Abstract
Many proteins are translocated through the SecY channel in bacteria and archaea, and the related Sec61 channel in eukaryotes1. The channel has an hourglass shape with a narrow constriction approximately halfway across the membrane, formed by a pore ring of amino acids2. While the cytoplasmic cavity of the channel is empty, the extra-cellular cavity is filled with a short helix, the plug2, which moves out of the way during protein translocation3,4. The mechanism by which the channel transports large polypeptides and yet prevents the passage of small molecules, such as ions or metabolites, has been controversial2,5–8. Here, we have addressed this issuein intact E. coli cells by testing the permeation of small molecules through wild-type and mutant SecY channels, which are either in the resting state or contain a defined translocating polypeptide chain. In the resting state, the channel is sealed by both the pore ring and the plug domain. During translocation the pore ring forms a gasket-like seal around the polypeptide chain, preventing the permeation of small molecules. The structural conservation of the channel in all organisms suggests a universal mechanism by which the membrane barrier is maintained during protein translocation.
Bacteria offer a unique opportunity to test the permeation of small molecules through the protein translocation channel, as the channel is located in the plasma membrane and is therefore accessible in intact cells. To test the permeability of the resting channel, we compared E. coli wild-type SecY, expected to be sealed, with a plug-deletion mutant(ΔP), which should be constitutively open(Fig. S1); although a new plug may form from neighboring polypeptide segments9, it likely blocks the channel only transiently8. Wild-type and ΔP mutant SecY channels were expressed under an inducible promoter at about the same level as the endogenous protein (Fig. S2). Expression of the ΔP mutant caused only a moderate growth defect (Fig. S2).
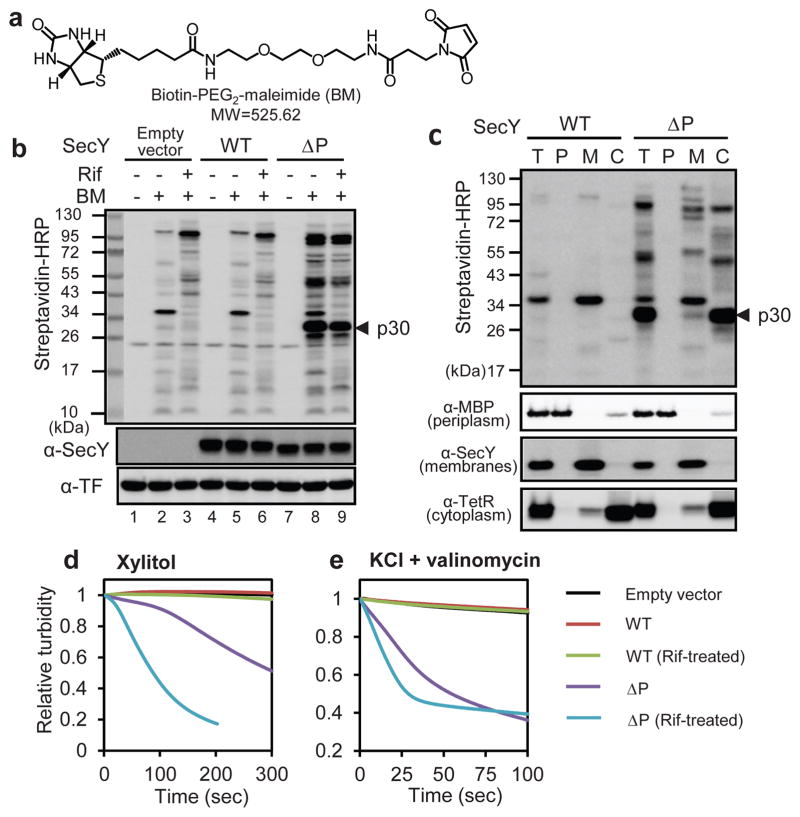
We first studied the permeation of a relatively large(525 Da), uncharged cysteine-modifying reagent (biotin-PEG2-maleimide(BM); Fig. 1a), which can cross the outer membrane through porins, but not the inner membrane10 (Fig. S1). When BM was added to wild-type E. coli cells, few proteins were biotinylated(Fig. 1b, lane 5). In contrast, with the ΔP mutant, several proteins were strongly modified, particularly a protein of about 30 kDa (Fig. 1b, lane 8). The majority of the modified proteins are located in the cytosol, as demonstrated by cell fractionation (Fig. 1c). The extent of modification was about the same after treatment with the transcription inhibitor rifampicin(Rif)(Fig. 1b, lane 9), which clears all SecY channels of translocating polypeptides(see Fig. 2c). Thus, permeation of BM occurs primarily through resting ΔP channels. We also found that many signal sequence suppressor (prl) SecY mutants allow the permeation of BM, although to a lesser extent than the ΔP mutant (Fig. S3). Channel opening in the absence of a translocation substrate explains why these mutants translocate proteins with defective or missing signal sequences11–13.
