Abstract
BACKGROUND/AIMS
Liver X receptor-α (LXRA) is a nuclear receptor that regulates genes important in cholesterol homeostasis and inflammation. Several single nucleotide polymorphisms (SNPs) in the LXRA gene have been previously associated with metabolic phenotypes (dyslipidemia and elevated BMI). Metabolic dysregulation is a major contributor to coronary disease; therefore, we assessed LXRA in INVEST-GENES, a genetic-substudy of a large clinical trial in patients with hypertension and coronary artery disease.
METHODS
Seven tag SNPs in the LXRA gene region (NR1H3) were selected for study: rs11039149, rs12221497, rs2279238, rs7120118, rs326213, rs11039159 and rs10501321. 1059 subjects were genotyped from the INVEST-GENES case-control set (Verapamil-SR or Atenolol based treatment strategies), comprised of 297 cases frequency matched approximately 2.5:1 with event-free controls by sex and race. The primary outcome was defined as first occurrence of all-cause death, nonfatal MI, or nonfatal stroke. Adjusted odds ratios (OR) were calculated using logistic regression.
RESULTS
Three of the seven SNPs were associated with significant effects on the primary outcome in Non-Blacks. The variant G allele of rs11039149 and the variant A allele of rs12221497 were associated with reduced risk of experiencing the primary outcome (OR: 0.62, CI: 0.45-0.85, P=0.003 and OR: 0.60, CI: 0.39-0.91, P=0.016 respectively). The rs2279238 genotype was associated with a significant increase in risk for the primary outcome (OR: 1.42, CI: 1.03-1.95, P=0.03). Furthermore, there was a significant genotype-treatment strategy interaction for carriers of the variant T allele of rs2279238 (OR for Verapamil SR strategy compared to Atenolol: 2.86, CI: 1.50-5.46, P=0.0015). Diplotype analyses revealed that the SNPs are rarely co-inherited and support the directionally opposite effects of the SNPs on the primary outcome.
CONCLUSIONS
LXRA genotypes were associated with variable risk for cardiovascular outcomes and pharmacogenetic effect in INVEST-GENES. These novel findings suggest LXRA is a genetic/pharmacogenetic target that should be further explored.
Keywords: LXRA, Nuclear Receptor, Pharmacogenetics, Polymorphisms, Cardiovascular Disease, Hypertension, Atenolol, Verapamil
Introduction
Cardiovascular disease (CVD) is the leading cause of mortality in industrialized countries, accounting for approximately 50% of all deaths [1]. Risk factors for the development of CVD are attributed to both genetic and environmental causes. Epidemiological studies have consistently identified decreased HDL cholesterol and increased LDL cholesterol and triglycerides as major contributors to atherogenesis [2, 3]. Furthermore, over the past decade it has been shown that inflammation plays a prominent role in CVD and its complications [4, 5]. Even with aggressive cardiovascular treatment strategies, residual risks remain[6]. Biomarkers and pathways central to lipids and inflammation are being investigated to understand what contributes to the residual risk for CVD [7, 8].
The liver X receptors (LXRs) are a class of nuclear receptors that regulate pathways central to lipid homeostasis, glucose homeostasis, vascular homeostasis and inflammation [9-12]. The two LXRs, LXRA and LXRB, are encoded by the NR1H3 and NR1H2 genes, respectively. While LXRB has ubiquitous tissue expression, LXRA is predominately expressed in the liver, heart, adrenal glands, adipose tissues, intestines and macrophages [10-12]. LXRs function as whole body cholesterol sensors and their natural ligands include cholesterol metabolites, such as oxysterols[10]. LXRs are partnered with retinoid X receptors (RXR) in the nucleus, where they are bound to LXR response elements in target genes[10]. Upon binding of endogenous (and presumably exogenous) ligands to the LXR/RXR complex, LXRs undergo conformational changes that induce the expression of a number of genes that protect cells from excess cholesterol[13]. Furthermore, ligands of LXRs decrease the activation of the renin-angiotensin-aldosterone system which adds to their appeal as contributors in cardiovascular homeostasis [14-16].
