Abstract
In C. elegans, the domains of Hox gene expression are controlled by the novel global regulatory gene sop-2. We identified a region located 3 prime of the Hox gene egl-5 that promotes ectopic expression of an egl-5 reporter gene in a sop-2 mutant. SOP-2 could directly block positive regulatory factors acting in this region, or it could block their expression. We identified three possible miRNA binding sites within the egl-5 3 prime UTR. Cognate microRNAs are expressed in relevant tissues and can block egl-5 expression when expressed from a transgene. Mutation of the putative binding sites in the egl-5 3 prime UTR resulted in a modest degree of misexpression of a minimal egl-5 reporter gene, suggesting that microRNAs may contribute to the tight restriction of egl-5 expression to particular cell lineages.
Keywords: gene regulation, microRNA, polycomb protein, transcription, Hox gene
INTRODUCTION
Hox genes play a key role in patterning the animal body (Lemons and McGinnis, 2006). They perform this function through largely cell-autonomous effects that direct the fates of cells into specific differentiation pathways appropriate to particular body regions. Understanding how the region-specific expression pattern of Hox genes is established is therefore important for understanding how animal morphology develops (Pearson et al., 2005).
In the nematode C. elegans, the posterior Hox gene egl-5 specifies the fates of cells in the tail region (Chisholm, 1991; Aboobaker and Blaxter, 2003). Its function is particularly significant in the male, where egl-5 specifies the development of multiple male-specific mating structures. In accordance with this role, egl-5 expression is found in a subset of cells and cell lineages throughout the tail region (Ferreira et al., 1999). Analysis of the egl-5 promoter has shown that the complex egl-5 expression pattern is established under the direction of a number of conserved cis regulatory elements distributed through large 5’ and 3’ intergenic domains (Teng et al., 2004).
In spite of their conserved developmental role across phyla, the modes of regulation of Hox genes are diverse. In vertebrates, the genes are organized in tight genomic clusters collinear with the region of the body where they act. Such clusters may reflect the presence of shared and intermingled transcriptional regulatory elements (Lemons and McGinnis, 2006). In nematodes, echinoderms and urochordates, on the other hand, the Hox genes are disordered or dispersed to separate genomic sites (Aboobaker and Blaxter, 2003; Spagnuolo et al., 2003; Seo et al., 2004; Cameron et al., 2006). In many animal phyla, polycomb group (PcG) genes are essential regulators in establishment of the regional pattern of Hox gene expression (Schuettengruber et al., 2007). However, of the two widely conserved classes of PcG genes, while C. elegans contains members of the PRC2 complex, it lacks the components of the PRC1 complex (Schuettengruber et al., 2007). Instead, C. elegans Hox genes are under the global regulation of novel PcG-like genes not found in other organisms (Zhang et al., 2003; Zhang et al., 2006)
Here we investigate regulation of egl-5 by one of these Polycomb-like genes, sop-2. sop-2 is required for correct temporal and sexual as well as spatial regulation of gene expression during C. elegans development. In mutants of sop-2, as well as of its interacting partner sor-1, Hox genes are expressed in inappropriate body regions in a manner similar to the phenotypes of PcG mutants in other animals (Zhang et al., 2003; Zhang et al., 2006). The regulatory role of sop-2, an essential gene, is not restricted to Hox genes and is required for the correct expression of many genes across multiple developmental axes. One hypothesis is that sop-2 acts by disrupting miRNA regulatory pathways (Grishok et al., 2005; Parry et al., 2007; Yang et al., 2007; Cai et al., 2008). Remarkably, neither sop-2 nor sor-1 are found in the genomes of the most closely-related nematode species, C. briggsae and C. remanei, nor have orthologs been reported to date in any available genome sequence, making their origin and role in C. elegans development particularly intriguing.
In order to gain insight into the mechanism by which sop-2 restricts expression of certain genes to specific domains, we focused on egl-5, comparing the expression of egl-5 transgenes in wild type and in a sop-2 mutant background. We found that the expression domain of a minimal reporter gene driven by an egl-5 lineage-specific cis regulatory element is restricted by sop-2 in a manner similar to the endogenous egl-5 gene. We found that sop-2 restricts expression by inhibiting the activity of a novel cis regulatory element in the egl-5 3’ DNA which induces ectopic expression in a sop-2 mutant background. We investigated the possible role of miRNA pathways in regulation of egl-5 and identified putative miRNA binding sites in the egl-5 3’UTR. These sites have a weak effect on reporter gene expression and appear to function independently of regulation by sop-2.
MATERIALS AND METHODS
Nematodes and culture methods
Nematodes were cultured following standard methods (Brenner, 1974). The N2 Bristol-derived strain used as wild type (CB4088) carried the him-5(e1490)V mutation so that it spontaneously generated males. Other strains used were: EM574 (pha-1(e2123ts)III;him-5(e1490)V), EM1081 (sop-2(bx91)II;pha-1(e2123ts)III;him-5(e1490)V); EM1080 (bxIs13;pha-1(e2123ts)III;him-5(e1490)V).
