Summary
Bacteria rapidly adapt to nutritional changes via the stringent response, which entails starvation-induced synthesis of the small-molecule, ppGpp, by RelA/SpoT homolog (Rsh) enzymes. Binding of ppGpp to RNA polymerase modulates the transcription of hundreds of genes and remodels the physiology of the cell. Studies of the stringent response have primarily focused on copiotrophic bacteria such as E. coli; little is known about how stringent signaling is regulated in species that live in consistently nutrient limited (i.e. oligotrophic) environments. Here we define the input logic and transcriptional output of the stringent response in the oligotroph, Caulobacter crescentus. The sole Rsh protein, SpoTCC, binds to and is regulated by the ribosome, and exhibits AND-type control logic in which amino acid starvation is a necessary but insufficient signal for activation of ppGpp synthesis. While both glucose and ammonium starvation upregulate the synthesis of ppGpp, SpoTCC detects these starvation signals by two independent mechanisms. Although the logic of stringent response control in C. crescentus differs from E. coli, the global transcriptional effects of elevated ppGpp are similar, with the exception of 16S rRNA transcription, which is controlled independently of spoTCC. This study highlights how the regulatory logic controlling the stringent response may be adapted to the nutritional niche of a bacterial species.
Keywords: Rsh, stringent response, IS3 insertion element, ribosome, L11, SpoT, Caulobacter, oligotroph, ppGpp
INTRODUCTION
Bacteria must coordinate growth rate with nutrient availability so as to balance survival under starvation conditions with reproduction in nutrient replete conditions. The stringent response is a bacterial signaling system that plays a critical role in this coordination of growth with nutrient availability (Potrykus & Cashel, 2008). This system is conserved in all bacterial species except obligate intracellular pathogens and obligate symbionts (Mittenhuber, 2001). In this response, RelA/SpoT homolog (Rsh) enzymes synthesize guanosine tetra- or penta-phosphate ((p)ppGpp) upon starvation. In gram-negative species, (p)ppGpp binds RNA polymerase (RNAP) and alters its activity at different promoters such that genes required for translation and cell building are downregulated while stress response genes are upregulated (Barker et al., 2001a, Barker et al., 2001b, Traxler et al., 2006, Traxler et al., 2008). (p)ppGpp also modulates the activity of certain DNA replication factors (Schreiber et al., 1995, Ferullo & Lovett, 2008, Lesley & Shapiro, 2008). Although the stringent response was initially defined in E. coli as the downregulation of stable RNA transcription in response to amino acid starvation (Cashel, 1969), several decades of research have demonstrated that (p)ppGpp is central in the vast network of bacterial sensory/signaling systems: this small molecule is rapidly synthesized upon multiple types of stress and alters transcription of large swathes of the genome (Potrykus & Cashel, 2008). We note that in the older literature the term “stringent response” refers specifically to the production of stable RNA after amino acid starvation. However, this term has come to encompass all aspects of cell physiology regulated by elevated ppGpp levels (Potrykus & Cashel, 2008). For the purposes of this study we use the term stringent response to refer to all effects of cytosolic ppGpp accumulation.
(p)ppGpp plays a central role in adaptation to changing environments in many bacterial species. For example, several mammalian pathogens use (p)ppGpp signaling to upregulate virulence genes upon host entry (Nakanishi et al., 2006, Dozot et al., 2006), to survive upon elimination from the host (Das et al., 2009), and to activate infection (Kim et al., 2005, Dalebroux et al., 2009, Edwards et al., 2009) or persistence (Dahl et al., 2003) factors. In E. coli the transition from the rich environment of the mammalian gut to the comparatively poor environment of the water table (Savageau, 1983) is mediated by (p)ppGpp signaling (Traxler et al., 2006). These species are all classified as copiotrophs, which experience an existence of feast and famine (Koch, 1971, Poindexter, 1981) and exhibit a “classic” stringent response whereby amino acid starvation induces the synthesis of (p)ppGpp. However, there are examples of species that exhibit a “non-classic” stringent response; the study of these cases provides important insight into the variable and conserved aspects of (p)ppGpp signaling, and highlights how the nutritional niche of a species can affect the structure of its molecular signaling systems.
Examples of species that do not produce ppGpp upon amino acid starvation include Rhodobacter sphaeroides, Rhizobium meliloti strain 41, Rhizobium tropici, Azomonas agilis and Azobacter vinelandii (Acosta & Lueking, 1987, Belitsky & Kari, 1982, Eccleston Jr. & Gray, 1973, Howorth & England, 1999). Both R. meliloti 41 and R. sphaeroides restrict rRNA transcription without production of ppGpp in amino acid starvation (Acosta & Lueking, 1987, Belitsky & Kari, 1982, Eccleston Jr. & Gray, 1973), but produce ppGpp in other types of starvation: R. meliloti 41 in carbon and ammonium starvation (Belitsky & Kari, 1982), and R. sphaeroides, a photosynthetic bacterium, upon downshift of light intensity (Eccleston Jr. & Gray, 1973). These species still downregulate rRNA synthesis during amino acid starvation and are thus “stringent” in the original sense of the word.
There are reports of “relaxed” species, including Helicobacter pylori and Caulobacter crescentus, which neither reduce rRNA transcription nor synthesize ppGpp in a few tested conditions of amino acid starvation (Scoarughi et al., 1999, Chiaverotti et al., 1981). Yet these species still synthesize ppGpp: C. crescentus in glucose (Lesley & Shapiro, 2008) and ammonium (Chiaverotti et al., 1981) starvation, and H. pylori in carbon and serum starvation and acid stress (Wells & Gaynor, 2006, Zhou et al., 2008). Both species also restrict rRNA transcription in conditions which activate ppGpp accumulation: carbon starvation in H. pylori (Wells & Gaynor, 2006) and nutrient downshift in C. crescentus (data not shown and (Amemiya, 1989)). To date, it has not been shown that ppGpp is responsible for rRNA transcriptional control in either species. Thus, while (p)ppGpp signaling is nearly ubiquitous in bacteria, it is clearly cued by different signals in different species; in some species other factors may be responsible for rRNA transcriptional control.
When studying diversity in the regulation of (p)ppGpp synthesis, it is necessary to consider the Rsh enzymes. The most well-studied of these are the paralogous enzymes RelAEC and SpoTEC of E. coli. RelAEC functions solely as a (p)ppGpp synthase (Aravind & Koonin, 1998); it binds directly to the ribosome and is activated by uncharged tRNA in the acceptor (A) site (Haseltine & Block, 1973). The C-terminal regulatory domains of RelAEC (Schreiber et al., 1991) and the L11 protein of the 50S ribosome (Friesen et al., 1974, Parker et al., 1976) function together to detect this uncharged tRNA signal. SpoTEC, on the other hand, both synthesizes and hydrolyzes (p)ppGpp (Xiao et al., 1991) and is regulated by starvation for phosphate (Spira et al., 1995), carbon (Xiao et al., 1991), iron (Vinella et al., 2005), and lipids (Seyfzadeh et al., 1993) and by deprivation for multiple amino acids in combination. Unlike RelAEC, starvation of single amino acids does not activate SpoTEC (Murray & Bremer, 1996). SpoTEC associates with the 50S subunit of the ribosome, but does not apparently associate with the translating ribosome (Jiang et al., 2007). It is not clear how SpoTEC is activated, except in the case of lipid starvation (Battesti & Bouveret, 2006).
In this work we examine the regulation of stringent signaling in C. crescentus, a bacterium that lives in nutrient poor (i.e. oligotrophic) freshwater environments, and exhibits a non-classical stringent response that is regulated by a single Rsh enzyme - annotated SpoTCC. Our results confirm that C. crescentus synthesizes ppGpp in response to glucose and ammonium starvation, but not amino acid starvation. We show that SpoTCC associates with the ribosome, and exhibits AND-type signaling logic in which detection of an uncharged tRNA at the acceptor site is a necessary but insufficient signal for activation of ppGpp synthesis by SpoTCC. Our work with SpoTCC and a C. crescentus ribosomal mutant provides evidence that glucose and ammonium starvation are detected via different mechanisms. A genome-scale transcriptional analysis delineates the global effects of increased cytosolic ppGpp on gene expression during carbon starvation. Quantification of 16S rRNA transcription by qRT-PCR shows that rRNA levels are restricted independently of spoTCC, and only in select amino acid starvation conditions. This study defines regulatory features of the stringent response in an oligotrophic bacterium, and provides an example of how the signaling logic of a broadly conserved signaling system may be tailored to a particular environmental niche.
RESULTS
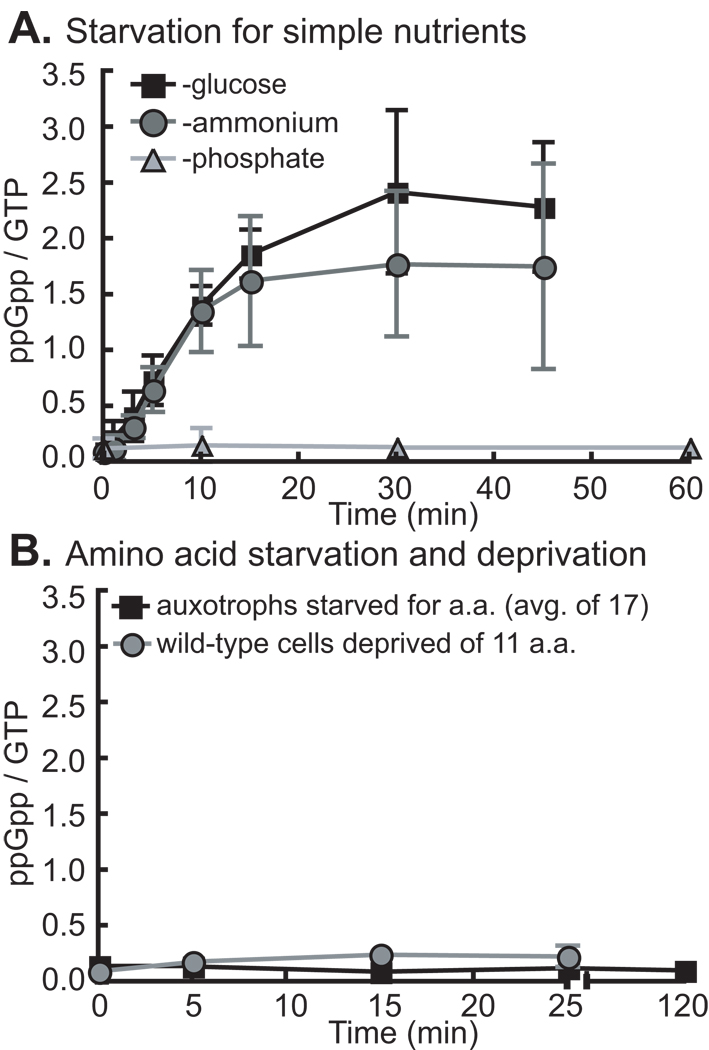
ppGpp synthesis in Caulobacter crescentus is regulated by glucose and ammonium, but not phosphate or amino acid starvation
To determine the types of starvation to which the stringent response of wild-type C. cresecentus strain NA1000 (Evinger & Agabian, 1977, Marks et al., 2010) is sensitive, we measured ppGpp accumulation by thin-layer chromatography (TLC) after selective removal of nutrients from glucose defined medium (M2G) supplemented with KH232PO4. It is known that C. crescentus synthesizes ppGpp under glucose (Lesley & Shapiro, 2008) and ammonium (Chiaverotti et al., 1981) starvation conditions. Our data confirm these results and show that SpoTCC does not respond to phosphate starvation (Figure 1A). Previous work has shown that starvation for arginine, proline, isoleucine, serine and lysine failed to induce ppGpp accumulation at short time points in C. crescentus. To more comprehensively examine the response of SpoTCC to amino acid starvation, we used strains that were auxotrophic for 17 amino acids – all except alanine, glutamine and asparagine. These strains have transposon insertions within, or deletions of, amino acid biosynthetic genes and thus require specific amino acids for growth. We measured ppGpp accumulation in these auxotrophs when starved for their respective required amino acid at short and long time points. Our results show that C. crescentus only accumulates very low levels of ppGpp in any of the tested amino acid starvation conditions (Figure 1B, S1).
Figure 1. SpoTCC responds to glucose and ammonium, but not amino acid or phosphate starvation.
A) ppGpp accumulation in wild-type cells in glucose, ammonium and phosphate starvation. For glucose and ammonium starvation, n=4; for phosphate starvation, n=2. B) ppGpp production in a series of auxotrophic strains starved for their respective amino acids at 5 minute and 120 minute time points. Data for separate auxotrophic strains starved individually for 17 different amino acids are similar and have been averaged (black line). ppGpp starvation curves for individual amino acid auxotrophs are presented in Figure S1. Also, presented, ppGpp accumulation in wild-type cells cultured in the presence of all 11 non-inhibitory amino acids and then simultaneously deprived of these amino acids (gray line; N=2). Error bars refer to standard deviation for all experiments.