It has been observed in recent epidemiological studies that polymorphisms and haplotypes of NR1H3 are associated with body mass index (BMI)[17], cholesterol levels[18, 19], and at-risk metabolic phenotypes[20]. It is possible that NR1H3 gene variants may contribute to variable risk of CVD morbidity and mortality in patients with documented coronary disease that are otherwise treated for the risk factors (e.g., hypertension, dyslipidemia, diabetes). We therefore hypothesized that NR1H3 gene polymorphisms are associated with adverse cardiovascular outcomes in the INternational VErapamil SR Trandolapril STudy GENEticSubstudy (INVEST-GENES). INVEST was a clinical outcomes trial that compared beta blocker and calcium channel blocker based antihypertensive treatment strategies. The treatment strategies were equally efficacious in controlling blood pressure and preventing adverse events [21], thus CVD risk associated with NR1H3 gene variants could be determined in the absence of confounding by blood pressure response. Since the treatment strategies work via distinct pathways, we also performed exploratory pharmacogenetic analyses of the relationship of NR1H3 and risk based on treatment strategy in INVEST.
Methods
Study Participants
The design and findings from INVEST have been described in detail elsewhere [21, 22]. In brief, INVEST was a prospective, randomized, open-label, blinded-endpoint trial of beta blocker versus calcium channel blocker-based antihypertensive therapy in 22,576 patients with hypertension and documented coronary artery disease (CAD). The primary outcome was the first occurrence of all-cause mortality, nonfatal MI, or nonfatal stroke. The treatments strategies were equivalent in extent of blood pressure control at 24 months and the incidence of the primary outcome [21].
The INVEST-GENES Case-Control Population
Informed consent for genetic studies was obtained and DNA samples were collected from 5,979 patients participating in INVEST in the United States and Puerto Rico. Within this cohort, 297 patients experienced a primary outcome event after a mean (SD) of 2.8 (0.7) years. All events constituting the primary outcome were adjudicated by an endpoint committee blinded to treatment strategy. Controls were selected from patients not experiencing a primary outcome event during the follow-up period, and were selected at a ratio of approximately 2.5 controls per case. Controls were frequency-matched to cases by sex and race/ethnicity. We have previously demonstrated that, for INVEST, the nested case-control approach provides equivalent similar statistical power as genotyping the entire cohort [23-27]. Furthermore, power analyses conducted using the methods described by Skol et al. predict that sufficient power is obtained using this case control approach (Table S1)[28].
DNA Collection and Genotyping
Buccal tissue samples were obtained by mouthwash and genomic DNA was isolated as previously described[29]. We selected the following seven polymorphisms in the LXRA gene region (NR1H3 ±5kb) using a tag SNP approach in Blacks and Whites (CEU/CEPH and Yoruban) from the International HapMap project (Haploview software): rs11039149, rs12221497, rs2279238, rs7120118, rs326213, rs11039159 and rs10501321. The minor allele frequency was set at 10% and r2 was set at 80% for general SNP selection. Additionally, SNPs were selected for entry if putative functionality was predicted. Genotyping was performed using Taqman (Applied Biosystems, 7900, Foster City, CA). Genotype accuracy was verified by blind duplicate re-genotyping of 5% of the samples and concordance was 100%.
Statistics
Hardy-Weinberg equilibrium was tested within each racial/ethnic group using χ2 analysis. Linkage disequilibrium (LD; r2) between the SNPs in each racial/ethnic group was estimated using Haploview. Demographic and baseline clinical characteristics were compared by genotype using χ2 tests for categorical data, and analysis of variance or a nonparametric equivalent for continuous data. Logistic regression was performed to estimate odds ratios (OR) and 95% confidence intervals (95%CI) for carriers of the variant allele relative to wild-type homozyogtes. Associations with the primary and secondary outcomes (the individual components of the primary outcome) were tested using stepwise logistic regression models. The models adjusted for age, sex, and race/ethnicity, with additional covariates selected from the following baseline characteristics using the stepwise procedure (P<0.1 for entry, P<0.05 for retention): history of heart failure, previous myocardial infarction (MI), diabetes, stroke or transient ischemic attack (TIA), renal insufficiency, dyslipidemia, left ventricular hypertrophy, peripheral vascular disease, stable angina, unstable angina, arrhythmia, cancer, ever-smoking, BMI, and baseline systolic and diastolic blood pressures.
