Summary
Multi-body potentials have been of much interest recently because they take into account three dimensional interactions related to residue packing and capture the cooperativity of these interactions in protein structures. Our goal was to combine long range multi-body potentials and short range potentials to improve recognition of native structure among misfolded decoys. We optimized the weights for four-body non-sequential, four-body sequential and short range potentials in order to obtain optimal model ranking results for threading and have compared these data against results obtained with other potentials. (Twenty six different coarse-grained potentials from the Potentials ‘R’Us web server have been used.) Our optimized multi-body potentials outperform all other contact potentials in the recognition of the native structure among decoys, both for models from homology template-based modeling and from template-free modeling in CASP8 decoy sets. We have compared the results obtained for this optimized coarse-grained potentials, where each residue is represented by a single point, with results obtained by using the DFIRE potential, which takes into account atomic level information of proteins. We found that for all proteins larger than 80 amino acids our optimized coarse-grained potentials yield results comparable to those obtained with the atomic DFIRE potential.
Introduction
Knowledge-based potential functions are used in many different types of computational protein studies, including protein structure prediction1–5, protein design6–9, docking applications10–13 and protein folding mechanism studies14–17. Many atomistic potential functions18–20 and coarse-grained potential functions21–24 have been developed. The use of these potentials has grown significantly, and they are of interest because their use can significantly reduce the computational cost of modeling and prediction of protein structures. A major challenge in computational biology is to derive better coarse-grained potentials that are able to perform as well as atomistic potentials, yet are computationally much less expensive.
Many different coarse-grained potentials have been extensively applied in the assessment of protein models and the native structure recognition. One of the most widely used two-body potentials are the Miyazawa-Jernigan potentials22. Betancourt and Thirumalai 25 suggested that pair-wise potentials are not likely to be sufficient for threading applications. The alternative multi-body potentials, in principal, are able to take account of more complex three dimensional interactions, revealing the effects of dense residue packing. In particular, they can capture the strong cooperativity operative within protein structures. Three-body potentials were proposed and developed by Munson and Singh26 and also by Li and Liang27 and they all showed improvements over two-body potentials. Four-body potentials were first derived in the context of Delaunay tessellation by Krishnamoorthy and Tropsha28 and they demonstrated that these potentials also perform better than two-body potentials.
The four-body contact potentials developed by our group29 incorporated sequence information and considered in detail the interactions between backbones and side chains through a simple geometric construction (see Methods for the model description). We also developed them to distinguish between different levels of solvent accessibility of the residues.
These four-body potentials (both sequential and non-sequential) have been successful in recognizing the native structure among most of the misfolded decoy sets from Decoys ‘R’Us data set. However these potentials fail to recognize the native structures of some significant number of proteins.
In this paper we have improved the performance of the four-body contact potentials by combining the four-body sequential29 with the four-body non-sequential potentials30 and with short range potentials. For the short range knowledge-based potentials, we consider the identity for two consecutive amino acids along the sequence, and the pairwise couplings between their virtual torsion and bond angles31. The results for the rankings of the best models are obtained by combining these three sets of potentials, and optimizing globally the weights for each component in the sum.
Different measures of the quality of model selection predictions such as: rankings of the native structure for the decoy sets, RMSD values of the best ranked model and correlation coefficients all show that both the four-body sequential and the four-body non-sequential potentials on average perform better than or as well as two-body coarse-grained potentials. After optimization, however, the resulting residue-level coarse-grained potentials, i.e. the weighted sum of four-body sequential, non-sequential potentials and short range potentials performs better than all other coarse-grained potentials and almost as good as much more detailed (but computationally more costly) atomistic empirical potentials.