In E. coli, SpoTEC does not synthesize ppGpp upon starvation for single amino acids, but rather upon depletion of multiple amino acids (Murray & Bremer, 1996). To test whether SpoTCC responds similarly to depletion of multiple amino acids for which it is prototrophic, we cultured wild-type C. crescentus with the 11 amino acids that do not inhibit growth of this species (Ferber & Ely, 1982) at 100 µg/ml in M2G with KH232PO4. We then washed all amino acids out of the medium and measured ppGpp accumulation. Our results show that simultaneous removal of multiple amino acids from the medium does not induce significant ppGpp accumulation (Figure 1B); thus SpoTCC differs from SpoTEC in its response to amino acid deprivation and phosphate starvation (Spira et al., 1995).
SpoTCC is physically associated with the ribosome
E. coli RelAEC associates with the ribosome (Ramagopal & Davis, 1974, Gentry & Cashel, 1995); this interaction is presumed to be required for regulation of RelAEC activity by uncharged tRNAs (Haseltine & Block, 1973). SpoTEC associates only with the 50S subunit (Jiang et al., 2007). C. crescentus SpoTCC differs from its E. coli Rsh homologs in that it is unresponsive to amino acid starvation (Figure 1B). We sought to test whether an Rsh protein that does not respond directly to amino acid starvation may still associate with the ribosome.
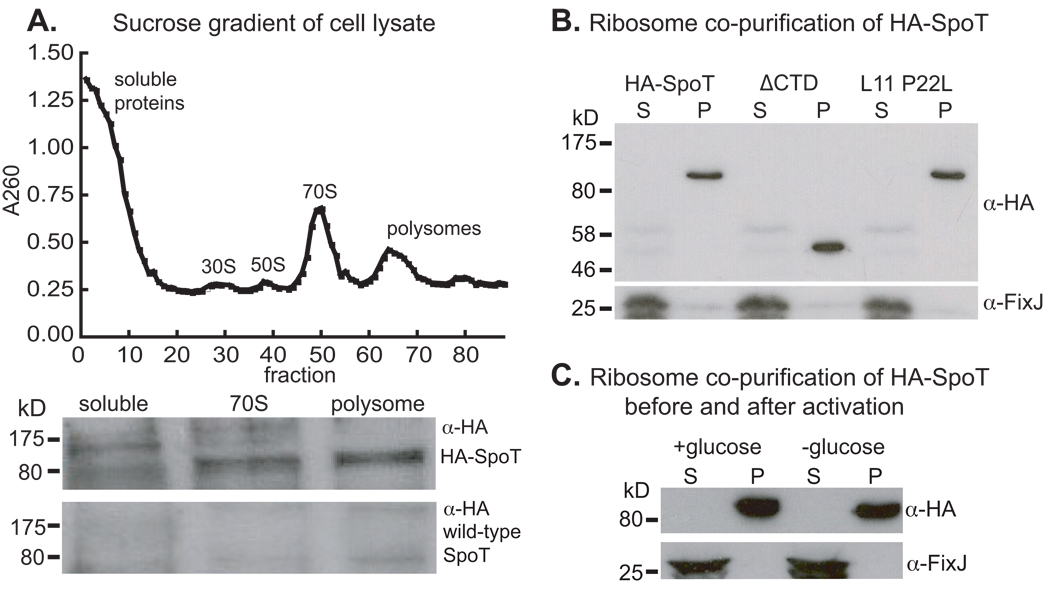
To assess ribosome association of SpoTCC, we built a C. crescentus strain in which spoTCC is fused to an N-terminal HA tag (HA-spoTCC). This tag does not interfere with SpoTCC activity or cause spurious ribosome association in a cytoplasmic control protein (Figure S2). Cell lysate from this strain was resolved on a sucrose gradient and ribosomal peaks were analyzed by Western blot with an α-HA antibody. Data from this experiment demonstrate that HA-SpoTCC is enriched in the 70S and polysome fractions of the lysate, while little or no HA-SpoTCC is present in the soluble fraction (Figure 2A). From this, we conclude that SpoTCC associates with the translating ribosome.
Figure 2. SpoTCC associates with the ribosome.
A) sucrose-gradient polysome profile (top) and western blot (bottom) of pooled fractions from the soluble, 70S and polysome peaks. The top blot strip is from the HA-SpoT strain, the bottom strip is from the wild-type strain, which serves as a negative control. B) Western blots of supernatant (S) and pellet (P) fractions from cell lysates spun through a 1M sucrose cushion to separate the ribosomes from other cell constituents (See Figure S2). HA-SpoT co-purifies with the ribosome when the C-terminal domains are removed (ΔCTD) and in the L11 P22L ribosomal mutant. C) HA-SpoT co-purifies with the ribosome in M2G and after 15 minutes of glucose starvation in M2. FixJ is presented as a non-ribosome-associated control.
It is known that deletions of C-terminal regions of RelAEC and a P22L mutation in ribosomal protein L11 prevent RelAEC from detecting uncharged tRNA in the ribosomal A site (Jenvert & Schiavone, 2007, Schreiber et al., 1991). To test whether ribosome-association of SpoTCC is affected by similar mutations, we isolated C. crescentus ribosomes by sucrose cushion centrifugation (Cross, 1970, Spedding, 1996) and analyzed the ribosome pellet and soluble fractions by Western blot. HA-SpoTCC is present in the ribosome pellet fractions in both the C-terminal truncation of SpoTCC, and in an L11 P22L ribosomal mutant (Figure 2B, Figure S3), demonstrating that these mutations do not affect ribosome association. The effects of these mutations on SpoTCC activity will be discussed below.
In vitro, RelAEC is thought to have a lower affinity for ribosomes during active ppGpp synthesis (Wendrich et al., 2002). To test whether ribosome association of SpoTCC is altered during ppGpp synthesis in vivo, we isolated ribosomes from unstarved and glucose-starved cells. SpoTCC is found only in the ribosome fraction, regardless of whether it is actively synthesizing ppGpp (Figure 2C).
Ribosome activity affects the regulation of cytosolic ppGpp levels by SpoTCC
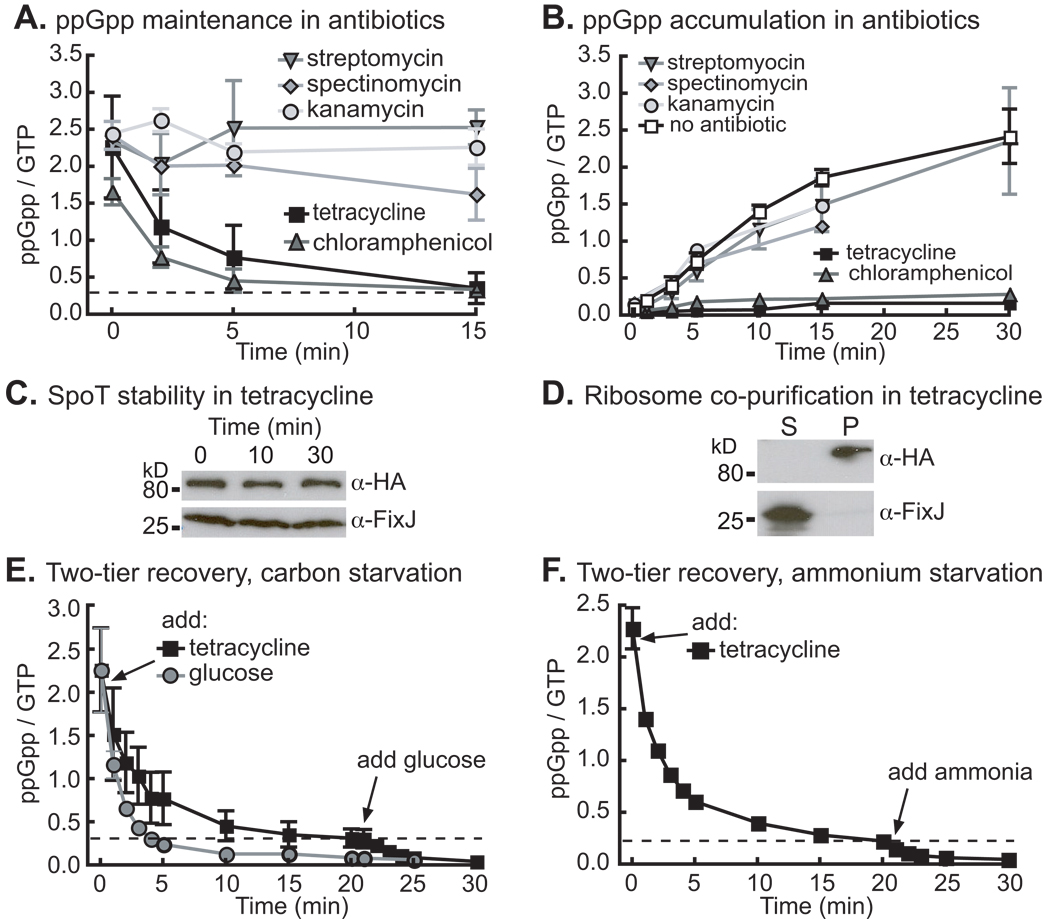
Given that amino acid starvation alone is not sufficient to induce robust ppGpp synthesis in C. crescentus, the functional role of SpoTCC ribosome association was not immediately evident. Antibiotics that inhibit different aspects of ribosome activity have been used to explore the mechanism of RelAEC regulation (Lund & Kjeldgaard, 1972, Sokawa & Sokawa, 1978). To test whether inhibition of the C. crescentus ribosome affects SpoTCC activity, we measured ppGpp decay during glucose starvation upon addition of the ribosome antibiotics tetracycline, chloramphenicol, streptomycin, spectinomycin and kanamycin. When added to cells pre-starved for glucose, tetracycline and chloramphenicol induce rapid ppGpp decay; the other tested antibiotics do not induce decay (Figure 3A). The converse experiment yields consistent results: tetracycline and chloramphenicol prevent ppGpp accumulation upon glucose starvation, while streptomycin, spectinomycin and kanamycin do not (Figure 3B). Chloramphenicol inhibits peptidyltransferase activity and tetracycline blocks tRNAs from entering the A site: these drugs both bind to the 50S subunit (Gale et al., 1981). Chloramphenicol and tetracycline may prevent ppGpp accumulation either by inhibiting a ribosome function that is required for SpoTCC activity (see Discussion), or by some direct effect on SpoTCC. However, SpoTCC is stable (Figure 3C) and co-purifies with the ribosome (Figure 3D) in the presence of tetracycline, ruling out antibiotic effects on SpoTCC stability or ribosome association.
Figure 3. SpoTCC response to starvation is abrogated by certain ribosome poisons.
A) ppGpp decay in wild-type cells starved for glucose for two hours and then treated with chloramphenicol (50 µg/ml), tetracycline (50 µg/ml), kanamycin (250 µg/ml), spectinomycin (1250 µg/ml) or streptomycin (50 µg/ml). B) ppGpp accumulation in wild-type cells grown in M2G, and then washed and resuspended in M2 without glucose and with an antibiotic at the same concentration as in A. C) Western blot of HA-SpoT culture aliquots taken before (0) and after 50 µg/ml tetracycline was added to the culture. D) Western blots of supernatant (S) and pellet (P) fraction from an HA-SpoT culture that was treated with 50 µg/ml tetracycline for 15 minutes before being lysed and separated through a sucrose cushion. FixJ is a non-ribosome-associated control. E) ppGpp decay in wild-type cells starved for glucose for two hours and then treated with 50 µg/ml tetracycline or 0.2% glucose for 20 minutes: the tetracycline-treated cells were then treated with 0.2% glucose and analyzed for ppGpp content for an additional 10 minutes. F) ppGpp decay in wild-type cells starved for ammonium for two hours, treated with 50 µg/ml tetracycline for 20 minutes, then treated with 9.3 mM NH4Cl and analyzed for ppGpp content for an additional 10 minutes. N=2 for each experiment, error bars refer to the standard deviation.
As carbon starvation entails amino acid starvation (Ballesteros et al., 2001), uncharged tRNAs are expected to accumulate during glucose limitation and can function as a potential starvation signal. Our antibiotic data provide evidence that peptidyltransferase activity and binding of tRNA to the A site are necessary for SpoTCC activation. We therefore propose that, like RelAEC, activation of SpoTCC requires an uncharged tRNA to occupy the A site. However, unlike RelAEC, this signal is insufficient (Figure 1B, S1) and an additional signal induced by carbon or nitrogen starvation is required for full SpoTCC activation. This regulatory model is further tested below.
A two-tiered regulatory response of C. crescentus to ribosome inhibition and starvation
We observed that ppGpp levels did not completely decay to non-starvation levels when tetracyline and chloramphenicol were added to starved cultures (Figure 3A). This may be because the antibiotics directly, but incompletely, affect SpoTCC activity, or because they are interfering with only one of multiple signals sensed by SpoTCC during glucose starvation. Based on analogy to RelAEC regulation, we propose that tetracycline interferes with the signal for amino acid starvation. We therefore tested whether tetracycline-treated cells could still respond to relief of starvation by addition of glucose. We starved wild-type cells for glucose in the presence of KH232PO4 for two hours to induce ppGpp accumulation, then added tetracycline and measured ppGpp decay for 20 minutes. We then added glucose to these cells to relieve starvation (Figure 3E). We also measured ppGpp decay in a glucose-starved culture after the addition of glucose alone. These data show that when glucose is added to a starved culture, ppGpp levels quickly decay back to non-starvation levels. When tetracycline is added to a starved culture, ppGpp decays to a ppGpp/GTP ratio of ∼0.3–0.5. Only when glucose is added to this tetracycline-treated culture does ppGpp return to non-starvation levels (Figure 3E). This two-tiered response implies that there are two signals controlling SpoTCC activity: tetracycline interferes with one signal while glucose removes both. We conducted the same experiment with ammonium-starved cells, and observed the same two-tiered response. These results imply that amino acid starvation is also a necessary (Figure 3F) but insufficient (Figure 1B, S1) signal for SpoTCC activation in ammonium starvation.