The threshold for screening SNPs in the overall population was set at P<0.2. Subsequently, SNPs with P<0.2 were analyzed for significance within each racial group. Previous studies have found four of the seven selected SNPs (rs12221497, rs2279238, rs7120118, and rs11039149) to be associated with conditions where the sequelae are CVD morbidity and mortality [17, 18, 20, 30]. Therefore, P<0.05 was considered appropriate for confirmation of the previous findings for a role in CVD for these SNPs. We selected an adjusted p-value of 0.007 as significant for the remaining SNPs that would produce novel findings (p=0.05/7; rs326213, rs11039159 and rs10501321). Stratified logistic regression was used to estimate genotype risks in each treatment stratum. Furthermore, analyses were performed using racial stratification and ancestral informative markers (AIMs) to control for population structure. All statistical analyses were performed using SPSS 17.0 (SPSS, Chicago, IL).
Results
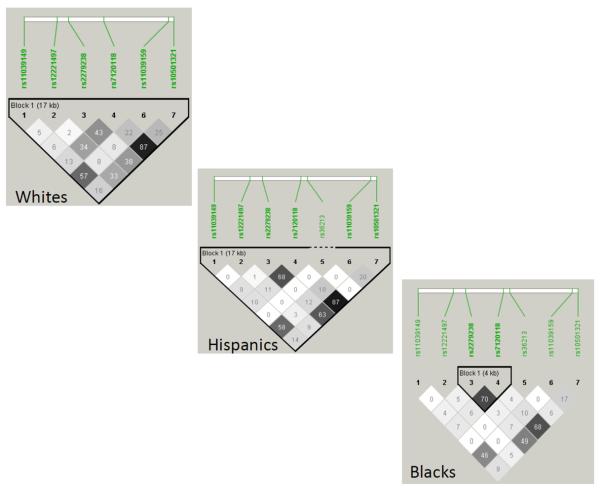
Pertinent baseline characteristics are listed in Table 1. Because age, systolic blood pressure, BMI, and histories of myocardial infarction, stroke or TIA, heart failure, peripheral vascular disease, diabetes, and renal impairment differed comparing the case and control groups, they were included as covariates in the analyses. Successful genotyping for all loci was achieved in over 90% of the individuals in the data set. Minor allele frequencies are listed in Table 2. The linkage disequilibrium plots for the selected tag SNPs in each population are shown in Figure 1.
Table 1.
Demographics of INVEST-GENES Case Control Data Set
| Characteristic (N, % unless otherwise noted) |
Cases (N=297) |
Controls (N=762) |
|---|---|---|
| Age, mean (SD), years* | 71.4 (9.8) | 70.2 (9.3) |
| Women | 148 (49.8) | 387 (50.8) |
| Race/ethnicity | ||
| White | 185 (62.3) | 462 (60.6) |
| Hispanic | 70 (23.6) | 197 (25.9) |
| Black | 41 (13.8) | 100 (13.1) |
| Other/multiracial | 1 (0.3) | 3 (0.4) |
| BP, mean (SD), mm HG | ||
| Systolic* | 150.6 (18.7) | 147.4 (18.9) |
| Diastolic | 83.3 (11) | 83.4 (11.1) |
| BMI, mean (SD), kg/m2* | 27.5 (4.8) | 28.9 (5.5) |
| Medical history | ||
| Prior myocardial infarction* | 107 (36) | 225 (29.5) |
| Stable angina | 181 (60.9) | 476 (62.5) |
| Prior stroke/TIA* | 44 (14.8) | 67 (8.8) |
| Left Ventricular Hypertrophy | 56 (18.9) | 132 (17.3) |
| Heart failure (class I-III)* | 31 (10.4) | 27 (3.5) |
| Peripheral vascular disease* | 50 (16.8) | 86 (11.3) |
| Smoking Ever | 154 (51.9) | 349 (45.8) |
| Diabetes*@ | 90 (30.3) | 170 (22.3) |
| Hypercholesterolemia@ | 184 (62) | 475 (62.3) |
| Renal Impairment* | 15 (5.1) | 17 (2.2) |
| Cancer | 25 (8.4) | 44 (5.8) |
Abbreviations: BP, blood pressure, BMI, body mass index, TIA, transient ischemic attack.
Indicates p<0.05.
Diabetes/Hypercholesterolemia defined as history or currently taking anti-diabetic or lipid-lowering medications
Table 2.