egl-5 reporter genes
A 306 nucleotide region (genomic coordinates 7810910-7811215) surrounding the 181 nucleotide V6 lineage specific enhancer element V6CRE (Teng et al., 2004) and the 2680 nucleotide region downstream of egl-5 protein coding sequence (genomic coordinates 7816269-7818949) were amplified from genomic DNA by PCR (Expand Long Template or Expand High Fidelity PCR Systems from Roche Molecular Biochemicals). The V6CRE sequence was inserted upstream of the Δpes-10 promoter of plasmid pPD122.53, which contains GFP coding sequence terminated by the 3’ UTR of the unc-54 myosin gene (A. Fire laboratory vector, 1999). The cloned plasmid (EM#320) was microinjected into nematodes to generate the extrachromosomal transgenic array bxEx145 (strain EM1063). A segment of 113 nucleotides from the egl-5 3’UTR (genomic coordinates 7817076-7817189) was inserted into EM#320 by fusion PCR and the construct was injected into worms to generate bxEx146. To generate the starting plasmid for deletion analysis, the unc-54 3’ UTR in EM#320 was replaced by the 2680 nucleotide PCR product from the egl-5 3’ DNA, generating plasmid EM#321 and extrachromosomal transgenic array bxEx139 (EM1057). We call this starting reporter gene V6CRE-egl-5-3’REG after the two regulatory elements it contains, the enhancer V6CRE and egl-5 gene 3’ regulatory DNA. Single, double and triple deletion of the three potential miRNA binding sequences were generated from EM#321 by PCR and cloned in E. coli and injected into worms. Other deletion and point mutation derivatives were generated from EM#321 by PCR and directly injected into worms in the stitching method (Hobert, 2002). The identity of each construct was verified by restriction digestion and sequencing; DNA was prepared from multiple independent isolates and a mixture was used for injections.
Fragments of C. elegans egl-5 gene included in the 3’ regions of constructs were as follows (genomic nucleotide number): bxEx139: 7816269–7818949; bxEx140: 7816269–7817279; bxEx141: 7816269-7817189; bxEx142: 7816269–7817076; bxEx143: 7816269–7817055; bxEx144: 7816269–7816379.
miRNA reporter genes and misexpression constructs
mir-247prom::gfp and mir-249prom::gfp were generated by joining about 5 and 6 kb, respectively, upstream sequence up to the start of the precursor miRNA of the mir-247 (genomic coordinates 4752037-4756998) and mir-249 gene (genomic coordinates 3006528-3012389) to the Δpes-10 promoter driving GFP-coding sequence in pPD122.53 by PCR fusion and directly injected into worms (giving bxEx158 and bxEx156, respectively). All other miRNAprom::gfp constructs were made by introducing restriction sites into PCR primers and then cloning in E. coli. 3kb upstream sequence (genomic coordinates 11770136-11773453) was used for mir-61 (bxEx157). Since mir-54, 55 and 56 are clustered in the genome, about 6 kb 5’ region was taken as common promoter for all three miRNAs (genomic coordinates 13145008-13150692) (bxEx155).
The miRNA misexpression constructs were made by fusing the V6CRE-Δpes-10 promoter with PCR-amplified genomic sequence covering the hairpin sequence of each miRNA. Transgenic worms with misexpression constructs were generated by coinjecting PCR products (20 ng/μl) and pBX1 (100 ng/μl) into strain EM1080, bxIs13;pha-1(e2123ts);him-5(e1490), following the stitching method. The genomic coordinates amplified were: mir-54: 13144915-13145013 (bxEx159); mir-56: 13144610-13144706 (bxEx160); mir-249: 3006431-3006527 (bxEx161); mir-247: 4756939-4757036 (bxEx162); mir-61: 11770038-11770134 (bxEx163).
Generation of transgenic nematodes
Transgenic nematodes were generated by microinjection as described by Mello et al. (1991). PCR products or plasmids (20-50 ng/μl) are coinjected with pBX1 (100 ng/μl) (Granato et al., 1994) into strain pha-1(e2123ts);him-5(e1490) (EM574). Multiple independent lines were examined for consistency of expression patterns. Transgenes were introduced into a sop-2 mutant background by crossing the wild-type transgenic animals to strain sop-2(bx91);pha-1(e2123ts);him-5(e1490) (EM1081). At 25°C, sop-2(bx91) mutant animals arrest at early larval stages. In order to obtain older individuals for observation, we picked adult hermaphrodite worms and allowed them to lay eggs for one hour at 20°C, kept the eggs at 20°C for 5-6 hours and then transferred the eggs to 25°C.
Microscopy of transgenic animals
Animals from a minimum of two lines per construct were mounted on 5% agarose/noble agar pads and examined under Nomarski optics at 400× or 1000×. Fluorescence was observed at 1000× with a Zeiss 487905 filter set/Chroma High Q GFP LP filter set.