SpoTCC functions as a synergistic ‘AND’ logic gate
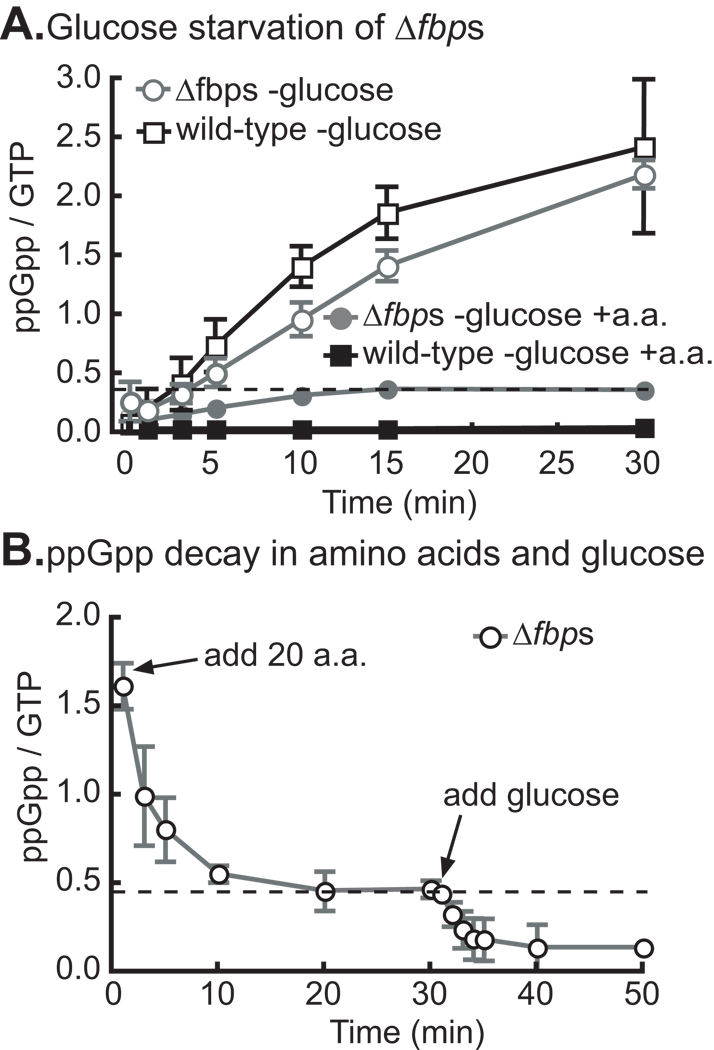
To genetically test whether glucose starvation in the absence of amino acid starvation is sufficient to activate SpoTCC, we built strain Δfbps in which both genes for the gluconeogenic enzyme fructose 1–6 bisphosphatase have been deleted (Table 1). This strain is a glucose auxotroph that cannot grow with amino acids as the sole carbon source (data not shown), and which synthesizes ppGpp normally upon glucose starvation (Figure 4A). We cultured Δfbps in M2G defined medium with KH232PO4 and all 20 amino acids (100 µg/ml), then washed glucose from the medium and measured ppGpp accumulation. Our results show that glucose starvation during amino acid supplementation induces only a low level of ppGpp (Figure 4A), and is therefore not sufficient to induce a full ppGpp response in the Δfbps strain. As a control, we show that wild-type cells starved for glucose in medium supplemented with amino acids do not accumulate detectable ppGpp (Figure 4A); these cells are able to synthesize glucose from the amino acids and are therefore not starving for any nutrient.
Table 1.
Strains
| FC # | Genotype | CCNA#, Gene altered | Reference |
|---|---|---|---|
| 20 | Caulobacter crescentus NA1000 | (Evinger & Agabian, 1977) | |
| 769 | NA1000 ΔspoT | CCNA_01622 | This work |
| 1220 | NA1000 ΔpheA | CCNA_03028 | This work |
| 1145 | NA1000 CCNA0559∷hMu | CCNA_00559 | (Huitema et al, 2006) |
| 1165 | NA1000 lysA∷mar414 | CCNA_02296 | (Huitema et al, 2006) |
| 1166 | NA1000 serA∷mar414 | CCNA_03322 | (Huitema et al, 2006) |
| 1164 | NA1000 CCNA1419∷mar414 | CCNA_01419 | (Huitema et al, 2006) |
| 1250 | NA1000 ΔgltB | CCNA_03722 | This work |
| 1183 | NA1000 cysE∷hMu | CCNA_02734 | (Huitema et al, 2006) |
| 1167 | NA1000 asp∷mar414 | CCNA_01603 | (Huitema et al, 2006) |
| 329 | NA1000 thrB∷mar414 | CCNA_03475 | (Huitema et al, 2006) |
| 316 | NA1000 proC∷hMu | CCNA_00528 | (Huitema et al, 2006) |
| 1163 | NA1000 argB∷hMu | CCNA_00285 | (Huitema et al, 2006) |
| 1144 | NA1000 ilvB∷tn5 | CCNA_02185 | (Huitema et al, 2006) |
| 314 | NA1000 leuC∷hMu | CCNA_00196 | (Huitema et al, 2006) |
| 325 | NA1000 trpE∷mar414 | CCNA_01972 | (Huitema et al, 2006) |
| 323 | NA1000 hisD∷mar414 | CCNA_02431 | (Huitema et al, 2006) |
| 943 | NA1000 HA-SpoT | CCNA_01622 | This work |
| 1105 | NA1000 HA-SpoTΔCTD | CCNA_01622 | This work |
| 1283 | NA1000 HA-SpoT L11 P22L | CCNA_01622,CCNA_00678 | This work |
| 1095 | NA1000 SpoTΔCTD | CCNA_01622 | This work |
| 1010 | NA1000 SpoTΔACT | CCNA_01622 | This work |
| 772 | NA1000 SpoTΔACT(668–719) | CCNA_01622 | This work |
| 1019 | NA1000 SpoTΔTGS | CCNA_01622 | This work |
| 1068 | NA1000 HA-SpoTΔACT | CCNA_01622 | This work |
| 1155 | NA1000 HA-SpoTΔACT(668–719) | CCNA_01622 | This work |
| 1083 | NA1000 HA-SpoTΔTGS | CCNA_01622 | This work |
| 1334 | NA1000 L11 P22L | CCNA_00678 | This work |
| 1257 | NA1000 xylX∷pP16Slacz | This work | |
| 1258 | NA1000 ΔspoT xylX∷pP16Slacz | CCNA_01622 | This work |
| 1304 | NA1000 ΔdksA xylX∷pP16Slacz | CCNA_02663 | This work |
| 1259 | NA1000 ΔpheA xylX∷pP16Slacz | CCNA_03028 | This work |
| 1261 | NA1000 ΔgltB xylX∷pP16Slacz | CCNA_03722 | This work |
| 1397 | NA1000 Δfbp ΔglpX, i.e., Δfbps | CCNA_01448, CCNA_01449 |
This work |
Figure 4. Glucose starvation in the presence of amino acids does not induce significant ppGpp accumulation.
A) ppGpp accumulation in Δfbps (glucose auxotroph) and wild-type cells starved for glucose with all 20 amino acids supplemented. Cells were grown in M2G with all 20 amino acids supplemented at 100 µg/ml each. Cells were then washed and resuspended in media with all 20 amino acids but no glucose, and ppGpp measured at times after glucose removal. The data for both strains starved for glucose without amino acid supplementation is included for comparison. B) ppGpp decay in Δfbps cells starved for glucose for two hours and then treated with addition of all 20 amino acids, after 30 minutes 0.2% glucose was added and further time points were taken. N=2 for each experiment, error bars refer to the standard deviation.
The converse experiment showed consistent results: glucose-starved Δfbps cells synthesize high levels of ppGpp, induce partial ppGpp decay upon addition of all 20 amino acids, and full decay upon addition of glucose (Figure 4B). These results provide evidence for a regulatory model in which SpoTCC functions as a synergistic ‘AND’ logic gate that requires both amino acid and glucose starvation signals for full activation. Individual starvation for either glucose or amino acids results only in very low levels of accumulated ppGpp (Figure 1B, 4A,B).
C-terminal truncations of SpoTCC decouple glucose and ammonium starvation responses
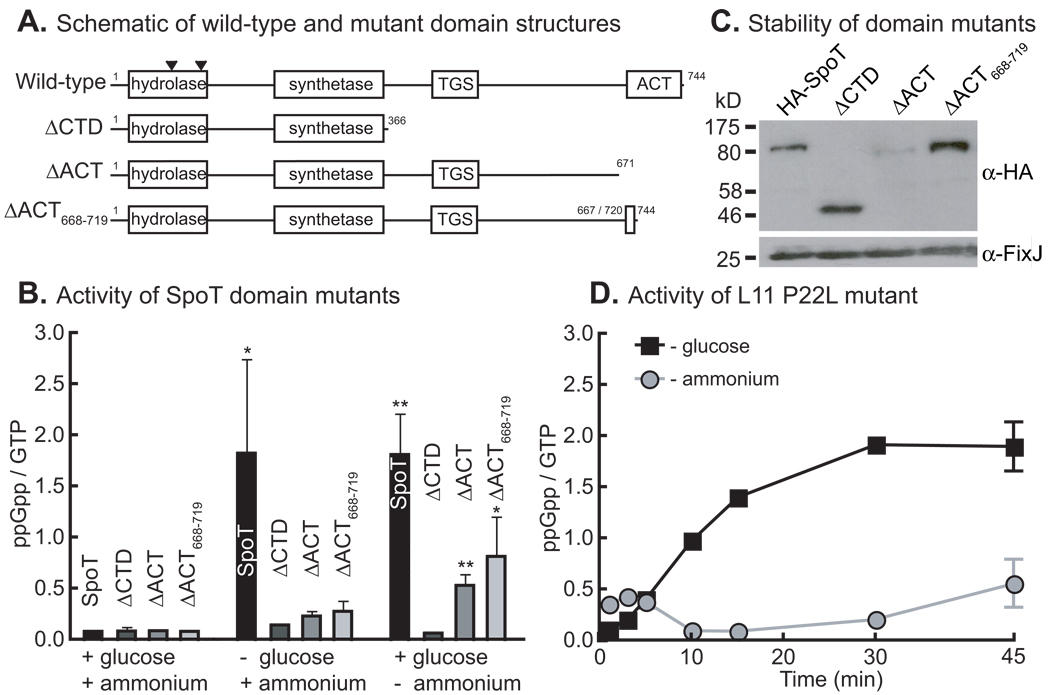
Rsh proteins have two conserved C-terminal domains, TGS and ACT, that are involved in detection of starvation signals (Battesti & Bouveret, 2006, Schreiber et al., 1991). To test the roles of these domains in starvation signal detection in SpoTCC, we constructed strains in which the wild-type allele of SpoTCC was replaced with alleles carrying truncations within the C-terminus (Figure 5A).
Figure 5. Glucose and ammonium starvation are sensed by different mechanisms.
A) Schematic showing domain structure of the full length SpoT protein and domain-deletion alleles. Locations of IS insertion elements found in hydrolase suppressor mutants are indicated by black triangles. (See last section of Results). B) In vivo ppGpp accumulation by the wild-type and mutant SpoT proteins in M2G, or after 2 hours of glucose or ammonium starvation. The stars indicate statistically significant difference, by student’s t-test, from the ΔCTD strain for each condition where *= p<0.01 and **=p<0.001. N=3. C) Western blot showing the stability of each domain-deletion protein in cells. N-terminal HA-tagged versions of each mutant were used. FixJ was used as a loading control. D) ppGpp accumulation in an L11 P22L mutant starved for glucose or ammonium. N=2. Error bars refer to standard deviation for all experiments
These mutant strains were incubated for two hours in the presence of KH232PO4 in replete M2G defined medium, M2 without glucose, or M2G without ammonium, and then analyzed by TLC to assess ppGpp levels. All of the mutants have impaired ppGpp accumulation in glucose and ammonium starvation (Figure 5B). The ΔCTD mutant fails to accumulate ppGpp under any condition. Notably, while all of the mutants fail to accumulate significant ppGpp under glucose starvation conditions, the ACT domain mutants do accumulate ppGpp under ammonium starvation. The stability of each of these SpoTCC truncations was assessed by Western blot in strains with N-terminal HA tags on each. These data show that all of the mutants are stable except ΔACT, which is present at very low levels (Figure 5C). The relatively high ppGpp accumulation observed in ammonium starvation by the unstable ΔACT mutant implies that it is hyper-responsive under this starvation condition.
