Abstract
Tuberculosis remains a serious global health threat, with the emergence of multidrug-resistant strains highlighting the urgent need for novel antituberculosis drugs. The enzyme 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS) catalyzes the first step of the shikimate pathway for the biosynthesis of aromatic compounds. This pathway has been shown to be essential in Mycobacterium tuberculosis, the pathogen responsible for tuberculosis. DAH7PS catalyzes a condensation reaction between P-enolpyruvate and erythrose 4-phosphate to give 3-deoxy-d-arabino-heptulosonate 7-phosphate. The enzyme reaction mechanism is proposed to include a tetrahedral intermediate, which is formed by attack of an active site water on the central carbon of P-enolpyruvate during the course of the reaction. Molecular modeling of this intermediate into the active site reported in this study shows a configurational preference consistent with water attack from the re face of P-enolpyruvate. Based on this model, we designed and synthesized an inhibitor of DAH7PS that mimics this reaction intermediate. Both enantiomers of this intermediate mimic were potent inhibitors of M. tuberculosis DAH7PS, with inhibitory constants in the nanomolar range. The crystal structure of the DAH7PS-inhibitor complex was solved to 2.35 Å. Both the position of the inhibitor and the conformational changes of active site residues observed in this structure correspond closely to the predictions from the intermediate modeling. This structure also identifies a water molecule that is located in the appropriate position to attack the re face of P-enolpyruvate during the course of the reaction, allowing the catalytic mechanism for this enzyme to be clearly defined.
Keywords: Enzyme Catalysis, Enzyme Inhibitors, Enzyme Structure, Enzymes, Protein Structure, Aromatic Amino Acid Biosynthesis, Shikimate Pathway
Introduction
Tuberculosis remains a serious global health threat with over a million deaths per year. The recent emergence of multidrug-resistant strains of Mycobacterium tuberculosis, the pathogen that causes the lung disease, highlights the need for rapid development of new antibacterial drugs to combat tuberculosis (1–4).
3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS)4 is an important enzyme in M. tuberculosis and other pathogens. It catalyzes the first committed step in the shikimate pathway, which is responsible for the biosynthesis of aromatic amino acids and other essential aromatic metabolites in microorganisms, plants, and apicomplexan parasites (5–7). This pathway is absent in humans, and inhibitors of amino acid biosynthesis have been shown to be effective antimicrobial and herbicidal agents (8, 9). Gene disruption studies have demonstrated that M. tuberculosis is not viable if the shikimate pathway is not operational (10). These findings make DAH7PS an attractive target for drug development.
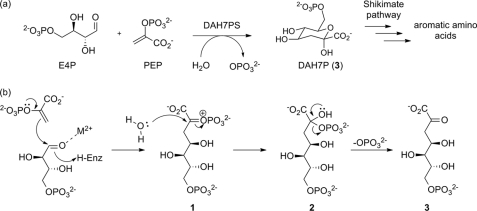
DAH7PS catalyzes the aldol-like condensation of P-enolpyruvate and d-erythrose 4-phosphate (E4P) to yield 3-deoxy-d-arabino-heptulusonate 7-phosphate (Fig. 1a). The reaction mechanism has been subject to extensive study, and many of the key details of the mechanism have been elucidated (11–16). The reaction occurs stereospecifically with respect to both substrates, with the si face of P-enolpyruvate attacking the re face of E4P. A divalent metal ion present in the active site is essential for activity. The reaction takes place with cleavage of the C–O bond of P-enolpyruvate rather than the O–P bond, requiring water to attack C2 of P-enolpyruvate at some stage during the reaction.
FIGURE 1.
Catalysis by DAH7PS. a, overall reaction catalyzed by DAH7PS. b, proposed mechanism of condensation of P-enolpyruvate (PEP) with E4P. H-Enz, H-enzyme.
A mechanism consistent with the data published to date starts with nucleophilic attack of P-enolpyruvate at the E4P aldehyde moiety, resulting in the formation of oxocarbenium species 1 (Fig. 1b). This oxocarbenium ion 1 can be attacked by an active site water to form phosphohemiketal 2. It is noteworthy that water can potentially attack from either face of 1, giving rise to two possible diastereoisomers of tetrahedral intermediate 2, differing in their absolute configuration at C2. Although this stereogenic center is transient and the stereochemical information is lost by elimination of phosphate in the final step to generate the product DAH7P (3), the geometry of the enzyme active site is likely to favor stereoselective attack of water to form one diastereoisomer of 2 preferentially. In this way, DAH7P is formed in its acyclic form and cyclizes into its cyclic pyranose form following release from the enzyme.
3-Deoxy-d-manno-octulosonate 8-phosphate synthase (KDO8PS), an enzyme involved in the synthesis of the cell-wall lipopolysaccharide of Gram-negative bacteria, is structurally and evolutionary related to DAH7PS. KDO8PS catalyzes an analogous reaction between P-enolpyruvate and the five-carbon sugar arabinose 5-phosphate (17). Studies of the reaction mechanism of KDO8PS suggest that water is activated by the active site metal prior to attack at the reaction intermediate (18). Computational and structural studies of KDO8PS indicate that after activation, water or hydroxide attacks from the si face of P-enolpyruvate, resulting in the overall syn addition of arabinose-5-phosphate and a hydroxyl group to the double bond of P-enolpyruvate (19). Due to lack of comparable data for the reaction catalyzed by DAH7PS, it is unclear whether these findings also apply to the reaction catalyzed by DAH7PS. Despite the similarities in their reaction chemistry, a number of key structural and mechanistic differences of DAH7PS and KDO8PS such as divalent metal ion requirement and substrate specificity have also been identified (20).
