Abstract
To identify interaction sites we measured the rotational motion of a spin label covalently bound to the side chain of a cysteine genetically incorporated into rabbit skeletal muscle tropomyosin (Tm) at positions 13, 36, 146, 160, 174, 190, 209, 230, 271, and 279. Upon the addition of F-actin, the mobility of all the spin labels, especially at position 13, 271, or 279, of Tm was inhibited significantly. Slow spin-label motion at the C-terminus (at the 230th and 271st residues) was observed upon addition of troponin. The binding of myosin-head S1 fragments without troponin immobilized Tm residues at 146, 160, 190, 209, 230, 271, and 279, suggesting that these residues are involved in a direct interaction between Tm and actin in its open state. As immobilization occurred at substoichiometric amounts of S1 binding to actin (a 1:7 molar ratio), the structural changes induced by S1 binding to one actin subunit must have propagated and influenced interaction sites over seven actin subunits.
Introduction
A skeletal muscle thin filament consists of actin, tropomyosin (Tm), and troponin (Tn). Conformational changes in Tn, induced by the binding of Ca2+, and in association with Tm, act as a switch that regulates striated muscle contraction (1–4). The crystal structures of actin, tropomyosin, and the Tn core domain have been resolved. However, it is not known at the atomic level how these molecules assemble to form the thin filament. As there is no high-resolution structure of the thin filament available, the molecular mechanism of this regulated process remains uncertain. A well-known steric blocking model, in which binding of Ca2+ to Tn triggers the movement of Tm to a different position on the actin filament (5–7), was based on x-ray diffraction and electron microscopy (8–15). In the three-state model, the movement of Tm occurs upon transition from the blocked to the closed state and proceeds further upon binding of myosin heads to actin, resulting in the open state (16–19). However, fluorescence resonance energy transfer (FRET) measurements did not detect any significant change in transfer efficiency accompanying a change in the position of Tm on the actin filament (20–24).
Tm is a two-chain α-helical coiled coil that binds along the length of the actin filament and regulates its function. There is no high-resolution structure available to indicate where Tm binds to the actin filament. The purpose of the research presented herein was to determine how the dynamics and position of Tm on the actin filament are affected in response to Ca2+ and myosin-head binding. We have used site-directed spin-labeling electron paramagnetic resonance (EPR) spectroscopy, a technique exceedingly sensitive to tertiary contacts or steric hindrance (25–27), to study the rotational motion of a spin label attached to side chains at positions e and f along the entire coiled coil of Tm. The results support previous mutation studies (28) in which Tm's flexible end-to-end complexes were fixed weakly on an actin surface. Ca2+ binding to the thin filament did not change, or only slightly changed, spin-label mobility in accordance with previous studies of the spin-label mobility of native cysteines 36 and 190 at the a position (29) and with FRET measurements (23–27). However, substantial changes in spin-label mobility were observed when the thin filament was partially saturated with S1. The interaction sites of Tm with Tn, actin, and myosin were different from each other. These results suggest that Tm adjusts its position in different ways, depending on whether the thin filament is activated by Ca2+ (in the closed state) or by myosin binding (in the open state).
Materials and Methods
Protein preparation
Skeletal muscle proteins were prepared from rabbit back and leg muscles according to conventional methods: Tm and Tn were prepared according to Ebashi et al. (30–32), actin was prepared according to Spudich and Watt (33), and myosin S1 was prepared as described by Weeds and Taylor (34). Single-cysteine Tm mutants (13, 146, 160, 174, 190, 209, 230, 271, or 279) were constructed by following the methods laid out by Kimura-Sakiyama et al. (35).
Spin labeling
Tm was labeled at its cysteine residues with spin labels 4-maleimido-2,2,6,6-tetramethyl-1-piperidinyloxy (MSL) or (1-oxyl-2,2,5,5-tetramethylpyrrolidin-3-yl)methyl methanethiosulfonate (MTSL) (Fig. 1). The reaction proceeded as follows: a solution of 20–30 μM Tm was incubated with a four-molar excess of MSL or MTSL for 2 h at 37°C, in 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 4 M guanidine hydrochloride, where all coiled double strands dissociate into single chains. The solution was then dialyzed against 100 mM KCl and 20 mM Tris-HCl (pH 7.5) to remove unreacted spin label and to renature the protein.
Figure 1.
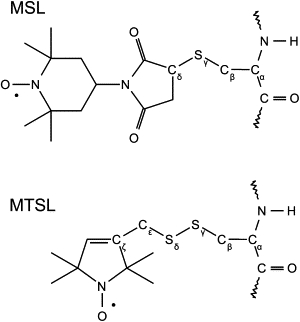
Structures of the spin labels used in this study, with cysteine disulfide bonds: MSL and MTSL.
The ratio of spin label to protein was determined by double integration of the EPR signal, which indicates the number of spins, whereas protein concentration was measured by the BCA protein assay (Thermo Fisher Scientific, Rockford, IL).
Reconstitution of thin filaments
Reconstitution of thin filaments with spin-labeled Tm was performed as described previously (31,32). G-actin was incubated in 20 mM Tris-HCl (pH 7.5), 100 mM KCl, and 4 mM MgCl2 for 30 min. Spin-labeled Tm and Tn were added to a G-actin solution (∼100 μM, 0.3–0.5 ml) and further incubated for 1 h in the presence of either 0.1 mM CaCl2 (+Ca2+ state) or 5 mM EGTA (-Ca2+ state). The molar ratio was set at 1:1:3 for the spin-labeled Tm and Tn complex and actin. Then, the mixture was centrifuged at 130,000 g for 30 min, and the pellet cosedimented with spin-labeled Tm was finally resuspended in a small volume (20–100 μl) of 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 4 mM MgCl2, and either 0.1 mM CaCl2 or 1 mM EGTA. The amounts of TnC, TnI, TnT, Tm, and actin were assessed by SDS-PAGE. The suspension, containing 20–40 μM of spin-labeled Tm, was used for EPR measurements. The same conditions were used for the preparation of Tm-actin. Before centrifugation, the Tm-actin or thin filaments reconstituted with spin label were mixed with various molar ratios of myosin S1 over actin.