RESULTS
A minimal egl-5 reporter gene is regulated by sop-2 along spatial, temporal, and sexual developmental axes
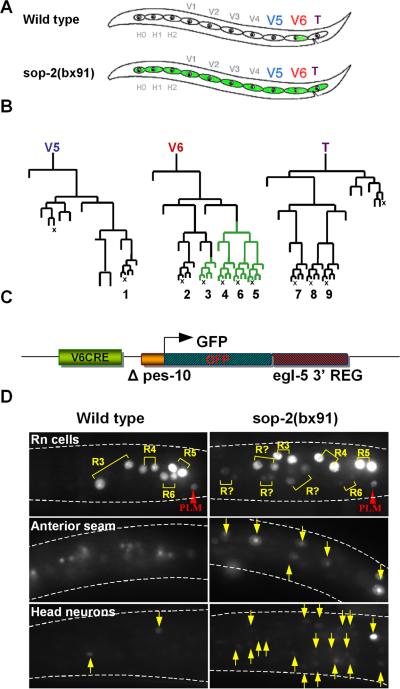
In order to investigate regulation of gene expression by sop-2, we focused on its action in controlling the expression of an egl-5 reporter gene in a single tissue, the hypodermal seam. The seam consists of a row of 10-13 blast cells arrayed in the hypodermis from head to tail along each side of the animal (Fig. 1A). Seam cells divide during postembryonic development to generate additional hypodermal cells and neurons (Sulston and Horvitz, 1977). In males, the three most posterior seam cells, V5, V6, and T, undergo additional rounds of division to generate the rays, three-celled male-specific sensilla used in mating (Sulston et al., 1980). In wild type, egl-5 is expressed in the posterior branches of the V6 seam cell lineage, where it specifies the development and properties of rays 3-6 (Chow and Emmons, 1994; Ferreira et al., 1999; Lints et al., 2004) (Fig. 1B).
Fig. 1.
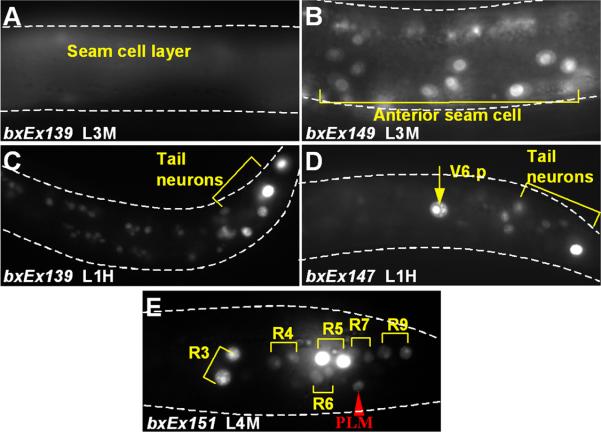
The expression domain of a minimal egl-5 seam cell reporter gene is restricted by sop-2. A. Seam cells in an L1 larva. Seam cells form a row of hypodermal blast cells that generate additional hypodermal cells and neurons during postembryonic development. The three posterior seam cells V5, V6, and T generate the male rays. The wild type egl-5 gene is expressed in the posterior lineage branches of V6. In a sop-2 mutant, it is expressed in more anterior seam cell lineages as well. B. Postembryonic cell lineages of seam cells V5, V6 and T in the male. egl-5 is expressed in the posterior branches of the V6 lineage beginning in the L2 larval stage (green lineage branches). The terminal sublineages labeled 1-9 generate the rays (X, programmed cell death). C. Structure of the minimal egl-5 seam cell reporter, V6CRE-egl-5-3’REG. In this reporter, GFP transcribed from the C. elegans promoter Δpes-10 is controlled by the egl-5 V6 lineage cis regulatory element V6CRE and egl-5 3’ DNA. D. Expression of V6CRE-egl-5-3’REG in the anterior seam is repressed by sop-2. In wild type, shown is GFP fluorescence in the first two daughters of the ray precursor cells (Rn cells), R3, R4, R5, and R6, which generate the ray sublineages leading to rays 3-6. Flurosecence is also present in the posterior touch cell PLM. The reporter is not expressed in the anterior seam (background fluorescence is due to gut autofluorescence), and has weak expression in a few head neurons (arrows). In sop-2(bx91), additional ray sublineages express the reporter, as well as anterior seam cells and additional head neurons (arrows).
Restriction of egl-5 expression in the seam to the posterior branches of the V6 lineage requires the function of sop-2. In the temperature-sensitive mutant sop-2(bx91) at restrictive temperature (25°C), the expression domain of egl-5 expands to include anterior seam cells (Zhang et al., 2003) (Fig. 1A). Thus sop-2 represses egl-5 expression outside of its normal posterior expression domain in the seam.
Expression of egl-5 in the V6 lineage is directed by a conserved 180 nucleotide cis-regulatory element termed V6CRE (Teng et al., 2004). To understand how sop-2 represses expression of egl-5 in the anterior seam, we utilized a minimal reporter gene (V6CRE-egl-5-3’REG) in which V6CRE drives transcription of a GFP gene in the seam (Fig. 1C). In this reporter, GFP coding sequence is transcribed from a naïve promoter (Δpes-10) and is terminated by 2680 nucleotides of egl-5 3’ DNA sequence including the entire egl-5 3’ UTR. V6CRE-egl-5-3’REG recapitulates the wild type pattern of egl-5 expression in the V6 lineage (Fig 1D). Expression begins during the L2 stage in V6.ppp, remains on in V.6ppp descendants, and in addition comes on in V6.pappp such that by mid L3 it is expressed in the precursor cells of rays 3-6 (R3-R6) and their descendants (Fig. 1B). V6CRE-egl-5-3’REG also recapitulates normal expression of egl-5 in the bilateral pair of PLM neurons, and is weakly expressed in a small number of head neurons, where endogenous egl-5 gene expression has not been reported.