LXRA gene (NR1H3) polymorphism frequencies in INVEST-GENES Case Control Data Set
| Polymorphism | Position | Genotype Success Rate |
Minor Allele Frequency | |||
|---|---|---|---|---|---|---|
| Overall | Whites | Hispanics | Blacks | |||
| rs11039149 (A>G) | 5′ UTR/Promoter | 97% | 0.23 | 0.28 | 0.20 | 0.06 |
| rs12221497 (G>A) | 5′ UTR /Promoter | 94% | 0.11 | 0.13 | 0.07 | 0.07 |
| rs2279238 (C>T) | Exon 5 | 97% | 0.22 | 0.15 | 0.26 | 0.42 |
| rs7120118 (C>T) | Intron 7 | 90% | 0.33 | 0.28 | 0.35 | 0.52 |
| rs326213 (C>A) | Intron 7 | 93% | 0.01 | 0.01 | 0.02 | 0.05 |
| rs11039159 (G>T) | 3′ UTR | 91% | 0.33 | 0.39 | 0.29 | 0.13 |
| rs10501321 (G>T) | 3′ UTR | 92% | 0.36 | 0.30 | 0.37 | 0.59 |
Figure 1. Linkage disequlibrium plots for the seven NR1H3 tag SNPs.
The plots are shown with R2 values for the three major populations of INVEST-GENES (Whites, Hispanics, and Blacks).
Associations of LXRA Genotypes and Primary Outcomes in the Overall INVEST-GENES Case Control Group
Results for the SNP associations with primary outcome in the entire case-control group are shown in Table 3. Three of the four SNPs selected for validation met the criteria for further analysis in the group stratified by race (P<0.2; rs11039149, rs12221497 and rs2279238). However, none of the three SNPs selected to provide novel CVD risk data (rs326213, rs11039159, and rs10501321) met this criterion (P<0.2).
Table 3.
Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for primary outcomes by carrier status
| Adjusted OR (95% CI) | Adjusted p-value | |
|---|---|---|
| rs11039149 (Variant G allele carriers vs. Wild Type)* | 0.64 (0.47-0.87) | 0.004 |
| rs12221497 (Variant A allele carriers vs. Wild Type)* | 0.69 (0.47-1.02) | 0.064 |
| rs2279238 (Variant T allele carriers vs. Wild Type)* | 1.34 (0.99-1.81) | 0.060 |
| rs7120118 (Variant T allele carriers vs. Wild Type) | 1.16 (0.85-1.58) | 0.351 |
| rs326213 (Variant A allele carriers vs. Wild Type) | 1.21 (0.41-3.58) | 0.737 |
| rs11039159 (Variant T allele carriers vs. Wild Type) | 0.82 (0.60-1.11) | 0.200 |
| rs10501321 (Variant T allele carriers vs. Wild Type) | 1.19 (0.87-1.62) | 0.271 |
Odds Ratios (OR) from logistic regression adjusted for: age, sex, race/ethnicity, BMI, INVEST treatment strategy, history of CHF, history of MI and history of diabetes at baseline
Indicates p meets pre-specified significance level for subsequent testing in racial strata
Associations of LXRA Genotypes and Primary/Secondary Outcomes Stratified by Race/Ethnicity
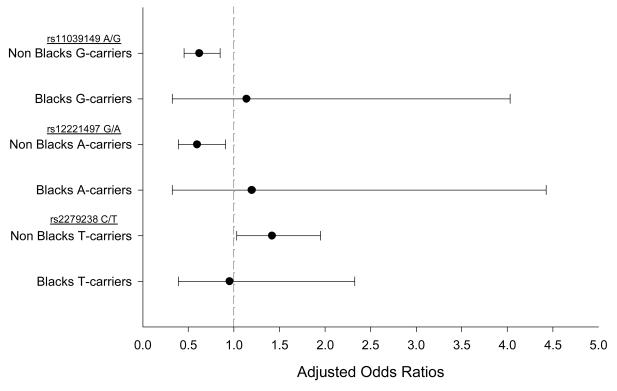
The results for the effects of the three SNPs that met the criteria to be tested in the racially stratified subsets are represented in Table 4. Whites and Hispanics were analyzed together as Non-Blacks because they shared similar point estimates and LD block structures (Figure 1). Each of the three SNPs (rs11039149, rs12221497 and rs2279238) was significantly associated with the primary outcome in Non-Blacks (Figure 2). The variant G allele of rs11039149 was associated with a significant reduction of risk of the primary outcome (OR: 0.62, 95% CI: 0.45-0.85, P=0.003). Furthermore, Non-Black carriers of the variant A allele of rs12221497 also experienced a reduction in risk for the primary outcome (OR: 0.60, 95% CI: 0.39-0.91, P=0.016). However, among Non-Blacks the variant T allele of rs2279238 was associated with a 1.42-fold increase in risk of having the primary outcome (95% CI: 1.03-1.95, P=0.031) (Figure 2).