Methods
Geometric construction for considering interactions
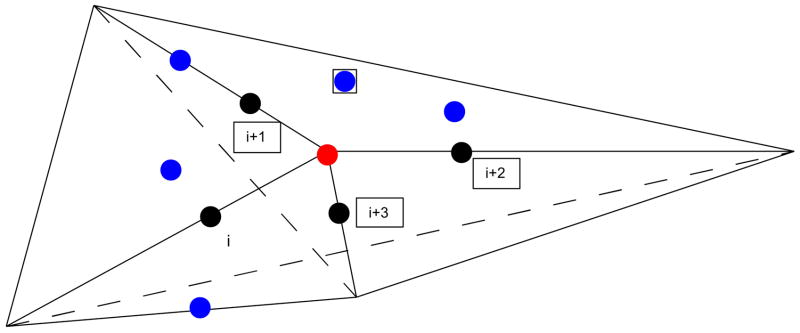
For each four consecutive amino acids (i, i+1, i+2, i+3) along the sequence (in black in Figure 1), we calculated the geometrical center (red) of their four side chain centers (Cα for Gly). Blue residues are residues in close proximity to the geometrical center. Six planes can be defined by the combinations of all possible pairs of these four points and the red center point, and these planes subdivide the space surrounding the red point into four tetrahedra., Each tetrahedron has a common vertex, which is the geometrical center of four side chain centers. Each of the four contacting bodies for our four body potentials are obtained as follows. One triplet of amino acids from a tetrahedron is taken along the sequence with another amino acid which is not along the sequence but within a cutoff distance from the quartet’s geometrical center (blue residue in Figure 1). This amino acid is considered to be in contact with the triplet within a cutoff distance of 8 Å. The cutoff distance 8 Å was selected because it gives the best threading results compared to other values of cutoff distances that we considered. One example of four-bodies is marked in Figure 1 by the four residues in black boxes. We use tetrahedra to capture long-range interactions between non-bonded side chains and groups of backbone residues. In case of these sequential four-body potentials we require, the triplet of amino acids to be sequential, but for the nonsequential four-body potentials this requirement is no longer necessary. Optimized potentials in this paper combine both the sequential and non-sequential four-body potentials along with short ranged pair-wise potentials mentioned earlier.
Figure 1.
Model description of the four-body potentials; Black points represent four sequential residues and the red point is the geometric center of these residues. Blue residues are in close proximity with the geometric center. Six plans can be defined by all the possible combinations of pairs of black points and the central point which subdivide the space around the red point into four tetrahedra. Four-body sets are selected such that a triplet of residues is selected from sequential residues (black nodes) and the fourth node is a residue which is not along the chain but within 8 Å from the center point (blue node). One example of the four contacting bodies is shown by the four residues in black boxes.
Extensive studies have been carried out, where the performance of different knowledge-based potential functions was compared20,33,38 on large data sets of protein models. The way the evaluations have been done is by finding the success in the ranking of the native structure as the conformation with the lowest energy and also by computing average Z-score between the energy of the native structure and the next most favorable structure (the larger the average Z-score the better the evaluation).
We have used CASP8 models as decoy sets (see supplementary materials) for the evaluations of how well two-body and four-body potential functions perform in identifying native (or near native) protein structures. Twenty-three different two-body (more details about these potentials can be found in Pokarowski et al.34 ) and sequential29 and non-sequential30 four-body potentials were used. The targets were divided into two subsets according to the method used to generate decoys for each target. One set is comprised of models that were obtained using homology (template-based) modeling (153 cases) and the other set of models is obtained from template-free modeling approaches (12 cases).
The four-body sequential, the four-body non-sequential and the short ranged potentials were combined in simple linear way by using the following formula:
| (1) |
Optimization of the weight of each term was performed to find an optimized potential for computational applications.
The optimization was carried out using Particle Swarm Optimization32 (PSO) technique. We set the weight of the four-body sequential term to 1.0 (w4-body-seq = 1) and vary the weight coefficients for the other two terms w4-body-nonseq and wSR by using PSO. The main philosophy behind PSO lies in the observation of swarms of birds or bees. The optimal solution is searched for by maintaining a population of candidate solutions (also called particles) and the best found positions for each particle and the whole population are remembered by the algorithm. Particles scan the search-space according to a simple movement formula which takes into account the best found solution by individual particles and the whole population. For the case of optimizing only two parameters, there are other possible methods to optimize them and get similar results. However, in the case of optimization of a function in a higher dimensional space, this method has significant advantage over the other, because in comparison to, for example, grid methods, it is computationally more efficient, and in comparison to simulated annealing methods it does not require any arbitrary assumptions. For each combination of terms we calculated the average RMSD for the best ranked model and the Z-scores for all CASP8 targets. Heat maps for average best ranked models RMSD and Z-score were computed for varying weights w4-body-nonseq and wSR of the optimized potentials for proteins modeled using (homology) template-based methods, and using template-free modeled targets. The native structure rankings obtained for the optimized potentials were compared to those obtained using other coarse-grained potentials and for the atomistic DFIRE potentials20. The Decoys ‘R’Us dataset 33 was used for comparison with atomistic potentials. Both single and multiple decoy sets were used in this assessment. A single decoy set consists of a pair of structures: native structure and decoy structure, and multiple decoys set contains many decoys for each target structure. We have excluded the multiple loop set from our assessment because of the poor amino acid packing in loop regions, and also excluded the ifu decoys set, because multi-body potentials do not perform well for small structures. (For small proteins there are problems with proper tessellation. Residues at the surface cannot be tessellated correctly without taking into account neighboring solvent molecules.)