In sop-2(bx91), V6CRE-egl-5-3’REG is mis-expressed in the seam in a manner similar to the endogenous egl-5 gene. At permissive temperature (20°C), there was no difference from the wild type expression pattern. However, at restrictive temperature (25°), 83% of sop-2(bx91) transgenic animals containing V6CRE-egl-5-3’REG as an extrachromosomal array (bxEx139) had an average of 7 (range: 1-30; n = 201) ectopic seam cell nuclei that were positive for GFP expression (Fig. 1D, Fig. 2). Ectopic seam cell nuclei included nuclei in the anterior seam, additional descendants of V6, and descendants of seam cell T. Ectopic expression was first observed during early L2 and increased until late L2. These results show that V6CRE-egl-5-3’REG expression was spatially mis-regulated. Expression of the transgene was also temporally mis-regulated. In the wild type male V6 lineage, expression is first seen in V6.ppp (100%, n = 93). In sop-2(bx91), expression often began in V6.p or V6.pp (48%, n = 204). Finally, expression was also sexually misregulated. Ectopic anterior seam cell as well as V6 lineage seam cell expression of V6CRE-egl-5-3’REG was also observed in hermaphrodites (39/49 animals scored). Thus sop-2 is required for correct expression of V6CRE-egl-5-3’REG along three developmental axes — spatial, temporal and sexual — as has been observed for the endogenous egl-5 gene (Zhang et al., 2003; Cai et al., 2008).
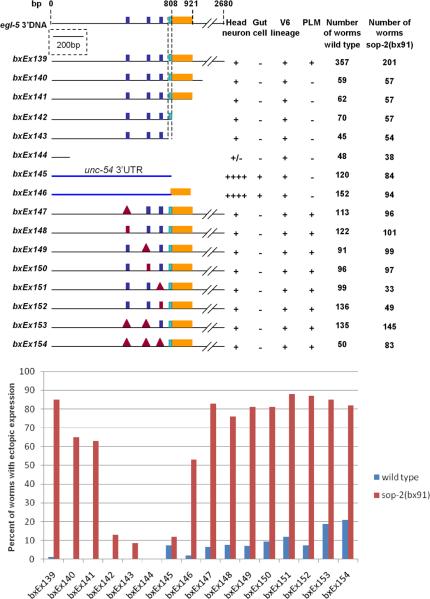
Fig. 2.
Effect of egl-5 3’ DNA sequences on expression of the minimal seam cell reporter. Top: the structure of deletion derivatives of V6CRE-egl-5-3’REG and their expression in several tissues. Bottom: the percent of worms with ectopic seam cell expression in wild type and in a sop-2 mutant background. Dark blue boxes indicate the sites of three potential miRNA binding sequences. Red box indicates the introduction of point mutations within the sequence, red triangle indicates the sequence was deleted. The light blue box indicates the 3’ end of an egl-5 cDNA and the position of a short conserved region (F5, see Figure 3). The yellow box indicates the 113 nucleotide region that causes ectopic expression in a sop-2 background.
Ectopic reporter gene expression requires a sequence in egl-5 3’ DNA
Since evidence suggests sop-2 may act by affecting miRNA pathways, we examined the influence of the egl-5 3’ DNA sequence on the expression of V6CRE-egl-5-3’REG in wild type and in sop-2(bx91). In a wild type background, expression of the reporter was not affected by a series of deletions that removed DNA up to within 111 nucleotides of the end of egl-5 coding sequence (bxEx144), with the exception that PLM expression was lost (Fig. 2). The results map a PLM enhancer to the region 1011-2680 nucleotides downstream of egl-5 coding sequence (compare bxEx139 and bxEx140). However, in the seam there was no ectopic expression, while in the V6 lineage expression was normal. Therefore, considering the role of sop-2, this gene evidently does not repress reporter gene activity via a sequence in egl-5 3’ DNA up to within 111 nucleotides of the end of the gene. Deletion of such a sequence would have resulted in ectopic seam cell expression in a wild type background, similar to that seen in sop-2(bx91).
However, unlike in wild type, in a sop-2(bx91) mutant background expression of the reporter was affected by these deletions (Fig. 2). Ectopic expression was lost when a 113 nucleotide region was deleted between 808 and 921 nucleotides downstream from the egl-5 stop codon (compare bxEx141 and bxEx142). This region is immediately downstream of the 3’ end of a documented egl-5 cDNA (C08C3.1b isoform, www.WormBase.org). Therefore, ectopic expression of the reporter in a sop-2 mutant background is promoted by this 113 nucleotide region. This region may either contain a novel seam cell enhancer element or a sequence that increases productive transcripts initiated by the V6CRE enhancer. The activity of this sequence is inhibited by sop-2.
We next attempted to determine where sop-2 acted to repress the activity of this novel 3’ regulatory sequence. We replaced the egl-5 3’ DNA sequence in V6CRE-egl-5-3’REG with the 3’ UTR sequence of the muscle myosin gene unc-54 (bxEx145). This reporter has normal V6 lineage expression and, as expected, since it lacks the novel regulatory sequence, seam cell expression is not significantly increased in a sop-2 mutant background (Fig. 2; bxEx145). In addition to being expressed in the V6 lineage, this reporter is strongly expressed in gut cells (Teng et al., 2004) and in additional head neurons in both wild type and in sop-2(bx91) (data not shown), indicating that the unc-54 3’ UTR contains a sequence that increases transcription or productive processing of reporter gene transcripts in these tissues. When the 113 nucleotide novel regulatory region from egl-5 was added to this reporter (bxEx146), the reporter was expressed ectopically in the seam in sop-2(bx91) but not in wild type (Fig. 2). Expression in gut cells and head neurons was unaffected. This confirms that the 113 nucleotide region promotes ectopic seam cell expression in a sop-2 mutant background. It shows further that the DNA site necessary for sop-2-mediated repression of the 113 nucleotide region does not map within egl-5 3’ DNA sequence outside of the 113 nucleotide region. This site could lie within the 113 nucleotide region itself, possibly because it contains a SOP-2 binding sequence, or elsewhere in the transgene, for example within V6CRE. Alternatively, sop-2 might repress ectopic expression of the transgene indirectly by repressing an activator encoded elsewhere in the genome that acted within the 113 nucleotide region, inducing expression in the anterior seam.