Ribosomal protein L11 is involved in transduction of ammonium starvation signals to SpoTCC
In E. coli, a P22L mutation of L11 abrogates the ability of RelAEC to respond to amino acid starvation but has little effect on protein synthesis (Jenvert & Schiavone, 2007). To assess whether L11 is involved in the response of SpoTCC to starvation signals, we constructed an allelic replacement strain with a P22L mutation in L11 and tested its ability to accumulate ppGpp upon glucose and ammonium starvation. The data show that this mutant exhibits a defect in ppGpp accumulation under ammonium starvation but not glucose starvation (Figure 5D). This demonstrates that though the signals perceived by RelAEC and SpoTCC are different, there are conserved features of signal transduction through the ribosome. Moreover, the result that L11 P22L and SpoTΔACT alleles decouple the ammonium and glucose starvation responses provides evidence for independent mechanisms of carbon and nitrogen starvation signal detection by SpoTCC.
Activated SpoTCC downregulates growth genes and upregulates stress and starvation genes
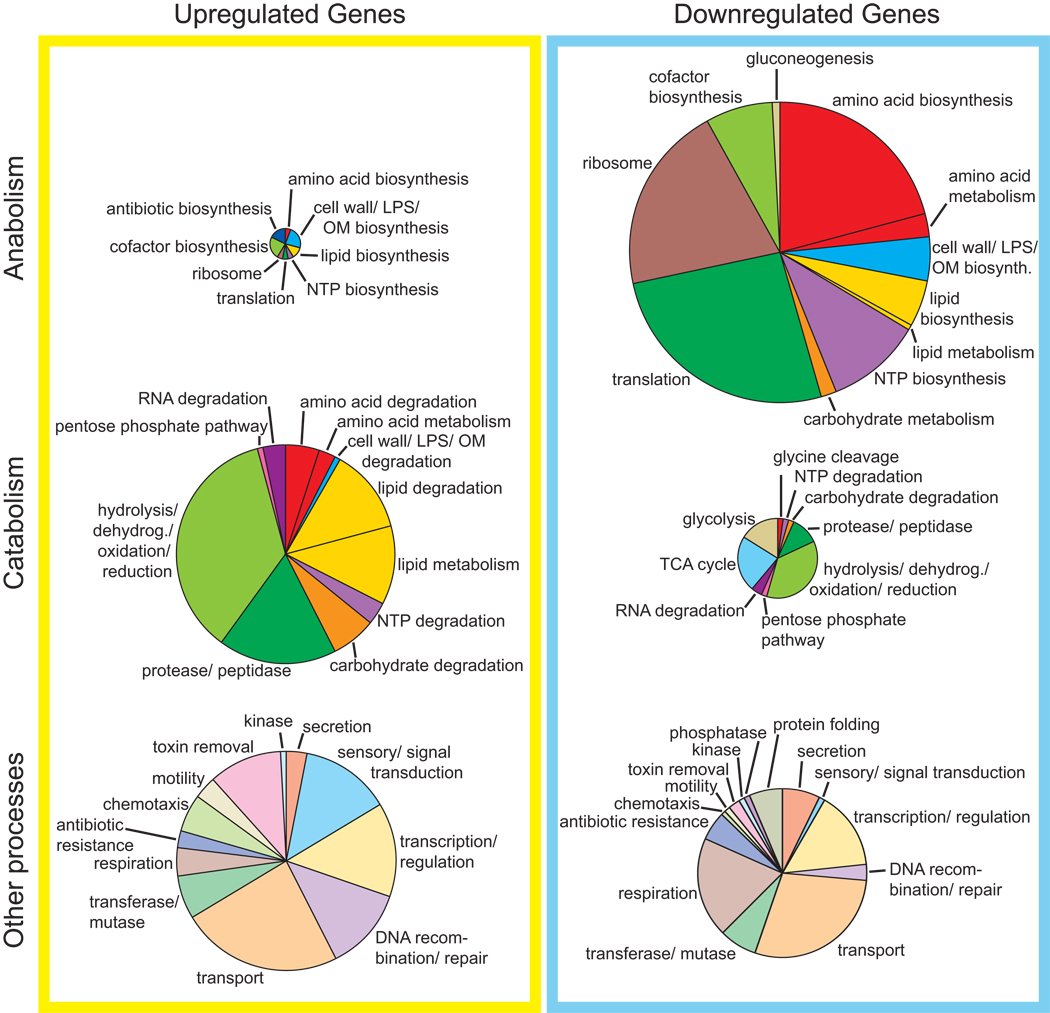
We sought to identify the genes that are regulated by C. crescentus SpoTCC upon carbon starvation. To quantify gene expression during the stringent response we grew wild-type strain NA1000 and NA1000 ΔspoTCC in M2G, washed glucose out of the medium and starved the cells for five minutes before extracting RNA and hybridizing labeled cDNA to Affymetrix GeneChip, CauloHi1 (GEO microarray accession number GSE21206). The results from this experiment show large-scale downregulation of anabolic genes and upregulation of catabolic, sensory, chemotaxis and DNA recombination genes (Figure 6, Table S1). These results are generally consistent with the stringent transcriptional response described in other species, with the notable exception of amino acid biosynthetic genes, which are downregulated during carbon starvation by spoTCC. This exception is likely due to our experimental protocol in which cells are starved for glucose, not amino acids. Several other genes of interest are regulated in carbon starvation by spoTCC, including genes involved in cell division and nutrient granule metabolism (Table 2).
Figure 6. Microarray analysis of genes regulated by C. crescentus SpoTCC.
The genes included in this figure are protein-coding open reading frames that were regulated at least 3-fold in the wild-type versus ΔspoTCC Affymetrix data set. The size of each pie is proportional to the number of genes in that category. In total, 379 genes were upregulated at least 3-fold (17-anabolic, 120-catabolic, 126-other processes, 8-cell cycle/nutrient granule, 108-unknown function), and 382 genes were downregulated at least 3-fold by spoT (166-anabolic, 44-catabolic, 94-other processes, 6-cell cycle/nutrient granule, 71-unknown function) (see Table S1). Genes of unknown function and cell cycle/nutrient granule genes in Table 2 are not included in the pie charts
Table 2.
Regulated cell cycle and nutrient granule genes
| Upregulated genes | ||||
|---|---|---|---|---|
| Function | Gene # | Gene name | WT/ΔspoT | References |
| cell division | CCNA_02635 | Cell division protein ftsW | 4.23 | (Mercer & Weiss, 2002) |
| PHB degradation | CCNA_00250 | Poly(3-hydroxyalkanoate) depolymerase |
3.04 | (Karp et al., 1999, Romero et al., 2001) |
| PHB metabolism | CCNA_00544 | Acetyl-CoA acetyltransferase | 4.32 | (Karp et al., 1999, Romero et al., 2001) |
| PHB metabolism | CCNA_00545 | acetoacetyl-CoA reductase | 3.45 | (Karp et al., 1999, Romero et al., 2001) |
| PHB metabolism | CCNA_01444 | Poly(3-hydroxyalkanoate) polymerase | 3.03 | (Karp et al., 1999, Romero et al., 2001) |
| PHB metabolism | CCNA_03293 | Enoyl-CoA hydratase | 5.28 | (Karp et al., 1999, Romero et al., 2001) |
| polyphosphate synthesis | CCNA_03529 | Polyphosphate kinase | 7.86 | (Karp et al., 1999, Romero et al., 2001) |
| replication inhibition | CCNA_01451 | Cold shock protein cspD | 3.78 | (Yamanaka et al., 2001) |
| Downregulated genes | ||||
|---|---|---|---|---|
| Function | Gene # | Gene name | WT/ΔspoT | References |
| cell division | CCNA_03792 | Intracellular septation protein | 0.33 | (Siomoin et al., 1996) |
| cell division inhibition | CCNA_02427 | Septum formation protein Maf | 0.26 | (Butler et al., 1993) |
| DNA methylation/ cell cycle |
CCNA_00382 | modification methylase CcrMI | 0.16 | (Collier et al., 2007, Reisenauer & Shapiro, 2002) |
| polyphosphatase | CCNA_01780 | Exopolyphosphatase | 0.06 | (Karp et al., 1999, Romero et al., 2001) |
| replication | CCNA_01596 | DNA helicase II | 0.04 | (Lesley & Shapiro, 2008) |
| replication | CCNA_03701 | Integration host factor beta-subunit | 0.19 | (Lesley & Shapiro, 2008) |
SpoTCC does not regulate rRNA transcription during glucose starvation
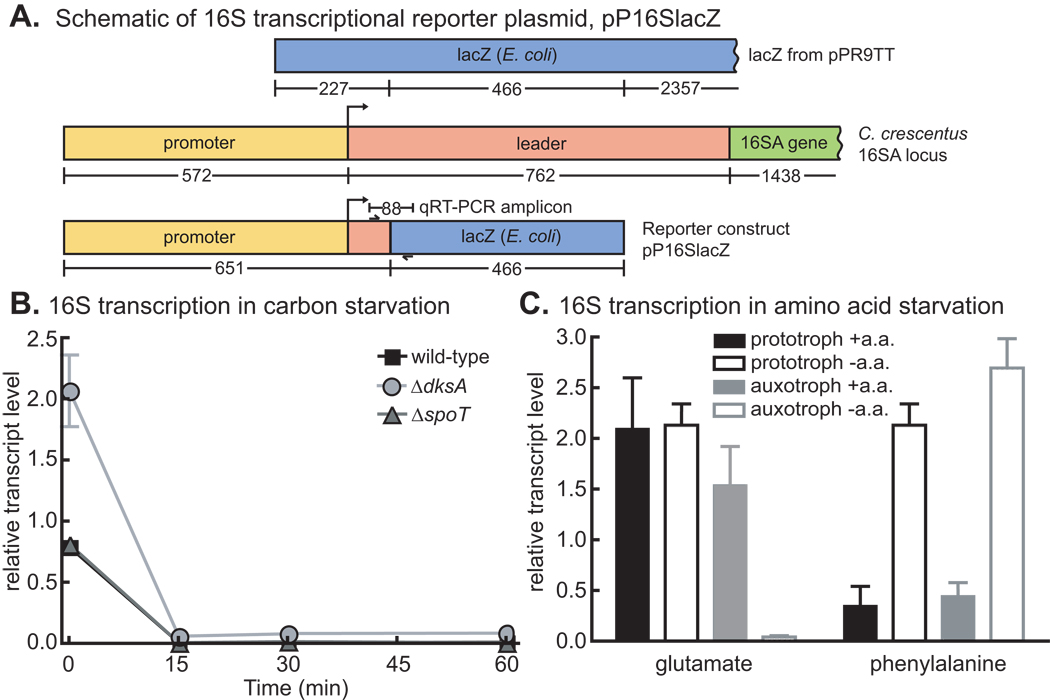
The first function assigned to RelAEC was that of a negative regulator of rRNA transcription during amino acid starvation (Stent & Brenner, 1961). To assess the role of SpoTCC in rRNA transcriptional control, we constructed a 16S rRNA transcriptional reporter plasmid that contains the 16SA promoter and the first 81 nucleotides of the 16SA leader followed by a segment of the lacZ gene from E. coli. The unstable transcript from this reporter can be quantified by qRT-PCR to yield a measure of 16S rRNA transcription (Figure 7A). Strains containing this reporter plasmid were grown in M2G medium and then starved by washing out glucose; RNA samples were collected from cells before and after starvation. For each sample, data from the 16S amplicon were normalized to data from an amplicon of the ruvA DNA helicase gene. ruvA was chosen as a normalization control because ruvA transcript levels did not change under starvation conditions as assessed using multiple unique probes on the Affymetrix microarray. The qRT-PCR data show that 16S transcription is quickly downregulated after glucose deprivation, but that this response is not dependent on spoTCC (Figure 7B). DksA is required for regulation of rRNA transcription in E. coli (Paul et al., 2004). We repeated the rRNA transcriptional assay in a ΔdksA strain: this mutation affects the basal level of 16S transcription but is not responsible for downregulation of transcription during glucose starvation (Figure 7B) in C. crescentus. These data provide evidence for a SpoT/DksA-independent mechanism of rRNA transcriptional control during starvation in C. crescentus.
Figure 7. rRNA transcriptional control.
A) Schematic of 16S transcriptional reporter plasmid, which produces an unstable transcript from the 16S A rRNA promoter. Numbers below genes represent the number of nucleotides. Location of qRT-PCR amplicon primers are indicated by small arrows. B) 16S rRNA promoter activity in wild-type, ΔspoT and ΔdksA strains before (0 minutes) and during an hour of glucose starvation. N=3. C) 16S rRNA promoter activity in the prototrophic strain (NA1000 pxyl∷pP16Slacz) and the auxotrophic strains (NA1000 ΔgltB pxyl∷pP16Slacz and NA1000 ΔpheA pxyl∷pP16Slacz) in M2G with and without added glutamate or phenylalanine. The cells were grown up in M2G + glu or M2G + phe + ala, and then washed and resuspended in M2G or M2G + ala and grown in those conditions for two hours before the starvation samples were taken. All data were normalized to the signal from the ruvA amplicon on the same biological sample (N=3). Error bars refer to standard deviation for all experiments.