Figure 1. Testing the permeability of the resting SecY channel.
a, Structure of the modification reagent BM. b, Wild-type(WT) SecY or the ΔP mutant were expressed under the inducible Tet promoter. Cells were incubated with BM, and the proteins separated by SDS-PAGE, followed by blotting with streptavidin-HRP conjugate, with SecY-antibodies, or with trigger factor (TF)-antibodies (loading control). Where indicated, rifampicin (Rif) was added before BM. Endogenous SecY was tagged at its C-terminus, abolishing recognition by SecY antibodies (Fig. S2). p30, a prominent biotinylated protein. c, As in b, but after incubation with BM, cells(T)were fractionated into periplasm (P), membranes (M), and cytosol (C). Fractionation was controlled by immunoblotting for the indicated marker proteins. MBP, maltose-binding protein, TetR, tetracycline repressor. d, Spheroplasts were diluted into an iso-osmotic solution of xylitol and the change in turbidity followed over time. f, As in e, but with dilution into iso-osmotic KCl containing valinomycin.
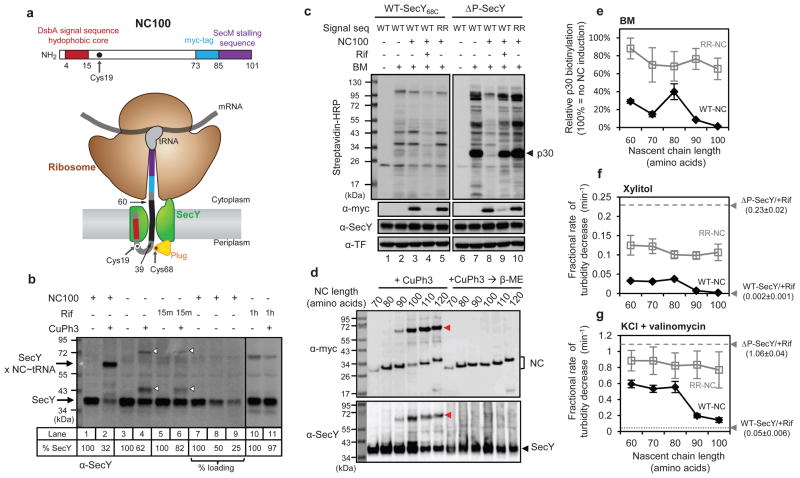
Figure 2. Testing the permeability of a translocating SecY channel.