ATPase assay
The ATPase activities of reconstituted systems comprised of S1, Tn, Tm, and F-actin were measured in the presence and absence of Ca2+. Measurements were performed at 25°C in 10 mM KCl, 4 mM MgCl2, 20 mM Tris-HCl (pH 7.6), and either 50 μM CaCl2 (+Ca2+ state) or 1 mM EGTA (–Ca2+ state). Protein concentrations were 4 μM F-actin, 0.7 μM Tm, 0.8 μM Tn, and 1 μM S1.
The ATPase reaction was coupled with a regeneration system, as described by Matsuo et al. (36), in which 2 mM phosphoenolpyruvic acid, 0.3 mM NADH, 38 U/ml pyruvate kinase, and 50 U/ml lactate dehydrogenase were mixed. The reaction was initiated by the addition of 2 mM ATP, and a time course of NADH absorption at 340 nm was followed for 10 min.
EPR spectroscopy and analysis
EPR spectroscopy was performed as described previously (31,37,38), using a Bruker ELEXSYS E500 spectrometer equipped with a dielectric resonator. Sample solutions (∼15 μl) were loaded into capillaries (inside diameter 1.0 mm) and inserted into the resonator, and EPR spectra were acquired using a 1-G field modulation amplitude at 100 kHz and 5 mW incident microwave power at 25°C. Overlays of spectra were made after experimental spectra were corrected for baseline and normalized to the same spins by double integration. The difference spectrum produced by computer subtraction between the spectra before and after complex formation was used for calculation of effective rotational correlation time. The effective rotational correlation time (τ) was determined using the equations for isotropic rotational motion as described previously (31,39), although the observed spectrum reflected anisotropic rotational motion. In the fast motion regime (τ = 10−9 s), the correlation time was calculated from the spectrum consisting of a single motion component using the equation of Redfield (40): τ = −1.22 × 10−9 ΔH0{(30.5/4)(V0/V+1)0.5 − (V0/V−1)0.5}, where ΔH0 is the peak-to-peak line width of the central line and Vm (m = −1, 0, +1) is the peak-to-peak height. The effective rotational correlation time in the slow motion regime (τ = 10−9 ∼ 2 × 10−7 s) was calculated from a spectrum consisting of single or multiple motion (fast and slow) component(s), according to an equation of Goldman et al. (41): τ = a(1 − Teff/Tmax)b, where a = 5.4 × 10−10 s and b = −1.36, Tmax (= 35 G) is the rigid limit for a particular spin, and 2 Teff is the width between the low-field and high-field absorption peaks. Flat peak and wide splitting were difficult to accurately estimate the values of Teff and rotational correlation time. Although difference spectra provided a more accurate estimation of Teff, we measured at least three spectra from different samples and determined average values of correlation time. Teff had at least 2−3% error for faint peaks, and correlation time then had ∼20–30% error for an average correlation time of <10−20 ns, respectively. However, when Teff was close to the rigid limit, the error exceeded 50% at an average correlation time of >50 ns.
Results and Discussion
The central aim of our research is to understand the topology and conformational dynamics of Tm on muscle thin filaments. We have used site-directed spin-labeling EPR, which allowed us to determine the side-chain mobility of the specific amino acid residue. We can use this technique to determine residual steric hindrance from the binding or contact of Tm with actin, Tn, or myosin, and to obtain a topological model of the thin filaments.
Spin labeling
The labeling of rabbit skeletal Tm with MSL or MTSL (Fig. 1) resulted in the covalent modification of Cys residues 13, 36, 146, 160, 174, 190, 209, 230, 271, and 279 in the Tm molecule. We have not examined the residues between 36 and 146 at periods 2 and 3, since the residues at periods 1, 5, and 7 of Tm were thought to be primarily important for actin binding from the mutational analysis of Tm (28). We have already reported that fluorescent labeling at these 10 residues does not impair calcium regulation of the actomyosin S1 ATPase rate (35). However, we still checked the calcium sensitivity of myosin S1 ATPase upon activation by thin filaments reconstituted from spin-labeled Tm. All activities were inhibited 80–90% in the absence of Ca2+, similar to the behavior of intact actomyosin ATPase, indicating that spin-labeling did not significantly affect the regulatory activity of Tm. As determined from the concentrations of protein and spin label the labeling efficiency of rabbit tropomyosin was >0.8 mol/mol for tropomyosin cysteine residues. Therefore, EPR spectra may detect that two spin labels at one region of the Tm dimer move differently.
Tm
The EPR spectra of MSL- and MTSL-Tm in solution are shown in Figs. 2 and 3, respectively, and the calculated rotational correlation times are shown later (see Figs. 7 and 8), respectively. Most spectra from MSLs or MTSLs bound to two canonical sites at the e or f position (residues 13, 146, 160, 174, 209, 230, 271, or 279) of the coiled-coil double strand showed a sharp absorption-line shape. The spectra of MTSL-labeled Tm residues had only a fast component. The spectra of MSL-labeled Tm residues had a fast component or a composite of poorly separated fast and weakly immobilized components at residues 146 and 279. The effective rotational correlation times were calculated from the inverse of the amplitude of absorption lines at lower and higher magnetic fields, V0/V+1 and V0/V−1, and the width, ΔH0, and then were 0.5–2.0 ns, although these values were somewhat ambiguous, because two motional components coexist. This indicated that these residues were solvent-exposed or only slightly immobilized by adjacent residues. However, there was large splitting of the slow component and relatively broad absorption lines in the spectra of MSL and MTSL labeled Tm residues 36 or 190 (τ = 2–7 ns), compared with other residues. It should be noted that these residues are located in the a position opposite the α-helical coil. Therefore, it seems that MSL or MTSL was immobilized at the a position in the coil. The same result has been reported previously (29). This is highly consistent with the structural configuration, wherein residues 13, 146, 160, 174, 209, 230, and 271 are all at the f position and residue 279 is at the e position in the α-helical coiled coil.
