Telomerase, the enzyme dedicated to the synthesis of telomere repeat units, has attracted considerable attention owing to its critical function in maintaining chromosome ends and extending cellular life span (Artandi and Cooper, 2009). Though telomerase was referred to as telomere terminal transferase upon its initial discovery, it soon became clear that the enzyme uses an integral RNA component (TER) as the template for DNA synthesis, and is thus by definition a reverse transcriptase (RT) (Greider and Blackburn, 1989). Its evolutionary kinship to other RTs, however, was not resolved until more than a decade later, when the catalytic protein component (TERT) was cloned (Lingner et al., 1997). The initial TERT sequences from yeast and a ciliated protozoon, as well as from numerous homologues subsequently identified, reveal a core RT domain that clearly shares common ancestry with other prototypical RTs. The ensuing biochemical analyses and the recent crystal structures a TERT homologue from Tribolium castaneum (TcTERT) further reinforce the notion that telomerase utilizes similar chemical mechanisms as other RTs to catalyze the nucleotidyl transfer reaction (Autexier and Lue, 2006; Gillis et al., 2008). A central question for devotees of this “special” RT then shifted to how a core RT domain can be elaborated and joined with other protein and RNA domains to perform its dedicated biochemical function. Considerable “tweaking” of the basic RT reaction is evidently necessary given that telomerase (1) captures and extrudes single stranded DNA, (2) repetitively reverse transcribes the same limited template region within a much larger RNA molecule (Fig. 1). Through the efforts of many groups working on disparate systems, the outline of the answer to the central question is coming into closer focus; the RNA and protein domains necessary for telomere repeat synthesis are reasonably well defined and in some cases, their contributions to specific steps of the reaction cycle characterized. The dissection of different systems was productive because of the vagaries of expressing and manipulating telomerase components and the distinct genetic and cell biological tools available for each organism. It also yielded a greater appreciation of the variability and diversity of telomerase structures and properties. For instance, the Tetrahymena telomerase has a greater propensity to reverse transcribe the template iteratively, thus producing long DNA products, whereas others generate mainly short products (Cohn and Blackburn, 1995; Greider, 1991). Having data from multiple systems, though, makes the task of integrating the findings and deriving common themes become all the more challenging. In this regard, the work by Robart and Collins (2011) in the current issue of Molecular Cell helps to resolve a number of uncertainties and yield a more unifying picture of how telomerase works.
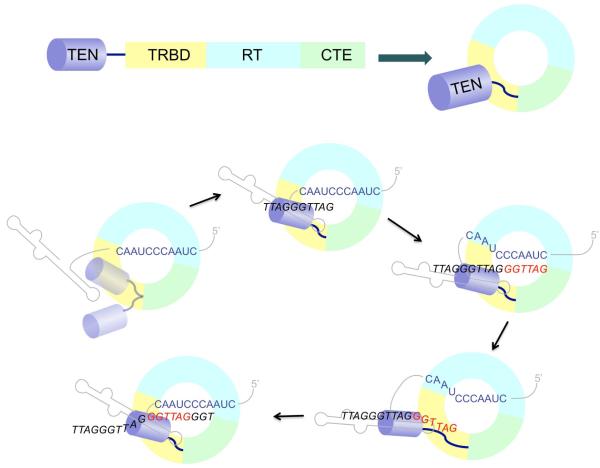
Fig. 1. The architecture of TERT and the telomerase reaction cycle.
(Top) The TERT polypeptide consists of four widely conserved domains: TEN, telomerase essential N-terminal; TRBD, TERT RNA-binding domain; RT, reverse transcriptase, and CTE, C-terminal extension. The TEN domain is connected through a flexible linker to the rest of the protein, which adopts a toroidal shape. (Bottom) The TEN domain is hypothesized to be a flexible appendage of the core enzyme. In the presence of DNA, a network of TEN-DNA, TEN-RNA, TEN-RT and RNA-DNA interactions help to trap the DNA substrate/product within the RNP and allow the DNA to be processively extended.
The authors set out to define the interactions between human TERT and TER (telomerase RNA) domains and assess the roles of these interactions in promoting telomere repeat synthesis. Their basic strategy was to express tagged human TERT and TER fragments in human cell lines, prepare cell extracts, and then examine the interactions between fragments by affinity purification, as well as determine the activities of the isolated fragments or complexes. Similar issues were addressed in earlier studies, but primarily through in vitro expression and reconstitution of telomerase fragments. An important finding by Robart and Collins was the detection of a robust interaction between the TRBD and CTE domain of TERT (Fig. 1). In a sense, this interaction was not unexpected; the crystal structure of TcTERT revealed a ring-shaped structure involving an extensive interface between precisely these two domains. However, many of the residues at the interface are not well conserved. Uncertainty as to the general applicability of the TcTERT structure is also stoked by the lack of information on insect telomerase RNA and the fact that TcTERT is devoid of a widely conserved domain known as the TEN domain (see below). Viewed in this light, the interaction between the TRBD and CTE domains of human TERT is quite reassuring, and bolsters the case for a conserved architecture for TERTs in different organisms.
