Abstract
Objectives
To determine the protein expression of TNFAIP3 in synovium and to show the capability of 6q23 intergenic SNPs, associated with rheumatoid arthritis (RA) susceptibility, to influence TNFAIP3 gene transcription.
Methods
Immunohistochemistry for TNFAIP3, NF-κB p65 and phosphorylated NF-κB p65 protein expression was performed in 6 RA knee joint synovium samples compared to 9 osteoarthritis (OA) samples.
Luciferase reporter gene assays were used to examine the regulatory ability of RA associated SNP variants on TNFAIP3 promoter activity. Sense and antisense constructs were prepared for rs6920220 alleles, together with each of the 4 SNPs in r2=1 with it (rs6933404, rs2327832, rs6927172 and rs17264332), coupled to the TNFAIP3 promoter. Transient transfections were performed in a human T lymphoblastoid (CEMC7A) cell line. Bioinformatic software was utilised to prioritise SNPs for further investigation. Electrophoretic mobility shift assays (EMSA), using CEMC7A nuclear extracts, were conducted for the rs6927172 SNP alleles.
Results
TNFAIP3 protein expression was seen in the synovium samples and differential TNFAIP3 protein expression between RA vs. OA synoviocytes observed. Within RA synoviocytes TNFAIP3 expression is predominately cytoplasmic, whereas in OA its expression is strongly nuclear and cytoplasmic.
For 3 of the 5 SNPs investigated (rs6920220, rs6933404, rs6927172) evidence of repressor activity of TNFAIP3 transcription was seen and EMSA data showed evidence of differential transcription factor binding to rs6927172 alleles.
Conclusion
This is the first observation of TNFAIP3 protein expression in RA and OA synovium. In vitro analysis of 6q23 intergenic SNPs supports the possibility of the functional regulation of TNFAIP3.
Keywords: TNFAIP3, A20, synovium, transcriptional regulation, rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is a common systemic autoimmune inflammatory disease characterised by chronic inflammation and destruction of synovial joints leading to progressive disability. The aetiology of RA remains to be understood, but recent advances in defining the genetic basis to the condition have been made post genome wide association studies (GWAS). Following the well described genetic associations with HLA-DRB1 and PTPN22 alleles, the next most strongly associated genetic variant, within UK RA datasets, has emerged as rs6920220 (1). This SNP maps to an intergenic region of 6q23. A subsequent fine mapping study of the region revealed that multiple SNPs are independently associated with RA in this locus (2). One of them, rs5029937, is located in TNFAIP3 intron 2, and is in linkage disequilibrium (LD) with rs2230926, a non-synonymous variant also associated with systemic lupus erythematosus that affects the ability of TNFAIP3 to inhibit TNF-induced NF-κB signalling (3). However, the SNP showing the strongest association with RA in our previous fine mapping was rs6920220. This SNP is not in LD with rs2230926, and conditional logistic regression showed that its effect is independent of that provided by the non synonymous SNP. Four SNPs genotyped by the HapMap consortium (rs6933404, rs2327832, rs6927127 and rs17264332) have a pairwise r2=1 with rs6920220. All five SNPs map to a single linkage disequilibrium block spanning a 60kb region (4), which lies between the genes OLIG3 and TNFAIP3 (also known as A20). OLIG3 is important in the development and differentiation of neuronal cells, whereas TNFAIP3 negatively regulates the transcription factor NFκB responses to tumour necrosis factor alpha (TNF-α), toll-like receptor and NOD2 signaling (5). Mice null for TNFAIP3 have multi-organ inflammation, including inflammation of synovial joints (6), therefore TNFAIP3 is an attractive candidate RA susceptibility gene.
One microarray expression study observed TNFAIP3 transcripts in RA synoviocytes (7). To date, no characterisation of TNFAIP3 protein has been described in human synovium. Documenting TNFAIP3 protein expression and distribution within the main active site of the disease pathogenesis, the synovium, would enhance its feasibility as a key RA candidate locus.