Figure 2.
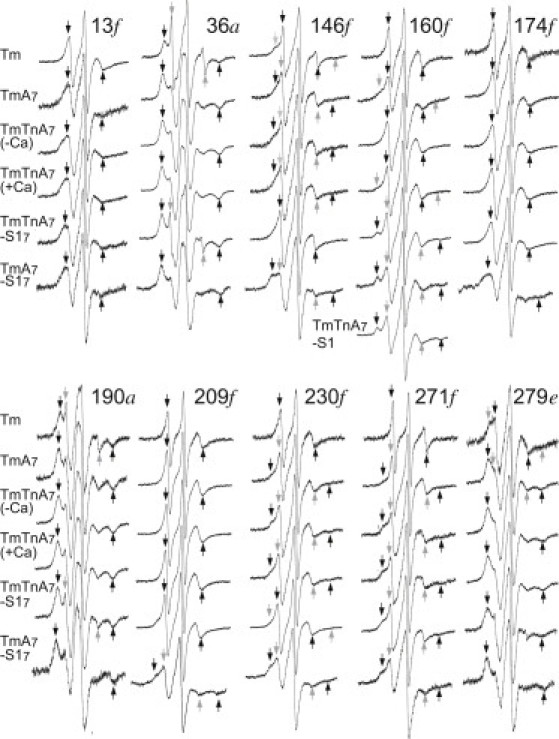
EPR spectra of MSL-labeled Tm in reconstituted filaments. Tm (first row), Tm-actin(A)7 (second row), Tm-Tn-actin(A)7 (–Ca2+) (third row), Tm-Tn-actin(A)7 (+Ca2+) (fourth row), Tm-Tn-actin(A)7-S17 (fifth row), and Tm-actin(A)7-S17 (sixth row). The scan width is 100 G. EPR spectra were taken from 13C, 36C, 146C, 160C, and 174C mutants (upper) and 190C, 209C, 230C, 271C, and 279C mutants (lower). Positions a, e, and f of the residues in the coiled coil are indicated at the end of the residue number. The dark and gray arrows indicate outer peaks for major (or significantly changed) and minor motional components, respectively. The molar ratio of S1/actin for Tm-Tn-actin(A)7-S17 and Tm-actin(A)7-S17 (fifth and sixth rows) was usually over unity. The spectrum from Tm labeled at the 160th residue shows a strongly immobilized component in both Tm-Tn-actin(A)7-S17 and Tm-actin(A)7-S17, comparable to the spectrum obtained by adding S1 to Tm-Tn-actin7 at molar ratio 1:7 (shown in the seventh row as Tm-Tn-A7-S1).
Figure 3.
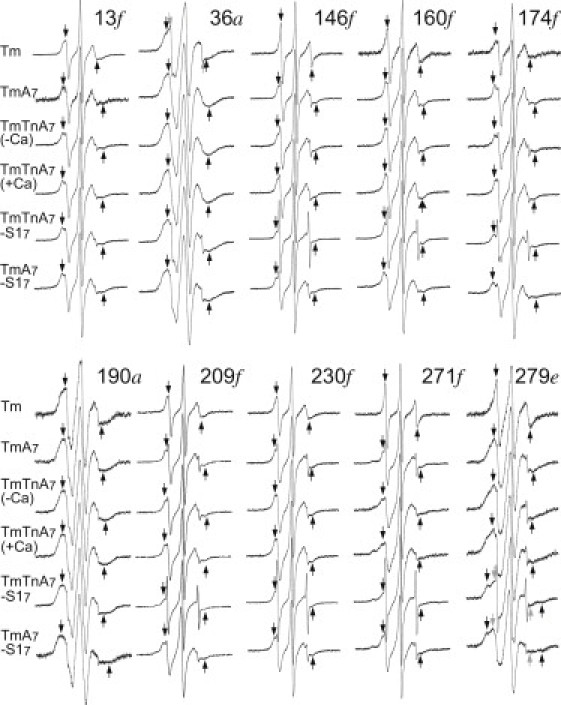
EPR spectra of MTSL-labeled Tm in reconstituted filaments. The conditions are as described for Fig. 2, except that Tm was labeled with MTSL.
Figure 7.
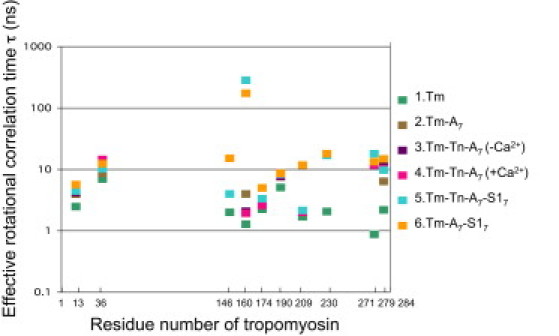
Profile of effective rotational correlation time as a function of the number of MSL-labeled Tm residues. Effective rotational correlation time was estimated from the spectra shown in Fig. 2 and the difference spectra obtained as shown in Figs. 4–6 and indicated by the average value obtained from at least three preparations (see Materials and Methods). When the spectrum consisted of fast and slow components, the major peak (Fig. 2, dark arrow) was used for estimation of effective rotational correlation time. On the right side of upper figure, each color of markers indicates the order of rows and the sample preparation shown in Fig. 2.
Figure 8.
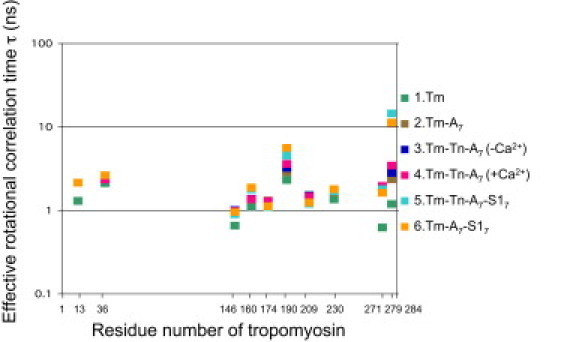
Profile of the effective rotational correlation time as a function of the number of MTSL-labeled Tm residues. The conditions are as described for Fig. 7, except that Tm was labeled with MTSL and the spectra shown in Fig. 3 were used for estimation of the effective rotational correlation time.