Another striking and unifying observation relates to the function of the aforementioned TEN domain. A review the telomerase reaction cycle is worthwhile for understanding the remarkable activity of this domain. In order for telomerase to engage in repetitive copying of the template RNA, it has to first capture the DNA 3′ end, anneal the end to the RNA template region, and shuttle the hybrid to the active site of TERT to commence reverse transcription (Fig. 1). After the 5′ end of the RNA template is reached, the template/product hybrid must unpair to allow re-alignment and another cycle of reverse transcription. Classic biochemical analyses suggest the existence of a separate DNA-binding domain (anchor site) within the telomerase complex that allows the complex to retain the DNA substrate/product during the unpairing and re-alignment steps. Indeed, subsequent experiments generally point to the TEN (telomerase essential N-terminal) domain as being the main provider of anchor site function (Autexier and Lue, 2006). This widespread, though not universally conserved domain, is connected to the core RT domain through a flexible linker, and has been shown to bind both DNA and RNA with low affinity (Jacobs et al., 2006). Ascribing anchor site function to the TEN domain would place it physically and functionally part from the catalytic site, and predicts preferential loss of long extension products as a result of TEN domain mutations. However, other studies have suggested a direct involvement of this domain in the polymerization reaction (Jurczyluk et al., 2010). Robart and Collins showed in their new study that a “TEN-less” telomerase can indeed catalyze a single round of reverse transcription, but not processive DNA synthesis. Moreover, they were able to restore multiple repeat addition by supplying the TEN domain in trans. Coupled with previous studies in yeast and the fact that some insect TERTs are naturally TEN-less, these observations re-emphasize a conserved function for the TEN domain in promoting processivity rather than catalysis (Gillis et al., 2008; Lue, 2005).
Two observations by Robart and Collins speak to intriguing aspects of TEN domain mechanisms. First, as a separate polypeptide, it binds to the rest of the TERT protein in an RNA-stimulated manner. Second, whereas the TEN-less telomerase is unable to retain a DNA substrate stably through affinity purification, it acquires this ability when the TEN domain is supplied in trans. Such findings imply a complex set of interactions (aptly characterized by the authors as constituting network) that provides telomerase with increased versatility for engaging single stranded telomeric DNA (Fig. 1). That the TEN domain is able to help capture the DNA is at first glance somewhat surprising. Direct analyses of isolated polypeptide fragments have generally shown this domain to have lower affinity for DNA than the rest of TERT (Wyatt et al., 2007). However, when the TEN domain is supplied in trans, it does not simply add a surface for DNA-binding, but rather becomes tethered to the rest of the RNP through both protein-protein and protein-RNA interactions. These interactions are likely to reinforce the TEN-DNA interaction to allow DNA capture and retention.
One may naturally wonder as to whether such an elaborate scheme for engaging substrate is really necessary. One plausible advantage, as emphasized by the authors, is the greater opportunity for regulatory control of telomerase action in vivo: each of the myriad of interactions could in principle be targeted by regulatory factors. But another intriguing possibility is that having multiple weak interactions confers the system with a dynamic flexibility that is beneficial. Perhaps a high affinity DNA binding anchor site or a rigidly positioned anchor site will actually hamper the movement of the DNA substrate through the RNP and make the enzyme less proficient at making telomeric DNA. With more efficient systems for reconstituting and analyzing telomerase in place, these and other interesting issues should now be amenable to experimental tests and are likely to keep devotees of telomerase and nucleic acid enzymes occupied for years to come.
Footnotes
[SC1]The synthesis of telomeric DNA by telomerase entails repeated cycles of reverse transcription on a short RNA template. In this issue of Molecular Cell, Robart and Collins (2011) describe a set of interactions between human telomerase RNA, protein domains, and the substrate DNA that drives the intricate reaction cycle.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Artandi SE, Cooper JP. Reverse transcribing the code for chromosome stability. Mol Cell. 2009;36:715–719. doi: 10.1016/j.molcel.2009.11.030. [DOI] [PubMed] [Google Scholar]
- Autexier C, Lue NF. The Structure And Function Of Telomerase Reverse Transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Greider C. Telomerase is processive. Mol. Cell. Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, Parker MW, Cohen SB, Bryan TM. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2010;39:1774–1788. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lue N. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol Cell Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]