The RMSD values between the native structure and the best fitting decoy for each decoy set was computed with the TM-score algorithm39. Spearman’s, Pearson’s and Kendall’s correlation coefficients were calculated for all the target-decoy pairs by using potential energies and RMSD values to the native conformation. All incomplete decoys were removed from the sets. Z-scores were also calculated for decoys to evaluate the separation between the native structure and other structure sets in energy space. Pearson’s correlation coefficient is expressed as the covariance of two variables normalized by their standard deviations:
| (2) |
Because Pearson’s correlation coefficient assumes linearity between the two variables (in the context of this paper: energy and RMSD), it would be more suitable to use alternative correlation measures. In particular it seems appropriate to use rank order correlation coefficients. Spearman’s rank correlation coefficient is a non-parametric measure of the statistical dependence between two ranked variables. In the case of existence of tied ranks (when two different observations have the same value - in case of this study, when two structures with different RMSD have the same energy) ρS is computed from the same formula as for ρP. In the case where there are no tied rankings Spearman’s correlation coefficient is computed from the simpler formula:
| (3) |
with di = xi − yi being the difference between the ranks on the two variables for the same structure model.
Kendall’s τ coefficient is a measure of rank correlation, i.e. the similarity of the ordering of the data when ranked by different quantities, defined as:
| (4) |
where nc is the number of concordant pairs, nd is the number of discordant pairs, and the denominator is the total number of pairs. We call the two pairs of variables [Ei, RMSDi] and [Ej, RMSDj] concordant with each other, if Ei > E j; then RMSDi > RMSDj (or vice versa), otherwise we consider them to be discordant.
The three correlation coefficients are calculated for each target using energy and RMSD values away from the native target structure for each target decoy. Then all coefficient values are averaged over all targets in each of the two categories to obtain average values for each potential function.
Results
Performances of different individual potential functions for model ranking
Tested potentials are all knowledge-based coarse-grained potentials and they usually capture the statistics of contacts based on the coordinates of Cα (sometimes Cβ) atoms. Therefore they do not take into account the atomic details of proteins. We observe that for template-based modeled targets, the BT potential derived by Betancourt and Thirumalai25 performs best in comparisons with other two-body potentials and the two four-body potentials individually (in terms of correlation coefficients, average Z-score and average RMSD). The best RMSD values are in the range of 4 Å to 5 Å (See Table 1). Four-body potentials perform well in the identification of native structures and there are a few other two-body potentials which show similar performances with RMSD in the 4 Å range.
Table 1.
Model ranking results for CASP8 template-based homology modeled targets. We show averages computed for all homology modeled CASP8 targets.(Abbreviations and definitions of potentials are explained on our Web-Server (http://gor.bb.iastate/potential) and in Pokarowski et al. 34).