Tm-Actin7
Most spectra of MSL in the residues of Tm on the actin filament resulted in a reduction of the mobile component and an increase in the slow component, as compared with the corresponding components of Tm alone (Fig. 2). This may indicate that two side chains at one region of the Tm dimer move differently. The spectrum of MSL at the 13th residue of Tm showed marked changes when Tm was complexed with actin (Fig. 4 e). The binding of Tm to actin filaments resulted in increases in the inverse of the amplitude of absorption lines at lower and higher magnetic fields, V0/V+1 and V0/V−1, from 2.2 to 2.7 and from 9 to 16, respectively, that coincided with an increase in the width, ΔH0, of the central absorption line from 3.1 to 4.1 G. The calculated effective rotational correlation time of the MSL-labeled 13th residue of Tm increased from 2.4 ns to 4.6 ns when Tm was complexed with an actin filament (see Fig. 7). The slow component was enhanced and the mobile component almost disappeared in the spectrum of MSL-labeled residue 279 (Fig. 4, d). The mobile component was produced by subtracting the actin-bound Tm spectrum from the free Tm spectrum (Fig. 4 d). The average value of the correlation time of the slow component was 6.4 ns (see Fig. 7), which was calculated from the widths between the low-field and high-field absorption (2Teff = 58 G) in the actin-bound Tm spectra of three separate experiments, as described in Materials and Methods. A slow component also appeared in the spectra of MSL-labeled 146, 160, 230, and 271 residues of Tm. The slow spectral components were produced by subtracting 40–80% of the free Tm spectrum from the actin-bound Tm spectrum (Fig. 4). The average values of the correlation times of resultant slow components of MSL-labeled residues 146, 160, 230, and 271 of Tm were calculated from the widths between the low-field and high-field absorption and increased more than twofold compared with those of mobile components; from 2.0 to 4.0 ns, from 1.3 to 4.0 ns, from 2 to17 ns, and from 0.9 to 12 ns, respectively. On the other hand, when Tm was bound to actin, the absorption lines appeared with a wider outer-splitting in all the spectra from MTSL-labeled residues, especially at residues 13, 271, and 279 (Fig. 3). Fig. 4 g shows the overlaid spectra from MTSL at residue 271 of Tm in solution and on actin filament. The average values of the correlation times for residues 13, 271, and 279 increased from 1.3 to 2.2 ns, from 0.6 to 1.8 ns, and from 1.2 to 2.4 ns, respectively, indicating a significant immobilization of MTSL when Tm was bound to actin (see Fig. 8). These results support the previously postulated view (28) that a very short region in the C-terminus of Tm, where residues 271 and 279 are located, forms a head-to-tail overlap with the N-terminal region of the next Tm, where residue 13 is located. A middle portion of Tm also interacts weakly with actin, because all the residues are weakly immobilized (as compared to strongly immobilized residues 160 or 279 of Tm-Tn-actin7-S17).
Figure 4.
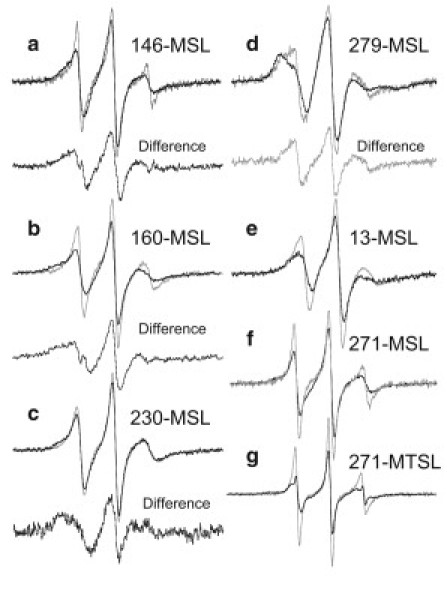
Overlay of EPR spectra from MSL and MTSL of Tm in solution (Tm) (gray line) and Tm on actin filament (Tm-actin7) (dark line). (a) Spectra from MSL at residue 146. Difference spectrum was produced by subtracting 50% of the Tm spectrum from Tm-actin7 spectrum. (b) Spectra from MSL at residue 160. The difference spectrum was produced by subtracting 40% of Tm spectrum from Tm-actin7 spectrum. (c) Spectra from MSL at residue 230. The difference spectrum was produced by subtracting 78% of Tm spectrum from Tm-actin7 spectrum. (d) Spectra from MSL at residue 279. The difference spectrum was produced by subtracting 60% of Tm-actin7 spectrum from Tm spectrum. (e) Spectra from MSL at residue 13. (f) Spectra from MSL at residue 271. (g) Spectra from MTSL at residue 271.
Tm-Tn-Actin7
In the reconstituted thin filament, the spectra of MSL-labeled residues 271 of Tm resulted in a reduction of the mobile component (with the effective rotational correlation time of ∼2 ns), and the slow component (with a correlation time of 10–20 ns, calculated from 2Teff = 64–65 G) was enhanced (Fig. 2 upper). Fig. 5, f and h, shows spectral differences in the overlaid spectra from MSL and MTSL, respectively, at residue 271 of Tm on actin filament without and with Tn. Difference spectra between those with and without Tn showed a clear slow component (Fig. 5 f). A small reduction of mobile and an enhancement of slow components were observed for MSL at residue 230 (Fig. 5 e). No obvious changes were found for MSL and MTSL at other residues, such as MSL-labeled residues 146 and 174 and MTSL-labeled residue 174 of Tm (Fig. 5). The enhancement of these slow components indicated that steric hindrances or contacts had been generated (indicating that the side chains of residues 230 and 271 in Tm are located close to actin or the Tn surface). A part of Tn (TnT) might actually bind only to limited residues of the C-terminal region of Tm, though it is established that Tn lies in the C-terminal half of Tm on the thin filament (1,7,35). It should be noted that the increase in the mobile component was found for MSL at residue 160 (Fig. 5 c). This suggests that Tm moves at residue 160 when Tn binds to Tm-actin7. Moreover, the MTSL at residue 230 did not sense the binding of Tn with Tm (Fig. 3 lower, and see Fig. 8). The orientation or structure of MTSL on the side chain may fail to sense the tertiary contact between Tn and Tm.