a, Schematics of the model nascent chain (NC100) (upper panel)and its insertion into the SecY channel (lower panel). The indicated cysteines in NC100 and SecY form a disulfide bridge. Residues 39–60 are inside the channel. b, NC100 with Cys19 was expressed under an arabinose-inducible promoter together with SecY68C under the endogenous promoter. The start-codon of SecY68C was changed from AUG to GUG. Where indicated, cells were treated with the oxidant copper phenanthroline (CuPh3). Rifampicin(Rif) was added prior to oxidation for 15 min or 1 hr, as indicated. %SecY, percentage of non-crosslinked SecY. c, NC100 with either a wild-type(WT) or defective (RR) signal sequence was expressed together with SecY68C or the ΔP mutant. Where indicated, cells were pretreated with Rif before addition of BM. The samples were analyzed by SDS-PAGE, followed by blotting with streptavidin-HRP conjugate or myc-(detects NC100), SecY-, and TF-antibodies. d, Nascent chains (NC) of different lengths, all with Cys19, were expressed together with SecY68C, and crosslinked with CuPh3. The samples were analyzed by SDS-PAGE with or without prior reduction with β-mercaptoethanol (β-ME), followed by immunoblotting. Red arrowhead, crosslinked product between SecY and NC-tRNA. e, Nascent chains (NC) of different lengths with either wild-type(WT-NC) or defective (RR-NC) signal sequence were expressed together with ΔP channel. As a control, NC100 synthesis was not induced (−NC). BM was added to the cells and biotinylated proteins detected by SDS-PAGE, followed by blotting. The modification of p30 was quantified (error bars, s.d.; n=3). f, As in e, but spheroplasts were diluted into an iso-osmotic solution of xylitol and the initial, linear rate of turbidity decrease was determined (error bars, s.d.; n=3). Spheroplasts were also analyzed after treatment with Rif. The ΔP-SecY/+Rif sample showed an initial lag phase, which was ignored. g, As in f, but with dilution into iso-osmotic KCl containing valinomycin.
Next we used osmotic swelling/bursting of cells (Fig. S1) to test the permeability of the resting SecY channel for xylitol, an uncharged sugar of 152 Da. E. coli cells were converted into spheroplasts and diluted into an iso-osmotic solution of xylitol. Spheroplasts containing wild-type SecY channel did not take up xylitol and therefore the turbidity of the sample changed little over time (Fig. 1d). In contrast, the ΔP mutant allowed xylitol to permeate, particularly when the channel was cleared of translocating chains by rifampicin(Fig. 1d). Finally, we used osmotic swelling/bursting to test the permeation of charged and small (35 Da) Cl− ions. Spheroplasts were diluted into an iso-osmotic solution of KCl in the presence of valinomycin, an ionophore that allows the K+ counterions to directly move through the lipid bilayer. The data show that wild-type SecY does not conduct Cl− ions, in contrast to the ΔP mutant (Fig. 1e). We conclude that the resting wild-type channel is impermeable to the small molecules tested, and that the plug domain of SecY is required for the seal. It should be noted that the ΔP mutant did not allow passage of K+ (Fig. S4), Na+, or SO42− ions (not shown). Thus, in agreement with previous results14, the open channel still provides a barrier to some molecules(see Supplementary Discussion), which may explain the relatively minor growth defect of the ΔP mutant (Fig. S2).
To study the permeability of the active SecY channel, we developed a method to occupy the channels in vivo with a defined co-translational translocation intermediate. The model substrate (NC100) contains 100 amino acids(Fig. 2a), including the signal sequence of DsbA, which targets it to the signal recognition particle (SRP)-dependent co-translational translocation pathway15, a sequence from an unrelated protein, a myc-tag for detection, and the SecM-stalling sequence16–18. After synthesis of the SecM sequence, the ribosome stallson the mRNA, with the nascent chain associated as peptidyl-tRNA(Fig. 2a). The construct was synthesized from an inducible promoter in cells expressing the SecY channel from a constitutive promoter. The insertion of the nascent chain into the channel was verified by in vivo disulfide crosslinking; addition of an oxidant to the cell culture led to efficient crosslinking of a single cysteine at position 19 in NC100 to a single cysteine at position 68 of the plug domain of SecY(SecY68C; Fig. S5). Channel insertion was dependent on the hydrophobicity of the signal sequence and was strongly reduced in the absence of SRP (Fig. S6). When the expression of SecY68C was diminished by changing the start-codon, about 70% of SecY68C was crosslinked to NC100, as judged by the reduction of the non-crosslinked SecY band upon addition of the oxidant (Fig. 2b, lane 2 versus 1; quantification was confirmed by loading different amounts; lanes 7–9). When NC100 expression was not induced, a lower percentage of SecY68C crosslinked to endogenous proteins (lane 4 versus 2; white arrowheads); these crosslinks disappeared over time in the presence of rifampicin(lanes 6 and 11). These results indicate that most of the SecY molecules can be occupied by NC100. Given the almost 1:1 molar ratio of nascent chain and SecY, a single SecY copy may be sufficient for cotranslational translocation of a nascent chain, as proposed before19,20.