miRNAs with seed matches to three potential binding sequences in the egl-5 3’UTR are expressed in the seam
To investigate whether miRNA regulation played a role in limiting egl-5 expression to the posterior seam, or to a particular time during development or a particular sex, we identified potential miRNA binding sites in the egl-5 3’UTR and tested their function. Through sequence alignment we identified three phylogenetic footprints conserved across C. elegans, C briggsae, and C. remanei that contain seed matches respectively to the mir-56 group, which includes mir-51, mir-52, mir-53, mir-54, mir-55, and mir-56; to mir-249; and to mir-44, mir-45, mir-61, and mir-247 (Fig. 3). The region with a match to mir-249 was previously identified as the conserved region CR15 in Teng et al. (2004). The other two conserved sequences were not previously described.
Fig 3.
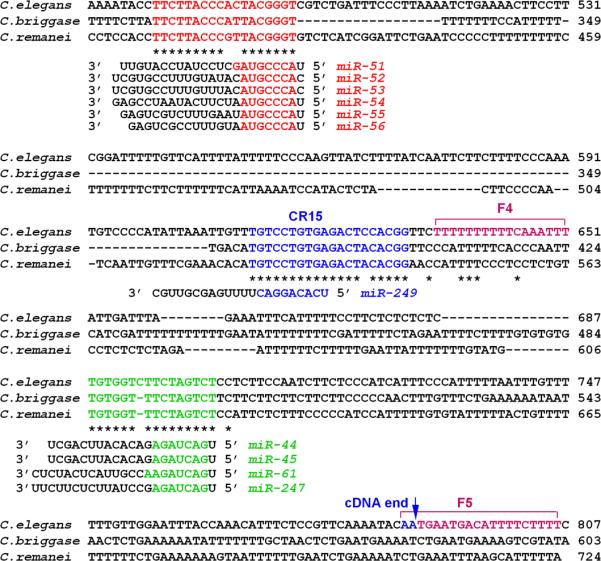
Three conserved sequences in the egl-5 3’UTR have seed matches to C. elegans miRNA's. The coordinates shown are the number of nucleotides downstream of the egl-5 stop codon, the third nucleotide of which is at genomic position 7816268*. The positions are shown of three conserved regions described by Teng at al. (2004), CR15, F4 and F5. The position of the end of an egl-5 cDNA is also shown.
To determine whether egl-5 might be regulated by these miRNA's, we examined their expression patterns during wild type development. Reporter genes were made for mir-54/55/56, 249, 247 and 61, by fusing a region of genomic sequence immediately upstream of the miRNA encoding sequence to the green fluorescent protein gene. Several independent transgenic lines carrying the mirprom::gfp extrachromosomal array displayed GFP expression in some cells and tissue types of the worm, including seam cells.
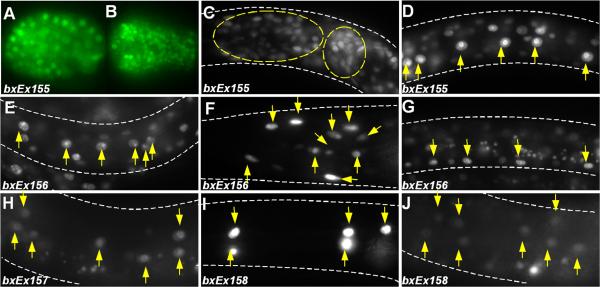
mir-54/55/56::gfp was detected in the cell nuclei of essentially all somatic cells when driven by a region extending 6.0 kb upstream from the mir-54 coding sequence. Expression was first observed in the 28-cell stage embryo, weakly and diffusely within nuclei of all somatic cells. The expression then became strong from ~200-cell stage onwards and persisted throughout embryogenesis, larval stages and adulthood (Figure 4A-D).
Fig. 4.
miRNA's defined by seed matches to conserved egl-5 3’UTR sequences are expressed in the seam. A-D: mir-54/55/56::gfp (bxEx155) is ubiquitously expressed at several developmental stages. (A) about 80 cell stage embryo; (B) bean stage embryo; (C) head neuron expression; (D) seam cell expression. mir-249::gfp (bxEx156) is expressed in many seam cells (E), several head neurons (F), and P lineage cells (G). mir-61::gfp (bxEx157) is expressed transiently in seam cells in worms at L3 stage (H) and older. mir-247::gfp (bxEx158) is expressed transiently in several head neurons (I) and seam cells in L3 stage (J) and older worms.