Figure 5.
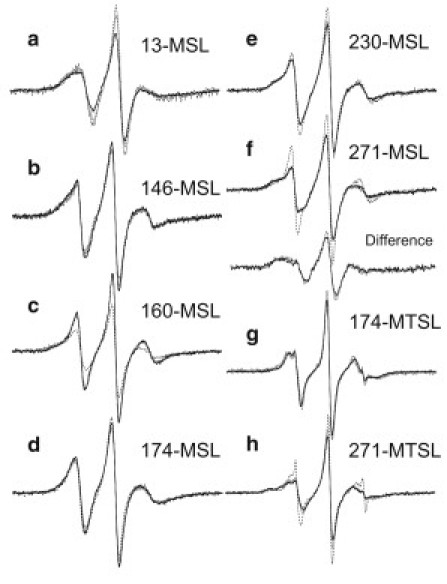
Overlay of spectra from MSL and MTSL of Tm on actin filament (Tm-actin7) (dashed line), and with Tn on actin filament (Tm-Tn-actin7 (-Ca) (gray line)), plus Ca2+ (Tm-Tn-actin7 (+Ca) (dark line)). (a–f) Spectra from MSL at residues 13 (a), 146 (b). 160 (c). 174 (d), 230 (e), and 271 (f). Difference spectra (gray or dark lines) were produced by subtracting 40% or 45% of the Tm-actin7 spectrum from Tm-Tn-actin7 (-Ca) or Tm-Tn-actin7 (+Ca) spectra, respectively. (g) Spectra from MTSL at residue 174. (h) Spectra from MTSL at residue 271.
It is very important to examine the effect of Ca2+, because Tm is thought to play an essential role in the steric blocking model. Evidence supporting this model comes from electron microscopy data that illustrate Tm shifts on the actin filament upon transition from the blocked state to the closed state when Ca2+ concentration is elevated. However, in the presence of Ca2+ (the closed state), none of the spectra from any of the MSL- or MTSL-labeled Tm residues changed markedly (Figs. 2 and 3, respectively). Fig. 5 shows overlaid spectra from MSL at residues 13, 146, 160, 174, 230, and 271 and from MTSL at residues 174 and 271 of Tm and Tn on actin filament with (dark line) and without (gray line) Ca2+. The orientation or structure of the spin label on the side chain may be accidentally unsuitable for sensing Ca2+-mediated changes in the tertiary contact between Tn and Tm. The straightforward interpretation of a lack of Ca2+ effect is that Tm does not shift azimuthally on the thin filament. Consistent with our findings, an azimuthal movement of Tm on the actin filament could not be detected by FRET from two fluorescent labels attached to Tm and actin (20–24). It is very difficult to imagine a case in which Tm would have two azimuthally different positions (corresponding to blocked and closed states in the steric blocking model) on actin without showing any significant change in the transfer efficiency or in the interacting residues between Tm and actin. If Tm localizes to the same average position, exhibiting thermally driven fluctuations with altered flexibilities and/or having different affinities for actin in the blocked and closed states, then FRET and EPR may not detect such Ca2+-mediated conformational changes. It seemed possible, however, that the overlaid spectra of MSL-labeled Tm in the thin filaments showed a very small decrease (at residue 13) and increases (at residues 146, 160, and 230) in the mobile components upon addition of Ca2+ in accordance with original or modified steric blocking model (Fig. 5, a–c and e).
Tm-Tn-Actin7-S17 and Tm-Actin7-S17
The most strongly immobilized component (Teff > 68 G and effective rotational correlation time τ = ∼200 ns) appeared in the spectrum of MSL-labeled Tm residue 160, together with a mobile component (rotational correlation time τ = ∼2 ns) when S1 was added to the reconstituted thin filaments (Fig. 2). In fact, the strongly immobilized spectrum was produced by subtracting the spectrum (80%) of the reconstituted thin filament (Tm-Tn-actin7) from the spectrum of S1-bound filament (Tm-Tn-actin7-S17) (see Fig. 6 b). We found similar immobilized and mobilized components of MSL at Tm residue 160 when S1 was added to the Tm-actin7 complex (Fig. 2 upper and Fig. 7). The strongly immobilized spectra generated by subtraction were identical within experimental error between Tm-Tn-actin7-S17 and Tm-actin7-S17 complexes (data not shown). It was suggested that one of two MSLs of the Tm coiled-coil dimer may be completely fixed because a small fraction (∼20%) of MSL was immobilized. MSL at residues 209 and 230 showed a reduction of the mobilized component when S1 was added to the Tm-Tn-actin7 complex (Fig. 2 lower), and the rotational correlation times of the immobilized component were estimated from the widths between the low and high field absorption to be 10–15 ns (see Fig. 7). Enhancement of the immobilized component of MSL at residues 146, 190, 209, 230, 271, and 279 was also found when S1 was added to the Tm-actin7 complex (Fig. 2). The effective rotational correlation time of immobilized component was estimated from the spectra produced by subtraction of actin-bound Tm spectrum from S1-bound Tm-actin7 spectrum (Fig. 6). The average values of the correlation times for MSL at residues 146, 209, 230, and 271 were 15.2, 11.8, 18.1, and 13.3 ns, respectively. The mobilized component was significantly enhanced in the spectra of MSL-labeled residues 146, 174, 209, and 230, as compared with the corresponding component of these residues in the Tm-Tn-actin7-S1 complex, when S1 was added to the Tm-actin7 complex (Fig. 2). If the surrounding structures of these residues change, or Tm itself moves at these residues when Tn binds to Tm in the absence of Ca2+, then S1 induces distinct mobility changes of these residues in the presence and absence of Tn. The immobilized component in spectra from MTSL at residues 160 and 279 was enhanced significantly by S1 binding to thin filament or Tm-actin filament (Fig. 3). The immobilized spectrum of MTSL at residue 279 was produced by subtracting the spectrum (80%) of Tm-actin from the spectrum of S1-bound Tm-actin (Fig. 6 f). The average values of the rotational correlation time of immobilized component increased from 1.3 to 1.8 ns and from 3 to 11 or 14 ns for MTSL at residues 160 and 279, respectively (see Fig. 8). These results suggest that conformational changes induced by S1 binding to actin occur on a relatively large portion (C-terminal half at least) of Tm. S1-mediated conformational changes of Tm were also found by an earlier EPR study, which showed a significant increase in the mobility of MSL at native residues 36 and 190 of Tm (29). The spectrum of MSL at residue 160 reached almost maximal perturbation at a 1:7 molar ratio of S1 to actin (Fig. 2 upper, row 7); the spectrum did not change further when the ratio increased to 1:1. It is therefore suggested that S1 binding to one actin subunit induces a conformational change of the thin filament that propagates over seven actin subunits or the entire length of Tm and causes residue 160 to contact the actin surface. Such S1-mediated cooperative change in Tm was recently reported from FRET measurements between Cys374 of actin and residues 190, 206, 247, or 282 of cardiac Tm (24).