Table 4.
Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the primary/secondary outcomes within Non-Black variant allele carriers
| Main Effect | rs11039149 (G carriers) | rs12221497 (A Carriers) | rs2279238 (T Carriers) |
|---|---|---|---|
| Primary Outcome | |||
| INVEST-GENES case control | 0.64 (0.47-0.87) | 0.69 (0.47-1.02) | 1.34(0.99-1.81) |
| Non-Blacks (n=914) | 0.62 (0.45-0.85) | 0.60 (0.39-0.91) | 1.41 (1.03-1.95) |
| Atenolol Based Strategy (n=459) | 0.63 (0.39-0.99) | 0.81 (0.43-1.51) | 0.85 (0.53-1.36) |
| Verapamil Based Strategy (n=455) | 0.64 (0.41-0.99) | 0.46 (0.26-0.84) | 2.13 (1.37-3.32) |
| Secondary Outcomes | |||
| All Cause Death | 0.85(0.53-1.31) | 0.72 (0.39-1.34) | 0.95 (0.61-1.48) |
| Non Fatal Stroke | 0.52 (0.30-0.90) | 0.63 (0.32-1.25) | 1.61 (0.96-2.68) |
| Non Fatal MI | 0.74 (0.44-1.24) | 0.71 (0.36-1.39) | 1.25 (0.74-2.13) |
Odds Ratios (OR) from logistic regression adjusted for: age, sex, race/ethnicity, BMI, INVEST treatment strategy, history of CHF, history of MI and history of diabetes at baseline
Figure 2. Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the primary outcome within variant allele carriers stratified by racial groups.
Reference is the wild type allele for the respective SNPs. Non-Blacks indicates the White and Hispanic subjects that were grouped together based on the similar haplotype structures in Figure 1. Primary outcomes are a composite of the first occurrence of all-cause death, nonfatal MI or nonfatal stroke. Odds Ratios (OR) from logistic regression adjusted for: age, sex, race/ethnicity, BMI, INVEST treatment strategy, history of CHF, history of MI and history of diabetes at baseline.
In the analysis of secondary outcomes in Non-Blacks, the findings for variant carriers of rs11039149, rs12221497 and rs2279238 were similar in directional trends to the effects observed on the primary outcome, although strongest for nonfatal stroke (Table 4).
Exploratory Pharmacogenetic Associations with LXRA Genotypes in Non-Blacks
The protective effects associated with the variant G allele of rs11039149 were observed similarly in participants randomized to both treatment strategies (Table 4). However, there were variable risks for variant allele carriers of rs12221497 and rs2279238 based on treatment strategy assignment. The observed protective effects associated with the variant A allele of rs12221497 were only significant in patients stratified to the verapamil-SR strategy (OR: 0.46, 95% CI: 0.26-0.84, P=0.011). Similarly, the observed risk associated with the variant T allele of rs2279238 was associated with participants randomized to the verapamil-SR strategy (OR: 2.13, 95% CI: 1.37-3.32, P=0.001) (Table 4). Of note, the protective association of the variant A allele of rs12221497 and the excess risk associated with the variant T allele of rs2279238 were not present in participants treated in the atenolol strategy (Table 4). Furthermore, a gene-treatment strategy interaction was observed for the variant carriers of the rs2279238 polymorphism being treated with verapamil-SR strategy compared to atenolol strategy (OR: 2.86, 95% CI: 1.50-5.46, P=0.0015).
LXRA Diplotypes and Primary Outcomes by Treatment Strategy in Non-Blacks
PHASE software was used to form diplotypes for SNPs rs11039149 (A>G), rs12221497 (G>A) and rs2279238 (C>T) for risk of primary outcome. The frequencies of the diplotypes are listed in Supplementary Table S2. There were nine diplotypes (D1-D9) with frequencies above two percent in Non-Blacks. D1, which contained the wild type alleles for each of the three SNPs, was selected as the reference for the logistic regression analyses. Diplotype analyses supported the individual SNP associations, as the variant alleles associated with directionally opposite effects on risk were not predicted to be commonly co-inherited (Supplemental figure S1).