Expression of mir-249::gfp driven by a 6.0 kb upstream region was first observed in most, if not all seam cells in L1 stage worms in both hermaphrodites (Figure 4E) and males. Some head neurons (Figure 4F) and P lineage cells (Figure 4G) also consistently expressed gfp.
mir-247::gfp with 3kb promoter was observed transiently in seam cells in L3 and L4 stage worms (Figure 4H) and also some neurons (data not shown). mir-61::gfp, driven by 3kb upstream region, is also expressed in several head neurons (Figure 4I) and transiently and weekly in some seam cells (Figure 4J) in L3 and L4 stage worms, in addition to vulva precursor cells and gonadal cells as reported before (Yoo and Greenwald, 2006). Since all the miRNAs investigated are expressed consistently or transiently in all or some of the seam cells, they could play a role in regulating gene expression in the seam.
Driving expression of miRNAs in the seam blocks egl-5 expression and function
To test the ability of miRNAs to block egl-5 expression, for several of the miRNA's we asked whether expressing them ectopically from a transgene in the V6 lineage would inhibit expression of an egl-5 reporter gene. We joined miRNA genomic coding sequence covering the entire inverted repeat structure of the miRNA primary transcript to the V6 lineage cis-regulatory element V6CRE and the Δpes-10 naïve promoter. We introduced these constructs into a strain containing the integrated egl-5 reporter transgene bxIs13, which consists of the intact egl-5 chromosomal locus from 7 kb upstream of the gene to 2.7 kb downstream, with GFP coding sequence inserted in frame at the 3’ end of egl-5 coding sequence. The region included contains V6CRE and several additional tissue-specific regulatory elements, and bxIs13 is expressed in the corresponding tissues (Teng et al., 2004).
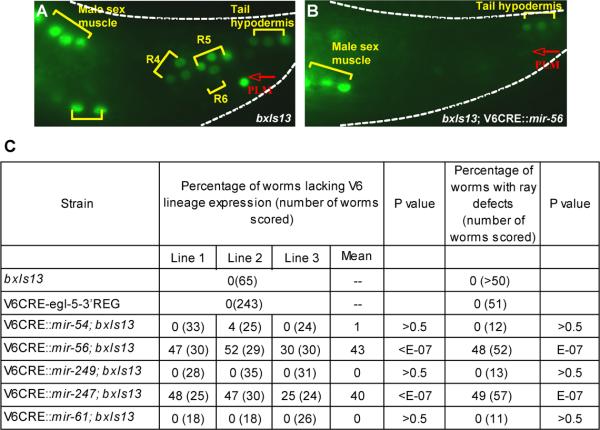
Transgenes containing the coding sequences of mir-56 and mir-247 driven by V6CRE eliminated GFP expression from bxIs13 in the V6 lineage in some animals, but did not affect expression in male sex muscles or tail hypodermis (Fig. 5). Therefore, it appears that mir-56 and mir-247 can repress egl-5 gene expression. No affect on the GFP expression pattern of bxIs13 was observed in strains carrying transgenes for mir-54, mir-249, or mir-61. Since we did not monitor expression of the miRNA from these transgenes, no conclusion can be drawn from this negative result.
Fig. 5.
Driving miRNA expression in the V6 lineage can repress egl-5 expression. A. The integrated egl-5 reporter gene bxIs13 is expressed in ray sublineages, male sex muscles, tail hypodermis, and PLM. B. A transgene containing the mir-56 gene under the control of the V6 lineage cis-regulatory element V6CRE blocks bxIs13 expression in the ray sublineages and PLM but not in sex muscles or tail hypodermis. C. Transgenes containing mir-56 and mir-247 under the control of V6CRE can block bxIs13 expression in the V6 lineage and also generate a ray-defective phenotype consistent with absence of chromosomal egl-5 gene expression. Transgenes designed to express three other mir genes did not. Expression data is given for three independently transformed lines. P values are for comparison to bxIs13.
Elimination of egl-5 gene expression from bxIs13 in the V6 lineage by transgenes expressing mir-56 and mir-247 suggested that these transgenes ought to also block expression of the endogenous, chromosomal egl-5 gene in this lineage, producing an Egl-5 phenotype. We examined the adult morphology of males and found that this was indeed the case. egl-5 is necessary for the identities of sensory rays 3-5 and for generation of ray 6 (Chisholm, 1991; Chow and Emmons, 1994; Lints et al., 2004). In the absence of egl-5 function, rays 3-5 take the identity of ray 2 and fuse with it (Chow and Emmons, 1994; Lints et al., 2004). For strains expressing mir-56 and mir-247, we found a significant level of abnormalities affecting rays 3-6 (Fig. 5). These abnormalities included missing rays and morphologically abnormal rays including fused rays. The fraction of animals exhibiting these abnormalities was similar to the fraction of animals with absent bxIs13 GFP expression in V6 lineage cells. Strains expressing mir-54, mir-249, and mir-61, as well as the strain containing V6CRE-egl-5-3’REG, did not exhibit such abnormalities, suggesting that the abnormal morphology phenotype did not result from titration of egl-5 regulatory factors by the transgenic arrays.
