Abstract
The importance of mRNA localization and localized protein synthesis to spatially modulate protein levels in distinct subcellular domains has increasingly been recognized in recent years. Axonal and dendritic processes of neurons represent separate functional domains of the cell that have shown the capacity to autonomously respond to extracellular stimuli through localized protein synthesis. With the vast distance often separating distal axons and dendrites from the neuronal cell body, these processes have provided an appealing and useful model system to study the mechanisms that drive mRNA localization and regulate localized mRNA translation. Here, we discuss the methodologies that have been used to isolate neuronal processes to purity, and provide an in-depth method for using a modified Boyden chamber to isolate axons from adult dorsal root ganglion neurons for analyses of axonal mRNA content. We further show how this method can be utilized to identify specific mRNAs whose transport into axons is altered in response to extracellular stimuli, providing a means to begin to understand how axonal protein synthesis contributes to the proper function of the neuron.
Keywords: mRNA localization, RNA transport, axonal protein synthesis, local translation
1. INTRODUCTION
The ability to amplify very small quantities of nucleic acids has provided new possibilities to query the RNA profiles of subcellular compartments, at least when the compartment or region can be isolated to purity (1). Neurons have provided a particularly appealing experimental model for analyses of RNA localization, since primary neurons can extend processes for hundreds of microns to a few centimeters in culture. The possibility that proteins can be synthesized in the efferent processes of neurons, referred to as dendrites, was suggested by ultrastructural studies showing that ribosomes concentrate adjacent to post-synaptic regions of these cells (2). Subsequent studies looking at this localized protein synthesis have supported the early hypotheses that the proteins generated from dendritically localized mRNAs play a role in synaptic plasticity (3). Synaptic plasticity is thought to underlie learning and memory, so stimuli regulating mRNA transport into and translation within dendrites could have profound effects on brain function. Protein synthesis in the axonal compartment has been more closely linked to growth of this process, both during development and after injury (4,5). As mentioned in other chapters herein and in recent reviews (6,7), subcellular localization of mRNAs is a common feature of polarized eukaryotic cells, and neurons provide a convenient experimental system for profiling the populations of localized mRNAs and for determining how mRNA transport is regulated. In this chapter, we will detail methodology that we have developed for analyses of axonal mRNA content, both for profiling the populations of mRNAs that localize and for quantifying stimulus-dependent transport of mRNAs into the axonal compartment.
Investigators have used several different approaches to isolate subcellular domains of neurons. From in vivo preparations, density gradient ultracentrifugation has been used to fractionate growth cones and synaptic structures (synaptosomes and post-synaptic densities) from cell bodies and non-neuronal cells (8–10). The growth cone is the terminal portion of the growing axon where new membrane and cytoskeleton is laid down for extension of this process; actively growing dendrites also have growth cones, but dendrites only extend for a few millimeters while axons can grow for hundreds of millimeters to many centimeters. The synaptosome preparations contain both pre-synaptic (axonal) and post-synaptic (dendritic) components, while the post-synaptic density is only the dendritic. Though mRNAs have been detected and protein synthesis has been demonstrated using the synaptosome-type preparations (10–13), contamination from other cellular elements, particularly glial cells, is a major concern with this approach. Proteomics analyses of synaptosome and post-synaptic density preparations clearly show that a small complement of glial proteins co-fractionate with these structures (14,15). Thus, most analyses of localized protein synthesis and mRNA transport have used cultured neurons.
In vitro, neurons isolated from the central and peripheral nervous systems (CNS and PNS, respectively) will extend processes, axons and dendrites, for several hundred microns away from the cell body. For rodent preparations, culture of CNS neurons is largely restricted to embryonic and the early post-natal period. In contrast, PNS neurons can be cultured from adult animals. With the large geographic separation of the ends of the neuronal processes from the cell body, one simply needs to determine a means to compartmentalize the growing processes away from the cell body and any non-neuronal cells that are included in the preparation. Since neurons are post-mitotic, the non-neuronal cells (typically astrocytes in CNS cultures and Schwann cells in PNS cultures) can be minimized by inclusion of mitotic inhibitors. However, in our hands this requires several days to effectively kill off these glial cells and the glia are known to exert varying trophic effects on neurons (16–18).