Starvation for glutamate - but not phenylalanine - results in downregulation of rRNA transcription
While the data presented above show that ppGpp is not involved in stringent control of rRNA transcription (Figure 7B), we wished to test whether C. crescentus restricts rRNA synthesis upon amino acid starvation. It has been reported that net RNA synthesis in C. crescentus is not restricted upon the addition of chemical inhibitors of isoleucine, serine and lysine synthesis or utilization; however, controls demonstrating that these chemicals induced starvation were not described (Chiaverotti et al., 1981). With respect to C. crescentus growth, there are two classes of amino acids: nine which inhibit growth when added to minimal medium (Cys, His, Ile, Leu, Met, Phe, Ser, Thr, Val) and 11 which do not (Ferber & Ely, 1982). We hypothesized that the rRNA transcriptional response to starvation of amino acids in these two classes may be different. To study a representative from each class, we built phenylalanine (ΔpheA) and glutamate (ΔgltB) auxotrophs. These auxotrophic strains and a prototrophic strain were grown in M2G with phenylalanine or glutamate added. We then washed out the amino acids, starved the cells for two hours, and measured 16S rRNA transcription by qRT-PCR as described above. Our results show that glutamate deprivation results in a reduction of rRNA transcription only in the ΔgltB auxotroph, which experiences starvation (Figure 7C). Addition of phenylalanine, however, results in a reduction of 16S transcription in the prototroph and auxotroph. Starvation for phenylalanine in the auxotroph does not result in reduced rRNA transcription. The growth inhibition observed in C. crescentus when phenylalanine is added to growth media (Ferber & Ely, 1982) may indirectly cause rRNA transcriptional repression.
ppGpp accumulation activates IS3 elements in the C. crescentus genome
We observed an increase in transcription of insertion sequence (IS) elements in our global transcriptional analysis (Figure 6, Table S1). This result was functionally confirmed in a strain with SpoTCCH67A, which carries a point mutation that ablates ppGpp hydrolase activity (Hogg et al., 2004). Initial characterization of the SpoTCC H67A strain indicated slow growth, as expected for a strain unable to hydrolyze ppGpp. However, suppressors with wild-type growth rates appeared quickly, and we determined that they were no longer able to synthesize ppGpp (data not shown). Sequencing of the spoTCC locus in three independent suppressors revealed IS3 insertion at codon 115 in one and at codon 161 in the other two (Figure 5A). The transcriptional data and isolation of multiple IS3-mediated H67A suppressors provides evidence that increased cellular ppGpp, independent of starvation stress, is sufficient to activate IS3 insertion elements.
DISCUSSION
Prokaryotes inhabit a diverse range of ecological niches; the structure of environmental sensing and signaling mechanisms is correspondingly diverse. The stringent response has been most thoroughly studied in a few copiotrophic species including E. coli (Potrykus & Cashel, 2008), B. subtilis (Ochi et al., 1982), and Legionella pneumophila (Dalebroux et al., 2009, Edwards et al., 2009, Wells & Gaynor, 2006, Zhou et al., 2008). However, there is evidence that species living in different ecological niches have stringent response systems that are structured and regulated differently (Chiaverotti et al., 1981, Acosta & Lueking, 1987, Belitsky & Kari, 1982, Howorth & England, 1999, Scoarughi et al., 1999, Wells & Gaynor, 2006, Zhou et al., 2008). In this work we illuminate several regulatory features of the stringent response in the model oligotroph C. crescentus.
Amino acid starvation is not a sufficient signal to induce ppGpp accumulation in the oligotroph C. crescentus
For E. coli, amino acids are a constant and important nutrient source in the intestine. In the transition from intestine to the environment outside its commensal host, amino acid concentration scales with total nutrient availability (Savageau, 1983). Thus measuring amino acid levels is a convenient proxy for E. coli to assess environmental nutrient status. In contrast, C. crescentus lives in oligotrophic environments where amino acid concentrations are consistently low (Poindexter, 1981, Thomas, 1997) and may have little correlation with total nutrient availability. C. crescentus is able to respond to low cytoplasmic concentrations of amino acids by upregulating expression of amino acid biosynthetic genes (Tarleton et al., 1994), but, in contrast to E. coli and other copiotrophs, it does not activate the stringent response upon amino acid starvation alone.
SpoTCC is bound to the ribosome
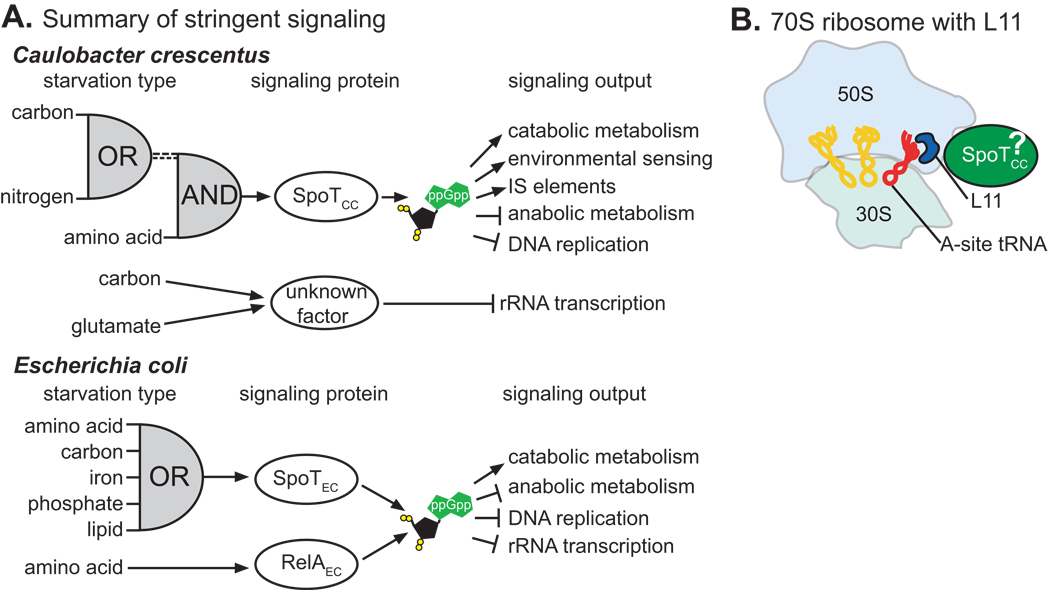
The E. coli bifunctional ppGpp hydrolase/synthetase SpoTEC binds to the 50S ribosomal subunit, but not the translating ribosome (Jiang et al., 2007). Bifunctional Rsh proteins that are regulated solely by amino acid starvation and bind the ribosome have been previously identified (Avarbock et al., 2000, Avarbock et al., 1999, Martinez-Costa et al., 1998). Our data are the first to demonstrate association of a bifunctional Rsh protein that does not respond solely to amino acid starvation with the translating ribosome (Figure 2A). Deletion of the C-terminal domain of SpoTCC does not prevent ribosome association (Figure 2B), indicating that either the N-terminus alone mediates association, or that both halves of the protein independently associate with the ribosome. In the case of E. coli RelAEC, it is not known if the C-terminal domains bind to the ribosome, only that they receive a signal through the ribosome (Schreiber et al., 1991, Gropp et al., 2001). It is noteworthy that on the ribosomal side, association with RelAEC is mediated by L10 (Howard et al., 1976) but signal transduction is mediated by L11 (Friesen et al., 1974, Schreiber et al., 1991): thus binding and signal transduction are spatially separated.
An L11 P22L ribosome mutation perturbs SpoTCC regulation (Figure 5D), but does not abrogate ribosome association (Figure 2B). This mirrors the observation in E. coli that L11, though important for signaling, does not bind directly to RelAEC (Yang & Ishiguro, 2001). SpoTCC does not appear to modulate its association with the ribosome during glucose starvation (Figure 2C), although this experiment cannot rule out small changes in SpoTCC-ribosome affinity. Antibiotics that bind to the 50S but not the 30S subunit interfere with SpoTCC activity (Figure 3A,B). The L11 protein and the amino-acylation portion of the A-site tRNA are near each other on the 50S subunit (Voorhees et al., 2010). We therefore propose that SpoTCC binds near these signals (Figure 8B).
Figure 8. Summary of the regulatory logic of the stringent response in a copiotrophic and an oligotrophic species, and a ribosome model.
A) Comparison of the signaling proteins, signaling logic, and signaling output of the stringent response in the oligotroph, C. crescentus, and the copiotroph, E. coli. The dashed lines from the OR gate of C. crescentus represent the different signals that activate the stringent response during carbon and nitrogen starvation. B) Cartoon of the bacterial 70S ribosome. SpoTCC is in green. SpoTCC is presumed to be near the 70S A site and ribosomal protein L11 based on regulatory data presented herein.
Combinatorial signal integration by SpoTCC: Evidence for ‘AND’ logic
The ribosome-association result reported herein was initially surprising given the observation that SpoTCC does not respond to amino acid starvation in the same way as RelAEC or SpoTEC (Figure 1, S1). However, our functional data demonstrate a role for ribosomal protein L11 in SpoTCC regulation (Figure 5D) and pharmacological data show that certain ribosome functions are required for ppGpp accumulation during glucose starvation (Figure 3). Together, these data provide support for a model in which the ribosome plays an important role in SpoTCC regulation.
Chloramphenicol and tetracycline both appear to interfere with the signal of the uncharged tRNA at the A site of the ribosome, which is required for RelAEC activation (Sokawa & Sokawa, 1978, Lund & Kjeldgaard, 1972, Gale et al., 1981, Haseltine & Block, 1973). Both chloramphenicol and tetracyline bind the 50S subunit of the ribosome (Gale et al., 1981), where RelAEC has been shown to bind (Howard et al., 1976). These same drugs also interfere with SpoTCC function (Figure 3). While it is possible that chloramphenicol and tetracycline inhibit Rsh protein activity by affecting binding to the ribosome (Gale et al., 1981), our data show no effect of chloramphenicol (data not shown) or tetracycline on SpoTCC association with the ribosome (Figure 3D) or stability (Figure 3C). The fact that these drugs both prevent ppGpp accumulation (Figure 3B) and induce ppGpp decay in pre-starved cultures (Figure 3A,E,F) also supports the argument that they do not inhibit the SpoTCC enzyme but rather remove the signal required for its activation toward synthesis.
Three antibiotics did not interfere with ppGpp accumulation or maintenance (Figure 3A,B): these all bind to the 30S subunit. Streptomycin causes mistranslation and inhibits initiation (Gale et al., 1981). Kanamycin binds to the decoding region of the A site and prevents discrimination between cognate and non-cognate tRNAs (Magnet & Blanchard, 2005). Spectinomycin binds to the head domain and blocks translocation (Borovinskaya et al., 2007). Our pharmacological data therefore suggest that an active peptidyltransferase site and binding of tRNA to the acceptor site are required for SpoTCC activation, while translational accuracy, initiation, and translocation are not. In E. coli, ppGpp synthesis by RelAEC is inhibited by chloramphenicol, tetracycline and spectinomycin but not by kanamycin and streptomycin (Cortay & Cozzone, 1983), indicating that tRNA binding, peptidyltransferase and translocation are essential for RelAEC activity, but initiation and fidelity are not. Experiments with spectinomycin show that RelAEC and SpoTCC are different with respect to translocation. This and the data with the mutated L11 protein (Figure 5C) show that detection of amino acid starvation at the ribosome occurs via subtly different mechanisms in SpoTCC and RelAEC. It is not clear why SpoTCC should be insensitive to inhibition of translocation.
Both glucose and ammonium starvation lead to accumulation of uncharged tRNAs and an increase in ppGpp (Figure 1A). Our genetic studies using amino acid auxotrophs demonstrate that amino acid starvation alone is not sufficient to induce ppGpp accumulation (Figure 1B, S1). Using a glucose auxotroph, we show that glucose starvation in the presence of amino acids is also not sufficient to induce full accumulation of ppGpp (Figure 4A). In ppGpp decay assays there is a two-tiered response to relief of these two types of starvation. When glucose or ammonium starved cells are treated so as to solely remove the amino acid starvation signal - by adding tetracycline (Figure 3E,F) or by adding amino acids (Figure 4B) – ppGpp levels are partially reduced but level off at a ppGpp/GTP ratio of ∼0.3 – 0.5. When these cells are subsequently treated with glucose or ammonium, full ppGpp decay is induced (Figure 3E,F, 4B).
Our data support a model in which SpoTCC has regulatory properties of a synergistic ‘AND’ logic gate in which the presence of one activating signal results in low levels of ppGpp accumulation (Figure 1B, S1, 3, 4A). Only when both amino acid AND glucose OR ammonium starvation signals are present do high levels of ppGpp accumulate (Figure 1A, 4A, 8A). In glucose starvation it is possible that a glycolytic intermediate could be a molecular signal for SpoTCC activity, as may be the case in E. coli (Schneider & Gourse, 2003). The Δfbps strain would have abnormal levels of some glycolytic intermediates during glucose starvation and amino acid supplementation due to the block in gluconeogenesis, and this could prevent full ppGpp accumulation in this condition (Figure 4A,B). However, we think this an unlikely explanation of the results in Figure 4 because the analogous experiments with tetracycline show similar levels of ppGpp accumulation (Figure 3A, 3E, 3F).
The ‘AND’ gate signaling logic we propose for the C. crescentus stringent response entails a stricter definition of starvation than is observed in E. coli, which has ‘OR’ signaling logic in its stringent response (Figure 8A). This higher threshold for stringent response activation is likely an adaptation to the near constant starvation conditions encountered by C. crescentus in its oligotrophic niche. A regulatory structure in which multiple starvation cues are required to fully trigger the stringent response is likely better suited to oligtrophic niches where organisms must utilize their substantial metabolic capacity to convert nutrients and continue growth whenever possible.