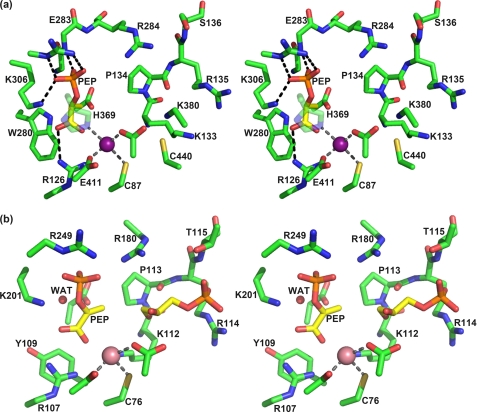
M. tuberculosis DAH7PS (MtuDAH7PS) is the only member of the DAH7PS type II family that has been structurally characterized (21–23). Type II DAH7PS enzymes show very little sequence similarity with their type I DAH7PS counterparts, which are relatively well characterized and are found in organisms such as Escherichia coli (14, 15) and Saccharomyces cerevisiae (16, 24). Both type I and type II DAH7PS enzymes share the common triosephosphate isomerase (TIM barrel) fold, and mechanistic studies have suggested that the key details of the reaction chemistry are similar for enzymes of both DAH7PS types (25). Despite the low sequence similarity, the active site architecture of MtuDAH7PS shows remarkable correspondence to that of type I enzymes (Fig. 2). P-enolpyruvate is held in place by a tightly knit network of interactions. The P-enolpyruvate phosphate forms salt bridges to Lys306 (MtuDAH7PS numbering) and Arg337 and forms a hydrogen bond to the backbone N–H of Glu283, whereas the P-enolpyruvate carboxylate forms a salt bridge to Arg126. The metal ion is coordinated by His369, Glu411, Cys87, and Asp441 in a trigonal pyramidal fashion, leaving one coordination site potentially free for the carbonyl moiety of the E4P aldehyde moiety, thereby activating this functionality to nucleophilic attack. The proposed E4P binding site is constituted mostly by a 133KPRS136 motif that is highly conserved in the type II subfamily, whereas members of the type I DAH7P synthase subfamily display a very similar, also highly conserved KPRT motif at the equivalent position. The best indication of how E4P is bound to the enzyme can be found in the crystal structure of S. cerevisiae type I DAH7PS (SceDAH7PS) in complex with the E4P analog, glycerol 3-phosphate (G3P) (16). Arginine and threonine from the KPRT motif interact with the G3P phosphate moiety, whereas the primary hydroxyl group of G3P forms a hydrogen bond to the backbone carbonyl of the proline residue of the KPRT motif, and the secondary hydroxyl interacts with the metal-coordinating aspartate.
FIGURE 2.
Active site architecture of DAH7PS. a, stereo view of the active site of M. tuberculosis DAH7PS (PDB code 3NV8) (22) showing P-enolpyruvate (PEP, yellow carbons) in its binding site. The manganese iron is represented as a purple sphere. Major interactions between P-enolpyruvate and the enzyme and the metal coordination sphere are shown as dashed lines. b, stereo view of the active site of S. cerevisiae DAH7PS in complex with P-enolpyruvate, G3P (yellow carbons), and a Co2+ ion (magenta sphere)(PDB code 1OF8) (16). A water molecule (WAT) on the re side of P-enolpyruvate (left in this orientation), which is proposed to play the role of the substrate water, is shown as a red sphere.
Despite the many similarities in active site architecture and reaction chemistry between DAH7P synthases from different organisms and families, there are some significant distinctions of type II DAH7P synthases. For example, Trp280 forms part of the P-enolpyruvate binding site in MtuDAH7PS, whereas in the E. coli and S. cerevisiae enzymes, this residue is replaced by Ala. However, in the type I enzymes, the space occupied by Trp280 is occupied by a Tyr residue from another part of the structure. Another significant distinction between MtuDAH7PS and all other DAH7P synthases characterized to date is its complex mechanism of allosteric regulation, which we have recently investigated in detail (22). Furthermore, the recent discovery that M. tuberculosis chorismate mutase is activated in complex with DAH7PS suggests a key role of DAH7PS in the regulatory network of aromatic metabolism of M. tuberculosis (26).
In vitro studies with mechanism-based inhibitors have been previously reported for E. coli DAH7PS (27, 28). Recent results in our laboratory have shown that extension of inhibitors targeting the P-enolpyruvate binding site to pick up interactions in the E4P binding site leads to increased inhibitor potency (49). In addition, carbohydrate-derived compounds, designed as mechanism-based inhibitors of both KDO8PS and DAH7PS, were shown to have mild antibacterial properties when tested against E. coli, Yersinia enterocolitica, Pseudomonas aeruginosa, Staphylococcus aureus, and Bacillus subtilis, although further in vitro studies were suggested to confirm that these compounds indeed target KDO8PS and/or DAH7PS (29). However, to date, no inhibitors for the DAH7PS from the pathogen M. tuberculosis have been reported. Herein we report the design, synthesis, and testing of an intermediate mimic as the first potent inhibitor of MtuDAH7PS in both its racemic and its enantiomerically pure forms. The crystal structure of the target enzyme in complex with the inhibitor shows that the simplified intermediate analog binds in a fashion in accordance with its design and thus provides valuable insight into both the DAH7PS reaction mechanism and a lead structure for novel potent antimycobacterial drugs targeting MtuDAH7PS.
EXPERIMENTAL PROCEDURES
Analytical Methods
Optical rotations [α]D20 were measured at room temperature on a PerkinElmer Life Sciences Model 341 polarimeter; analyte concentrations are given in g/100 ml. Specific rotations are reported. NMR spectroscopy was carried out on a Varian UNITY 300 MHz or Varian INOVA 500 MHz NMR spectrometer. 1H and 13C NMR spectra are referenced to external tetramethylsilane; 31P NMR spectra are referenced to external 85% phosphoric acid. Mass spectrometry was performed on Bruker maXis 3G or Micromass LCT by electrospray ionization in positive or negative ionization modes.
UV-visible spectrophotometry was carried out on a Varian Cary® One UV-visible spectrophotometer, in stoppered quartz cells. The temperature was continuously controlled at 303 K (30 °C) by the use of a jacketed multicell holder, connected to an external Varian Peltier temperature controller filled with either water or ethylene glycol.
Expression, Purification, and Crystallization of MtuDAH7PS
M. tuberculosis DAH7PS was overexpressed and purified as described previously (21–23). A reservoir solution containing 400 μm inhibitor was prepared by dissolving an appropriate amount of solid rac-4 in crystallization buffer (0.1 m Tris-HCl buffer at pH 8, 1.5 m ammonium sulfate, and 12% v/v glycerol). 500 μl of enzyme stock solution was concentrated to ∼50 μl using centrifugation with a 10-kDa cutoff membrane, and the resulting solution was diluted to ∼500 μl using the inhibitor solution. This process was repeated twice to buffer-exchange the solution as completely as possible. 1 μl of the buffer-exchanged enzyme solution (concentration 3–5 mg/ml) was mixed with 1 μl of crystallization buffer. Crystals were grown by sitting drop vapor diffusion over 300 μl of reservoir solution and appeared within 24 h at 25 °C. Crystals were flash-frozen in liquid nitrogen in a cryoprotectant solution consisting of crystallization buffer and 20% v/v glycerol.