| Potential | Spearman ρ | Pearson ρ | Kendall τ | Z-score | Top Ranked RMSD |
|---|---|---|---|---|---|
| 4B OPT POT | 0.36 | 0.4 | 0.24 | 1.33 | 3.7 |
| BT | 0.46 | 0.49 | 0.33 | 1.5 | 4.1 |
| 4B POT | 0.33 | 0.38 | 0.23 | 1.29 | 4.6 |
| SKJG | 0.44 | 0.43 | 0.31 | 1.41 | 4.6 |
| MJ3 | 0.40 | 0.4 | 0.28 | 1.29 | 4.6 |
| VD | 0.41 | 0.43 | 0.29 | 1.4 | 4.6 |
| 4BG POT | 0.31 | 0.36 | 0.21 | 1.1 | 4.7 |
| TEl | 0.43 | 0.46 | 0.31 | 1.41 | 4.7 |
| SKOb | 0.43 | 0.44 | 0.3 | 1.48 | 4.8 |
| MJ3h | 0.46 | 0.48 | 0.33 | 1.4 | 4.9 |
| BFKV | 0.45 | 0.48 | 0.33 | 1.45 | 4.9 |
| Qm | 0.39 | 0.37 | 0.27 | 1.25 | 5 |
| SKOa | 0.42 | 0.4 | 0.29 | 1.42 | 5.2 |
| MS | 0.38 | 0.4 | 0.27 | 1.25 | 5.2 |
| Qa | 0.38 | 0.36 | 0.26 | 1.09 | 5.4 |
| TD | 0.44 | 0.45 | 0.32 | 1.27 | 5.4 |
| RO | 0.24 | 0.26 | 0.16 | 0.46 | 5.9 |
| TEs | 0.42 | 0.45 | 0.3 | 1.39 | 6.1 |
| GKS | 0.3 | 0.31 | 0.21 | 1.16 | 6.3 |
| Qp | 0.41 | 0.39 | 0.29 | 1.22 | 6.5 |
| HLPL | 0.39 | 0.38 | 0.28 | 1.18 | 6.7 |
| SR | 0.21 | 0.25 | 0.15 | 0.77 | 6.9 |
| MJ2h | 0.32 | 0.3 | 0.23 | 0.81 | 8.1 |
| MSBM | 0.07 | 0.05 | 0.05 | 0.02 | 8.6 |
| MJPL | 0.3 | 0.26 | 0.22 | 0.75 | 9.3 |
| TS | 0.28 | 0.24 | 0.2 | 0.66 | 9.4 |
For the targets from template-free modeling, the performance (in terms of correlation coefficients or average values of Z-score or RMSD) is worse than that for the homology-based modeled proteins (See Table 2). Potentials that perform best for template-free modeled targets also perform best for homology template-based modeled targets but do not yield results that are as good as the latter. This is due to the fact that the template-free modeled structures submitted to CASP8 deviated significantly more from the native structures than template-based homology models, and were usually poorly packed and/or poorly folded. Therefore empirical potentials, which are derived based on real globular proteins interactions, cannot be applied well to these cases.
Table 2.
Model ranking results for CASP8 template-free modeled targets. We show averages computed for all template-free modeled CASP8 targets. (Abbreviations and definitions of potentials are explained on our Web-Server (http://gor.bb.iastate/potential) and in Pokarowski et al.34.
| Potential | Spearman ρ | Pearson ρ | Kendall τ | Z-score | Top Ranked RMSD |
|---|---|---|---|---|---|
| 4B OPT POT | 0.19 | 0.17 | 0.13 | 1.3 | 7.5 |
| BT | 0.19 | 0.16 | 0.14 | 2.14 | 7.7 |
| MJ3h | 0.15 | 0.12 | 0.11 | 2.02 | 8.4 |
| 4B G POT | 0.14 | 0.14 | 0.09 | 1.2 | 9.1 |
| BFKV | 0.17 | 0.13 | 0.13 | 1.98 | 9.2 |
| Qm | 0.19 | 0.14 | 0.13 | 1.7 | 9.3 |
| MJ3 | 0.22 | 0.18 | 0.15 | 1.66 | 9.6 |
| Qp | 0.16 | 0.04 | 0.13 | 1.43 | 9.7 |
| TD | 0.16 | 0.1 | 0.13 | 1.78 | 9.9 |
| 4B POT | 0.17 | 0.19 | 0.12 | 1.29 | 10.3 |
| HLPL | 0.16 | 0.03 | 0.13 | 1.32 | 10.3 |
| MS | 0.22 | 0.18 | 0.14 | 1.56 | 10.3 |
| SKOa | 0.21 | 0.15 | 0.14 | 2.01 | 10.4 |
| TEl | 0.15 | 0.12 | 0.11 | 1.7 | 10.6 |
| SKJG | 0.2 | 0.16 | 0.13 | 1.88 | 10.8 |
| MSBM | 0.1 | 0.06 | 0.07 | 1.05 | 10.8 |
| TEs | 0.15 | 0.11 | 0.1 | 1.59 | 10.9 |
| VD | 0.16 | 0.14 | 0.12 | 1.58 | 10.9 |
| SR | 0.15 | 0.13 | 0.1 | 0.91 | 11.1 |
| GKS | 0.16 | 0.12 | 0.11 | 1.33 | 11.1 |
| SKOb | 0.19 | 0.13 | 0.13 | 1.94 | 11.4 |
| MJPL | 0.15 | −0.02 | 0.12 | 0.88 | 11.4 |
| TS | 0.15 | −0.02 | 0.12 | 0.81 | 11.6 |
| MJ2h | 0.16 | −0.003 | 0.13 | 1 | 11.8 |
| RO | 0.12 | 0.13 | 0.08 | 0.46 | 12.3 |
| Qa | 0.19 | 0.16 | 0.12 | 1.52 | 16.2 |
Rankings, RMSDs and correlation coefficients results all show that the four-body sequential and four-body non-sequential potentials on average perform better than or as well as two-body potentials.