Putative miRNA binding sites play a limited role in restricting V6CRE-EGL-5-REG expression to the posterior seam
In view of the evidence that miRNA's cognate to potential binding sequences within the egl-5 3’UTR were expressed in the anterior seam and could repress egl-5 gene expression if ectopically expressed from an extrachromosomal transgene, we examined the roles of the putative miRNA binding sequences in the egl-5 3’UTR in regulation of gene expression. We modified each of these sequences in V6CRE-egl-5-3’REG either by deleting it or by substituting an unrelated sequence of equal length (from tacgggt to cgtaaca, from gtcctgtg to acttcaca, and from tctagtc to ctcgact). Deletions and substitutions had similar effects. Individually, mutation of each of these putative binding sequences resulted in a small increase in ectopic seam cell expression in wild type animals (Fig. 2). In addition to expression in anterior seam cells, expression was observed prematurely in V6.p, in T seam cell descendants and in hermaphrodites (Fig. 6). Thus there was evidence for less precise regulation of gene expression along three developmental axes. The effects of single mutations were additive. In a transgene in which all three sequences were deleted, ectopic seam cell expression was observed in 21% of animals (Fig. 2). We conclude that miRNA may contribute to bringing about a tight restriction of egl-5 gene expression to specific V6 lineage branches in the male.
Fig. 6.
Mutation of putative miRNA binding sites in the egl-5 3’UTR results in ectopic seam cell expression. A. V6CRE-egl-5-3’REG is not expressed in the seam in the midbody. B-F. Examples of ectopic expression patterns of several transgenes with mutated miRNA target sequences (see Fig. 2). Expression is shown in (B) the midbody seam of an L3 male; (C) posterior neurons of the L1 hermaphrodite; (D) hermaphrodite V6.p cell; (E) ray 7 and 9 precursor descendants of T seam cell in an L4 male.
Confirming the evidence given above that sequences in the egl-5 3’UTR outside of the 113 nucleotide region that enhanced ectopic seam cell expression in sop-2(bx91) were not required for sop-2 to repress reporter gene expression, mutation of putative miRNA binding sequences did not affect regulation of the transgene by sop-2. Transgenes containing mutations in putative miRNA binding sequences were expressed ectopically at a high level in sop-2(bx91) compared to wild type (Fig. 2). Thus neither repression of ectopic seam cell expression by sop-2, nor ectopic expression in the absence of sop-2 function, seemed to require the function of miRNA acting at any of the three putative miRNA binding sequences. However, the absence of any indication of additivity of the effects of putative miRNA binding site mutation and sop-2 mutation raises the possibility that regulation of gene expression via these sites requires sop-2 function.
Discussion
In search of a sop-2 response element
Global regulation of gene expression in C. elegans by sop-2 resembles in many ways gene regulation by the repressive PRC1 Polycomb Group (PcG) genes of other animals. In particular, in both PcG mutants and in sop-2 mutants, Hox genes are expressed outside of their normal spatial domains. PcG genes repress gene activity via a diverse class of cis-regulatory sequences known collectively as Polycomb Response Elements (PRE) to which they are tethered by other proteins with DNA-binding activity (Muller and Kassis, 2006; Ringrose and Paro, 2007; Schuettengruber et al., 2007). To determine whether sop-2 acted in a similar manner, we searched for a sop-2 Response Element in flanking DNA of the Hox gene egl-5.
We found that a minimal egl-5 reporter gene, consisting only of a lineage-specific enhancer element from egl-5, a GFP coding sequence driven by a naïve promoter and followed by 2680 nucleotides of egl-5 3’ DNA sequence, contained an element that conferred regulation of the transgene by sop-2. Like the chromosomal egl-5 gene, expression of this transgene in the anterior seam is repressed by sop-2. We mapped the responsible element to a 113 nucleotide region in egl-5 3’ DNA. This region promotes transgene expression in a sop-2(-) background, but has no observable effect in wild type.
Grishok et al. (2005) showed that transgene silencing in the C. elegans soma could be brought about by dsRNA with homology to plasmid sequences that are outside of the transgene transcriptional unit. This silencing required the function of sop-2 and the RNAi pathway. Grishok et al. (2005) suggest that this effect is due to chromosomal transcriptional gene silencing targeted against non-specific double-stranded transcripts generated by read-through of the complex transgenic arrays. The finding in our case of regulation of an egl-5 reporter gene by sop-2 in a tissue-specific manner similar to sop-2 regulation of the endogenous gene, via a sequence in egl-5 DNA, appears to rule out such a non-specific transgene silencing mechanism here.
We could not identify a sequence outside of the 113 nucleotide region where sop-2 acted to repress anterior seam cell expression. One possibility is that both a positively-acting factor and SOP-2 act within the 113 nucleotide region. Prior work on PREs has shown they are complex sequences with binding sites for multiple DNA-binding factors that can have both positively and negatively acting effects on gene expression (Muller and Kassis, 2006; Ringrose and Paro, 2007; Schuettengruber et al., 2007; DeVido et al., 2008). Blast search revealed no conserved motifs within the 113 nucleotide region, and this region was not conserved in other nematode species. Alternatively, sop-2 could act within V6CRE or by repressing a trans-activator gene located elsewhere in the genome. SOP-2 contains multiple RNA-binding domains but has not been demonstrated to bind DNA (Zhang et al., 2004).
Regulation of egl-5 by miRNA
We identified three conserved regions in the egl-5 3’ UTR that contain seed matches to C. elegans miRNA's. We demonstrated that the cognate miRNAs are expressed in the seam and, when expressed from an ectopic promoter in a particular cell lineage, are capable of repressing egl-5 expression from an integrated transgene. Ectopic expression also caused defects in male sensory structures that suggested expression of chromosomal egl-5 was also blocked. Thus egl-5 is capable of being regulated by miRNAs with cognate sites in the egl-5 3’UTR.