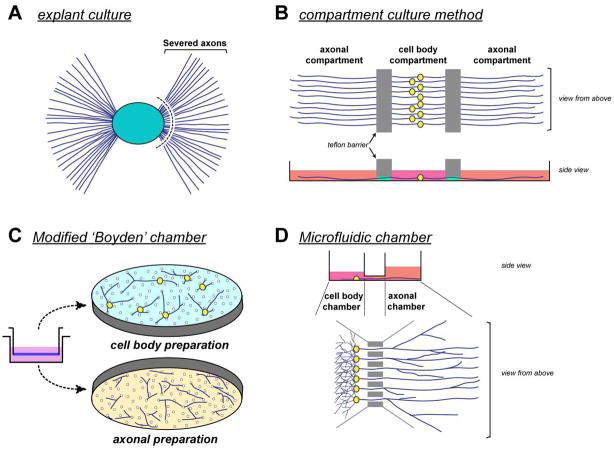
Several methods have been used to physically separate neuronal processes of PNS and CNS neurons from their cell bodies in culture (Figure 1). In the following section, we detail use of the ‘modified Boyden’ chamber approach for isolation of axonal processes from dissociated cultures of adult DRG neurons. Torre and Steward (1996) initially used this approach to show glycosylation of dendritically synthesized proteins in cultures of rodent hippocampal neurons (19). In their method, Matrigel™ was used in the lower compartment, providing a substrate for dendrites to grow into and facilitating separation of the processes. While this was effective for these glycosylation studies, the proteins within the Matrigel are too concentrated to allow for reasonable fractionation of any metabolically labeled proteins from lysates. The high protein content of Matrigel does not allow for electrophoretic methods for detection and, in our hands, some lots of matrigel have generated non-specific amplifications by reverse transcriptase-coupled polymerase chain reaction (RT-PCR). In searching for a more versatile method, we found that DRG axons would traverse pores of the laminin-coated membrane and adhere to the undersurface to provide an effective separation of cell bodies and distal processes (20). Other groups have used an analogous method to purify axonal processes from developing DRG neurons (21), dendrites from hippocampal neurons (22), and pseudopodia from fibroblasts (23). In the sections below, we outline the use of this culture method to profile axonal mRNA levels. The reader should take note that the methods for RNA analysis provided here could readily be adapted to the other culture methods outlined in Figure 1, particularly the microfluidics approach that Taylor et al. (2009) have recently used for profiling RNA transport in cortical neurons (24).
Figure 1. Culture methods for isolation of neuronal processes.
A variety of methods have been developed to isolate axons and dendrites from cell bodies and non-neuronal cells. A, The simplest approach is to physically dissect away the neurites as Olink-Coux and Hollenbeck (1996) did for explant cultures of rodent PNS ganglia (36). The much larger size of some invertebrate neurons allows for such dissections even in dissociated cultures (37). B, The compartmented or ‘Campenot’ culture approach has been used for sympathetic and developing DRG cultures (31). Vogelaar et al (2009) recently modified this chamber method for explant DRG cultures from adult rodents (38). This is an appealing approach since the Teflon barrier not only blocks passage of glial cells but also provides physically separate media compartment where processes can be treated directly with trophic/tropic and pharmacologic agents (39). The separate media compartments also facilitate preparation of axonal versus cell body lysates. A drawback for this approach is the distance that processes must extend to reach the ‘axonal compartment’, since the barrier is typically 1 mm thick and cell body compartment can add a few mm more depending on where the neuron initially adheres. Consequently, these cultures are maintained for several days to weeks. C, Several groups, including the authors, have used a modified Boyden chamber to isolate axonal or dendritic processes (19–22). The porous membrane used in these preparations is only a few microns thick (8 μm for Falcon PET membrane with 8 μm diameter pores), so axons can traverse the membrane pores to reach the undersurface within hours of plating. This short distance allows isolation of dendrites from neurons that are more polarized than the DRGs and sympathetic neurons (19,22). Neuronal processes are isolated from cell bodies and non-neuronal cells by physically scraping, which requires care to reach rigorous RT-PCR based standards of purity that we and others have typically used (see section 3.1.3). Notably the Macara lab has used this approach to isolate pseudopodia from cultured fibroblasts (23). D, Taylor et al. (2005) developed a microfluidic device for isolation of axonal processes (24,40). Neuronal processes extend through grooves in this device to reach a separate chamber, and similar to the Campenot chamber cultures, media of the axonal and cell body compartments are separate providing a means to uniquely stimulate the neuronal processes (27). In contrast to the Campenot chambers, the cell body and axonal compartments can be separated by only a few hundred microns in these microfluidic devices. Beyond the methods outlined in this schematic, a wafer chip based method was very recently published for generating directed growth of neuronal processes and also allows for axonal isolation and fluorescence-based imaging (41). Additionally, a pure axoplasm preparation from peripheral nerve preparations was recently published that holds promise for analysis of axonal RNA transport and localized protein synthesis in vivo (42).
