head is bound and the other tethered.
Our findings seem to support a two-head-bound conformation for NcKin433 kinesin in the ATP waiting state. Such a conformation was also suggested by Yildiz et al. (16) who tracked fluorescently labeled heads. Later work from the same group (17) confirmed that in the waiting state both heads are 8 nm apart but do not necessarily have to be firmly bound to the MT. The latter was also proposed by Asenjo and Sosa (14), who showed one head to be mobile, but suggested it might be weakly bound to the MT. The studies reviewed above were all performed with cysteine-light mutants of human or Drosophila Kinesin-1. Our experiments, on the other hand, were performed with the fungal Kinesin-1 NcKin (31), and it may be possible that this particular type binds more firmly with both heads to the MT as compared to other members of the Kinesin-1 family. The interaction of both heads with the MT surface is at least strong enough to withstand the scanning by the AFM tip. We cannot exclude an increased mobility of one of the heads, which might have been suppressed by the tip-sample interaction, or that we select for motors with both heads bound to the MT. Furthermore, it should be noted that for most of our experiments AMP-PNP was used. Although we washed our sample and replaced the buffer with an ATP solution, one cannot exclude that traces of AMP-PNP may have in some cases affected the observed binding mode.
In conclusion, we have shown that the Kinesin-1 Nckin433 binds and moves along a single protofilament and can be found with both heads spaced 8 nm apart and bound to the MT while it waits for ATP in between steps. Our approach opens the way to study dynamics of kinesins and other proteins, exploring collective phenomena, traffic in crowded conditions or around obstacles, with even more details revealed by further increasing time resolution.
Acknowledgments
We thank Claudia Veigel, Erwin Peterman, Julio Gómez-Herrero, Lukas Kapitein, Marileen Dogterom, Sander Verbrugge, and Stefan Lakämper for comments and discussions, and Günther Wöhlke for providing the kinesin.
This work was initiated as part of the research program of the Stichting voor Fundamenteel Onderzoek der Materie, which is financially supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek. Further support came from the EU through a Marie Curie Intra-European Fellowship (to I.A.T.S), and the Deutsche Forschungsgemeinschaft through the Research Center for Molecular Physiology of the Brain/Excellence Cluster 171 (to C.F.S. and I.A.T.S).
Supporting Material
References
- 1.Alberts B., Johnson A., Walter P. Garland Science; New York: 2008. Molecular Biology of the Cell. [Google Scholar]
- 2.Vale R.D., Milligan R.A. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 3.Veigel C., Schmidt C.F. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat. Rev. Mol. Cell Biol. 2011;12:163–176. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- 4.Kuo S.C., Gelles J., Sheetz M.P. A model for kinesin movement from nanometer-level movements of kinesin and cytoplasmic dynein and force measurements. J. Cell Sci. Suppl. 1991;14:135–138. doi: 10.1242/jcs.1991.supplement_14.27. [DOI] [PubMed] [Google Scholar]
- 5.Ray S., Meyhöfer E., Howard J. Kinesin follows the microtubule's protofilament axis. J. Cell Biol. 1993;121:1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnitzer M.J., Block S.M. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 7.Svoboda K., Schmidt C.F., Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 8.Kozielski F., Sack S., Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 9.Marx A., Müller J., Mandelkow E. Interaction of kinesin motors, microtubules, and MAPs. J. Muscle Res. Cell Motil. 2006;27:125–137. doi: 10.1007/s10974-005-9051-4. [DOI] [PubMed] [Google Scholar]
- 10.Hua W., Chung J., Gelles J. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 2002;295:844–848. doi: 10.1126/science.1063089. [DOI] [PubMed] [Google Scholar]
- 11.Kaseda K., Higuchi H., Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 12.Asbury C.L., Fehr A.N., Block S.M. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso M.C., Drummond D.R., Cross R.A. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asenjo A.B., Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc. Natl. Acad. Sci. USA. 2009;106:5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori T., Vale R.D., Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 16.Yildiz A., Tomishige M., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 17.Toprak E., Yildiz A., Selvin P.R. Why kinesin is so processive. Proc. Natl. Acad. Sci. USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asenjo A.B., Krohn N., Sosa H. Configuration of the two kinesin motor domains during ATP hydrolysis. Nat. Struct. Biol. 2003;10:836–842. doi: 10.1038/nsb984. [DOI] [PubMed] [Google Scholar]