Performance of the optimized potentials
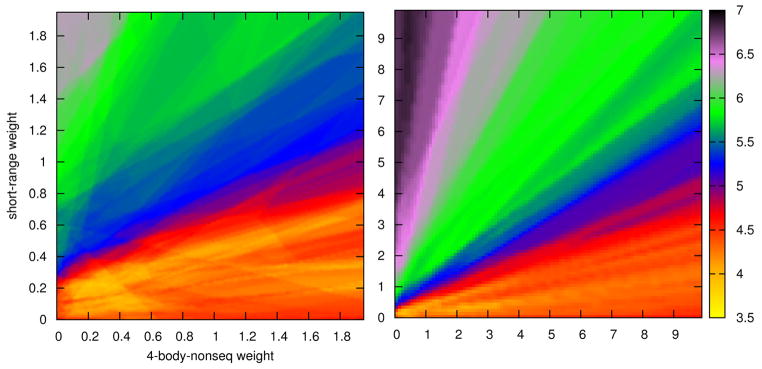
The heat map shows the average RMSD (expressed by color) from the native structure for best ranked homology models, where w4-body-nonseq is plotted on the x-axis and wSR on the y-axis, both in steps of 0.05 (see Figure 2). Additional heat maps are given in the Supplementary materials (Figures S1, S2, S3). The best weights in linear combination of four-body non-sequential, four-body sequential and short range potentials correspond to the yellow regions in Fig. 2. The weight for four-body sequential potentials is equal 1.0. It can be seen that all heat maps (see Supplementary materials Figures S1, S2, S3) show the same region of best weights and there can be several values that give similar results. The optimized weights obtained for the four-body non-sequential and short-range potentials are about 0.28 and 0.22 respectively for the template-based modeled (homology) targets. For template-free modeled targets the corresponding weights are different and equal 1.01 and 0.56, respectively. The weights obtained for homology modeled targets were used in assessing the quality of our optimized potential using Decoys ‘R’Us data set.
Figure 2.
Heat map for the average RMSDs for best ranked models, for homology modeled targets from CASP8, for different weights of the four-body non-sequential and short-range potentials where the color gives the value of RMSD (Å). Right: Heat map for the full range of parameters. Left: Enlarged heat map for the best range of parameters.
The four-body non-sequential potentials don’t necessarily perform better than the sequential potentials, but after optimization, the resulting potentials perform better than either of the two individually, better than all other coarse-grained potentials (with an average RMSD approaching ~3.7 Å for the homology modeled targets), and almost at the same level of performance as fully atomistic potentials. For template-free modeled targets the Betancourt-Thirumalai25 potentials perform almost as well as the optimized potentials but for template-based modeled targets the improvement of the RMSD for the optimized potentials is significantly better.
For the misfolded, asilmarh and Pdberr&sgpa data sets from the Decoys ‘R’Us database the optimized potentials identify all native structures from these datasets and thereby performs as well as the other atomistic potentials (data not shown) like RAPDF33 atomic KBP19 and DFIRE (in the case of the DIFIRE-B potential, there was one mismatch). In Table S1 (see Supplementary materials), the native structure ranks and the Z-scores are compared for the above atomistic potentials and for our optimized potentials using multiple decoy sets. Optimized potentials are able to predict all native structures in the lattice-ssfit decoy set and they fail to identify only two native states in the 4-state reduced decoy set. Average Z-scores for the optimized potentials for these decoys is 1.87. Multi-body potentials perform well if protein structures are large enough, sufficiently compact and well-packed with many multi-body contacts (see Discussion).
Discussion
Coarse-grained potentials cannot be expected to recognize protein native structures with 100% accuracy regardless of the type of modeling used to generate structural models. This limitation could be due to the sample of structures used to derive the knowledge-based potentials, the geometric characterization afforded by the models used and the optimization methods used to generate models or the importance of long distance ranges of interactions that are not considered in their derivations. Therefore in order to obtain better quality assessments it is reasonable to produce decoys using one potential and assess their quality using other scoring functions. Such an example can be found in McGuffin35.