Discussion
In this study, we used a tag SNP approach to select seven SNPs in the NR1H3 gene region to test whether risk of CVD is associated with LXRA in the INVEST-GENES case-control study. We observed variable CVD outcomes based on NR1H3 genotypes. Specifically, two SNPs located in the 5′ UTR/promoter region of NR1H3 (rs11039149 and rs12221497) were associated with decreased risk of experiencing the primary outcome, while another SNP located in exon 5 (rs2279238) was associated with an increased risk of experiencing the primary outcome in Non-Blacks. Additional analyses revealed similar directional trends in risk across all of the individual outcomes with the presence of the variant alleles for the respective SNPs in Non-Blacks. Interestingly, exploratory pharmacogenetic analyses revealed the detrimental effects of rs2279238 were primarily observed in the patients randomized to the verapamil-SR strategy.
LXRA is a biologically attractive candidate gene for cardiovascular risk. Recent experimental models suggest that modulating the LXRA pathway has beneficial cardiovascular effects. LXRA agonists used in the treatment of experimental stroke in animal models have shown some benefit. Saez et al. reported neuro-protection associated with the use of an experimental LXRA agonist in a rat model of acute brain ischemia[31]. Further support from animal models came when Chen et al. reported that treatment of experimental stroke in mice with the LXRA agonist TO90317 resulted in improved vascular maturation, increased angiogenesis, increased levels of HDL and increased eNOS phosphorylation in the ischemic portions of the brain [32]. Moreover, Kuipers et al. reported that the LXRA agonist TO90317 blunts RAAS activation (AT1R, ACE and Renin) induced by isoproterenol in mice hearts and kidney [15]. With these data supporting a potential role for LXRA agonists in improving the function of the vasculature of the brain and maintaining RAAS homeostasis, it may be plausible that gain of function alleles in NR1H3 may contribute similar protective effects. The recent data generated in basic science models correlate with the growing clinical observations suggesting a role for LXRA in cardiovascular homeostasis. Dahlman et al. reported that rs2279238 C/T carriers experienced an increased BMI in an observational study of 559 Swedish subjects[17]. Furthermore, the wild type C allele was carried in a NR1H3 haplotype that was also associated with a lower BMI[17]. Subsequently, Robitaille et al. reported that variant carriers of rs12221497, rs61896015, and rs3758674 had increased plasma total cholesterol levels in a cohort of 732 French-Canadian subjects [18]. In another population-based study comprised of two French cohorts (n=1130 and n=1160), Legry et al. reported that variant carriers of rs11039155 in the 5′ UTR/promoter region of NR1H3 had increased HDL and a 30% decrease in risk of having the metabolic syndrome[20]. Furthermore, Sabatti et al. recently published a genome-wide association study associating the variant alleles of two NR1H3 SNPs (rs2167079 and rs7120118) with increased HDL in a Finnish birth cohort of over 4000 subjects[19]. Moreover, a recent genetic study in the Copenhagen City Heart Study (CCHS, n=10,300) and the Copenhagen General Population Study (CGPS, n=51,500) reported that SNPs in the promoter region of NR1H3 (rs6189605 and rs12221497) predicted risk of developing a myriad of ischemic vascular diseases (Ischemic heart disease, myocardial infarction, ischemic cerebrovascular disease and ischemic stroke) in a general population[30]. The CCHS and CGPS subjects differ from our INVEST-GENES subjects that were enrolled with documented hypertension and CAD, and therefore direct comparisons of our findings are difficult. Nonetheless, the findings are supportive of a role of NR1H3 SNPs in CVD and provide additional support that our findings are not due to chance. Correlating with our data, some of the previous clinical observations with NR1H3 variability have been associated with favorable phenotypes, while other SNPs have been associated with cardiovascular risk phenotypes. Therefore, considering the data across the translational divide suggests modulating the LXRA pathway has cardiovascular effects.
In-silico analyses fail to suggest putative functionality for rs11039149, but suggest that rs12221497 may be located at an exon splicing enhancer. Furthermore, in-silico analyses suggest that rs2279238 is also located at an exon splicing enhancer (ESE) of NR1H3 where splicing factor SRp55 binds. Several splice variants of NR1H3 have been identified but their contributions to normal physiology or disease pathogenesis have not been fully explored[33]. Further functional studies may add clarity to the function of rs11039149, rs12221497 and rs2279238 in normal physiology and pathogenesis of cardiovascular disease.
The exploratory pharmacogenetic association observed with rs2279238 genotype and the risk of adverse outcomes in the verapamil-SR strategy warrant further studies to help to clarify the results of these findings. Future studies designed with NR1H3 genotypes, gene expression and clinical biomarkers associated with cardiovascular risk may be able to provide more functional insight.