2. MATERIALS
2.1 DRG Cell Culture and Axonal Treatment
8 μm polyethylene tetraphalate (PET) tissue culture inserts for 6-well plates (Falcon #353093; Franklin Lakes, NJ) – these membranes are translucent so they can also be used for imaging.
6-well tissue culture plates, deep-welled (Falcon #353046).
6-well tissue culture plates, shallow-welled (Greiner Bio-One #657160; Monroe, NC).
Cell culture tested poly-L-lysine, mol wt 70,000–150,000 (Sigma #P4707).
Mouse laminin (Millipore #CC095; Billerica, MA).
DMEB/F12 cell culture media – DMEM and Ham’s F-12 1:1 mix supplemented with 0.1% cell culture tested BSA, fraction V.
Complete Culture Media – DMEM and Ham’s F-12 1:1 mix, with N1 medium supplement to 1X (Sigma #N6530; St. Louis, MO) and 10% Horse Serum. This is made up in 50 ml batches and kept for up to one month at 4°C.
Phosphate buffered saline, pH 7.4 (PBS).
Collagenase, type XI (Sigma #C9697); stock solution of 500,000 units/ml in PBS is stored at −80°C in 100 μl aliquots.
5 3/4″ glass pipettes – Fire-polished to decrease cell damage during trituration. (see Note 1)
Cytosine arabinoside (Sigma #C1768).
5,6-dichlorobenzimidazole riboside (DRB; Sigma #D1916).
20 μm diameter carboxylated polystyrene microparticles (Polyscience, Inc. #24811-2; Warington, PA).
Mouse 2.5S Nerve Growth Factor (NGF; Harlan; Indianapolis, IN); 50 μg per experiment.
PolyLink protein kit for carboxylated microparticles (Polysciences, Inc. #24350-1).
2.2 RNA and Protein Isolation, Quantification and Normalization
RNaqueous-micro kit (Ambion #AM1931; Austin, TX).
RiboGreen RNA quantification reagent and kit (Invitrogen #R11490; Carlsbad, CA).
NanoOrange protein quantitation kit (Invitrogen #N6666).
VersaFluor fluorometer (BioRad #170-2402; Hercules, CA) with RNA and protein analyses filter sets (exitation 485–495 nm/emission 515–525 nm and excitation 470–490 nm/emission 585–595 nm, respectively).
Microcon centrifugal filter, 10 kilodalton (kDa) cutoff (Millipore).
2.3 Reverse Transcription and Real-time PCR
iScript cDNA synthesis kit (BioRad #170-8891).
HotStarTaq 2X Master Mix kit (Qiagen #203445-14; Valencia, CA).
2X SybrGreen Master Mix kit (Qiagen#204145-14).
Real-time primers for each transcript of interest optimized for comparative cycle threshold (Ct) method. (see Note 2).
7900HT real-time amplification and sequence detection system (Applied Biosystems; Foster City, CA).
Biomek2000 liquid handling robotic system (Beckman Coulter; Brea, CA).