Structure Determination
X-ray diffraction data were collected at the MX2 beamline at the Australian Synchrotron (see Table 1). The dataset was integrated using iMosflm (30). The space group and cell parameters were identical to those of previously reported structures of MtuDAH7PS allowing phases calculated from the original structure (Protein Data Bank (PDB) code 2B7O) (21) to be used to solve the structure. The structure was refined using the CCP4 software package (30) by methods analogous to the ones described for the native structure (21). Both enantiomers of inhibitor 4 were built using the Dundee PRODRG2 server (31). The two enantiomers of 4 were initially positioned manually into the electron density observed in the active site using COOT and then refined at half-occupancy. The model was optimized using repetitive cycles of model building with COOT and refinement with REFMAC5. Water molecules were added automatically in COOT and verified using |2Fo − Fc| and |Fo − Fc| maps and potential to hydrogen-bond to at least one protein atom or other water molecule. A bias-removed |Fo − Fc| omit map (see Fig. 5a) was generated by deleting the ligand and active site waters from the model and subjecting the model to simulated annealing and refinement in PHENIX. The |2Fo − Fc| map obtained by this procedure (see Fig. 5a) shows clear continuous electron density for ligand 4. The structure was validated using SFCHECK, with 97.5% of residues in the most favored region of the Ramachandran plot (Table 1). The final model included two protein chains each containing all 462 natural residues and two residues from His tag cleavage, 554 water molecules, two manganese ions, one sulfate ion, one chloride ion, and (R)- and (S)-4.
TABLE 1.
Data collection and refinement statistics for M. tuberculosis DAH7PS in complex with 4
| Data collection | |
| Crystal system | Trigonal |
| Space group | P3221 |
| Unit cell parameters | |
| a (Å) | 203.58 |
| b (Å) | 203.58 |
| c (Å) | 66.49 |
| Resolution range (Å) | 55.641–2.35 Å (2.48–2.35) |
| Measurements | 726,170 |
| Unique reflections | 65,888 |
| Redundancy | 11.0 (11.2) |
| Completeness (%) | 100 (100) |
| I/σ(I) | 5.0 (1.9) |
| Rmerge (%) | 13.0 (39.2) |
| Wilson B-value (Å2) | 26.5 |
| Refinement | |
| Resolution (Å) | 51.16–2.35 (2.41–2.35) |
| Rcryst (%) | 16.4 |
| Rfree (%) | 19.6 |
| Amino acids (chain length 464 residues) | 464 + 464 res.; 7136 atom sites |
| Water molecules | 554 |
| Other | 2 Mn2+a, 1 SO42−, 1 Cl−, (R)-4a, (S)-4a |
| Mean B (Å2) | |
| Protein | 25.13 |
| Water | 32.06 |
| Other | 12.12 |
| r.m.s.d.b from target values | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.214 |
| Dihedral angles (°) | 5.634 |
| Ramachandran | |
| Most favored (%) | 98.3 |
| Allowed (%) | 1.2 |
| Disallowed (%) | 0.5 |
| PDB code | 3PFP |
a Refined at reduced occupancy.
b r.m.s.d., root mean square deviation.
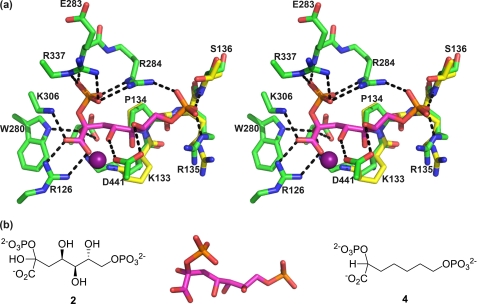
FIGURE 5.
Stereo view of the active site of the enzyme in complex with 4 (only the (R)-enantiomer is shown for clarity). a, |Fo − Fc| simulated annealing omit map contoured at 3σ showing the electron density for inhibitor 4 (blue carbons) in the active site. WAT, water. b, interactions of 4 with the active site residues (green carbons) and metal ion (purple sphere). c, overlay of P-enolpyruvate (yellow carbons)-DAH7PS structure 3NV8 (cyan carbons) with DAH7PS-4-complex (green carbons). d, overlay of the preferred pose of the linear intermediate (magenta carbons) and 4 (blue carbons; the oxygens on the C2-phosphate were omitted for clarity). The enzyme structure experimentally obtained from the DAH7PS-4-complex (green carbons) superimposes well onto the enzyme structure obtained by induced fit modeling of the linear intermediate (yellow carbons, peptide; magenta carbons, intermediate).
Inhibition Assays
Enzyme activity was monitored by following the loss of absorbance at 232 nm (ϵ = 2800 liters mol−1 cm−1) due to the consumption of P-enolpyruvate (32). Assays were carried out in 50 mm 1,3-bis(tris(hydroxymethyl)methylamino)propane buffer at pH 7.5 containing 1 mm tris(2-carboxyethyl)phosphine at 30 °C. The buffer solution was prepared using ultrapure water, which was stirred over Chelex resin (Bio-Rad) and filtered before use. Manganese(II) sulfate stock solution (10 mm) was made up in ultrapure water pretreated with Chelex resin. P-enolpyruvate and E4P substrate solutions were made up to ∼10 mm by dissolving the P-enolpyruvate monopotassium salt and E4P monosodium salt (Aldrich) in buffer solution. Accurate substrate concentrations were determined by enzymatic reaction; the absorbance changes of solutions containing one substrate in excess were determined before and after conversion of the limiting substrate by added DAH7PS had occurred, and the extinction coefficient of P-enolpyruvate was used to calculate the concentration of the limiting substrate. The absorbance change was corrected by the absorbance change caused when DAH7PS was added to a control solution that did not contain E4P. For inhibition studies, assay solutions containing manganese (II), varying concentrations of P-enolpyruvate and inhibitors, and buffer were equilibrated at 30 °C. 2 μl (1.3 mg/ml) of M. tuberculosis DAH7PS was added, and the mixture was equilibrated for 2 min before the reaction was initiated by the addition of E4P (11 μl). Final assay conditions were 10 μm Mn2+, 5.2 nm DAH7PS, 100 μm E4P, 57.6–115.2 μm P-enolpyruvate, 0–940 nm inhibitor with an appropriate amount of buffer to make up a final volume of 1 ml.
Lanzetta Phosphate Assay
Lanzetta reagent (33) was prepared fresh as required from the following components: 3 parts 0.045% w/v malachite green in water, 1 part 4.2% w/v ammonium molybdate in 4 m HCl, 0.1 parts 1.5% v/v Triton X-100 in water. The components were mixed in the dark and stirred for 1 h before the solution was filtered through a 0.45-μm syringe filter. For the qualitative detection of phosphate-containing fractions after anion exchange chromatography, a 20-μl sample of each fraction was mixed with 250 μl of Lanzetta reagent, and the color change was judged by optical inspection. For the quantitative determination of inhibitor concentration, 300 μl of the inhibitor solutions was incubated with 10 μl of calf alkaline phosphatase solution (5 units/ml in 4 mm MgCl2) for at least 2 h. To 100 μl of the digested sample was added 700 μl of Lanzetta reagent, and the absorbance at 630 nm was determined after 20 min. A calibration curve for the determination of phosphate concentration was obtained from analogous analysis of solutions of appropriate concentrations (6–150 μm) of KH2PO4, which had been dried in high vacuum for at least 3 h before use. As a control, a glucose-6-phosphate solution of known concentration was also digested with calf alkaline phosphatase and analyzed.