Deletion of the conserved potential miRNA binding sites in the egl-5 3’UTR resulted in a small but significant degree of mis-expression of our reporter gene in the seam. Regulation of C. elegans Hox genes by miRNA has not been studied previously. In mouse, HoxB8 mRNA is cleaved by miR-196, while HoxA11 is down-regulated by miR-181 (Yekta et al., 2004; Naguibneva et al., 2006). miRNA genes present in vertebrate Hox clusters appear to function in reinforcing the antero-posterior pattern of Hox gene expression (Yekta et al., 2008). Our results suggest that miRNAs may be one component of multi-factor regulation of egl-5 expression, making a contribution to the precise localization of this expression to the posterior branches of the V6 lineage in males. This supplementary or refining role of miRNAs in the case of egl-5 would be consistent with recent observations on the global role of miRNA, which appears to be that of a modulator with a modest effect on the level of expression of many target genes (Baek et al., 2008; Selbach et al., 2008)
Acknowledgements
We thank the C. elegans Genetics Center for strains; A. Fire for plasmids; and members of the Emmons laboratory for helpful discussions. C. Smith and J. Di Mele provided expert technical assistance. This research was supported by NIH Grant R01 GM39353 to S. W. E. S. W. E. is the Siegfried Ullmann Professor of Genetics.
References
- Aboobaker AA, Blaxter ML. Hox gene loss during dynamic evolution of the nematode cluster. Current Biology. 2003;13:37–40. doi: 10.1016/s0960-9822(02)01399-4. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Sun Y, Huang X, Guo C, Zhang Y, Zhu Z, Zhang H. The Caenorhabditis elegans PcG-like gene sop-2 regulates the temporal and sexual specificities of cell fates. Genetics. 2008;178:1445–1456. doi: 10.1534/genetics.108.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RA, Rowen L, Nesbitt R, Bloom S, Rast JP, Berney K, Arenas-Mena C, Martinez P, Lucas S, Richardson PM, Davidson EH, Peterson KJ, Hood L. Unusual gene order and organization of the sea urchin hox cluster. J. Exp. Zool. B Mol. Dev. Evol. 2006;306:45–58. doi: 10.1002/jez.b.21070. [DOI] [PubMed] [Google Scholar]
- Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- Chow KL, Emmons SW. HOM-C/Hox genes and four interacting loci determine the morphogenetic properties of single cells in the nematode male tail. Development. 1994;120:2579–2592. doi: 10.1242/dev.120.9.2579. [DOI] [PubMed] [Google Scholar]
- DeVido SK, Kwon D, Brown JL, Kassis JA. The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development. 2008;135:669–676. doi: 10.1242/dev.014779. [DOI] [PubMed] [Google Scholar]
- Ferreira HB, Zhang Y, Zhao C, Emmons SW. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene-transfer in C. elegans. Nucleic Acids Research. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes and Development. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Lemons D, McGinnis W. Genomic evolution of hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Lints R, Jia L, Kim K, Li C, Emmons SW. Axial patterning of C. elegans male sensilla identities by selector genes. Dev Biol. 2004;269:137–151. doi: 10.1016/j.ydbio.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb DT, Ambros VR. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Current Opinion in Genetics and Development. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nature Cell Biology. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Parry DH, Xu J, Ruvkun G. A whole-genome RNAi screen for C. elegans miRNA pathway genes. Current Biology. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nature Reviews Genetics. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chaourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008 doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Seo H-C, Edvardsen RB, Maeland AD, Bjordal M, Jensen MF, Hansen A, Flaat M, Weissenbach J, Lehrach H, Wincker P, Reinhardt R, Chourrout D. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- Spagnuolo A, Ristoratore F, Di Gregorio A, Aniello F, Branno M, Di Lauro R. Unusual number and genomic organization of Hox genes in the tunicate Ciona intestinalis. Gene. 2003;309:71–79. doi: 10.1016/s0378-1119(03)00488-8. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Teng Y, Girard L, Ferreira HB, Sternberg PW, Emmons SW. Dissection of cis-regulatory elements in the C. elegans Hox gene egl-5 promoter. Dev Biol. 2004;276:476–492. doi: 10.1016/j.ydbio.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Yang Y, Sun Y, Luo X, Zhang Y, Chen Y, Tian E, Lints R, Zhang H. Polycomb-like genes are necessary for specification of dopaminergic and serotonergic neurons in Caenorhabditis elegans. PNAS. 2007;104:852–857. doi: 10.1073/pnas.0610261104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih I, Bartel D. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nature Reviews Genetics. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Azevedo RB, Lints R, Doyle C, Teng Y, Haber D, Emmons SW. Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev Cell. 2003;4:903–915. doi: 10.1016/s1534-5807(03)00136-9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cristoforou A, Aravind L, Emmons SW, van den Heuvel S, Haber DA. Polycomb group proteins directly bind to RNA. Molecular Cell. 2004;14:841–847. doi: 10.1016/j.molcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang T, Sun Y, Tian E, Deng H, Zhang Y, Luo X, Cai Q, Wang H, Chai J, Zhang H. RNA-binding proteins SOP-2 and SOR-1 form a novel PcG-like complex in C. elegans. Development. 2006;133:1023–1033. doi: 10.1242/dev.02275. [DOI] [PubMed] [Google Scholar]