Diplotype analyses did not provide further insights regarding risks not evident from the single SNP analyses. However, the analysis did reveal that the variant alleles of rs11039149 and rs12221497 are rarely co-inherited with rs2279238. This provides support for the directionally opposite findings observed in the single SNP analyses.
This analysis within INVEST GENES does have some limitations. There were considerably more Whites and Hispanics in the study than Blacks and thus it is unclear whether this association is absent in Blacks or we do not have sufficient power to detect it. Ancestral informative markers (AIMs) were selected to help delineate Native American, European and African ancestry for patients in the INVEST-GENES study. AIMs were used to control for population stratification to reduce the possibility of spurious associations. AIMs were used to adjust ORs for each of the ancestries, but had minimal effects on the results observed.
Conclusions
In this study we observed differential CVD risk based on NR1H3 genotypes. We identified protective associations with the variant alleles of NR1H3 (rs11039149 and rs12221497) and an increased risk of adverse cardiovascular outcomes with another variant allele (rs2279238). Furthermore, we observed a pharmacogenetic interaction between the rs2279238 variant allele and treatment strategy. To our knowledge, this is the first evaluation of NR1H3 polymorphisms and drug response. Collectively, these findings support the exploration of nuclear receptors/transcription factors that are essential to cardiovascular homeostasis as potential contributors to development of CVD and response to pharmacotherapy for CVD risk reduction. Specifically, this work provides support for additional prospective studies to confirm the contribution of NR1H3 gene variants in cardiovascular homeostasis, cardiovascular disease pathogenesis and pharmacological response.
Supplementary Material
Acknowledgments
Funding sources
This project was funded by National Institutes of Health (HL074730, HL69758, to J.J. and C.P.), T-32-DK007518-21S1 (EP), 1-F31-HL091710-01(EP), RR017568 and a grant from Abbott Pharmaceuticals (to J.J. and C.P.),
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lloyd-Jones D, et al. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121(7):e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 2.Castelli WP, et al. Incidence of coronary heart disease and lipoprotein cholesterol levels. The Framingham Study. JAMA. 1986;256(20):2835–8. [PubMed] [Google Scholar]
- 3.Gordon DJ, et al. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 1989;79(1):8–15. doi: 10.1161/01.cir.79.1.8. [DOI] [PubMed] [Google Scholar]
- 4.Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–74. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 5.Libby P. Inflammation and cardiovascular disease mechanisms. Am J Clin Nutr. 2006;83(2):456S–460S. doi: 10.1093/ajcn/83.2.456S. [DOI] [PubMed] [Google Scholar]
- 6.Pepine CJ. Residual risk for secondary ischemic events in patients with atherothrombotic disease: opportunity for future improvements in patient care. Ann Med. 2010;42(1):19–35. doi: 10.3109/07853890903260898. [DOI] [PubMed] [Google Scholar]
- 7.Peters SA, et al. C-reactive protein lowering with rosuvastatin in the METEOR study. J Intern Med. 2010;268(2):155–61. doi: 10.1111/j.1365-2796.2010.02230.x. [DOI] [PubMed] [Google Scholar]
- 8.Ridker PM, et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med. 2008;359(21):2195–207. doi: 10.1056/NEJMoa0807646. [DOI] [PubMed] [Google Scholar]
- 9.Barish GD, Evans RM. PPARs and LXRs: atherosclerosis goes nuclear. Trends Endocrinol Metab. 2004;15(4):158–65. doi: 10.1016/j.tem.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–29. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- 11.Joseph SB, Tontonoz P. LXRs: new therapeutic targets in atherosclerosis? Curr Opin Pharmacol. 2003;3(2):192–7. doi: 10.1016/s1471-4892(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 12.Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol Endocrinol. 2003;17(6):985–93. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 13.Zambon A, et al. Modulation of hepatic inflammatory risk markers of cardiovascular diseases by PPAR-alpha activators: clinical and experimental evidence. Arterioscler Thromb Vasc Biol. 2006;26(5):977–86. doi: 10.1161/01.ATV.0000204327.96431.9a. [DOI] [PubMed] [Google Scholar]