We used the new method to ask whether the open SecY channel, represented by the ΔP mutant (see Fig. 1), can be blocked for small molecules by NC100. The ΔP mutant expressed from a constitutive promoter was leaky for the modification reagent BM, but induction of NC100 abolished permeation (Fig. 2c, lane 8 versus 7). Inhibition of transcription by rifampicin released NC100 from ribosomes and restored leakiness for BM(lane 9). NC100 with a defective signal sequence (RR; two arginines in the hydrophobic core)did not block BM permeation (lane 10). With wild-type SecY channel, no leakage was observed, regardless of whether or not NC100 was expressed (lanes 1–5). Thus, the open pore of the ΔP mutant is sealed upon binding of the ribosome-nascent chain complex to the SecY channel.
To test whether the seal is provided by channel insertion of the nascent polypeptide, we expressed chains of different lengths. Nascent chains of 90 amino acids or longer were inserted into the SecY channel, as demonstrated by disulfide crosslinking (Fig. 2d). These chains almost completely blocked the permeation of BM through the ΔP channel (Figs. 2e and S7). Some reduction of BM permeability was also observed with chains of 60 to 80 residues, which were not inserted into SecY. These chains are still likely targeted to the SecY channel, as signal sequence mutants (RR) did not prevent BM permeation. Thus, the formation of a ribosome-channel junction may provide a barrier for BM, but complete blockage of permeation requires channel insertion of the nascent chain. Similar results were obtained when the permeation of xylitol was analyzed by osmotic swelling/bursting of spheroplasts(Fig. 2f). The permeation of Cl− ions through the ΔP channel was inhibited only moderately by short nascent chains(Fig. 2g), but was reduced by ~90% by channel-inserted nascent chains, although not quite to the level seen with wild-type channel. Chains with a defective signal sequence did not significantly block Cl− permeation. Consistent with our observation that short chains do not efficiently block the permeation of the smallest molecules through the ΔP channel, the cells grew significantly slower than those expressing channel-inserted chains (Fig. S8). Wild-type SecY did not cause chain-length dependent growth behavior (Fig. S8) and it did not allow significant Cl− uptake into spheroplasts regardless of nascent chain length, even when potentially counteracting pump activity was abolished by energy depletion (Fig. S9). Collectively, these results show that the nascent chain itself provides an effective seal. Consistent with this notion, expression of the post-translational, SecA-dependent substrate proOmpA in the ΔP mutant significantly reduced BM permeation, whereas a signal sequence mutant did not (Fig. S10). The level of channel blockage was lower than with a stalled ribosome-nascent chain complex, probably because proOmpA only transiently occupies the SecY channel. It should be noted that some Cl− permeation was observed in vitro with wild-type SecY channel engaged in SecA-mediated translocation14,21 (see Supplementary Discussion).