Previous studies have identified functional SNPs that are capable of altering gene expression or protein degradation, leading to variations in RA susceptibility and severity (8, 9) therefore it is possible that rs6920220 and its correlates could modulate TNFAIP3 protein levels. However, interrogation of data available from public databases shows the rs6920220 genotype not to be correlated to TNFAIP3 expression in EBV transformed B cell lines (10), though this may be due to lack of power. In order to establish if rs6920220, or one of the correlated SNPs, could alter TNFAIP3 expression, luciferase reporter gene assays in a T lymphoblast cell line (CEMC7A) have been performed with all 5 SNPs to see if they show evidence of regulating TNFAIP3 transcription.
Materials and methods
Immunohistochemistry
As TNFAIP3 expression is induced by TNF-α, anti-TNF treatment could influence the protein levels. Therefore, we used samples collected pre the anti-TNF era (pre 1970) to be certain that anti-TNF drugs had not been used, as treatment history is not routinely documented on the synovium sample archive we have access to.
Tissue samples of histologically confirmed RA (n=6, 5 female, 1 male, mean age 48.7 years, range 21-84) and OA (n=9, 6 female, 3 male, mean age 38.6 years, range 23-71) cases, as diagnosed by a Consultant histopathologist were available. Paraformaldehyde-fixed, paraffin-embedded tissue sections were dewaxed in xylene and rehydrated through graded alcohols. The sections were subjected to heat-induced antigen retrieval in citrate buffer solution (10mM, pH 6.0) and incubated with 3% H2O2 to block endogenous peroxidase activity. Non-specific binding sites were blocked by incubating the sections with 10% normal goat serum (Abcam) for 1-2 hours at room temperature.
Sections were incubated overnight at 4°C with chicken anti-human-TN-FAIP3 polyclonal antibody (3.5 μg/mL) (Abcam), rabbit anti-human-NF-κB p65 monoclonal antibody (0.3 μg/mL) and rabbit anti-human-Phospho-NF-κB p65 (Ser276) polyclonal antibody (0.3 μg/mL) (Cell Signaling Technologies), respectively. For control sections, the primary antibodies were omitted or irrelevant isotype-matched antibodies were applied at the same working concentration as the corresponding primary antibody. The secondary antibodies used were biotinylated anti-rabbit IgG, made in goat (BA-1000, Vector laboratories, Burlingame, CA), for the anti-NF-κB p65 and anti phospho-NF-κB p65 primary antibodies and biotinylated anti-chicken IgG, made in goat (BA-9010, Vector laboratories, Burlingame, CA), for the anti-TNFAIP3 primary antibody. Indirect immunoperoxidase staining was performed using a Vector ABC Kit and Vector SG as a chromogen (Vector laboratories, Burlingame, CA). Counterstaining was performed using Nuclear Fast Red (Vector laboratories, Burlingame, CA).
In order to determine TNFAIP3 synoviocyte expression quantifiable image analysis was performed using the Leica Qwin Pro V2.4 software (Leica, Cambridge, UK). For each specimen we measured protein levels in the synovial lining by defining an constant region, 63 μm thick from the synovial surface, magnification 600x.
Expression levels were compared between cases and controls by logistic regression using STATA ver 9.2 (STAT corporation, USA).
Construction of reporter gene plasmids
1Kb DNA sequences encompassing the SNPs of interest were amplified using the extensor high fidelity PCR master-mix (Abgene) with genomic DNA isolated from an individual homozygous at the given SNP locus and specific primers containing 5′BamHI sites. Only one of the SNPs under investigation was contained within a single the 1Kb constructs. Products were then BamHI digested and ligated into BamHI linearised pGL3 SV40 vector (Promega). Sequencing of transformants was performed to confirm sequence insertion and orientation, SNP allele and to exclude possible errors. Site directed mutagenesis was used to alter the SNP allele (Quikchange site directed mutagenesis kit (Stratagene)). To change the promoter driving luciferase expression in this plasmid, the extensor high fidelity PCR mastermix (Abgene) and specific primers with HindIII BglII 5′ ends were used to amplify a 405bp product encompassing the TNFAIP3 promoter from genomic DNA (11). This product was then HindIII BglII digested and ligated into the HindIII BglII linearised pGL3 backbone. Sequencing was performed to confirm sequence insertion and to exclude possible errors. All constructs used in transfections were prepared using the Endofree Maxi prep kit (Qiagen). Primer sequences are available on request.