Figure 6.
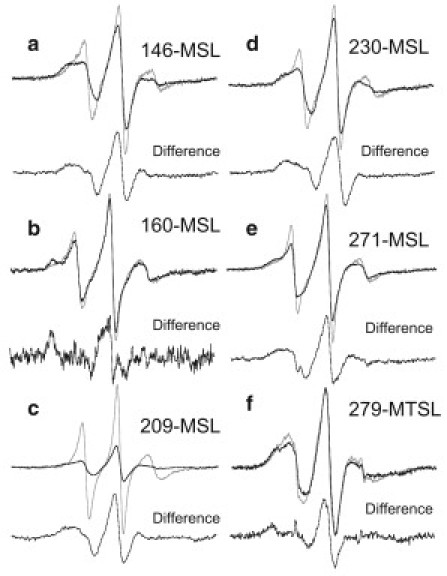
Overlay of spectra from MSL and MTSL of Tm on actin filament with Tn or on actin filament (Tm-Tn-actin7 or Tm-actin7, gray line), plus myosin S1 (Tm-Tn-actin7-S17 or Tm-actin7-S17, dark line). (a) Spectra from MSL at residue 146. The difference spectrum was produced by subtracting 30% of Tm-actin7 spectrum from Tm-actin7-S17. (b) Spectra from MSL at residue 160. The difference spectrum was produced by subtracting 80% of Tm-Tn-actin7 spectrum from Tm-Tn-actin7-S17 spectrum. (c) Spectra from MSL at residue 209. The difference spectrum was produced by subtracting 10% of Tm-actin7 spectrum from Tm-actin7-S17. (d) Spectra from MSL at residue 230. The difference spectrum was produced by subtracting 40% of Tm-actin7 spectrum from Tm-actin7-S17. (e) Spectra from MSL at residue 271. The difference spectrum was produced by subtracting 50% of Tm-Tn-actin7 spectrum from Tm-Tn- actin7-S17 spectrum. (f) Spectra from MTSL at residue 279. The difference spectrum was produced by subtracting 80% of Tm-actin7 spectrum from Tm-actin7-S17.
The data presented here are not enough to obtain a high-resolution model of Tm on actin or thin filament. To elucidate such a model, we will consolidate spectra from equivalent positions by the pattern of mobility changes with continuous scanning of single-Cys mutants along Tm. In fact, the spectra from160 and 209 MTSL, the 146, 174, and 230 MTSL, and the 174, 209, 230, and 271 MSL appear very similar (Figs. 2 and 3).
Conclusions
We have measured the mobility of spin labels (MSL or MTSL) bound to the residues of Tm under various conditions. In general, most EPR spectra appeared similar to each other and had weakly immobilized components whose relative populations were changed under various conditions. The mobility versus residue-number profiles from two spin labels were similar, although changes in MTSL were smaller than those in MSL (Figs. 7 and 8). The results suggest the possibility that the entire length of Tm is fixed weakly on the actin filament, and that residues 13, 271, and 279 of Tm are immobilized by end-to-end association of Tm. The Tm molecule is weakly bound to F-actin when assayed at substoichiometric levels (42). Stronger binding was seen under stoichiometric binding conditions that involve overlapping interactions of Tm at the N- and C-termini. If we assume that Tm has sevenfold periods that are quasiequivalent for actin binding (28), residues 13, 146, 174, 190, and 230 are consensus actin-binding residues of periods 1, 4, 5, 5, and 6, respectively. However, strong stereospecific binding is not involved, because spin-labeled side chain of these residues is not severely immobilized. A middle portion of Tm may be flexible and unstructured and bind weakly to actin filament. NMR (43), EPR (29), and x-ray crystallographic (44) studies showed that Tm is a destabilized coiled coil.
Second, slow spin-label motion of the C-terminus (the 230th and 271st residues) was observed upon further addition of Tn. We therefore conclude that residues 230 and 271 are the interaction sites of Tm with Tn. It is also suggested that the insensitivity of spin-label mobility resulted from no or small Ca2+-induced movements of Tm. This strongly supports previous studies that showed no Ca2+-induced changes in the mobility of spin label at residue 36 or 190 of Tm (29) and in the FRET between Tm and actin residues (20–24). This would support a speculative mechanism in which Tm moves only a few Ångstroms inward to bind at the actin surface or changes its thermally driven flexibility upon transition from the blocked to the closed state.