- 19.Verbrugge S., Lansky Z., Peterman E.J. Kinesin's step dissected with single-motor FRET. Proc. Natl. Acad. Sci. USA. 2009;106:17741–17746. doi: 10.1073/pnas.0905177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Block S.M., Svoboda K. Analysis of high resolution recordings of motor movement. Biophys. J. 1995;68:2305S–2395S. [PMC free article] [PubMed] [Google Scholar]
- 21.Skiniotis G., Surrey T., Hoenger A. Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehr A.N., Asbury C.L., Block S.M. Kinesin steps do not alternate in size. Biophys. J. 2008;94:L20–L22. doi: 10.1529/biophysj.107.126839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giessibl F.J. Atomic resolution of the silicon (111)-(7x7) surface by atomic force microscopy. Science. 1995;267:68–71. doi: 10.1126/science.267.5194.68. [DOI] [PubMed] [Google Scholar]
- 24.Ohnesorge F., Binnig G. True atomic resolution by atomic force microscopy through repulsive and attractive forces. Science. 1993;260:1451–1456. doi: 10.1126/science.260.5113.1451. [DOI] [PubMed] [Google Scholar]
- 25.Gittes F., Schmidt C.F. Thermal noise limitations on micromechanical experiments. Eur. Biophys. J. Biophys. Lett. 1998;27:75–81. [Google Scholar]
- 26.Weisenhorn A.L., Drake B., Gaub H.E. Immobilized proteins in buffer imaged at molecular resolution by atomic force microscopy. Biophys. J. 1990;58:1251–1258. doi: 10.1016/S0006-3495(90)82465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ando T., Kodera N., Toyoshima Y.Y. A high-speed atomic force microscope for studying biological macromolecules in action. ChemPhysChem. 2003;4:1196–1202. doi: 10.1002/cphc.200300795. [DOI] [PubMed] [Google Scholar]
- 28.Kodera N., Yamamoto D., Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 29.Schaap I.A.T., de Pablo P.J., Schmidt C.F. Resolving the molecular structure of microtubules under physiological conditions with scanning force microscopy. Eur. Biophys. J. 2004;33:462–467. doi: 10.1007/s00249-003-0386-8. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence C.J., Dawe R.K., Wordeman L. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallipolitou A., Deluca D., Woehlke G. Unusual properties of the fungal conventional kinesin neck domain from Neurospora crassa. EMBO J. 2001;20:6226–6235. doi: 10.1093/emboj/20.22.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinberg G., Schliwa M. Characterization of the biophysical and motility properties of kinesin from the fungus Neurospora crassa. J. Biol. Chem. 1996;271:7516–7521. doi: 10.1074/jbc.271.13.7516. [DOI] [PubMed] [Google Scholar]
- 33.Schaap I.A., Carrasco C., Schmidt C.F. Elastic response, buckling, and instability of microtubules under radial indentation. Biophys. J. 2006;91:1521–1531. doi: 10.1529/biophysj.105.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villarrubia J.S. Algorithms for scanned probe microscope image simulation, surface reconstruction, and tip estimation. J. Res. Natl. Inst. Stand. Technol. 1997;102:425–454. doi: 10.6028/jres.102.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Legleiter J., Park M., Kowalewski T. Scanning probe acceleration microscopy (SPAM) in fluids: mapping mechanical properties of surfaces at the nanoscale. Proc. Natl. Acad. Sci. USA. 2006;103:4813–4818. doi: 10.1073/pnas.0505628103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horcas I., Fernández R., Baro A.M. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007;78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi K., Ishiwata S. Nucleotide-dependent single- to double-headed binding of kinesin. Science. 2001;291:667–669. doi: 10.1126/science.291.5504.667. [DOI] [PubMed] [Google Scholar]
- 38.Merkel R., Nassoy P., Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 39.de Pablo P.J., Colchero J., Baro A.M. Jumping mode scanning force microscopy. Appl. Phys. Lett. 1998;73:3300–3302. [Google Scholar]
- 40.Schaap I.A., Hoffmann B., Schmidt C.F. Tau protein binding forms a 1 nm thick layer along protofilaments without affecting the radial elasticity of microtubules. J. Struct. Biol. 2007;158:282–292. doi: 10.1016/j.jsb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Yildiz A., Tomishige M., Vale R.D. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nogales E., Whittaker M., Downing K.H. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 43.Guydosh N.R., Block S.M. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