Cell culture
CEMC7A cells (human T lymphoblast) obtained from ECACC and were cultured in RPMI1640 (Gibco BRL, Paisley, UK) 10 % FBS, at 37°C in 5% CO2.
Transfections
CEMC7A cells were transfected as described previously (12). Total protein was assayed by Bradford assay and used for normalisation of luciferase values. Cells were harvested 20 hours later and luciferase and renilla assays performed using the Dual Luciferase assay kit (Promega) as per manufacturers’ instructions. Experiments were performed in triplicate, on at least three separate occasions.
Bioinformatic and Transcription Factor Binding (TFB) prediction: cross species sequence homology was determined using UCSC (http://genome.ucsc.edu/). AliBaba, Match, and Matinspector softwares (http://www.gene-regulation.com/pub/programs.html; http://www.genomatix.de/products/MatInspector/) were used to determine TFB.
Statistical analysis
Analysis of variance (ANOVA) followed by Dunnett’s Multiple Comparison Test were performed using GraphPad Prism 4. p<0.05 was considered significant.
Preparation of nuclear extract
CEMC7A cells were grown to a density of 1×105cells/ml in 1L of RPMI1640 media in order to gain a sufficient final amount of nuclear extract. Cells were harvested and washed twice in PBS. Nuclear extract was then prepared as described previously (13) with a minor modification. Rather than Dounce homogenisation, cells were disrupted by passing the cell suspensions through a 23 gauge needle 5 times.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using radiolabelled double stranded DNA oligonucleotides corresponding to the rs6927172 C or G alleles. The oligonucleotides used were: Forward C allele 5′ TGG GGG AAA TCC AGG TCA ACT T 3′ Reverse C allele 5′AAG TTG ACC TGG ATT TCC CCC A 3′ Forward G allele 5′TGG GGG AAA TGC AGG TCA ACT T 3′ Reverse G allele 5′AAG TTG ACC TGC ATT TCC CCC A 3′ Ets1 5′GGGCTGCTTGAGGAAGTATAAGAAT 3′ Mutated Ets1 (mt Ets1) 5′GGGCTGCTTGAGAGAGTATAAGAAT 3′ Annealed oligonucelotides were end labelled with (γ-32P) ATP (Amersham) using T4 polynucleotide kinase (Promega) and purified using G50 columns (Amersham). Reactions contained 10μl Buffer C (10mM Tris pH 7.8, 50mM NaCl, 1mM DTT, 1mM EDTA 5% glycerol) 2μg BSA, 100ng Poly dI:dC, 0.1ng labelled probe and 5μg nuclear extract as indicated in the figure legends. Competition experiments were performed by adding 100× or 500× unlabelled oligonucelotide to the reaction mixes prior to addition of radiolabelled probe as indicated. Reactions were incubated for 1 hour (25°C), loaded onto 5% 0.5× TBE gel, run at 150V for 3 hours. Gels were then dried and autoradiographed.
Results
Immunohistochemical analysis of the expression of TNFAIP3, NF-κB (p65) and Phospho-NF-κB (phospho-p65) in synovial tissue
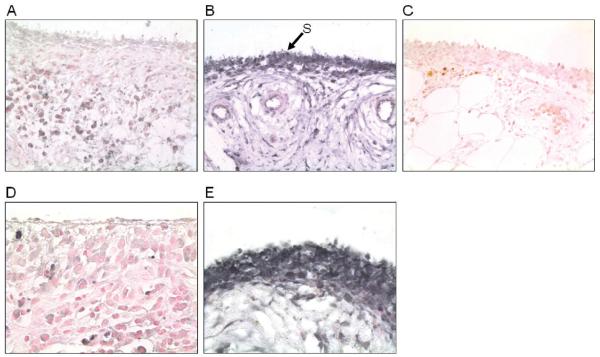
Expression of TNFAIP3 protein was detected in RA (n=6) and OA (n=9) synovium samples (Fig. 1), with weaker expression seen consistently throughout all the RA compared to the OA samples. Morphological examination showed positive TNFAIP3 expression in synoviocytes, lymphocytes, fibroblasts and muscle cells, and was especially strong in mast cells.