Third, the binding of myosin-head S1 subunits to actin filaments was accompanied by a significant inhibition in spin-label mobility at Tm residues 146, 160, 190, 230, 271, and 279. We can conclude that residues 146, 160, 190, 209, 230, 271, and 279 are involved in the direct interaction of Tm with actin in the open state induced by S1 binding. These results support those of previous spin-labeling (29) and FRET (24) studies, which showed conformational or positional change of Tm on the thin filament in response to S1 binding. The S1-mediated structural transition to the open state was shown to be highly cooperative and exhibited dependence similar to that of the S1-mediated changes in FRET from Tn-I or T (45,46) and cardiac Tm (24) to actin. We therefore suggest that the S1-mediated structural changes in the Tm-actin interface are a consequence of, and are directly coupled to, activation of the thin filament.
Acknowledgments
We thank Drs. M. Nakamura and S. Ueki for helpful discussions and advice.
This work was supported by Grants-in-Aid for the Special Coordination Funds (to T.A. and M.M.) and Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, and Core Research for Evolutional Science and Technology program from the Japan Science and Technology Cooperation (to T.A.).
Footnotes
Keisuke Ueda's present address is Institute for Protein Research, Osaka University, Yamadaoka 3-2, Suita, Osaka, Japan.
Chieko Kimura-Sakiyama's present address is Division of Biological Science, Graduate School of Science, Nagoya University, Furo, Nagoya, Japan.
Tomoki Aihara's present address is X-Ray Structural Analysis Research Team, RIKEN SPring-8 Center, RIKEN Harima Institute, Hyogo, Japan.
References
- 1.Ebashi S., Endo M., Otsuki I. Control of muscle contraction. Q. Rev. Biophys. 1969;2:351–384. doi: 10.1017/s0033583500001190. [DOI] [PubMed] [Google Scholar]
- 2.Gergely J. Molecular switches in troponin. Adv. Exp. Med. Biol. 1998;453:169–176. doi: 10.1007/978-1-4684-6039-1_20. [DOI] [PubMed] [Google Scholar]
- 3.Zot A.S., Potter J.D. Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annu. Rev. Biophys. Biophys. Chem. 1987;16:535–559. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]
- 4.Li M.X., Wang X., Sykes B.D. Structural based insights into the role of troponin in cardiac muscle pathophysiology. J. Muscle Res. Cell Motil. 2004;25:559–579. doi: 10.1007/s10974-004-5879-2. [DOI] [PubMed] [Google Scholar]
- 5.McLachlan A.D., Stewart M. The troponin binding region of tropomyosin. Evidence for a site near residues 197 to 127. J. Mol. Biol. 1976;106:1017–1022. doi: 10.1016/0022-2836(76)90349-1. [DOI] [PubMed] [Google Scholar]
- 6.Parry D.A.D. Movement of tropomyosin during regulation of vertebrate skeletal muscle: a simple physical model. Biochem. Biophys. Res. Commun. 1976;68:323–328. doi: 10.1016/0006-291x(76)91146-3. [DOI] [PubMed] [Google Scholar]
- 7.Phillips G.N., Jr., Fillers J.P., Cohen C. Tropomyosin crystal structure and muscle regulation. J. Mol. Biol. 1986;192:111–131. doi: 10.1016/0022-2836(86)90468-7. [DOI] [PubMed] [Google Scholar]
- 8.Haselgrove J. X-ray evidence for a conformational change in the actin containing filaments of vertebrate striated muscle. Cold Spring Harb. Symp. Quant. Biol. 1972;37:341–352. [Google Scholar]
- 9.Huxley H.E. Structural changes in the actin and myosin containing filaments during contraction. Cold Spring Harb. Symp. Quant. Biol. 1972;37:361–376. [Google Scholar]
- 10.Parry D.A., Squire J.M. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J. Mol. Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- 11.Wakabayashi T., Huxley H.E., Klug A. Three-dimensional image reconstruction of actin-tropomyosin complex and actin-tropomyosin-troponin T-troponin I complex. J. Mol. Biol. 1975;93:477–497. doi: 10.1016/0022-2836(75)90241-7. [DOI] [PubMed] [Google Scholar]
- 12.Xu C., Craig R., Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys. J. 1999;77:985–992. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig R., Lehman W. Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J. Mol. Biol. 2001;311:1027–1036. doi: 10.1006/jmbi.2001.4897. [DOI] [PubMed] [Google Scholar]
- 14.Narita A., Yasunaga T., Wakabayashi T. Ca2+-induced switching of troponin and tropomyosin on actin filaments as revealed by electron cryo-microscopy. J. Mol. Biol. 2001;308:241–261. doi: 10.1006/jmbi.2001.4598. [DOI] [PubMed] [Google Scholar]
- 15.Pirani A., Vinogradova M.V., Lehman W. An atomic model of the thin filament in the relaxed and Ca2+-activated states. J. Mol. Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 16.Lehrer S.S., Morris E.P. Dual effects of tropomyosin and troponin-tropomyosin on actomyosin subfragment 1 ATPase. J. Biol. Chem. 1982;257:8073–8080. [PubMed] [Google Scholar]
- 17.Lehrer S.S. The regulatory switch of the muscle thin filament: Ca2+ or myosin heads? J. Muscle Res. Cell Motil. 1994;15:232–236. doi: 10.1007/BF00123476. [DOI] [PubMed] [Google Scholar]
- 18.Maytum R., Lehrer S.S., Geeves M.A. Cooperativity and switching within the three-state model of muscle regulation. Biochemistry. 1999;38:1102–1110. doi: 10.1021/bi981603e. [DOI] [PubMed] [Google Scholar]
- 19.McKillop D.F., Geeves M.A. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao T., Lamkin M., Lehrer S.S. Excitation energy transfer studies of the proximity between tropomyosin and actin in reconstituted skeletal muscle thin filaments. Biochemistry. 1983;22:3059–3066. doi: 10.1021/bi00282a006. [DOI] [PubMed] [Google Scholar]