The RMSDs and Z-scores of the best predicted (by any potential) models using decoys for homology-based modeled targets and template-free modeled targets have been averaged over all targets. The results are shown in Table 3. This suggests that if we obtain RMSD and Z-score values that are not as good as these average values, then it might be possible to further improve the potentials used either by taking a linear combination of potentials or perhaps even by using a non-linear combination. For the results presented in Table 3, we knew the answer in advance, but in cases where there is not a large difference between results from single potentials, there is a chance that by combining potentials we might obtain a better performing combination. We recognize that there may be a significant opportunity for improvements in this field because for the template-free modeled targets there is a large gap between the best average prediction for a single (or optimized) potential, and those using sophisticated methods to combine them.
Table 3.
“Optimal” average Z-score and RMSD for best ranked decoys for hard (template-free) and easy (template-based modeled) targets from CASP8.
| Z-score | Top Ranked RMSD | |
|---|---|---|
| Easy | 2.21 | 1.24 |
| Hard | 2.75 | 2.12 |
Here we have combined two types of multi-body potentials along with the short range pair-wise potentials to obtain optimized potentials. The optimized potentials failed to identify the native structure for several cases of small protein from Decoys ‘R’Us data set (see Supplementary Materials), or in cases where the structure was stabilized by ions (Zn2+) or ligands (RNA). For proteins larger than about 80 amino acids and for those which are stable alone, our optimized potentials perform as well as the atomistic potentials. This simply reflects the fact that the correct packing is essential for protein stability, whether atomic or coarse-grained. In case when proteins are large, atomistic potentials in protein folding simulations are simply impractical. Thus, there is a need for efficient, well performing coarse-grained potentials. We believe that our optimized potentials will be helpful not only for threading and model ranking problems, but also in protein folding simulations.
It is also important to point out that this linear combination of three potential terms is robust. In Figure 2, where we show the average RMSD for the best ranked models for template-based (homology) modeled targets, a yellow island is observed within which the performances are nearly equal. It is interesting that the parameters set, which we received from optimization on template-free modeled targets (considered in the context of Figure 2), show no significant difference, to parameters optimized on homology template-based models. Thus these potentials can be considered to be universal and do not depend strictly on what type of modeling (homology or template-free) is being considered.
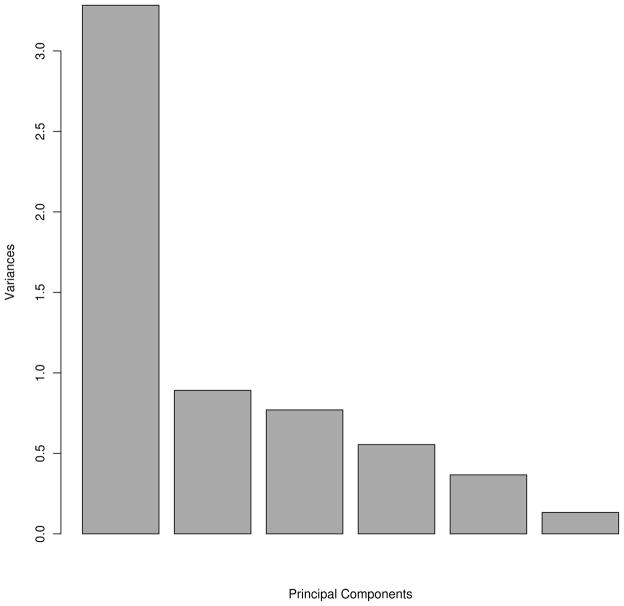
Principal component analysis (PCA) is a method to reduce the number of possibly correlated variables into a smaller number of uncorrelated variables. Li et al. carried out a PCA of Miyazawa-Jernigan potentials40. They used eigenvalue decomposition, which is the most commonly used method in PCA. By identifying the first principal component vector and finding a significant correlation with the vector of hydrophobicity indices of amino acids they showed that the dominant driving force for protein folding is the hydrophobic force. It is much more difficult and it requires more work to interpret major principal components in multi body combined potentials. We have carried out a principal component analysis using the four-body sequence dependent and non-sequence dependent, short-range, BT25, MJ336 and SKJG 37 for the case of the set 1sn3 from Decoys ‘R’Us. The variances of the principal components for the decoy energies with each potential are shown in Fig. 3. Each principal component is a combination of the above six potentials. It can be clearly seen that there is a major principal component that has the highest variance. The other five principal components are less important and, by definition, are orthogonal to the major principal component, and themselves. This tells that in energy model space there is a high redundancy of data (models usually capture common features of the system, and differ mostly in their details). Correlation coefficients between two-body potentials were calculated earlier by Pokarowski et al.34. Feng et al. found the correlations between sequential and non-sequential four-body potentials30. We presume that combining the best performing potentials that are less correlated should provide the best results. This is something that we will pursue in our future studies.