Modeling Procedure for Linear Intermediate and the Designed Inhibitors
Modeling studies were carried out using software packages from Schrödinger Suite 2006 software package (34–37); more detailed computational methods can be obtained from the supplemental material. The previously reported crystal structure of MtuDAH7PS (PDB code 2B7O) (21) was used as the receptor. As a starting point for docking studies, an ensemble of low-energy conformations of the ligands (R)- and (S)-4 and both epimers of the tetrahedral intermediate 2 were identified by conformational searches. The modeling of the tetrahedral intermediate epimers into the active site of MtuDAH7PS was carried out with the Schrödinger Suite 2006 Induced Fit Docking protocol (36). The induced fit docking procedure lead to an intermediate adapted protein receptor in which several residues in the active site adopted conformations different from the ones observed in the native crystal structure (PDB code 2B7O). This intermediate adapted structure of MtuDAH7PS was then used as a rigid receptor for docking studies of compounds (R)- and (S)-4, which were conducted using Glide (37).
Synthesis
All reactions were carried out under an inert atmosphere of nitrogen in flame-dried glassware unless otherwise stated. Organic solvents were dried before use by standard methods (38). Dess-Martin periodinane (39, 40) was prepared according to literature procedures. m-Chloroperbenzoic acid was recrystallized from dichloromethane before use, and all other reagents were purchased from Sigma-Aldrich and used without further purification.
Preparative procedures and characterization of previously unreported compounds can be found in the supplemental material. A representative procedure for the final step of the synthesis and characterization of inhibitor 4 is outlined in detail below.
Heptanoic Acid 2,7-Bisphosphate 4
Protected bisphosphate 11 (177 mg, 0.24 mmol) was dissolved in ethyl acetate (15 ml), and palladium on charcoal (10% palladium/carbon, 57 mg, 0.053 mmol of palladium) was added. The reaction flask was purged with hydrogen gas with three freeze-pump-thaw cycles. The reaction mixture was stirred for 16 h, after which TLC indicated complete consumption of starting material 11. The suspension was filtered through a Celite pad, the Celite was washed with ethyl acetate and methanol, and the solvent was evaporated under reduced pressure. The resulting residue was dissolved in 1 m aqueous potassium hydroxide solution (7 ml) and stirred vigorously for 2.5 h. The reaction mixture was passed down a column consisting of freshly regenerated DOWEX 50WX4-200 (H+) resin (∼5 × 1 cm), and the column was eluted with 50 ml of water. The eluate was neutralized with 1 m potassium hydroxide solution and lyophilized. The crude product was purified by anion exchange chromatography on SOURCE-Q resin eluting with a gradient of water/aqueous ammonium bicarbonate solution. Fractions containing the desired product were identified using the qualitative Lanzetta assay (see below), pooled, and lyophilized to give 42.5 mg of a white powder consisting of the product and residual ammonium bicarbonate. Lanzetta phosphate assay (see above) established that the purity of the white powder yielded was 80% by weight; the corrected yield for this step is thus 34%.
1H NMR (500 MHz, D2O) δ 4.34 (td, J = 5.6, 9.0 Hz, 1H), 3.73 (q, J = 6.4 Hz, 2H), 1.63–1.76 (m, 2H), 1.49–1.59 (m, 2H), 1.22–1.42 (m, 4H). 13C NMR (126 MHz, D2O) δ 24.0 (br), 25.2 (br), 30.1 (br), 33.7 (br), 65.4 (br), 76.1 (br), 180.5 (br). 31P NMR (121 MHz, D2O) δ ppm 3.31 (t, J = 6.0 Hz, 1P), 1.85 (d, J = 8.8 Hz, 1P). HRMS[ESI,neg] calculated for C7H14O10P2 321.0140, found 321.0131 [M-H]−. [α]D20 (R)-4, -0.9 (c 1.8, H2O), (S)-4, +0.8 (c 1.9, H2O).
RESULTS
Modeling of the Reaction Intermediate into MtuDAH7PS Active Site
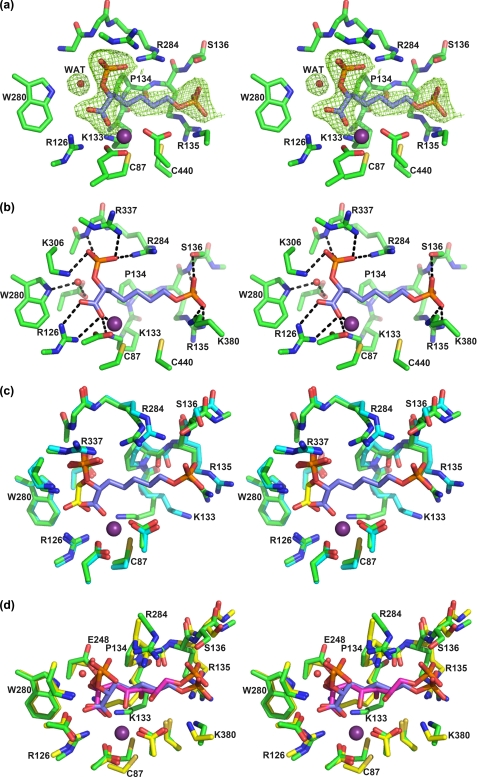
To inform inhibitor design, the predicted tetrahedral reaction intermediate (2, Fig. 3) was modeled into the active site of MtuDAH7PS. Because it is unknown from which side the water attacks the P-enolpyruvate substrate during the reaction, both possible epimers of the linear intermediate were modeled.
FIGURE 3.
Molecular modeling studies and inhibitor design. a, the modeled best pose of the tetrahedral open chain intermediate 2 (S-isomer) from induced fit docking to the active site of MtuDAH7PS superimposed on the crystal structure of SceDAH7PS (PDB code 1OF8). The active site carbons from the modeled structure are in green, and the active site residues of SceDAH7PS are shown with yellow carbons. The modeled linear intermediate molecule is shown with magenta carbons, and the C2-hydroxyl group is pointing behind the plane. The residues are labeled according to the numbering in MtuDAH7PS. The metal ion is displayed as a purple sphere. Hydrogen bonds are represented by dashed lines. b, left, linear intermediate 2. Center, molecular model of linear intermediate 2 showing the preferred 2S-configuration. Right, simplified intermediate analog 4, synthetic target for this study.
The modeling study produced a total of 30 possible poses for the two possible C2 diastereoisomers of the tetrahedral intermediate in the active site, out of which 17 retained the expected P-enolpyruvate binding interactions. All 17 poses with correct P-enolpyruvate orientation are from the S-isomer of the intermediate, consistent with water attacking the C3 carbon of P-enolpyruvate from its re face, which is the side opposite the active site Mn2+ ion.