- 21.Miki M., Miura T., Maéda Y. Fluorescence resonance energy transfer between points on tropomyosin and actin in skeletal muscle thin filaments: does tropomyosin move? J. Biochem. 1998;123:1104–1111. doi: 10.1093/oxfordjournals.jbchem.a022049. [DOI] [PubMed] [Google Scholar]
- 22.Bacchiocchi C., Lehrer S.S. Ca2+-induced movement of tropomyosin in skeletal muscle thin filaments observed by multi-site FRET. Biophys. J. 2002;82:1524–1536. doi: 10.1016/S0006-3495(02)75505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki M., Hai H., Wakabayashi T. Fluorescence resonance energy transfer between points on actin and the C-terminal region of tropomyosin in skeletal muscle thin filaments. J. Biochem. 2004;136:39–47. doi: 10.1093/jb/mvh090. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Mao S., Marriott G. Tropomyosin dynamics in cardiac thin filaments: a multisite forster resonance energy transfer and anisotropy study. Biophys. J. 2008;94:4358–4369. doi: 10.1529/biophysj.107.121129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mchaourab H.S., Lietzow M.A., Hubbell W.L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 26.Hubbell W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 27.Kweon D.H., Kim C.S., Shin Y.K. Regulation of neuronal SNARE assembly by the membrane. Nat. Struct. Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 28.Hitchcock-DeGregori S.E., Singh A. What makes tropomyosin an actin binding protein? A perspective. J. Struct. Biol. 2010;170:319–324. doi: 10.1016/j.jsb.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szczesna D., Fajer P.G. The tropomyosin domain is flexible and disordered in reconstituted thin filaments. Biochemistry. 1995;34:3614–3620. doi: 10.1021/bi00011a016. [DOI] [PubMed] [Google Scholar]
- 30.Ebashi S., Kodama A., Ebashi F. Troponin. I. Preparation and physiological function. J. Biochem. 1968;64:465–477. doi: 10.1093/oxfordjournals.jbchem.a128918. [DOI] [PubMed] [Google Scholar]
- 31.Aihara T., Ueki S., Arata T. Calcium-dependent movement of troponin I between troponin C and actin as revealed by spin-labeling EPR. Biochem. Biophys. Res. Commun. 2006;340:462–468. doi: 10.1016/j.bbrc.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 32.Aihara T., Nakamura M., Arata T. Switch action of troponin on muscle thin filament as revealed by spin labeling and pulsed EPR. J. Biol. Chem. 2010;285:10671–10677. doi: 10.1074/jbc.M109.082925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 34.Weeds A.G., Taylor R.S. Separation of subfragment-1 isoenzymes from rabbit skeletal muscle myosin. Nature. 1975;257:54–56. doi: 10.1038/257054a0. [DOI] [PubMed] [Google Scholar]
- 35.Kimura-Sakiyama C., Ueno Y., Miki M. Fluorescence resonance energy transfer between residues on troponin and tropomyosin in the reconstituted thin filament: modeling the troponin-tropomyosin complex. J. Mol. Biol. 2008;376:80–91. doi: 10.1016/j.jmb.2007.10.078. [DOI] [PubMed] [Google Scholar]
- 36.Matsuo N., Nagata Y., Yamamoto T. Coupling of calcium transport with ATP hydrolysis in scallop sarcoplasmic reticulum. J. Biochem. 2002;131:375–381. doi: 10.1093/oxfordjournals.jbchem.a003112. [DOI] [PubMed] [Google Scholar]
- 37.Ueki S., Nakamura M., Arata T. Site-directed spin labeling electron paramagnetic resonance study of the calcium-induced structural transition in the N-domain of human cardiac troponin C complexed with troponin I. Biochemistry. 2005;44:411–416. doi: 10.1021/bi048110w. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura M., Ueki S., Arata T. Calcium structural transition of human cardiac troponin C in reconstituted muscle fibres as studied by site-directed spin labelling. J. Mol. Biol. 2005;348:127–137. doi: 10.1016/j.jmb.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Yamada M.D., Maruta S., Arata T. Conformational dynamics of loops L11 and L12 of kinesin as revealed by spin-labeling EPR. Biochem. Biophys. Res. Commun. 2007;364:620–626. doi: 10.1016/j.bbrc.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 40.Redfield A. The theory of relaxation processes. Adv. Magn. Reson. 1965;1:1. [Google Scholar]
- 41.Goldman S.A., Bruno G.V., Freed J.H. Estimating slow motional rotational correlation times for nitroxides by electron spin resonance. J. Phys. Chem. 1972;76:1858–1860. [Google Scholar]
- 42.Hill L.E., Mehegan J.P., Tobacman L.S. Analysis of troponin-tropomyosin binding to actin. Troponin does not promote interactions between tropomyosin molecules. J. Biol. Chem. 1992;267:16106–16113. [PubMed] [Google Scholar]
- 43.Singh A., Hitchcock-DeGregori S.E. Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry. 2003;42:14114–14121. doi: 10.1021/bi0348462. [DOI] [PubMed] [Google Scholar]
- 44.Minakata S., Maeda K., Maéda Y. Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys. J. 2008;95:710–719. doi: 10.1529/biophysj.107.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hai H., Sano K.-I., Miki M. Ca2+- and S1-induced conformational changes of reconstituted skeletal muscle thin filaments observed by fluorescence energy transfer spectroscopy: structural evidence for three States of thin filament. J. Biochem. 2002;131:407–418. doi: 10.1093/oxfordjournals.jbchem.a003116. [DOI] [PubMed] [Google Scholar]
- 46.Kimura C., Maeda K., Miki M. Ca2+- and S1-induced movement of troponin T on reconstituted skeletal muscle thin filaments observed by fluorescence energy transfer spectroscopy. J. Biochem. 2002;132:93–102. doi: 10.1093/oxfordjournals.jbchem.a003204. [DOI] [PubMed] [Google Scholar]