3. METHODS
3.1 Culture method to isolate neuronal processes
Use of the modified Boyden chamber for culturing DRG neurons from adult rats to isolate axonal processes to purity is detailed here. Since the DRG neurons only extend axonal processes in culture (20), this system provides a pure preparation of axons. Neurons with full dendritic-axonal polarity (e.g., cortical or motor neurons) would generate a mixed preparation of axons and dendrites. However, with a single axonal process and many dendrites for each of these more polarized neurons, Poon et al. (2006) have shown that the majority of the processes reaching the lower membrane surface are dendritic when culturing hippocampal neurons on these porous membranes (22).
3.1.1 Preparation of Tissue Culture Inserts
The day before culturing, place 8 μm PET tissue culture inserts into deep-welled 6-well plates. (see Note 3) Add approximately 3 ml of poly-L-lysine (50 μg/ml) into each insert. (see Note 4)
Leave poly-L-lysine on plates for 30 to 60 minutes.
Remove poly-L-lysine and air-dry the inserts and plates in the hood for at least 30 minutes. Inspect to ensure that the plates are dry. If not, continue drying until no visible liquid remains.
Rinse the plates one time with sterile tissue culture grade dH2O. Aspirate the dH2O taking care to not disturb (i.e., puncture or tear) the membrane and air-dry for a minimum of 15 minutes.
Cover the plates and inserts with 3.0 ml of laminin (5 μg/ml in PBS). (see Note 5)
Incubate overnight at 4°C with gentle rocking to ensure complete coverage of the surface of the membranes.
3.1.2 DRG culture
Remove the L4 and L5 DRGs from the animals and place all of the isolated DRGs into 1 well of a 12-well plate containing 1 ml of DMEB/F12 + 1X N1 supplement. (see Note 6)
After all of the DRGs have been removed, wash them briefly by moving them from well to well through 6 wells containing 0.5 ml of DMEM/F12 plus 1X penicillin/streptomycin. (see Note 7)
After the penicillin/streptomycin rinse, put the DRGs into a well containing 0.5 ml of complete culture media, add collagenase to a final of 5000 units/ml and incubate at 37°C for 20 minutes.
After 20 minutes, triturate the DRG cell suspension 15–20 times by gently pipetting up and down using a fire-polished pipette to break apart the ganglia. Return to incubator for 5 minutes. (see Note 1)
Triturate again and remove DRGs from plate and put into 15 ml conical tube. Add 8.5 ml of DMEB/F12 media.
Pellet the cells at 160 × g for 5 minutes in a swinging bucket rotor at room temperature.
Aspirate media. Add 1 ml of DMEB/F12 media to the pellet and triturate 15–20 times with a fire polished Pasteur pipette until fully dissociated.
Add 8 ml of DMEB/F12 to the dissociated pellet, invert to mix, then centrifuge at 160 × g for 5 minutes.
Repeat steps 7–8 two additional times for a total of three washes.
After the final wash, resuspend the DRGs in complete culture medium plus 10 μM Cytosine arabinoside and 80 μM 5,6-dichlorobenzimidazole riboside. Use 2 ml of media per each filter. Add 2 ml of cells + media to the top of each filter. Add 2 ml of media containing no cells into the bottom of each well (for a total of 4 ml of media/well).
Culture overnight at 37°C, 5% CO2.
3.1.3 Isolation of axonal compartment
Rinse both the upper and lower surface of the tissue culture inserts with 1X PBS to remove media and any non-adherent cell material..
Gently scrape the upper surface of each insert with a cotton-tipped applicator. Use 4 applicators per insert, turning the insert 90 degrees after each swab to ensure that the entire surface of the insert is swabbed and that no areas are missed. It is imperative to remove all of the cell body side from the inserts used for axonal RNA preps without pressing so hard that material is pressed from the top surface through the filter. (see Note 8)
After shearing, rinse the upper surface of the inserts very briefly with 1X PBS. This will remove any cell body material that may have remained. Do this gently as the axons are now sheared from their cell bodies and will not tolerate being treated forcefully.
Remove the insert from its housing with a scalpel. Do not cut too close to the housing as this area is very hard to get clean on the upper surface and tends to be the biggest source of contamination. (see Note 9)
Place the excised insert axonal side down into a 35 mm dish containing 0.5 ml of RNA lysis buffer from the RNaqueous-micro kit.