Fig. 1.
Immunohistochemistry for the expression of TNFAIP3 in synovial tissue from patients with RA and OA. A: Expression of TNFAIP3 in RA synovium. B: Expression of TNFAIP3 in OA synovium. C: Chicken IgG negative control. (A-C: original magnification × 600).
D: Cytoplasmic expression of TNFAIP3 in RA synoviocytes. E: Nuclear and cytoplasmic expression of TNFAIP3 in OA synoviocytes. (D&E original magnification × 1200). Arrow S shows synoviocytes.
With regards to synoviocyte expression, TNFAIP3 localisation was seen as strongly nuclear, and also cytoplasmic in OA patients, but, in contrast, was predominately cytoplasmic in RA patients (Fig. 1).
Quantitative analysis of synoviocyte TNFAIP3 expression in RA patients compared to OA patients showed lower levels of TNFAIP3 in RA cases (mean normalised expression 0.23±0.15) compared to OA patients (mean normalised expression 0.34±0.17), however, this skewing did not reach statistical significance.
Expression of NFκB (p65) and its activated form phospho-p65 (Ser276) was determined. Expression of both was evident in RA and OA patients, being greater in those with RA, as previously described (14, 15). The RA cases (n=6) had significantly more expression of p65 and phospho-p65 (Ser276) than the OA group (n=9) (mean normalised expression 0.15±0.13 in RA cases vs. 0.04±0.04 in controls for p65 (p=0.05) and 0.30±0.18 in RA cases vs. 0.11±0.09 in controls for phospho-p65 (p=0.04)).
6q23 SNPs and regulation of TNFAIP3 transcription in CEMC7A cells
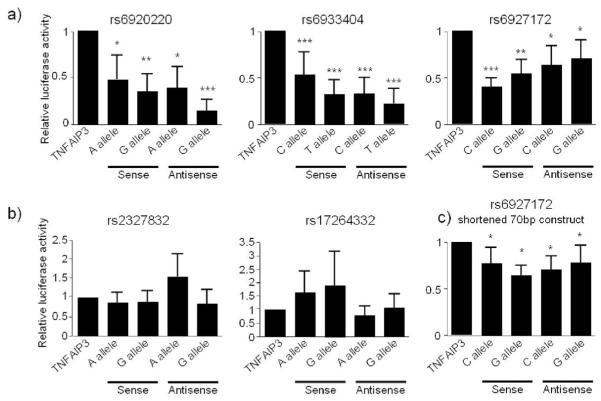
For each of the 5 SNPs under investigation 1kb sequences, with the allele variants at the centre, were cloned into the luciferase reporter gene construct pGL3, in both the sense and antisense direction, with reporter gene activity initiated from the TNFAIP3 promoter. Constructs were transfected into CEMC7A T lymphoblast cells. Figure 2 shows the results of the 5 SNPs tested. Three of the constructs (rs6920220, rs6933404, rs6927172) show orientation independent repression of TNFAIP3 transcription (Fig. 2a). rs2327832 and rs17264332 do not regulate TNFAIP3 (Fig. 2b).
Fig. 2.
Luciferase reporter gene assays for TNFAIP3. CEMC7A cells were transfected with the reporter gene constructs as indicated. Cells were harvested 20 hours later. Lysates were prepared, and luciferase content analysed. Luciferase values were normalised to total protein and the data presented graphically. Graph depicts mean±SD of triplicate wells, representative of 3 independent experiments. * p<0.01, **p<0.001, ***p<0.0001.