The best pose from the modeling study (Fig. 3a) was selected based on two criteria. The first is that the P-enolpyruvate part of the tetrahedral intermediate retains the same interactions as observed in the original crystal structure (PDB code 2B7O). The second is that the positioning of the E4P phosphate group should be as close as possible to the phosphate group of G3P in the crystal structure of SceDAH7PS (PDB code 1OF8), which is considered to provide the best prediction for the E4P phosphate position. In the best pose (Fig. 3a), the P-enolpyruvate part of the molecule interacts with Arg126, Lys306, Arg337, Arg284, and Glu283. Arg284 bridges the phosphate groups of P-enolpyruvate and E4P through salt bridges and hydrogen bonds. The E4P phosphate group interacts with Arg284, Arg135, and Ser136, which suggests that part of the KPRS motif (Arg135 and Ser136) plays a key role in positioning of the E4P phosphate, as expected from the G3P-bound SceDAH7PS structure. The E4P aldehyde group, now converted to a hydroxyl group in the linear intermediate, is at a distance of 2.2 Å from the metal ion. This indicates that the aldehyde of E4P may be held in place by coordination to the metal ion before reaction occurs. The hydroxyl group that corresponds to the incoming water molecule forms hydrogen bonds with Trp280, Glu248, and Lys133. Two of these residues, Glu248 and Lys133, are conserved across the entire DAH7PS family, whereas Trp280 is only conserved in the type II enzymes.
Inhibitor Design
Combining results from the intermediate modeling and previous work on mechanism-based inhibitors (27, 28), we designed bisphosphate 4 (Fig. 3b) as a simplified analog of the tetrahedral intermediate. The 2-phosphorylcarboxylic acid moiety targets the P-enolpyruvate binding site and resembles phospholactate, which has been reported as an efficient inhibitor of E. coli DAH7PS (27). Bisphosphate 4 represents a simplified analog of tetrahedral intermediate 2; the phosphoryl and carboxyl moieties of compound 4 are proposed to act as charged anchors arranged in a similar geometry to those of intermediate 2 and held together by a non-functionalized hydrocarbon linker. Given its low degree of functionalization, bisphosphate 4 may be used as a lead structure for specific introduction of functionality to enhance enzyme-ligand affinity. In light of the results for molecular modeling of the tetrahedral intermediate 2, which suggest a preference for the S-configuration at C2, we devised two syntheses leading to target compound 4 to assess possible enantio-discrimination: one racemic synthesis, which had already been proven useful in our laboratory, as well as a chiral pool synthesis, which allowed control of the absolute configuration at C2.
Synthesis of Racemic 4
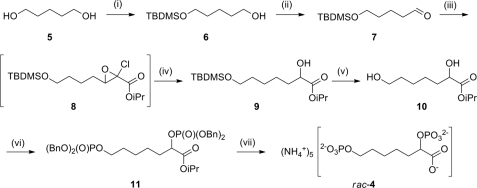
The synthetic strategy toward rac-4 relies on the establishment of the key α-hydroxyester moiety by a Darzens-type chain elongation of an aldehyde, a methodology that has been used for the synthesis of related compounds (Fig. 4) (29, 41). The final deprotection was achieved by hydrogenolysis of the benzyl groups followed by hydrolysis of the isopropyl ester. The final product, rac-4, was purified by anion exchange chromatography eluting with an ammonium bicarbonate gradient.
FIGURE 4.
Synthesis of racemic inhibitor 4. (i) TBDMSCl, imidazole, dichloromethane, 61%. (ii) Dess-Martin periodinane, dichloromethane, 70% (crude). (iii) isopropyl dichloroacetate, KOiPr, iPrOH, Et2O. iv, NaCNBH3, iPrOH, 60% (two steps). (v) tetrabutylammonium fluoride, tetrahydrofuran, 56%. (vi) 1) (BnO)2PN(iPr)2, 1H-tetrazole, dichloromethane; 2) meta-chloroperbenzoic acid, 50%. (vii) 1) H2, palladium/carbon; 2) KOH, H2O; 3) DOWEX-H+; 4) SOURCE-Q anion exchange chromatography, 42%.
Crystal Structure
The racemic inhibitor 4 was co-crystallized with DAH7PS from M. tuberculosis. Although generally both purification of the enzyme and crystal growth are difficult in the absence of P-enolpyruvate in the buffer solution, x-ray diffraction quality crystals were obtained by buffer exchange of the purified enzyme solution with a solution containing inhibitor 4 (Table 1). The crystal structure was solved by molecular replacement using the previously determined MtuDAH7PS structure (PDB code 2B7O) (21) as search model and was refined using x-ray diffraction to 2.35 Å (Table 1) The asymmetric unit contains two enzyme molecules as in other MtuDAH7PS crystal structures; tertiary and quaternary structures are essentially unaffected by binding of inhibitor 4. Superimposition on the wild-type protein (PDB code 3NV8) (22) showed that 5029 atoms could be matched with a root mean square positional difference of 0.22 Å.
Clear continuous electron density for inhibitor 4 was found in both active sites. The |Fo − Fc| omit map generated by removing the inhibitor and active site waters from the model and subjecting the resulting model to simulated annealing and refinement shows clear continuous density for inhibitor 4 and for a water molecule in the active site (Fig. 5a). Based on the electron density maps obtained, it could not be concluded whether there is preferential binding of one of the enantiomers from the racemic mixture. To represent this ambiguity, our final model includes both (R)-4 and (S)-4 at half-occupancy.
Inhibitor 4 binds as predicted with its 2-phosphoryl and carboxylate moieties in the P-enolpyruvate binding site and in an extended conformation so that the phosphate moiety on C7 is located near the proposed E4P phosphate binding site (Fig. 5b). Active site residues interacting with the P-enolpyruvate-mimicking portion of ligand 4 show little movement, consistent with this portion of the inhibitor being a good mimic of P-enolpyruvate. As expected, the conformations of the metal binding residues were unaffected by binding of compound 4. The 2-phosphate moiety of inhibitor 4 forms salt bridges with Arg284, Arg337, and Lys306 and a hydrogen bond to the backbone N–H of Glu283. The carboxyl group forms salt bridges with Lys133 and Arg126 and coordinates to the manganese ion present in the active site. The 7-phosphate moiety extends into the tentative E4P phosphate binding site, forming salt bridges with Lys380 and Arg135 and hydrogen bonds with the Ser136 hydroxyl group and backbone N–H. Conformational changes relative to the P-enolpyruvate-liganded structure were observed for the KPRS motif associated with inhibitor binding (Fig. 5c). Arg135 and Ser136 interact with the 7-phosphate moiety of inhibitor 4, moving closer into the active site pocket. The side chain of Lys133 shows the biggest conformational change of all active site residues, establishing a salt bridge to the carboxylate group of ligand 4. This big movement may also be partly due to increased electrostatic repulsion with the guanidinium group of Arg135, which has moved closer into the active site. Interestingly, the side chain movement of Lys133 correlates well with the movement that the induced fit molecular modeling results predict for this residue to accommodate the proposed reaction intermediate (Fig. 5d).