Continue with all 6 inserts, combining them into a single 35 mm dish.
Place on a rocker at 4°C for 20 minutes.
3.1.4 RNA isolation
Remove lysates from the 35 mm dish and place into 1.5 ml tubes. (see Note 10)
Vortex for 30 seconds to ensure lysis.
Add one half volume (~250 μl) 100% ethanol to the lysate and vortex briefly.
Load lysate/ethanol onto and RNaqueous-micro column (150 μl at time).
Centrifuge at max speed, 10 seconds. Repeat this several times to filter all of the axonal lysate.
Follow the RNaqueous-micro kit instructions to wash the columns.
After the final wash, transfer the columns to a new, RNase-free microcentrifuge tube and elute the RNA with the RNaqueous-micro elution solution (a total of 15 μl elution volume).
3.1.5 RNA quantification and reverse transcription
Quantify 1.0 μl of each RNA sample using the RiboGreen RNA quantification kit with a high-range working solution in a total volume of 200 μl. (see Note 11)
Read the RNA-RiboGreen fluorescence on a fluorometer such as a BioRad VersaFluor Fluorometer per quantification kit manfucturer’s protocol.
Use approximately 50 ng of total RNA for the reverse transcriptase (RT) reaction using the iScript cDNA synthesis kit. (see Note 12)
Dilute the RT product 1:5 with RNase-free water and use 1 μl for standard PCR to test for axonal purity. All axonal preparations must first be assessed for purity by standard RT-PCR for axonal, somatic and non-neuronal transcripts. (see Note 13)
3.2 Approach for quantitative analyses of axonal mRNA transport
Effects of neurotrophins and other growth regulating stimuli on general axonal and dendritic transport have long been known. However, it has only been in recent years that effects on RNA transport have been documented. Extracellular stimuli that modulate synaptic activity and axonal growth have been shown to trigger changes in transport of mRNAs from the cell body into neuronal processes (5). In cultures of developing CNS cortical neurons, neurotrophins were early on shown to modulate transport of axonal RNAs using SYTO® dyes (32). Bassell’s group went on to show that β-actin is among the mRNAs whose axonal transport is increased by neurotrophin 3 application to cortical neurons (33). The majority of studies of mRNA transport have been limited to single transcripts. We have used the culture method outlined above to broadly screen for ligand-dependent alterations in axonal RNA levels and the signaling mechanisms regulating these changes (25,26). The general scheme for this quantitative method is outlined below (see also Figure 2). Although we cannot entirely exclude the possibility that alterations in axonal mRNA stability contribute to the changes in axonal mRNA levels that we detect using this method, analysis of cell body mRNA levels in parallel experiments show an opposite trend in response to environmental stimuli compared to those seen in the axon (26). This strongly suggests that DRG sensory neurons alter mRNA delivery into the axons.
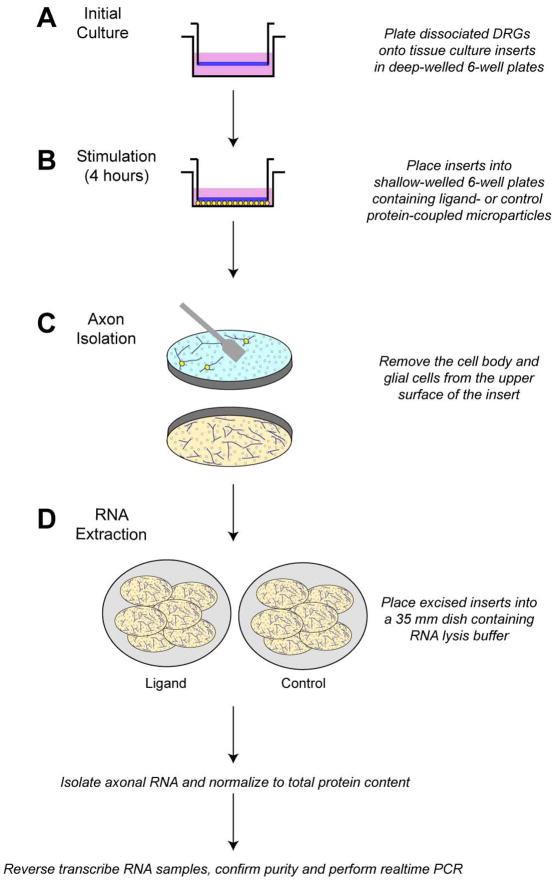
Figure 2. Method for analysis of stimulus-dependent axonal mRNA transport.