Figure 3.
Variances of principal components using four-body sequential, four-body nonsequential, short range, SKJG, BT and MT3 potentials.
Supplementary Material
Acknowledgments
We would acknowledge support from NIH Grants R01GM072014, R01GM081680 and R01GM081680-S1. We would like also to thank Yaping Feng for providing the server code.
References
- 1.Qu X, Swanson R, Day R, Tsai J. A Guide to Template Based Structure Prediction. Curr Protein Pept Sci. 2009;10:270–285. doi: 10.2174/138920309788452182. [DOI] [PubMed] [Google Scholar]
- 2.Kihara D, Chen, Yang YD. Quality Assessment of Protein Structure Models. Curr Protein Pept Sci. 2009;10:216–228. doi: 10.2174/138920309788452173. [DOI] [PubMed] [Google Scholar]
- 3.Skolnick J, Jaroszewski L, Kolinski A, Godzik A. Derivation and testing of pair potentials for protein folding. When is the quasichemical approximation correct? Protein Sci. 1997;6:676–688. doi: 10.1002/pro.5560060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skolnick J, Brylinski M. FINDSITE: a combined evolution/structure-based approach to protein function prediction. Brief Bioinform. 2009;10:378–391. doi: 10.1093/bib/bbp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryshtafovych A, Fidelis K. Protein structure prediction and model quality assessment. Drug Discov Today. 2009;14:386–393. doi: 10.1016/j.drudis.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellows ML, Floudas CA. Computational Methods for De novo Protein Design and its Applications to the Human Immunodeficiency Virus 1, Purine Nucleoside Phosphorylase, Ubiquitin Specific Protease 7, and Histone Demethylases. Curr Drug Targets. 2010;11:264–278. doi: 10.2174/138945010790711914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 8.Mandell DJ, Kortemme T. Backbone flexibilityin computational protein design. Curr Opin Biotechnol. 2009;20:420–428. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Gerlt JA, Babbitt PC. Enzyme (re)design: lessons from natural evolution and computation. Curr Opin Chem Biol. 2009;13:10–18. doi: 10.1016/j.cbpa.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vajda S, Kozakov D. Convergence and combination of methods in protein-protein docking. Curr Opin Struct Biol. 2009;19:164–170. doi: 10.1016/j.sbi.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Azevedo WF, Dias R. Computational Methods for Calculation of Ligand-Binding Affinity. Curr Drug Targets. 2008;9:1031–1039. doi: 10.2174/138945008786949405. [DOI] [PubMed] [Google Scholar]
- 12.Vakser IA, Kundrotas P. Predicting 3D Structures of Protein-Protein Complexes. Curr Pharm Biotechnol. 2008;9:57–66. doi: 10.2174/138920108783955209. [DOI] [PubMed] [Google Scholar]
- 13.Ritchie DW. Recent Progress and Future Directions in Protein-Protein Docking. Curr Protein Pept Sci. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- 14.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-timescale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Roccatano D. Computer Simulations Study of Biomolecules in Non-Aqueous or Cosolvent/Water Mixture Solutions. Curr Protein Pept Sci. 2008;9:407–426. doi: 10.2174/138920308785132686. [DOI] [PubMed] [Google Scholar]
- 16.Fawzi NL, Yap EH, Okabe Y, Kohlstedt KL, Brown SP, Head-Gordon T. Contrasting Disease and Nondisease Protein Aggregation by Molecular Simulation. Acc Chem Res. 2008;41:1037–1047. doi: 10.1021/ar800062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rumfeldt JAO, Galvagnion C, Vassall KA, Meiering EM. Conformational stability and folding mechanisms of dimeric proteins. Prog Biophys Mol Biol. 2008;98:61–84. doi: 10.1016/j.pbiomolbio.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J Mol Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- 19.Lu H, Skolnick J. A distance-dependent atomic knowledge-based potential for improved protein structure selection. Proteins. 2001;44:223–232. doi: 10.1002/prot.1087. [DOI] [PubMed] [Google Scholar]
- 20.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures: quasi-chemical approximation. Macromolecules. 1986;18:534–552. [Google Scholar]