- 14.Imayama I, et al. Liver X receptor activator downregulates angiotensin II type 1 receptor expression through dephosphorylation of Sp1. Hypertension. 2008;51(6):1631–6. doi: 10.1161/HYPERTENSIONAHA.107.106963. [DOI] [PubMed] [Google Scholar]
- 15.Kuipers I, et al. Activation of liver X receptor-alpha reduces activation of the renal and cardiac renin-angiotensin-aldosterone system. Lab Invest. 2010;90(4):630–6. doi: 10.1038/labinvest.2010.7. [DOI] [PubMed] [Google Scholar]
- 16.Morello F, et al. Liver X receptors alpha and beta regulate renin expression in vivo. J Clin Invest. 2005;115(7):1913–22. doi: 10.1172/JCI24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahlman I, et al. Liver X receptor gene polymorphisms and adipose tissue expression levels in obesity. Pharmacogenet Genomics. 2006;16(12):881–9. doi: 10.1097/01.fpc.0000236334.49422.48. [DOI] [PubMed] [Google Scholar]
- 18.Robitaille J, et al. The lipoprotein/lipid profile is modulated by a gene-diet interaction effect between polymorphisms in the liver X receptor-alpha and dietary cholesterol intake in French-Canadians. Br J Nutr. 2007;97(1):11–8. doi: 10.1017/S0007114507201722. [DOI] [PubMed] [Google Scholar]
- 19.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legry V, et al. Association between liver X receptor alpha gene polymorphisms and risk of metabolic syndrome in French populations. Int J Obes (Lond) 2008;32(3):421–8. doi: 10.1038/sj.ijo.0803705. [DOI] [PubMed] [Google Scholar]
- 21.Pepine CJ, et al. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA. 2003;290(21):2805–16. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- 22.Pepine CJ, et al. Rationale and design of the International Verapamil SR/Trandolapril Study (INVEST): an Internet-based randomized trial in coronary artery disease patients with hypertension. J Am Coll Cardiol. 1998;32(5):1228–37. doi: 10.1016/s0735-1097(98)00423-9. [DOI] [PubMed] [Google Scholar]
- 23.Beitelshees AL, et al. KCNMB1 genotype influences response to verapamil SR and adverse outcomes in the INternational VErapamil SR/Trandolapril STudy (INVEST) Pharmacogenet Genomics. 2007;17(9):719–29. doi: 10.1097/FPC.0b013e32810f2e3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beitelshees AL, et al. CACNA1C gene polymorphisms, cardiovascular disease outcomes, and treatment response. Circ Cardiovasc Genet. 2009;2(4):362–70. doi: 10.1161/CIRCGENETICS.109.857839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerhard T, et al. Alpha-adducin polymorphism associated with increased risk of adverse cardiovascular outcomes: results from GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST-GENES) Am Heart J. 2008;156(2):397–404. doi: 10.1016/j.ahj.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu Y, et al. Genetic variation in the beta2 subunit of the voltage-gated calcium channel and pharmacogenetic association with adverse cardiovascular outcomes in the INternational VErapamil SR-Trandolapril STudy GENEtic Substudy (INVEST-GENES) Circ Cardiovasc Genet. 3(6):548–55. doi: 10.1161/CIRCGENETICS.110.957654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacanowski MA, et al. beta-adrenergic receptor gene polymorphisms and beta-blocker treatment outcomes in hypertension. Clin Pharmacol Ther. 2008;84(6):715–21. doi: 10.1038/clpt.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skol AD, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 29.Andrisin TE, Humma LM, Johnson JA. Collection of genomic DNA by the noninvasive mouthwash method for use in pharmacogenetic studies. Pharmacotherapy. 2002;22(8):954–60. doi: 10.1592/phco.22.12.954.33598. [DOI] [PubMed] [Google Scholar]
- 30.Stender S, et al. Genetic Variation in Liver X Receptor Alpha Predicts Risk of Ischemic Vascular Disease in 60,000 Individuals from the General Population. Circulation. 2010;122(21 Supplement) p. Abstract 16602. [Google Scholar]
- 31.Sironi L, et al. Treatment with LXR agonists after focal cerebral ischemia prevents brain damage. FEBS Lett. 2008;582(23-24):3396–400. doi: 10.1016/j.febslet.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, et al. eNOS mediates TO90317 treatment-induced angiogenesis and functional outcome after stroke in mice. Stroke. 2009;40(7):2532–8. doi: 10.1161/STROKEAHA.108.545095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M, Beaven S, Tontonoz P. Identification and characterization of two alternatively spliced transcript variants of human liver X receptor alpha. J Lipid Res. 2005;46(12):2570–9. doi: 10.1194/jlr.M500157-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.