The superposition of structures of the DAH7PS-4-complex and the induced fit-modeling results (Fig. 5d) show clearly that the experimental binding mode of bisphosphate 4 closely matches the in silico modeling of the proposed reaction intermediate into the active site. The carboxylate and both phosphate groups of inhibitor 4 occupy positions that were predicted for the corresponding moieties of the intermediate model, with minor deviations found in the carbon chain. A crystallographically located water molecule occupies a position very close to the predicted position of the intermediate 2-hydroxyl group. This water is held in place by hydrogen bonds to Trp280 and Glu248. The distance of this water to C2 of the ligand is 3.4 Å when (R)-4 is modeled into the active site and 3.2 Å for the model containing (S)-4. Preliminary testing of rac-4 indicated that its Ki against MtuDAH7PS was in the submicromolar range. Encouraged by these results, and to assess whether one enantiomer of 4 is preferentially bound by the enzyme, we synthesized compounds (R)-4 and (S)-4 for inhibition assays.
Synthesis of (R)-4 and (S)-4
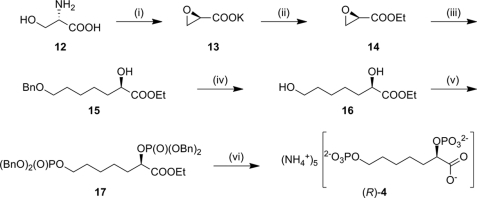
Because the previously described synthesis of rac-4 offers little potential with respect to stereoselective introduction of the α-hydroxyester moiety, another synthetic route was developed in which the chirality is derived from a cheap and readily available chiral pool starting material (Fig. 6). The chiral epoxyester 14 of appropriate configuration was prepared from d- or l-serine, respectively, following a previously reported method (42). Ethyl glycidate 14 was ring-opened regioselectively to yield α-hydroxyester 15. Hydrogenolysis of ester 15 liberated the 7-hydroxyl group to generate dihydroxyester 16, which could then be converted to (R)-4 or (S)-4 using the same procedures as described above for the racemic synthesis.
FIGURE 6.
Synthesis of enantiopure inhibitor 4. Yields are given for the synthesis of (R)-4; analogous procedures using l-serine as the starting material were used for the synthesis of (S)-4 in comparable yields. Please refer to the supplemental material for more detail. (i) HNO2, KBr, H2O then KOH/EtOH, 37% (two steps). (ii) EtBr, Bn(Et)3NCl, CH2Cl2, 66%. (iii) BnO(CH2)4Br, magnesium, tetrahydrofuran, room temperature, then CuBr·SMe2, −78 °C, then 14, 32%. (iv) H2, palladium/carbon, EtOAc, 66%. (v) (BnO)2P-N(iPr)2, 1H-tetrazole, dichloromethane, then meta-chloroperbenzoic acid 64%. (vi) H2, palladium/carbon, EtOAc, then KOH/H2O, then DOWEX-H+, anion exchange chromatography, 27%.
Inhibition Studies
(R)-4 and (S)-4 were tested for inhibition of MtuDAH7PS. Although inhibitor 4 was designed as a bisubstrate inhibitor, it is known that DAH7PS operates by a sequential ordered reaction mechanism in which P-enolpyruvate has to bind to the enzyme first (25). This means that although competition with P-enolpyruvate is most important to assess the inhibitory potency, the fixed concentration of the second substrate can influence the inhibition. The kinetic assays were carried out with P-enolpyruvate concentrations ranging from 57 to 115 μm at a fixed E4P concentration of 100 μm, which corresponds to four times the value determined for the Michaelis constant for E4P (Km(E4P)). The concentrations of (R)-4 and (S)-4 were varied from 0 to 940 nm. Even at these high levels of E4P, (R)-4 and (S)-4 gave inhibitory constants of 360 ± 50 and 620 ± 110 nm respectively (see supplemental Figs. S1 and S2). (R)-4 and (S)-4 represent the first potent inhibitors reported for MtuDAH7PS and the first inhibitors with substantially greater affinity than the substrate P-enolpyruvate (Km(PEP) = 29 μm for MtuDAH7PS) for any DAH7P synthase.
Modeling of (R)-4 and (S)-4 into MtuDAH7PS Active Site
Both (R)-4 and (S)-4 were modeled into the MtuDAH7PS active site, and the results showed that there is little preference for binding between the two isomers of inhibitor 4. Both isomers adopt similar binding modes, which are also very similar to that of the linear intermediate in the active site, with the R-isomer having a slightly better docking score than the S-isomer. The interactions found in the best poses from modeling of the two isomers are the same as those observed in the crystal structure of the MtuDAH7PS-4-complex (supplemental Fig. S3). These modeling results in conjunction with the inhibition data obtained for (R)- and (S)-4 and the tetrahedral intermediate modeling results suggest that a fully functionalized C2-carbon of the tetrahedral intermediate bearing carboxylate, phosphate, and hydroxyl substituents is necessary to observe stereoselective recognition in the enzyme active site.
DISCUSSION
The study presented here demonstrates the successful combination of synthetic, structural, and computational methods to inform the design of novel lead structures of inhibitors for MtuDAH7PS. Although both the substrate and the product of the enzymatic transformation, P-enolpyruvate and DAH7P, possess planar geometry at C2, mechanistic considerations suggest that the enzyme active site is also set up to accommodate preferentially a tetrahedral reaction intermediate. The high inhibitory potencies of (R)- and (S)-4 confirm that intermediate analogues with tetrahedral C2 are a promising scaffold for high-affinity inhibitors of DAH7PS.
The fact that the experimentally determined structure and the induced-fit model are in good agreement firstly demonstrates that inhibitor 4 is a good intermediate mimic, and secondly, verifies that our modeling approach is effective for predictions of enzyme-ligand interactions for MtuDAH7PS. The structural information from the enzyme-inhibitor complex can be used to inform future inhibitor design; based on the binding mode of inhibitor 4 in the active site, modifications to the lead structure allow the introduction of specific new interactions with the enzyme. For example, additional hydroxyl groups at C4 and C6 of inhibitor 4 with the appropriate absolute configuration are expected to form hydrogen bonds to Asp441, interactions that were observed in the modeling of the intermediate (Fig. 3). Introduction of a C4-hydroxyl or other metal-coordinating group such as an amine or thiol group could also lead to additional enzyme-inhibitor interactions through metal coordination.
Inhibitor 4 is a highly charged molecule, featuring two phosphate groups and a carboxylate moiety. This characteristic of this molecule is expected to significantly limit membrane permeability. Phosphates are abundant in naturally occurring molecules, making them highly interesting moieties in the development of drugs. Strategies to generate phosphate prodrugs containing suitable masking groups that facilitate membrane permeability and allow the release of the active drug inside the cell have been developed, and examples of these prodrugs are in clinical trials (43–45).