- 22.Miyazawa S, Jernigan RL. Residue-Residue Potentials with a Favorable Contact Pair Term and an Unfavorable High Packing Density Term, for Simulation and Threading. J Mol Biol. 1996;256:623–644. doi: 10.1006/jmbi.1996.0114. [DOI] [PubMed] [Google Scholar]
- 23.Sippl MJ. Calculation of conformational ensembles from potentials of mena force: An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka S, Scheraga HA. Medium- and Long-Range Interaction Parameters between Amino Acids for Predicting Three-Dimensional Structures of Proteins. Macromolecules. 1976;9:945–950. doi: 10.1021/ma60054a013. [DOI] [PubMed] [Google Scholar]
- 25.Betancourt M, Thirumalai D. Pair potentials for protein folding: choice of reference states and sensitivity of predicted native states to variations in the interaction schemes. Protein Sci. 1999;8:361–369. doi: 10.1110/ps.8.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munson P, Singh RK. Statistical significance of hierarchical multi-body potentials based on Delaunay tessellation and their application in sequence-structure alignment. Protein Sci. 1997;6:1467–1481. doi: 10.1002/pro.5560060711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Liang J. Geometric cooperativity and anticooperativity of three-body interactions in native proteins. Proteins. 2005;60:46–65. doi: 10.1002/prot.20438. [DOI] [PubMed] [Google Scholar]
- 28.Krishnamoorthy B, Tropsha A. Development of a four-body statistical pseudo-potential to discriminate native from non-native protein conformations. Bioinformatics. 2003;19:1540–1548. doi: 10.1093/bioinformatics/btg186. [DOI] [PubMed] [Google Scholar]
- 29.Feng Y, Kloczkowski A, Jernigan RL. Four-body contact potentials derived from two protein datasets to discriminate native structures from decoys. Proteins. 2007;68:57–66. doi: 10.1002/prot.21362. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y, Kloczkowski A, Jernigan R. Potentials ‘R’Us web-server for protein energy estimations with coarse-grained knowledge-based potentials. BMC Bioinformatics. 2010;11:92–5. doi: 10.1186/1471-2105-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bahar I, Kaplan M, Jernigan RL. Short-range conformational energies, secondary structure propensities, and recognition of correct sequence-structure matches. Proteins. 1997;29:292–308. doi: 10.1002/(sici)1097-0134(199711)29:3<292::aid-prot4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy J, Eberhart RC. Particle swarm optimization. Proceedings of IEEE International Conference on Neural Net-Works; 1995. pp. 1942–1948. [Google Scholar]
- 33.Samudrala R, Levitt M. Decoys “R” Us: a database of incorrect conformations to improve protein structure prediction. Protein Sci. 2000;9:1399–1401. doi: 10.1110/ps.9.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokarowski P, Kloczkowski A, Jernigan RL, Kothari NS, Pokarowska M, Kolinski A. Inferring ideal amino acid interaction forms from statistical protein contact potentials. Proteins. 2005;59:49–57. doi: 10.1002/prot.20380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuffin L. Benchmarking consensus model quality assessment for protein fold recognition. BMC Bioinformatics. 2007;8:345–15. doi: 10.1186/1471-2105-8-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyazawa S, Jernigan RL. Self-consistent estimation of inter-residue protein contact energies based on an equilibrium mixture approximation of residues. Proteins. 1999;34:49–68. doi: 10.1002/(sici)1097-0134(19990101)34:1<49::aid-prot5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 37.Skolnic J, Jaroszewski L, Kolinsk A, Godzik A. Derivation and testing of pair potentials for protein folding. When is the quasichemical approximation correct? Protein Sci. 1997:676–688. doi: 10.1002/pro.5560060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilis D. Protein decoy sets for evaluating energy functions. J Biomol Struct Dyn. 2004;21:725–736. doi: 10.1080/07391102.2004.10506963. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Tang C, Wingreen NS. Nature of driving force for protein folding: A result from analyzing the statistical potential. Phys Rev Letts. 1997;4:765–768. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.