A, Using a modified Boyden chamber, dissociated neurons are plated onto tissue culture inserts in 6-well plates with deep wells and allowed to grow until axons have extended through the pores. For injury-conditioned, adult DRG neurons, generally an overnight culture of 14–16 hours is sufficient to allow significant growth through the porous membrane. For cultures of neurons that have not been conditioned by injury, or ‘naive’ neurons, this initial culture period can be extended so that more robust axonal growth is seen along the undersurface. However, the smaller diameter pore membranes than 8 μm should be considered for longer term cultures. The deep wells are critical so that the bottom of the insert does not rest upon the bottom of the wells and hinder axonal growth onto the under surface of the insert. B, The tissue culture inserts are then transferred to a new 6-well plate with shallow wells with ligand- or control protein-coupled micoparticles placed at the bottom of each well. The shallow depth of the wells allows the bottom surface of the insert to rest on the surface of the well, facilitating contact between the axons and the microparticles. C, After stimulation (typically, 4 hours has been sufficient to see alterations in axonal mRNA transport in adult DRG neurons), the top surfaces of the inserts are scraped to remove the cell body and non-neuronal cells. Various methods for removing the material on the upper surface of the filter have been used. Most commonly, we used cotton-tipped applicators from a scientific supply vendors. However, we have found that over-the-counter cotton-tipped swabs that can be autoclaved to ensure sterility provide a more consistent result. D, After removal of the material on the top surface, the membrane insert is removed from it’s housing with a scalpel and all inserts treated with the same stimulus are combined into a 35 mm dish containing RNA lysis buffer. The isolated axonal RNA is quantified, normalized to total axonal protein content, reverse transcribed and checked for purity. Validated preparations are used for real-time PCR.
3.2.1 Exposure of axons to neurotrophins
Place 12.5 mg (approximately 0.5 ml) of 20 μm carboxylated polystyrene microparticles into a 1.5 ml centrifuge tube. (see Note 14)
Pellet at 1000 × g for 5 minutes and resuspend in 0.4 ml PolyLink Coupling Buffer.
Pellet at 1000 × g for 5 minutes and resuspend in 0.17 ml PolyLink Coupling Buffer.
Add 20 μl of freshly prepared 200 mg/ml EDAC solution to the resuspended microparticles and mix by gently vortexing.
Add 50 μg of NGF or BSA (i.e., control) dropwise to the microparticles suspension, mixing gently after each drop. (see Note 15)
Allow the protein to couple to the microparticles for 60 minutes at room temperature with end over end mixing.
Pellet at 1000 × g for 10 minutes and wash the microparticles 2 times with 0.4 ml PolyLink Coupling Buffer. Save the wash buffer after removal, as this will be used to calculate coupling efficiency to determine dosage of ligand on microparticles.
Quantify the amount of protein in the wash buffer by fluorometry using NanoOrange reagent to determine the amount of bound protein. Generally, 90–95% of the protein is bound to the microparticles. This calculation is based on the mass of protein added to coupling mixture less the mass of protein in the wash from # 7.
Add the bound microparticles to 12 ml of complete culture media and add 2 ml per well to a shallow 6-well plate; the density of microparticles can be varied based on the coverage desired and the protein concentration on the microparticles.
Remove the tissue culture inserts from the original 6-well plate after overnight culture, remove the media from the top of the filter and place each insert into the new, shallow 6-well plate containing the bound microparticles. Add 2 ml of complete culture media to the top of the inserts and return the plate to the incubator.
Incubate at 37°C, 5% CO2 for 4 hours. The time for exposure can be varied to account for transit of mRNAs. For example, if one wants to evaluate transport of newly transcribed mRNAs, then the exposure time should be extended to allow for transcriptional activation and DRB should be excluded from the culture preparations.