Although both enantiomers of inhibitor 4 displayed a high affinity to the enzyme active site, the small difference in affinity of the two enantiomers provides valuable insight into the enzyme mechanism and its inhibition. The observation that (S)-4 proved to be a slightly less potent inhibitor may be due to the fact that the C2-hydrogen of (S)-4 points in the direction of a water molecule also present in the active site, with interatom distances that suggest a steric clash between these moieties (supplemental Fig. S4). For the related enzyme KDO8PS, data from computational and crystallographic studies have led to the proposition that the substrate water attacks P-enolpyruvate from the si side during the course of the reaction (18, 19, 46). It, however, so far remains unclear whether this mechanistic proposal can be transferred to MtuDAH7PS or even DAH7P synthases in general. In the crystal structure reported in this study, a water molecule was located in close proximity to C2 of the inhibitor on what would be the re side of P-enolpyruvate. The resemblance of this arrangement of inhibitor and water to the location of the 2-hydroxyl group of the modeled tetrahedral reaction intermediate, combined with the configurational preference observed for the docking of the intermediate, suggest that in MtuDAH7PS, water attack proceeds from the re side of P-enolpyruvate. This is further supported by studies of type I DAH7P synthases from S. cerevisiae (16), E. coli (14), and Thermotoga maritima (47), which located a water molecule that was suggested to attack P-enolpyruvate during the course of the enzymatic reaction in a very similar environment. The glutamic acid residue has been proposed as a general base catalyst to facilitate nucleophilic attack of water. The highly conserved nature of the arrangement of water in the P-enolpyruvate binding site suggests a common catalytic mechanism for all DAH7P synthases that involves water attacking the re face of P-enolpyruvate during the course of the reaction.
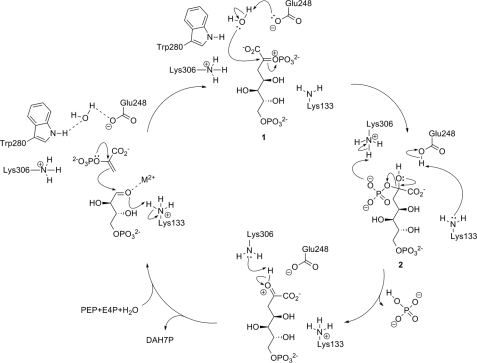
The combined results from intermediate modeling and the structural and kinetic study of intermediate analog 4 allow significant refinements of the proposed catalytic mechanism of MtuDAH7PS (Fig. 7). P-enolpyruvate and E4P binding has already been discussed in detail. We propose that the substrate water is bound on the re face of P-enolpyruvate through hydrogen-bonding interactions with Trp280 and Glu248. Nucleophilic attack of P-enolpyruvate at the E4P aldehyde moiety is facilitated by metal coordination, and the resulting oxyanion is protonated by Lys133. Both the intermediate modeling results and the crystal structure of the intermediate analog show significant movement of the Lys133 side chain upon ligand binding, positioning it in a suitable position to fulfill this role as a general acid. The substrate water is activated for attack at C2 of P-enolpyruvate by deprotonation with Glu248.
FIGURE 7.
Refined catalytic mechanism for MtuDAH7PS in accordance with the results obtained from this study.
A similar mechanism has been previously suggested in studies of S. cerevisiae and T. maritima type I DAH7PS (16, 47). The fact that a glutamate residue is found at the equivalent position for M. tuberculosis type II DAH7PS and the crystallographic localization of a suitable substrate water molecule in the MtuDAH7PS-4-complex in this study as well as our modeling results further support this mechanistic proposition. In the next step, the now neutral Lys133 can abstract the proton from Glu248, either by direct deprotonation or relayed via an active site water molecule. This step regenerates both the Lys133 ammonium and the Glu248 carboxylate group necessary for the next catalytic cycle. Lys306, which is positioned within hydrogen-bonding distance of the P-enolpyruvate phosphate, could aid the elimination of phosphate from tetrahedral intermediate 2 by protonating the phosphate, thus making it a better leaving group. Elimination of hydrogen phosphate yields DAH7PS protonated on the 2-keto moiety; this proton can then be removed by Lys306. After dissociation of the product DAH7P, all active site residues have returned to the protonation state that is required for the next catalytic turnover. This mechanism is the simplest that is in accordance with the data published to date, and it accounts for all protonation and deprotonation steps required on active site residues and intermediates. The only major side chain movement of an active site residue is required from the proton-shuttling Lys133, a residue that is not well defined or possesses high B-factor values in some crystal structures of DAH7P synthases, indicating mobility of its side chain (11, 21). Further support for the important role of both Lys133 and Lys306 comes from results in our laboratory that show that both residues are essential for catalytic activity.5
CONCLUSION
By combining molecular modeling-guided and mechanism-based design, we have synthesized the first potent inhibitor of MtuDAH7PS. The crystal structure of the enzyme-inhibitor complex shows that the structural features of the inhibitor designed to mimic features of the natural substrates and proposed reaction intermediate indeed lead to the predicted specific enzyme inhibitor interactions. Inhibitor 4 represents a promising lead structure for antimycobacterial drugs and offers ample opportunity for further modification to establish structure-activity relationships. The crystallographic location of an active site water molecule has clarified the enzyme catalytic mechanism.
Supplementary Material
Acknowledgments
This research was undertaken on the MX2 beamline at the Australian Synchrotron, Victoria, Australia (48). We acknowledge travel funding provided by the International Synchrotron Access Program (ISAP) managed by the Australian Synchrotron and the New Zealand Synchrotron Group.
This research was funded in part by the Maurice Wilkins Centre for Molecular Biodiscovery.
The atomic coordinates and structure factors (code 3PFP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. S1–S4.
L. R. Schofield and E. J. Parker, unpublished results.
- DAH7PS
- 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase
- MtuDAH7PS
- M. tuberculosis DAH7PS
- E4P
- d-erythrose 4-phosphate
- G3P
- glycerol-3-phosphate
- KDO8PS
- 3-deoxy-d-manno-octulosonate-8-phosphate synthase
- iPr
- isopropyl.