Bioinformatic analysis revealed the rs6927172 SNP, and its immediate flanking region, to show evidence of sequence conservation across mammalian species. This was in contrast to the other 4 SNPS (and flanking regions) which demonstrated no sequence conservation at all. We therefore shortened the rs6927172 insert in the TNFAIP3 constructs from 1Kb to 70bp, in order to confirm that the regulatory capacity of this sequence is centred around the SNP region. This shortened sequence is still able to inhibit TNFAIP3 promoter activity in CEMC7A cells in both orientations (Fig. 2c). However, we still did not observe any allele specific variation in the luciferase reporter gene data. Therefore, we next attempted to accurately predict TFB to each of the SNP allele variants as a way of demonstrating the possibility of differential allelic function of this SNP. TFB profiling was undertaken with three different prediction programs for the 5 SNPs under investigation. The only consistent result was observed with the rs6927172 SNP, where there was loss of a predicted Ets-1 site with the G allele variant of the SNP across two of the programmes.
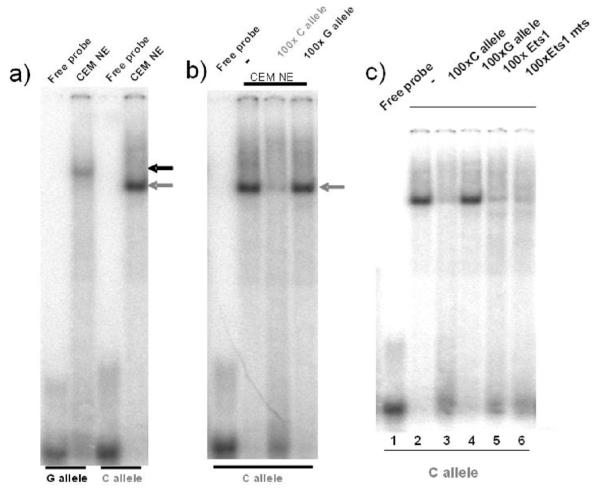
We looked at the ability of the alleles of rs6927172 to bind complexes in vitro using EMSA. CEMC7A nuclear extract was incubated with radiolabelled oligonucleotides corresponding to each of the rs6927172 SNP alleles, and the resulting complexes were resolved by native PAGE. As shown in Figure 3a, both alleles were capable of forming complexes. In order to determine whether these complexes are binding the allele variants in a sequence specific manner, competitions were performed by the addition of excess unlabelled oligonucleotide. This indicates that the sequence binding the C allele variant is distinct from that binding the G allele variant, since no competition is observed when excess unlabelled G allele oligonucleotide is added (Fig. 3b). Incubation with Ets1 consensus oligonucleotide causes an apparent reduction in complex binding to the C allele however, this is not sequence specific as mutated Ets1 has the same effect (Fig. 3c).
Fig. 3.
EMSA analysis shows differential transcription factor binding to rs6927172 alleles; lack of evidence for Ets1 binding. Nuclear extracts (NE) prepared from CEMC7A cells were incubated with radiolabelled oligonucleotides encompassing the rs6927172 C and G alleles and complexes were resolved by native gel electrophoresis. (a) The grey and black arrows indicate the C and G specific complexes, respectively. Competition experiments were performed by the addition of 100× unlabelled oligonucleotide as indicated (b and c). The C allele specific complex is indicated by the grey arrow. Gels are representative of 3 independent experiments.