Next we tested whether the specific sequence of the nascent chain inside the channel affects the permeability for small molecules. We determined that residues ~39 to 60 of NC100 are inside the central pore (Fig. 2a), based on the observation that the last ~40 residues are inside the ribosome and residues 19–34 are close to the plug domain on the periplasmic side of SecY (Fig. S11). We then varied the sequence of the nascent chain inside the SecY channel, making it more hydrophilic or hydrophobic than the original NC100 sequence, or replacing parts with stretches of glycines (Fig. S12). All variants completely blocked BM and xylitol permeation through the ΔP channel, like the original NC100 chain (Fig. S12; and data not shown). The more hydrophobic chains were somewhat more potent in blocking Cl− ion permeation (Fig. S12). With wild-type channel, little or no permeation was observed, regardless of which nascent chain was expressed. Thus, many different sequences of a translocating polypeptide can block the permeation of small molecules through the pore.
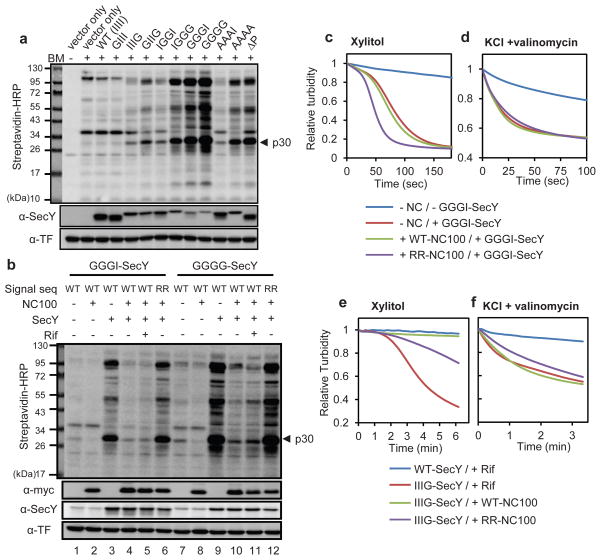
Next we tested the role of SecY’s pore ring in sealing the resting channel. Replacement of just one of the six isoleucines in the pore by a glycine caused only little BM permeation, but with increasing number of glycine substitutions the channel became progressively more leaky (Fig. 3a). Alanine substitutions had less severe effects. BM permeation occurs through the resting channel, as addition of rifampicin had no effect (Fig. S13a). The pore mutant channels were also permeable for xylitol (see below) and Cl− ions (Fig. S13b). In addition, the most severe pore mutants caused a strong growth defect (Fig. S13c); the cells died immediately after induction of the SecY mutants (Fig. S2). Cell death is likely caused by dissipation of the membrane potential: flow cytometry using a voltage-sensitive dye showed that the membrane potential was decreased to about the same extent as seen at 20 μM concentration of the ionophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Fig. S14). Taken together, these results show that the pore ring plays an important role in maintaining the seal of the resting SecY channel, and that a leaky channel is inconsistent with cell viability.
Figure 3. Permeability in pore ring mutants.
a, Wild-type SecY or pore mutants in which Ile86, Ile191, Ile278, or Ile408(IIII) were replaced by Gly or Ala were expressed under the Tet promoter. Cells containing the ΔP mutant oran empty vector were also analyzed. After treatment of cells with BM, the samples were analyzed by SDS-PAGE and blotting with streptavidin-HRP conjugate or SecY-and TF-antibodies. b, NC100 with wild-type (WT) or defective (RR) signal sequence was expressed from the inducible arabinose promoter together with SecY pore mutants under the Tet promoter. Where indicated, rifampicin (Rif) was added before BM. The samples were analyzed by SDS-PAGE, followed by blotting with streptavidin-HRP conjugate or myc-(detects NC100), SecY-, and TF-antibodies. Addition of Rif does not clear the mutant channels of nascent chains (lanes 5 and 11), likely because peptidyltransferase activity is compromised in these dying cells. c, As in b, but spheroplasts containing the GGGI pore mutant were diluted into an iso-osmotic xylitol solution, and the turbidity change followed over time. d, As in c, but spheroplasts were diluted into iso-osmotic KCl containing valinomycin. e, As in c, but with the IIIG pore mutant under the constitutive promoter with a GUG start-codon. The cells were treated with Rifor induced for NC100 expression. f, As in e, but spheroplasts were diluted into iso-osmotic KCl containing valinomycin.