Evidence for independent mechanisms of SpoTCC regulation by glucose and ammonium starvation signals
Rsh proteins have two conserved C-terminal domains: TGS and ACT, which are important for regulation. The TGS domain of SpoTEC binds to holo-acyl carrier protein during lipid starvation and induces synthesis of ppGpp (Battesti & Bouveret, 2006). The ACT domain and a region N-terminal to it are important in sensing amino acid starvation via ribosomal protein L11 in RelAEC (Schreiber et al., 1991, Durfee et al., 2008). We constructed strains in which the entire SpoTCC C-terminus or the ACT domain alone were deleted and tested them for ppGpp accumulation upon starvation. Our data show that SpoTCC with a missing ACT domain responds to ammonium but not glucose starvation. Thus signals perceived by SpoTCC are specific to the type of starvation and the ACT domain is not required for responding to ammonium starvation.
Further evidence for independent mechanisms of SpoTCC regulation by glucose and ammonium comes from our experiments with a ribosomal mutant. The L11 P22L mutant strain of C. crescentus was unable to accumulate ppGpp at a wild-type rate in ammonium starvation but showed no defect upon glucose starvation. This indicates that detection of an ammonium starvation signal by SpoTCC involves ribosomal protein L11 and that glucose starvation signals are either transduced differently through L11 or not transduced through this protein at all.
SpoTCC regulates large-scale transcriptional changes during carbon starvation
Experiments assessing global transcriptional change during the stringent response have been conducted in a number of species (Brockmann-Gretza & Kalinowski, 2006, Eymann et al., 2002, Gaynor et al., 2005, Kazmierczak et al., 2009, Nascimento et al., 2008, Traxler et al., 2008, Durfee et al., 2008). In these experiments ppGpp synthesis was induced by amino acid starvation. In general, many anabolic genes were downregulated while stress response genes and certain amino acid biosynthetic genes were upregulated. Work in E. coli, however, shows that the regulation of amino acid biosynthetic genes is variable and complex (Durfee et al., 2008). In E. coli grown under glucose-lactose diauxie, most anabolic genes are downregulated and stress response genes are upregulated. However, histidine and arginine biosynthetic genes were transiently upregulated (Chang et al., 2002), though this may not be dependent on ppGpp (Traxler et al., 2006). Overall, the general picture from microarray analyses of the stringent response is that anabolic genes are downregulated and stress response genes are upregulated. Amino acid biosynthetic genes are sometimes an exception to this trend, but this result is partially confounded by the fact that the tested cells were in most cases starved for amino acids.
The transcriptional effects of elevated ppGpp in glucose starvation in C. crescentus are similar to those reported in other species. We observe downregulation of genes encoding translation machinery, energy metabolism and anabolic metabolism (Figure 6, Table S1). In addition, almost all of the amino acid biosynthetic genes were downregulated; this is not consistently seen in other microarray experiments, but is consistent with adaptation to total growth arrest and nutrient deprivation. The one gene that is involved in amino acid biosynthesis that is upregulated in our data set encodes homoserine O-acetyltransferase (Figure 6, Table S1), which is required for biosynthesis of methionine as well as S-adenosyl-L-methionine, a universal methyl-donor molecule (Karp et al., 1999, Romero et al., 2001).
The majority of genes in the microarray data set involved in catabolic, or degradative processes, are upregulated. Thus SpoTCC activates the breakdown of cellular components during starvation, presumably for energy. We also observe that genes that function in motility, chemotaxis and environmental sensing are largely upregulated by SpoTCC, implying that the C. crescentus stringent response increases the capacity of the cell to respond to a wider range of nutrient sources. Such a response would be adaptive in an oligotrophic environment (Poindexter, 1981), and is not seen in E. coli in the amino-acid-starvation strigent response, where flagellar and chemotaxis genes are downregulated (Durfee et al., 2008, Lemke et al., 2009).
It has been predicted that oligotrophs should degrade storage polymers during starvation, and that cessation of growth should augment polymers of non-limiting nutrients (Poindexter, 1981). The PHB degradation enzyme Poly(3-hydroxyalkanoate) depolymerase is upregulated by spoTCC during carbon starvation, providing evidence that PHB is being used as a carbon source (Table 2). Additionally, SpoTCC downregulates the enzyme exopolyphosphatase, which degrades polyphosphate, while upregulating polyphosphate kinase (Table 2). Thus, SpoTCC regulates the ability of C. crescentus to increase its polyphosphate stores during stalled growth.
ccrM, a gene involved in cell division control, was downregulated by spoT (Table 2). CcrM methylates DNA, thereby controlling the transcription of genes with methylation-sensitive promoters. Normally ccrM is expressed just before cell division: its activity represses trancription of ctrA - a regulator responsible for expression of genes required for swarmer cell development and septation (Reisenauer & Shapiro, 2002) – and activates expression of DnaA, which initiates chromosome replication (Collier et al., 2007). Downregulation of ccrM during starvation should cause CtrA levels to remain high and DnaA levels to remain low, perhaps permitting completion of the ongoing cell division but repression of the next round of DNA replication (Lesley & Shapiro, 2008). The regulation of other genes involved in cell division hints at a complex integration of cell division and starvation signals (Table 2).
Rsh signaling diversity and an undetermined mechanism of rRNA regulation
Our data show that amino acid starvation is only conditionally linked to ppGpp synthesis (Figure 1B, 4A,B) and inconsistently linked to rRNA transcriptional control (Figure 7C). Moreover, we demonstrate that SpoTCC (and hence, ppGpp) is not required to downregulate 16SA rRNA transcription upon carbon starvation (Figure 7B). There are two rRNA operons in C. crescentus and we have only examined transcription of one of them, the A operon. We draw no conclusions about the regulation of the B operon, but it is clear that a spoT/dksA -independent mechanism regulates the A operon in carbon starvation. As described in the introduction, there are other examples of species in which rRNA transcriptional restriction occurs in the absence of ppGpp accumulation (Acosta & Lueking, 1987, Belitsky & Kari, 1982). Our study is the first to show ppGpp-independent downregulation of rRNA transcription in a condition that still induces ppGpp accumulation in the wild-type (Figure 7B). In E. coli the concentration of the initiating NTP (iNTP) controls rRNA transcription during outgrowth, and ppGpp controls it during nutrient shifts (Murray et al., 2003). It is possible that in C. crescentus the iNTP controls rRNA transcription under all conditions. The iNTP for the ribosomal A operon is GTP (Amemiya, 1989). We have data showing that GTP levels upon carbon starvation fall to similar extents in wild-type and spoT null strains, thus it is possible that iNTP levels alone could control rRNA transcription in C. crescentus (Figure S4). It is notable that in E. coli subjected to amino acid starvation, levels of the iNTPs fall in the wild-type strain but actually rise in the relA null strain (Edlin & Neuhard, 1967). Further study will be required to determine the mechanism of rRNA control in C. crescentus.
Activation of IS elements by the stringent response
Activation of the stringent response in C. crescentus results in increased transcription of IS transposases and DNA recombination enzymes (Figure 6, Table S1). The observation that strains carrying a hydrolase-deficient SpoTCC grow slowly initially and quickly acquire suppressors with IS3 elements interrupting the spoT locus provides a confirmation of our transcriptome data. It is likely that IS3 elements mobilized to many locations in the genome of the SpoT H67A strain; however, we only recovered strains with elements that interrupted the spoT locus, thereby relieving the growth inhibition by elevated ppGpp levels. These elements are a member of the IS3 family - the most prevalent family in bacteria (Mahillon et al., 1999). Our study shows that accumulation of ppGpp activates IS elements, independently of starvation. It is notable that a RelAEC null strain was isolated and shown to have an IS2 element within the RelA gene, disrupting its activity (Metzger et al., 1989). IS element activation may therefore be a conserved feature of ppGpp accumulation.
Concluding remarks
This work describes a stringent response signaling system in which a nearly universal bacterial signaling molecule, ppGpp, is used to effect transcriptional changes and growth arrest in a manner similar to that observed in E. coli and other species. C. crescentus is apparently adapted to its oligotrophic niche through divergence of its Rsh protein, SpoTCC, which has a high threshold for activation, requiring multiple starvation signals before it induces growth arrest via the stringent response.
MATERIALS AND METHODS
Bacterial strains and growth conditions
All strains used in this study are listed in Table 1. Experimental strains were derived from Caulobacter crescentus strain NA1000 (CB15N) (Evinger & Agabian, 1977, Marks et al., 2010). NA1000 strains were grown in M2G minimal medium (6.1mM Na2HPO4, 3.9mM KH2PO4, 9.3mM NH4Cl, 500µM MgSO4, 500µM CaCl2, 1X FeSO4/chelate (Sigma #F10518), 0.2% glucose) at 30°C. All auxotrophic strains except the glutamate auxotroph were grown in M2G supplemented with 100 µg/ml of the amino acid for which the strain was auxotrophic plus 200 µg/ml of alanine to relieve growth inhibition (Ferber & Ely, 1982). The glutamate auxotroph was grown in M2G supplemented with 500 µg/ml of glutamate. The concentration of kanamycin used for plasmid selection in C. crescentus was 5 µg/ml in liquid media and 25 µg/ml on solid media. E. coli strains TOP10 (Invitrogen, Carlsbad, CA) and Mach1 (Invitrogen) were used for cloning. E. coli strains were grown in Terrific Broth supplemented with 50 µg/ml kanamycin at 30°C.
Construction of strains
Strains in which genes were deleted, truncated, or fused to epitopes were constructed by homologous recombination using a two-step kanamycin selection/ sacB counterselection protocol with the suicide plasmid pNPTS138, as illustrated (Hinz et al., 2003, Fiebig et al., 2010). For gene deletions, ≈500 bp fragments upstream and downstream of the deletion target were PCR amplified, joined by triple ligation or gene stitching (Higuchi et al., 1988) and then cloned into pNPTS138. The ΔspoTCC strain encodes the first 29 codons and the last 24 codons of gene number CCNA_01622. The ΔdksA strain encodes the first 8 codons and the last 17 codons of gene number CCNA_02663. The ΔpheA strain encodes the first ten and last 17 codons of gene number CCNA_03028. The ΔgltB strain encodes the first 35 and last 30 codons of gene number CCNA_03722. The Δfbps strain encodes the first three codons of CCNA_01448 and the last 15 codons of CCNA_01449. The spoTΔACT668–719 strain encodes codons 1–667 and 720–744 of spoTCC. To create the SpoT domain truncation strains, ≈500 bp fragments surrounding the region to be mutagenized were PCR amplified and cloned into TOPO Blunt (Invitrogen); site-directed mutagenesis was used to change the appropriate codon to a stop codon. The mutagenized gene fragments were then cloned into pNPTS138 and used to replace the wild-type chromosomal allele using the two-step recombination protocol cited above. In the spoTΔCTD strain Y366 (TAC) was mutated to stop (TAG). In the spoTΔACT strain A671 (GCC) was mutated to stop (TAG).
To create N-terminal HA epitope fusions, mutagenic gene-stitching PCR (Higuchi et al., 1988) was used to introduce an HA tag into a DNA fragment containing the upstream and 5’ region of the target gene. This fusion allele was cloned into pNPTS138 and used to replace the wild-type chromosomal allele as above. The amino acid sequence of the beginning of the HA-SpoT protein is: MYPYDVPDYA followed by amino acids 2–744 of the wild-type protein. The p16Slac reporter plasmid was constructed by fusing the 16SA rRNA promoter region to a piece of E. coli lacZ using gene-stitching PCR, and cloning the resulting fusion into pMT585 (Thanbichler et al., 2007) using the AscI and NheI cut sites. The amplified region of the 16SA promoter starts 570 bp upstream of the transcription initiation site (Amemiya, 1989) and ends 80 bp after the initiation site. This chromosomal fragment was amplified using the following primers: 5’-atatggcgcgccaaacagctgatcgccaag-3’ and 5’-tatcggcctcaggaagtttctagcgaagcgtctgg-3’. The fragment of lacZ was amplified from pPR9TT (Santos et al., 2001) using the following primers: 5’-gcttcgctagaaacttcctgaggccgatactgtc-3’ and 5’-atatgctagccattaaagcgagtggcaaca-3’. All plasmids were introduced into C. crescentus by tri-parental conjugation using E. coli strain MT607 carrying the pRK600 helper plasmid (Finan et al., 1986).