Discussion
GWAS have resulted in several tens of new SNPs being found to be associated with complex diseases (16). In UK RA datasets, the strongest novel SNP association is with the rs6920220, which is intergenic on 6q23. Mechanistic advancement of aetiopathogenesis of RA could arise through understanding the functional affects of any given SNP association. This, however, is a difficult undertaking, especially when the SNP is not within a gene. Evidence from animal models proposes TNFAIP3 as a plausible candidate gene of importance in RA susceptibility. TNFAIP3 is a dual ubiquitin-editing enzyme, whose expression is induced by a large number of stimuli in a wide variety of cells. It is mainly involved in the negative feedback regulation of NF-κB, but it has been shown to have other functions, such as anti-apoptotic activity (5, 17). This is the first study showing TNFAIP3 protein expression in human synovium. Interestingly, TNFAIP3 expression was observed in several cell types that play important roles in the pathophysiology of RA, such as synoviocytes, lymphocytes and fibroblasts. The strong positive reaction found in mast cells should be considered with caution, since antibody binding has been shown to be influenced by ionic interaction with secretory granules in this cell type (18). We found lower TNFAIP3 expression levels between RA compared to OA, although this difference was not statistically significant, possibly due to small sample size. The protein showed a distinctly different pattern of localisation in RA and OA. In OA the synoviocytes showed both heavy nuclear and cytoplasmic expression of TNFAIP3, whereas, on the contrary, it was expressed predominantly in cytoplasm in RA. We have used formalin-fixed wax-embedded tissue for this histological evaluation. It was not therefore possible to extract any protein from these sections for further quantitative analysis. TNFAIP3 is generally considered a cytoplasmic protein. Nevertheless, it is a putative DNA-binding protein (UniProtKB/Swiss-Prot accession number P21580; http://www.uniprot.org), and it has been detected in the nucleus of an endothelial cell line (19). In view of the results from our immunohistochemistry study, it is tempting to hypothesise that this protein could be involved in the control of inflammation at the nuclear level in synoviocytes, and that this regulation could be impaired in RA due to reduced nuclear expression. The protein’s subcellular localisation could be altered by as yet unidentified genetic polymorphisms in the TNFAIP3 gene, which might be in LD with some of the previously identified RA associated SNPs. In this regard, a recent study has shown that mutations located in the zinc finger domains are able to disrupt the localisation of TNFAIP3 to an endocytic membrane compartment in HeLa cells, which results in reduced NF-κB inhibitory activity (20). Ongoing resequencing studies will help to identify new potentially functional variants in the TNFAIP3 region.
We studied rs6920220 and its perfect proxies using transient transfections in a human T lymphoblast cell line. Sequences containing 3 of the 5 SNPs studied significantly repressed TNFAIP3 expression. In vivo, such repression of TNFAIP3 transcription would limit the negative regulation of NF-κB, resulting in enhanced pro-inflammatory cytokine expression, as occurs in RA. No statistically significant allele specific differences were seen. This was the same regardless of induction with TNF-α (data not shown). This may be due to the lack of sensitivity of the reporter assay to identify small differences in SNP allele variation. Following bioinformatic and transcription factor binding analysis we prioritised investigation of the rs6927172 SNP. The repression of TNFAIP3, independent of orientation, remained even once the sequence was reduced from 1Kb to a 70bp, with the SNP at the centre. Additional support for the rs6927172 alleles being functionally different comes from our EMSA findings. Sequence specific complexes differentially binding to the C and to the G allele were consistently observed in nuclear extracts from CEMC7A cells. We do not, however, find evidence for an Ets1 specific binding site at the C allele, as predicted in silico. The future evaluation of the transcription factors binding to these alleles may help to elucidate their functional role in the potential regulation of TNFAIP3 gene transcription.
Most recently SNP data has been released from the sequencing of a 1000 genomes (http://www.1000genomes.org/page.php). This has identified two further SNPs, rs62432712 and rs928722, as r2=1 with rs6920220. Both these SNPs are in the same intergenic LD block as the other perfect proxies. The sequence flanking these SNPs is not conserved across species and the allele variants to not alter known TFB sites, suggesting a low priority for any functional evaluation.
In conclusion, we provide evidence for the protein expression of TNFAIP3 within synovium and show its cellular distribution to differ between RA and OA. This further supports the role of altered TNFAIP3 in RA aetiopathogenesis. We also provide in vitro evidence of altered TNFAIP3 transcription by 6q23 intergenic SNPs associated with RA. Further functional characterisation of the SNPs, in particular rs6927172, may provide mechanistic insight into TNFAIP3 gene regulation.
Acknowledgements
We thank Andy Berry, Donna Topping, Tom Eastell, Steve Eyre, Pauline Baird and Colin Allott for technical support.
This work was funded by the arc core grant support. The authors’ laboratories are supported by the Manchester Academic Health Sciences Centre (MAHSC) and by the NIHR Biomedical Research Centre.