Finally, we investigated the role of the pore ring in sealing the active channel. Expression of NC100 blocked the permeation of BM through even the most severe SecY pore mutants(Fig. 3b, lanes 4 versus 3 and 10 versus 9), while expressing a signal sequence mutant (RR) of NC100 did not (lanes 6 and 12). On the other hand, the permeation of the smaller molecules xylitol (Fig. 3c), Cl− ions (Fig. 3d), or K+ ions (Fig. S15) was not prevented by expression of NC100 in a pore mutant containing three glycines. With a mutant containing only one glycine in the pore ring, NC100 expression prevented xylitol (Fig. 3e), but not Cl− ions (Fig. 3f) from passing. The high permeability for Cl− ions was maintained regardless of the sequence inside the SecY channel (Fig. S16). Thus, a translocating channel requires isoleucines all around the pore ring to prevent small Cl− ions from passing, whereas pore defects are tolerated for larger molecules (Fig. S1).
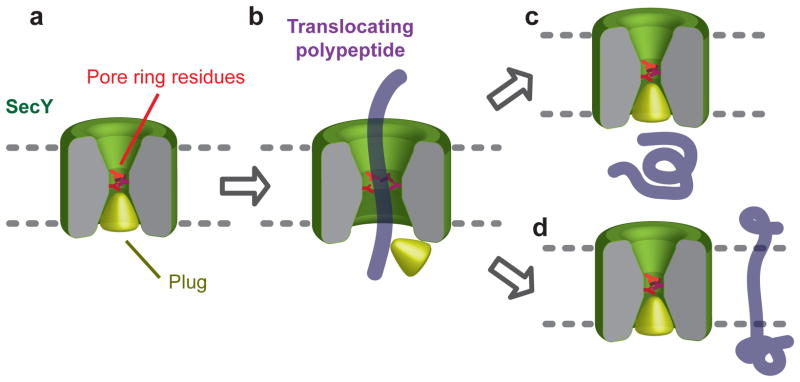
Our results show that the wild-type SecY channel is sealed for even the smallest molecules, both in its resting state and when translocating a polypeptide. A simple model explains how this is achieved(Fig. 4). In the resting state, the plug is located in the center of SecY, interacting with the pore ring residues and sealing the channel(Fig. 4a). During translocation the plug is displaced, while the pore ring forms a gasket-like seal around the translocating polypeptide chain to prevent the free flow of ions (Fig. 4b). The translocating chain itself serves as the major obstacle for small molecules; without it, the open pore allows many small molecules to pass. Whenever the polypeptide leaves the channel, either towards the extracellular side following termination of translocation (Fig. 4c), or sideways into lipid following the arrival of a hydrophobic trans-membrane sequence(Fig. 4d), the plug returns and re-seals the channel. This mechanism applies to both co-and post-translational translocation. However, in co-translational translocation, the ribosome-channel junction appears to provide an additional barrier for somewhat larger molecules, preventing metabolites and other cytosolic molecules from reaching the channel. Given the sequence conservation of SecY and Sec61 channels, these principles may be universal. However, in prokaryotes a tight seal is essential for cell viability, whereas the intracellular endoplasmic reticulum membrane may tolerate some leakiness22–24, which may explain why Sec61 pore mutants in S. cerevisiae have only minor growth defects25.
Figure 4. Model for the maintenance of the membrane barrier by the SecY channel.
For details, see text.