Measurement of cellular ppGpp levels
ppGpp was measured as described (Cashel, 1969) with protocol modifications outlined below. Cells were grown in M2G, then washed in either M2G-labeling (M2G with 12.2 mM NaCl and 3.9 mM KCl instead of Na2HPO4 and KH2PO4), M2-labeling (M2G-labeling without glucose) or M2G-N-labeling (M2G-labeling without NH4Cl) twice and then resuspended in the appropriate labeling medium. 100µCi/ml of KH232PO4 (PerkinElmer, Waltham, MA) was added and the cells were incubated at 30°C for 2 hours. In experiments with 2 hour time points, an equal volume of 2M formic acid was then added, and the formic acid extracts were placed on ice for at least 15 minutes. For shorter time points, cells were labeled in the appropriate labeling medium for 2 hours, spun for 1 minute in a microfuge at 12K rpm, the labeling media was pulled off, the cells were resuspended in 1 ml of the starvation or recovery medium, spun again, the washing medium was pulled off and then cells were resuspended in the appropriate starvation-labeling media with 20µCi/ml of KH232PO4. Aliquots were then taken and formic acid-extracted at time points after resuspension in the starvation media. The extracts were spun in a microfuge for three minutes and the cleared lysate was spotted on PEI-cellulose TLC sheets (EMD Chemicals) and developed in 1.5M KH2PO4 pH 3.4. TLC plates were imaged on a Typhoon Phosphoscanner and analyzed with QuantityOne software. The glutamate auxotroph strain FC1250 was cultured as described above, and starved in M2G-labeling media with KH232PO4, the other auxotrophs were starved in M2G-labeling media with KH232PO4 and 200 µg/ml of alanine. The Δfbps strain was starved in M2-labeling media with 100 µg/ml of each amino acid added. The concentrations of antibiotics used in the ppGpp assays was as follows: chloramphenicol, 50 µg/ml; tetracycline, 50µg/ml; streptomycin, 250µg/ml; kanamycin, 250 µg/ml; spectinomycin, 1250 µg/ml: in each case this is 50X the concentration of antibiotic normally used for selection.
Purification of ribosomes/polysomes by sucrose gradient centrifugation
800 ml of culture at OD660=0.2–0.3 was pelleted and resuspended in 3ml of buffer TM (20 mM Tris pH7.5, 15 mM MgCl2, 1mg/ml PMSF and EDTA-free Protease Inhibitor Cocktail (Roche, Indianapolis, IN)). 200 µl of 10 mg/ml lysozyme was added and the sample lysed by two successive freeze-thaw rounds in ethanol-dry ice / 10°C water bath. 60 µl of 10% deoxycholate and 40 µl of Rnase-free DnaseI (NEB, Ipswich, MA) was added and the sample was incubated on ice for 15 minutes, and then spun at 16,000 rpm for 15 minutes. The cleared lysate was removed and 50 OD260 was layered on top of a gradient of 10–40% sucrose in Buffer E (10 mM Tris, 10 mM MgCl2, 100 mM NH4Cl, 3mM β-mercaptoethanol) with 0.2 mg/ml PMSF and EDTA-free Protease Inhibitor Cocktail. The sucrose gradient was spun in a Beckman L8-M Ultracentrifuge at 27,000 rpm for 5 hours at 7°C. The gradient was manually fractionated by siphon into a UV-transparent 96-well plate and the OD260 of each fraction was read in a BioTek Synergy plate reader (BioTek, Winooski, VT). The fractions comprising the 70S and polysome peaks were combined and brought up to 30ml with Buffer E + 0.2 mg/ml PMSF, and then spun at 30,000 rpm for 12 hours. The supernatant was decanted and the pellets were resuspended in Buffer E. The soluble fractions from the gradient were combined and concentrated in an Amicon Ultra centrifugal filter device (Amicon, Billerica, MA). The concentrated ribosome and soluble fractions were then normalized to OD260 (1/5 of the OD260 of the ribosome fractions was used for the soluble fraction, so that background levels were comparable), separated by SDS-PAGE and immunoblotted to detect HA-SpoT.
Isolation of ribosomes by sucrose cushion centrifugation
To assay ribosome association of the various SpoT mutant proteins, we purified ribosomes using the sucrose cushion centrifugation method (Cross, 1970, Spedding, 1996). 150 ml of culture in M2G or M2 at OD660=0.2 was pelleted, lysed and cleared as described above. 0.5 ml of lysate was layered on top of a sucrose cushion (1M sucrose, 50mM Tris pH 7.6, 15mM MgCl2, 6mM BME, and 100mM NH4Cl) and spun in a Beckman Optima TLX Ultracentrifuge at 70,000 rpm for one hour. The top layer was removed and concentrated to a volume of 200 µl in an Amicon Ultra centrifugal filter device (Amicon). The sucrose cushion was decanted and the high-density ribosome pellet was resuspended in 200 µl of Buffer E. The soluble fractions and ribosome pellet fractions were then separated by SDS-PAGE and immunoblotted todetect HA-SpoT.
Detection of SpoT by immunoblot
Whole cells or samples from biochemical fractionation were boiled in SDS loading dye and proteins were separated on 12% SDS-PAGE gels. Proteins were transferred to a Immobilon polyvinylidene fluoride membrane (Millipore) by wet transfer. The membrane was then blocked in 5% milk for at least 30 minutes and then cut in half, when appropriate, so the FixJ loading control protein and the HA-SpoT proteins could be probed separately. HA-tagged proteins were hybridized by incubating for at least 40 minutes in a 1:5,000 dilution of purified monoclonal mouse anti-HA antibodies (Sigma-Aldrich), washing three times and then incubating for at least 40 minutes in a 1:2,500 dilution of goat anti-mouse antibodies conjugated to HRP (Thermo Scientific). FixJ was hybridized by incubating the membrane in 5% milk with a 1:2,000 dilution of polyclonal rabbit α-FixJ antibody for 40 minutes, washing twice, and then incubating with a 1:10,000 dilution of HRP-cojugated goat anti-rabbit antibodies (Thermo Scientific) for 40 minutes. All membranes were then washed five times, and the secondary antibodies were detected with SuperSignal West Femto Substrate (Thermo Scientific). Blots were exposed to film for 10 seconds to 5 minutes.
Transcriptional analysis of the C. crescentus stringent response by DNA microarray
Three independent replicate cultures each of wild-type strain NA1000 and NA1000ΔspoTCC were grown to OD660=0.2–0.3 in M2 with 0.2% glucose (M2G), washed once in glucose-free M2 medium, resuspended in 5ml of glucose-free M2, and rolled at 30°C for an additional 5 minutes to fully activate the stringent response. Cells were then pelleted and flash-frozen in liquid nitrogen. RNA was extracted using Trizol as previously described (Boutte et al., 2008) and RNA integrity assessed using an Agilent Bioanalyzer (Agilent, Santa Clara, CA). For each of the six RNA samples, 10 µg was processed to produce single-strand cDNA; RNA was removed by addition of 1N NaOH. cDNA was column-purified, fragmented using DNAseI (GE Life Sciences, Piscataway, NJ), and end labelled using GeneChip labelling reagent (Affymetrix, P/N 900542). Labelled cDNA was hybridized to GeneChip CauloHi1 (McGrath et al., 2007) according to GeneChip Expression analysis technical manual (Affymetrix, Santa Clara, CA). Hybridization proceeded for 16 hours at 50°C. Arrays were washed using protocol PRO-GE-W52-V3 and stained on a GeneChipFluidics Station (Affymetrix) according to the Genechip Expression analysis technical manual. The arrays were scanned using the Affymetrix Gene Chip Scanner 3000 7G and CEL intensity files were generated by GCOS (Gene Chip Operating Software) v. 1.4. Stringent response microarray data are deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under series accession GSE21206.
Data were analyzed by first calculating the mean expression for the wild-type and ΔspoT replicates and then calculating the wild-type / ΔspoT ratio. Only probes on the coding strand with at least a three-fold difference in expression were considered for further analysis (Table S1). In some cases there were multiple probes within a gene or its promoter region: the data for these probes were averaged. The standard deviation for the wild-type and ΔspoT replicates was calculated, and genes for which the standard deviation was greater than the mean for either sample set were discarded. Functional assignments for genes were made using BioCyc (Karp et al., 1999, Romero et al., 2001).
RNA isolation and qRT-PCR analysis
Strains for qRT-PCR analysis were grown in M2G + 5 µg/ml kanamycin plus appropriate amino acid supplements for the auxotrophic mutants. Cells for starvation samples were washed and resuspened in starvation media and rolled at 30°C for 15 minutes to one hour for glucose starvation and for two hours for amino acid starvation. 1.5ml of cells at OD660=0.2 were pelleted and resuspended in 1ml Trizol (Invitrogen), flash frozen in liquid nitrogen and stored at −80°C until purification. RNA was extracted according to the Trizol (Ambion, Austin, TX) protocol, the aqueous layer was extracted again with acid phenol-chloroform (Ambion) in phase-lock tubes (5 Prime). The aqueous layer from this extraction was decanted and precipitated with isopropanol. The RNA was then treated with TurboDNase (Ambion) for 4 hours at 37°C. Dnase Inactivating Reagent (Ambion) was used to remove DNase. 0.375 µg of RNA from each sample was used in a 15µl reverse transcription reaction with the iScript cDNA synthesis kit (BioRad) according to protocol. 4µl of the RT reactions was used in each 10µl qRT-PCR reaction. qRT-PCR was performed according to protocol using SsoFast EvaGreen Supermix (BioRad). The following primers were used to quantify 16S rRNA transcription from the pP16Slac plasmid: 5’-cgaaagggagttgcatcg-3’ and 5’cgtgcatctgccagtttg-3’. RuvA (gene number CCNA_03345) was used as an endogenous control, with the following primers: 5’-cgagtgaggaagccgtagag-3’ and 5’-gaccctgttgcacatcgag-3’. A relative starting quantity (SQ) of transcript was calculated by the Biorad CFX Manager software from a standard curve. The SQ value for the 16S amplicon was divided by the SQ value from the ruvA amplicon for each sample. The resulting ratio was the value recorded as “Relative transcript level” for each sample.
Supplementary Material
ACKNOWLEDGEMENTS
C.B. was supported by National Institutes of Health (NIH) Training Grant 5T32GM007183-34. S.C. acknowledges support for this project from the Arnold and Mabel Beckman Foundation (BYI), and the Mallinckrodt Foundation. We thank Ling Chan for advice and guidance with many of the experiments, and the Ira Wool Lab for the use of centrifuges. We also thank Melissa Marks for critical feedback and Emmanuelle Bouveret (LISM, CNRS-Marseille) and Aretha Fiebig for critical readings of the manuscript. Patrick Viollier (Université de Genève) generously provided a C. crescentus transposon insertion collection from which we isolated several amino acid auxotrophs.