Footnotes
Competing interests: none declared.
References
- 1.WELLCOME TRUST CASE CONTROL CONSORTIUM Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.OROZCO G, HINKS A, EYRE S, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23. Hum Mol Genet. 2009;18:2693–9. doi: 10.1093/hmg/ddp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MUSONE SL, TAYLOR KE, LU TT, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–4. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.THOMSON W, BARTON A, KE X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–3. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COORNAERT B, CARPENTIER I, BEYAERT R. A20: central gatekeeper in inflammation and immunity. J Biol Chem. 2009;284:8217–21. doi: 10.1074/jbc.R800032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LEE EG, BOONE DL, CHAI S, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–4. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GALLAGHER J, HOWLIN J, MCCARTHY C, et al. Identification of Naf1/ABIN-1 among TNF-alpha-induced expressed genes in human synoviocytes using oligonucleotide microarrays. FEBS Lett. 2003;551:8–12. doi: 10.1016/s0014-5793(03)00823-8. [DOI] [PubMed] [Google Scholar]
- 8.PALOMINO-MORALES R, GONZALEZ-JUANATEY C, VAZQUEZ-RODRIGUEZ TR, et al. Interleukin-6 gene -174 promoter polymorphism is associated with endothelial dysfunction but not with disease susceptibility in patients with rheumatoid arthritis. Clin Exp Rheumatol. 2009;27:964–70. [PubMed] [Google Scholar]
- 9.ASSMANN G, VOSWINKEL J, MUELLER M, et al. Association of rheumatoid arthritis with Mdm2 SNP309 and genetic evidence for an allele-specific interaction between MDM2 and p53 P72R variants: a case control study. Clin Exp Rheumatol. 2009;27:615–9. [PubMed] [Google Scholar]
- 10.STRANGER BE, NICA AC, FORREST MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.KRIKOS A, LAHERTY CD, DIXIT VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem. 1992;267:17971–6. [PubMed] [Google Scholar]
- 12.DONN R, ALOURFI Z, ZEGGINI E, et al. A functional promoter haplotype of macrophage migration inhibitory factor is linked and associated with juvenile idiopathic arthritis. Arthritis Rheum. 2004;50:1604–10. doi: 10.1002/art.20178. [DOI] [PubMed] [Google Scholar]
- 13.DIGNAM JD, LEBOVITZ RM, ROEDER RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.HANDEL ML, MCMORROW LB, GRAVALLESE EM. Nuclear factor-kappa B in rheumatoid synovium. Localization of p50 and p65. Arthritis Rheum. 1995;38:1762–70. doi: 10.1002/art.1780381209. [DOI] [PubMed] [Google Scholar]
- 15.MAROK R, WINYARD PG, COUMBE A, et al. Activation of the transcription factor nuclear factor-kappaB in human inflamed synovial tissue. Arthritis Rheum. 1996;39:583–91. doi: 10.1002/art.1780390407. [DOI] [PubMed] [Google Scholar]
- 16.HARDY J, SINGLETON A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–68. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.BEYAERT R, HEYNINCK K, VAN HS. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143–51. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 18.SCHILTZ PM, LIEBER J, GIORNO RC, CLAMAN HN. Mast cell immunohistochemistry: non-immunological immunostaining mediated by non-specific F(ab’)2-mast cell secretory granule interaction. Histochem J. 1993;25:642–7. doi: 10.1007/BF00157878. [DOI] [PubMed] [Google Scholar]
- 19.EVANS PC, TAYLOR ER, COADWELL J, HEYNINCK K, BEYAERT R, KILSHAW PJ. Isolation and characterization of two novel A20-like proteins. Biochem J. 2001;357(Pt 3):617–23. doi: 10.1042/0264-6021:3570617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LI L, HAILEY DW, SOETANDYO N, et al. Localization of A20 to a lysosome-associated compartment and its role in NFkappaB signaling. Biochim Biophys Acta. 2008;1783:1140–9. doi: 10.1016/j.bbamcr.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]