METHODS SUMMARY
All strains and plasmids used in this study are listed and described in Supplementary Tables 1 and 2. The subunits of the heterotrimeric SecY complex were expressed either from the Tet promoter in the pTet vector (Figs. 1, 3a–d), or from the endogenous rplN promoter on either the pACYC-SecYEG (Figs. 2c, 2e–g, 3e–f) or the pRSY (Figs. 2d) vectors. The latter also contained the SRP components (Ffh, 4.5S RNA, and FtsY) under their own promoters. NC100 or its variants were expressed from the arabinose promoter on the separate pBAD-NC100 plasmid, except in Fig. 2b, where both NC100(from the arabinose promoter)and SecY(from the rplN promoter)were expressed from the same plasmid and the SRP components from another. Except for Fig. 2d, a strain was used in which chromosomal SecY is tagged at the C-terminus with a calmodulin-binding peptide (CBP). To measure permeability of BM, cell cultures were incubated with 0.4 mM biotin-PEG2-maleimide, followed by quenching with 20 mM β-mercaptoethanol and lysis in SDS-sample buffer. For cell fractionation, cells were converted into spheroplasts by treatment with EDTA/lysozymein the presence of 18% sucrose. The spheroplasts were lysed by sonication, and the membranes separated from the cytosol by ultra-centrifugation. To measure permeability for other small molecules, spheroplasts were diluted 20-fold with iso-osmotic solutions and the absorbance was followed at 500 nm in a spectrophotometer. To disulfide-crosslink nascent chains with SecY, 0.25 mM CuPh3 was added directly to the culture, followed by quenching with 20 mM N-ethylmaleimide and lysis in SDS-sample buffer.
Supplementary Material
Acknowledgments
We thank P. Walter, H. Bernstein, and G. Phillips for materials, D. Boyd for advice, C. Akey for discussions, and C. Akey, A. Osborne, and A. Salic for critical reading of the manuscript. The work was supported by a grant from the NIH (GM052586). T.A.R. is a Howard Hughes Medical Institute investigator.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
E.P. performed the experiments, and E.P. and T.A.R. wrote the manuscript.
Author information
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 3.Harris CR, Silhavy TJ. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. Embo J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 6.Liao S, Lin J, Do H, Johnson AE. Bothlumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- 7.Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 8.Saparov SM, et al. Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell. 2007;26:501–509. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 9.Li W, et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 11.Bieker KL, Phillips GJ, Silhavy TJ. The sec and prl genes of Escherichia coli. J Bioenerg Biomembr. 1990;22:291–310. doi: 10.1007/BF00763169. [DOI] [PubMed] [Google Scholar]
- 12.Derman AI, Puziss JW, Bassford PJ, Beckwith J. A signal sequence is not required for protein export in prlA mutants of Escherichia Coli. EMBO Journal. 1993;12:879–888. doi: 10.1002/j.1460-2075.1993.tb05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalal K, Duong F. The SecY complex forms a channel capable of ionic discrimination. EMBO Rep. 2009;10:762–768. doi: 10.1038/embor.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schierle CF, et al. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J Bacteriol. 2003;185:5706–5713. doi: 10.1128/JB.185.19.5706-5713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 17.Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 18.Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Menetret JF, et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiebel E, Wickner W. Preprotein translocation creates a halide anion permeability in the Escherichia coli plasma membrane. J Biol Chem. 1992;267:7505–7510. [PubMed] [Google Scholar]
- 22.Le Gall S, Neuhof A, Rapoport TA. The endoplasmic reticulum membrane is permeable to small molecules. Mol Biol Cell. 2004;15:447–455. doi: 10.1091/mbc.E03-05-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heritage D, Wonderlin WF. Translocon pores in the endoplasmic reticulum are permeable to a neutral, polar molecule. J Biol Chem. 2001;276:22655–22662. doi: 10.1074/jbc.M102409200. [DOI] [PubMed] [Google Scholar]
- 24.Roy A, Wonderlin WF. The permeability of the endoplasmic reticulum is dynamically coupled to protein synthesis. J Biol Chem. 2003;278:4397–4403. doi: 10.1074/jbc.M207295200. [DOI] [PubMed] [Google Scholar]
- 25.Junne T, Kocik L, Spiess M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol Biol Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.