REFERENCES
- Acosta R, Lueking D. Stringency in the Absence of ppGpp Accumulation in Rhodobacter sphaeroides. J Bacteriol. 1987;169:908–912. doi: 10.1128/jb.169.2.908-912.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya K. Conserved Sequence Elements Upstream and Downstream from the Transcription Initiation Site of the Caulobacter crescentus rrnA Gene Cluster. J Mol Biol. 1989;210:245–254. doi: 10.1016/0022-2836(89)90327-6. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The HD domain defins a new superfamily of metal-depedent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- Avarbock D, Avarbock A, Rubin H. Differential Regulation of Opposing RelMtb Activity by the Aminoacylation State of a tRNA.Ribosome.mRNA.RelMtb Complex. Biochemistry. 2000;39:11640–11648. doi: 10.1021/bi001256k. [DOI] [PubMed] [Google Scholar]
- Avarbock D, Salem J, Li L, Wang Z, Rubin H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene. 1999;233:261–269. doi: 10.1016/s0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- Ballesteros M, Fredriksson A, Henricksson J, Nystrom T. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20:5280–5289. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of Regulation of Transcription Initiation by ppGpp. II. Models for Positive Control Based on Properties of RNAP Mutants and Competition for RNAP. J Mol Biol. 2001a;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanishm of Regulation of Transcription Initiation by ppGpp. I. Effects of ppGpp of Trancription Initiation in vivo and in vitro. J Mol Biol. 2001b;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- Belitsky B, Kari C. Absence of Accumulation of ppGpp and RNA during Amino Acid Starvation in Rhizobium meliloti. J Biol Chem. 1982;257:4677–4679. [PubMed] [Google Scholar]
- Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JHD. A Steric Block in Translation Caused by the Antibiotic Spectinomycin. Chem Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Srinivasan BS, Flannick JA, Novak AF, Martens AT, Batzoglou S, Viollier PH, Crosson S. Genetic and Computational Identification of a Conserved Bacterial Metabolic Module. PLoS Genet. 2008;4:e1000310. doi: 10.1371/journal.pgen.1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann-Gretza O, Kalinowski J. Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase. BMC Genomics. 2006;7:230. doi: 10.1186/1471-2164-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler YX, Abhayawardhane Y, Stewart GS. Amplification of the Bacillus subtilis maf Gene Results in Arrested Septum Formation. J Bacteriol. 1993;175:3139–3145. doi: 10.1128/jb.175.10.3139-3145.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M. The Control of Ribonucleic Acid Synthesis in Escherichia coli. J Biol Chem. 1969;244:3133–3141. [PubMed] [Google Scholar]
- Chang D, Smalley DJ, Conway T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol Microbiol. 2002;45:289–306. doi: 10.1046/j.1365-2958.2002.03001.x. [DOI] [PubMed] [Google Scholar]
- Chiaverotti TA, Parker G, Gallant J, Agabian N. Conditions that Trigger Guanosine Tetraphosphate Accumulation in Caulobacter crescentus. J Bacteriol. 1981;145:1463–1465. doi: 10.1128/jb.145.3.1463-1465.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci USA. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortay JC, Cozzone AJ. Effects of aminoglycoside antibiotics on the coupling of protein and RNA syntheses in Escherchia coli. Biochem Biophys Res Comm. 1983;112:801–808. doi: 10.1016/0006-291x(83)91688-1. [DOI] [PubMed] [Google Scholar]
- Cross GAM. Sedimentation properties of polyribosomes, ribosomes and ribosomal subunits from Crithidia oncopelti. Biochim. Biophys. Acta. 1970;204:470–477. doi: 10.1016/0005-2787(70)90167-x. [DOI] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HIM, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Barry III CE. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- Das B, Pal RR, Bag S, Bhadra RK. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol Microbiol. 2009;72:380–398. doi: 10.1111/j.1365-2958.2009.06653.x. [DOI] [PubMed] [Google Scholar]
- Dozot M, Boigegrain R, Delrue R, Hallez R, Ouahrani-Bettache S, Daese I, Letesson J, DeBolle X, Kohler S. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol. 2006;8:1791–1802. doi: 10.1111/j.1462-5822.2006.00749.x. [DOI] [PubMed] [Google Scholar]
- Durfee T, Hansen A, Zhi H, Blattner FR, Jin DJ. Transcription Profiling of the Stringent Response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston DE, JrM, Gray ED. Variations in ppGpp levels in Rhodopseudomonas spheroides during adaptation to decreased light intensity. Biochem Biophys Res Comm. 1973;54:1370–1376. doi: 10.1016/0006-291x(73)91138-8. [DOI] [PubMed] [Google Scholar]
- Edlin G, Neuhard J. Regulation of Nucleoside Triphosphate Pools in Escherchia coli. J Mol Biol. 1967;24:225–230. doi: 10.1016/0022-2836(67)90328-2. [DOI] [PubMed] [Google Scholar]
- Edwards RL, Dalebroux ZD, Swanson MS. Legionella pneumonophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol. 2009;71:1190–1204. doi: 10.1111/j.1365-2958.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associate nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymann D, Homuth G, Scharf C, Hecker M. Bacillus subtilis Functional Genomics: Global Characterization of the Stringent Response by Proteome and Transcriptome Analysis. J Bacteriol. 2002;184:2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber DM, Ely B. Resistance to Amino Acid Inhibitoin in Caulobacter crescentus. Mol Gen Genet. 1982;187:446–452. [Google Scholar]
- Ferullo DJ, Lovett ST. The Stringent Response and Cell Cycle Arrest in Escherchia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A, Castro-Rojas CM, Siegal-Gaskins D, Crosson S. Interaction specificity, toxicity and regulation of a paralogous set of ParE/RelE-family toxin-antitoxin systems. Mol Microbiol. 2010;77:236–251. doi: 10.1111/j.1365-2958.2010.07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan TM, Kunkel B, De Vos GF, Signer ER. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen JD, Fiil NP, Parker JM, Haseltine WA. A New Relaxed Mutant of Escherichia coli with an Altered 50S Ribosomal Subunit. Proc Natl Acad Sci USA. 1974;71:3465–3469. doi: 10.1073/pnas.71.9.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ. The Molecular Basis of Antibiotic Action. Bristol: John Wiley and Sons Ltd.; 1981. p. 646. [Google Scholar]
- Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol. 2005;56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Cashel M. Cellular Localization of the Escherichia coli SpoT protein. J Bacteriol. 1995;177:3890–3893. doi: 10.1128/jb.177.13.3890-3893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropp M, Strausz Y, Gross M, Glaser G. Regulation of Escherichia coli RelA Requires Oligomerization of the C-Terminal Domain. J Bacteriol. 2001;183:570–579. doi: 10.1128/JB.183.2.570-579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine WA, Block R. Synthesis of Guanosine Tetra- and Pentaphosphate Requires the Presence of a Codon-Specific, Uncharged Transfer Ribonucleic Acid in the Acceptor Site of Ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targetng of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational Antagonism between Opposing Active Sites in a Bifuctional RelA/SpoT Homolog Modulates (p)ppGpp Metabolism during the Stringent Response. Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- Howard GA, Gordon J, Farnung K, Richter D. Stringent Factor Binds to Escherichia coli Ribosome Only in the Presence of Protein L10. FEBS Lett. 1976;68:211–214. doi: 10.1016/0014-5793(76)80438-3. [DOI] [PubMed] [Google Scholar]
- Howorth SM, England RR. Accumulation of ppGpp in synbiotic and free-living nitrogen-fixing bacteria following amino acid starvation. Arch Microbiol. 1999;171:131–134. doi: 10.1007/s002030050689. [DOI] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial Birth Scar Proteins Mark Future Flagellum Assembly Site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Jenvert R, Schiavone LH. The Flexible N-terminal Domain of Ribosmal Protein L11 from Escherichia coli is Necessary for the Activation of Stringent Factor. J Mol Biol. 2007;365:764–772. doi: 10.1016/j.jmb.2006.10.065. [DOI] [PubMed] [Google Scholar]
- Jiang M, Sullivan SM, Wout P, Maddock JR. G-Protein Control of the Ribosome-Associated Stress Response Protein SpoT. J Bacteriol. 2007;189:6140–6147. doi: 10.1128/JB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Krummenacker M, Paley S, Wagg J. Integrated pathway-genome databases and their role in drug discover. Trends Biotech. 1999;17:275–281. doi: 10.1016/s0167-7799(99)01316-5. [DOI] [PubMed] [Google Scholar]
- Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol. 2009;72:590–611. doi: 10.1111/j.1365-2958.2009.06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Watanabe K, Suzuki H, Watarai M. Roles of Brucella abortus SpoT in morphological differentiation and intramacrophagic replication. Microbiology. 2005:151. doi: 10.1099/mic.0.27782-0. [DOI] [PubMed] [Google Scholar]
- Koch AL. The Adaptive Responses of Escherichia coli to a Feast and Famine Existence. Adv Microb Phys. 1971;6:147–217. doi: 10.1016/s0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley JA, Shapiro L. SpoT Regulates DnaA Stability and Initiation of DNA Replication in Carbon-Starved Caulobacter crescentus. J Bacteriol. 2008;190:6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Kjeldgaard KO. Metabolism of Guanosine Tetraphosphate in Escherichia coli. Eur J Biochem. 1972;28:316–326. doi: 10.1111/j.1432-1033.1972.tb01916.x. [DOI] [PubMed] [Google Scholar]
- Magnet S, Blanchard JS. Molecular Insights into Aminoglycoside Action and Resistance. Chem Rev. 2005;105:477–497. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Leonar C, Chandler M. IS elements as constituents of bacterial genomes. Res Microbiol. 1999;150:675–687. doi: 10.1016/s0923-2508(99)00124-2. [DOI] [PubMed] [Google Scholar]
- Marks ME, Castro-Rojas CM, Teiling C, L. D, Kapatral V, Walunas TL, Crosson S. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol. 2010;192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Costa OH, Fernandez-Moreno MA, Malpartida F. The relA/spoT-Homologous Gene in Streptomyces coelicolor Encodes Both Ribosome-Dependent (p)ppGpp-Synthesizing and -Degrading Activities. J Bacteriol. 1998;180:4123–4132. doi: 10.1128/jb.180.16.4123-4132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Lee H, Zhang L, Iniesta AA, Hottes AK, Tan MH, Hillson NJ, Hu P, Shapiro L, McAdams HH. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nature Biotech. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Mercer KLN, Weiss DS. The Escherichia coli Cell Division Protein FtsW is Required to Recruit Its Cognate Transpeptidase, FtsI (PBP3), to the Division Site. J Bacteriol. 2002;184:904–912. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 Mutation and a Comparison of relA1 with New relA Null Alleles in Escherichia coli. J Biol Chem. 1989;264:21146–21152. [PubMed] [Google Scholar]
- Mittenhuber G. Comparative Genomics and Evolution of Genes Encoding Bacterial (p)ppGpp Synthetases/Hydrolases (the Rel, RelA and SpoT Proteins) J Mol Microbiol Biotechnol. 2001;3:585–600. [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA Expression by Small Molecules Is Dynamic and Nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Murray KD, Bremer H. Control of spoT-dependent ppGpp Synthesis and Degradation in Escherchia coli. J Mol Biol. 1996;259:41–57. doi: 10.1006/jmbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. Role of RelA of Streptococcus mutans in Global Control of Gene Expression. J Bacteriol. 2008;190:28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K, Kandala J, Freese E. Evidence that Bacillus subtilis Sporulation Induced by the Stringent Response Is Caused by the Decrease in GTP or GDP. J Bacteriol. 1982;151:1062–1065. doi: 10.1128/jb.151.2.1062-1065.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Watson RJ, Friesen JD. A Relaxed Mutant with an Altered Ribosomal Protein L11. Mol Gen Genet. 1976;144:111–114. doi: 10.1007/BF00277313. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: A Critical Component of the Transcription Initiation Machinery that Potentiates the Regulaiton of rRNA Promoters by ppGpp and the Initiating NTP. Cell. 2004;188:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Poindexter JS. Oligotrophy: Fast and Famine Existence. Adv Microb Ecol. 1981;5:63–89. [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: Still Magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Ramagopal S, Davis BD. Localization of the Stringent Protein of Escherchia coli on the 50S Ribosomal Subunit. Proc Natl Acad Sci USA. 1974;71:820–824. doi: 10.1073/pnas.71.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A, Shapiro L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 2002;21:4969–4977. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Karp PD, Hottes A, Lee W, Laub M, Puniyani A, Green ML, Caspi R. BioCyc, Caulobacter crescentus CB15. 2001 [Google Scholar]
- Santos PM, DiBartolo I, Blatny JM, Zennaro E, Valla S. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol Lett. 2001;195:91–96. doi: 10.1111/j.1574-6968.2001.tb10503.x. [DOI] [PubMed] [Google Scholar]
- Savageau MA. Escherichia coli Habitats, Cell Types, and Molecular Mechanisms of Gene Control. Am Nat. 1983;122:732–744. [Google Scholar]
- Schneider DA, Gourse RL. Changes in the Concentrations of Guanosine 5’-Diphosphate 3'-Diphosphate and the Initiating Nucleotide Trisphosphate Account for Inhibition of rRNA Transcription in Fructose-1,6-Diphosphate Aldolase (fda) Mutants. J Bacteriol. 2003;185:6192–6194. doi: 10.1128/JB.185.20.6192-6194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G. Overexpression of the relA Gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- Schreiber G, Ron EZ, Glaser G. ppGpp-Mediated Regulation of DNA Replication and Cell Division in Escherichia coli. Curr Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- Scoarughi GL, Cimmino C, Donini P. Helicobacter pylori a Eubacterium Lacking the Stringent Response. J Bacteriol. 1999;181:552–555. doi: 10.1128/jb.181.2.552-555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfzadeh M, Keener J, Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherchia coli. Proc Natl Acad Sci USA. 1993;90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomoin RAM, Nakata N, Murai T, Yoshikawa M, Tsuji H, Sasakawa C. Identification and characterization of ispA, a Shigella flexneri chromosomal gene essential for normal in vivo cell division and intercellular spreading. Mol Microbiol. 1996;19:599–609. doi: 10.1046/j.1365-2958.1996.405941.x. [DOI] [PubMed] [Google Scholar]
- Sokawa J, Sokawa Y. Relaxation Effect of Chloramphenicol on the Stringent Control in Escherichia coli. J Biochem. 1978;83:1699–1705. doi: 10.1093/oxfordjournals.jbchem.a132083. [DOI] [PubMed] [Google Scholar]
- Spedding G. In: Ribosome preparation and protein synthesis techniques. In: Encyclopedia of Molecular Cell Biology and Molecular Medicine. Meyers RA, editor. Weinheim: VCH; 1996. pp. 327–337. [Google Scholar]
- Spira B, Silberstein N, Yagil E. Guanosine 3',5' -Bispyrophosphate (ppGpp) Synthesis in Cells of Escherichia coli Starved for Pi. J Bacteriol. 1995;177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stent GS, Brenner S. A Genetic Locus for the Regulation of Ribonucleic Acid Synthesis. Proc Natl Acad Sci USA. 1961;47:2005–2014. doi: 10.1073/pnas.47.12.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton JC, Malakooti J, Ely B. Regulation of Caulobacter crescentus ilvBN Gene Expression. J Bacteriol. 1994;176:3765–3774. doi: 10.1128/jb.176.12.3765-3774.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. doi:110.1093/nar/gkm1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD. The role of dissolved organic matter, particularly free amino acids and humic substances, in freshwater ecosystems. Freshw Biol. 1997;38:1–36. [Google Scholar]
- Traxler MF, Chang D, Conway T. Guanosine 3'-5'-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:2374–2379. doi: 10.1073/pnas.0510995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen H, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Ramakrishnan V. The Mechanism for Activation of GTP Hydrolysis on the Ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DH, Gaynor EC. Helicobacter pylor Initiates the Stringent Response upon Nutrient and pH Downshift. J Bacteriol. 2006;188:3726–3729. doi: 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the Mechanism for the Stringent Factor RelA. Mol Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yamanaka K, Zheng W, Crooke E, Wang YH, Inouye M. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherchia coli. Mol Microbiol. 2001;39:1572–1584. doi: 10.1046/j.1365-2958.2001.02345.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Ishiguro EE. Involvement of the N Terminus of Ribosomal Protein L11 in Regulation of the RelA Protein of Escherichia coli. J Bacteriol. 2001;183:6532–6537. doi: 10.1128/JB.183.22.6532-6537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YN, Coleman WG, Jr, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. Regulation of Cell Growth during Serum Starvation and Bacterial Survival in Macrophages by the Bifunctional Enzyme SpoT in Helicobacter pylori. J Bacteriol. 2008;190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.