REFERENCES
- 1. Sharma S. K., Mohan A. (2006) Chest 130, 261–272 [DOI] [PubMed] [Google Scholar]
- 2. Meya D. B., McAdam K. P. (2007) J. Intern. Med. 261, 309–329 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization (2009). Global Tuberculosis Control: Epidemiology, Strategy, and Financing, WHO Report 2009, pp. 6–33, WHO Press, Geneva [Google Scholar]
- 4. Barry C. E., 3rd, Blanchard J. S. (2010) Curr. Opin. Chem. Biol. 14, 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell S. A., Richards T. A., Mui E. J., Samuel B. U., Coggins J. R., McLeod R., Roberts C. W. (2004) Int. J. Parasitol. 34, 5–13 [DOI] [PubMed] [Google Scholar]
- 6. Roberts F., Roberts C. W., Johnson J. J., Kyle D. E., Krell T., Coggins J. R., Coombs G. H., Milhous W. K., Tzipori S., Ferguson D. J., Chakrabarti D., McLeod R. (1998) Nature 393, 801–805 [DOI] [PubMed] [Google Scholar]
- 7. Bentley R. (1990) Crit. Rev. Biochem. Mol. Biol. 25, 307–384 [DOI] [PubMed] [Google Scholar]
- 8. Sikorski J. A., Gruys K. J. (1997) Acc. Chem. Res. 30, 2–8 [Google Scholar]
- 9. Schönbrunn E., Eschenburg S., Shuttleworth W. A., Schloss J. V., Amrhein N., Evans J. N., Kabsch W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parish T., Stoker N. G. (2002) Microbiology 148, 3069–3077 [DOI] [PubMed] [Google Scholar]
- 11. Schofield L. R., Anderson B. F., Patchett M. L., Norris G. E., Jameson G. B., Parker E. J. (2005) Biochemistry 44, 11950–11962 [DOI] [PubMed] [Google Scholar]
- 12. Williamson R. M., Pietersma A. L., Jameson G. B., Parker E. J. (2005) Bioorg. Med. Chem. Lett. 15, 2339–2342 [DOI] [PubMed] [Google Scholar]
- 13. DeLeo A. B., Dayan J., Sprinson D. B. (1973) J. Biol. Chem. 248, 2344–2353 [PubMed] [Google Scholar]
- 14. Shumilin I. A., Bauerle R., Kretsinger R. H. (2003) Biochemistry 42, 3766–3776 [DOI] [PubMed] [Google Scholar]
- 15. Wagner T., Shumilin I. A., Bauerle R., Kretsinger R. H. (2000) J. Mol. Biol. 301, 389–399 [DOI] [PubMed] [Google Scholar]
- 16. König V., Pfeil A., Braus G. H., Schneider T. R. (2004) J. Mol. Biol. 337, 675–690 [DOI] [PubMed] [Google Scholar]
- 17. Ray P. H. (1980) J. Bacteriol. 141, 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kona F., Xu X., Martin P., Kuzmic P., Gatti D. L. (2007) Biochemistry 46, 4532–4544 [DOI] [PubMed] [Google Scholar]
- 19. Kona F., Tao P., Martin P., Xu X., Gatti D. L. (2009) Biochemistry 48, 3610–3630 [DOI] [PubMed] [Google Scholar]
- 20. Ahn M., Pietersma A. L., Schofield L. R., Parker E. J. (2005) Org. Biomol. Chem. 3, 4046–4049 [DOI] [PubMed] [Google Scholar]
- 21. Webby C. J., Baker H. M., Lott J. S., Baker E. N., Parker E. J. (2005) J. Mol. Biol. 354, 927–939 [DOI] [PubMed] [Google Scholar]
- 22. Webby C. J., Jiao W., Hutton R. D., Blackmore N. J., Baker H. M., Baker E. N., Jameson G. B., Parker E. J. (2010) J. Biol. Chem. 285, 30567–30576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Webby C. J., Lott J. S., Baker H. M., Baker E. N., Parker E. J. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 403–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartmann M., Schneider T. R., Pfeil A., Heinrich G., Lipscomb W. N., Braus G. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 862–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Webby C. J., Patchett M. L., Parker E. J. (2005) Biochem. J. 390, 223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sasso S., Okvist M., Roderer K., Gamper M., Codoni G., Krengel U., Kast P. (2009) EMBO J. 28, 2128–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walker S. R., Cumming H., Parker E. J. (2009) Org. Biomol. Chem. 7, 3031–3035 [Google Scholar]
- 28. Walker S. R., Parker E. J. (2006) Bioorg. Med. Chem. Lett. 16, 2951–2954 [DOI] [PubMed] [Google Scholar]
- 29. Grison C., Petek S., Finance C., Coutrot P. (2005) Carbohydr. Res. 340, 529–537 [DOI] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 31. Schüttelkopf A. W., van Aalten D. M. F. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 32. Schoner R., Herrmann K. M. (1976) J. Biol. Chem. 251, 5440–5447 [PubMed] [Google Scholar]
- 33. Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. (1979) Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]
- 34. Schrödinger, LLC (2005) MacroModel, Version 9.1, Schrödinger, LLC, New York [Google Scholar]
- 35. Schrödinger, LLC (2005) Maestro, Version 7.5, Schrödinger, LLC, New York [Google Scholar]
- 36. Schrödinger, LLC (2005) Schrödinger Suite 2006 Induced Fit Protocol, Prime version 1.5, Schrödinger, LLC, New York [Google Scholar]
- 37. Schrödinger, LLC (2005) Glide, Version 4.0, Schrodinger, LLC, New York [Google Scholar]
- 38. Armarego W. L., Chai C. (2003). Purification of Laboratory Chemicals, 5th Ed., Butterworth-Heinemann, Oxford [Google Scholar]
- 39. Dess D. B., Martin J. C. (1991) J. Am. Chem. Soc. 113, 7277–7287 [Google Scholar]
- 40. Ireland R. E., Liu L. (1993) J. Org. Chem. 58, 2899–2899 [Google Scholar]
- 41. Grison C., Coutrot F., Comoy C., Lemilbeau C., Coutrot P. (2000) Tetrahedron Lett. 41, 6571–6574 [Google Scholar]
- 42. Petit Y., Larcheveque M. (1998) Org. Synth. 75, 37–44 [Google Scholar]
- 43. Friis G. J., Bundgaard H. (1996) Eur. J. Pharm. Sci. 4, 49–59 [Google Scholar]
- 44. Schultz C. (2003) Bioorg. Med. Chem. 11, 885–898 [DOI] [PubMed] [Google Scholar]
- 45. Hecker S. J., Erion M. D. (2008) J. Med. Chem. 51, 2328–2345 [DOI] [PubMed] [Google Scholar]
- 46. Wang J., Duewel H. S., Woodard R. W., Gatti D. L. (2001) Biochemistry 40, 15676–15683 [DOI] [PubMed] [Google Scholar]
- 47. Shumilin I. A., Bauerle R., Wu J., Woodard R. W., Kretsinger R. H. (2004) J. Mol. Biol. 341, 455–466 [DOI] [PubMed] [Google Scholar]
- 48. McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) J. Synchrotron Rad. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 49. Walker S. R., Jiao W., Parker E. J. (2011) Bioorg. Med. Chem. Lett., 10.1016/j.bmcl.2011.03.071 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.