3.2.2 Isolation of axonal compartment
Rinse both the upper and lower surface of the tissue culture inserts with 1X PBS to remove media, any non-adherent cell material and any microparticles that might be stuck to the under surface.
Gently scrape the upper surface of each insert with a cotton-tipped applicator, rinse, excise the membran, and place in RNA lysis buffer as outlined in 3.1.3, steps 2–7.
3.2.3 RNA isolation
The RNA isolation for this approach follows the approach outlined in 3.1.4 except that the flow through from step 5 is saved for analyses of protein content in 3.2.4 below. This will be used to normalize axonal mass between the control and ligand stimulated axonal RNA preparation samples.
3.2.4 Quantification of protein for normalization
Place the 750 μl of flow-through sample generated in step 6 above into a Microcon centrifugal filter with molecular weight cut-off of 10 kDa.
Following the manufacturers instructions, centrifuge the filter to initially concentrate the samples.
Resuspend the concentrated protein in 0.5 ml 1 x PBS and repeat step 2.
Resuspend the concentrated protein in 0.5 ml 1 x PBS. (see Note 16)
Quantify the protein in each sample by fluorometry using the NanoOrange protein quantification kit.
Determine the normalization ratio between the NGF and BSA treated samples.
3.2.5 RNA quantification and reverse transcription
Quantify 1.0 μl of each RNA sample using the RiboGreen RNA quantification kit with a high-range working solution in a total volume of 200 μl as outlined in 3.1.5 steps 1–2. This is done as a confirmation for presence of RNA in the axonal samples and provides a confirmation of normalization; that is, the 4 hour stimulations used here do not seem to change overall axonal RNA content, but rather change the populations of mRNAs in transport.
Plan to use approximately 50 ng of total RNA for the RT reaction; however, the exact levels are normalized to the protein content of the lysates determined in 3.2.4 above. Using the iScript cDNA synthesis kit for RT, dilute the RT reactions by 1:5 with RNase-free water, and then test for purity as outlined in 3.1.5 steps 3–4 and Note 13.
3.2.6 Real-time PCR
The methods outlined below are based on comparative threshold (Ct) method for real-time analysis with SyberGreen reagent labeled PCR products. We have optimized this for our system with careful choice of controls for normalization as detailed below. This approach could also be adopted for multiplex analyses with other detection methodologies.
Primers used for Ct method real-time analysis with SyberGreen reagent are first run on a validation experiment to ensure that the efficiencies of the target and endogenous control amplifications are approximately equal. (see Note 2)
A robotic system (Biomek 2000) is used to standardize pipetting of samples and reagents into the 384-well plates. Automatic outlier calculations are performed using the ABI Prism SDS 2.3 software. (see Note 17)
Quantify the relative levels of the individual transcripts by normalizing to an endogenous control. The Ct for each transcript is determined using the automatic Ct algorithm of the SDS 2.3 software to calculate the optimal baseline range and threshold values. (see Note 18)
Calculate individual ΔCt values by subtracting the control RNA Ct value from the individual transcript Ct values. (see Note 19)
Determine the ΔΔCt value by subtracting the BSA calibrator ΔCt value from the NGF ΔCt value.
Express the fold difference as 2−ΔΔCt ± S, where ‘S’ is the standard deviation of the ΔΔCt values.
3.3 Concluding remarks
We consider the analysis method detailed above as a first step in assessing which mRNAs are present in the axon and how the transport of these mRNAs may be altered by extracellular signals. Subsequent techniques are then needed to more fully understand this regulated and dynamic process. For example, quantitative fluorescence in situ hybridization (FISH) can be used to confirm the results obtained from this method, as well as provide information on the spatial distribution of the mRNA along the axon shaft and in the growth cone, particularly relative to the source of the stimulus (26). Similarly, diffusion limited fluorescent proteins, such as myristolated GFP (34,35), can be used to generate reporter constructs useful for the identification of motifs that mediate axonal localization as well as the signaling mechanisms that drive alterations in axonal mRNA localization and localized translation.
Although we have outlined methods for profiling and quantitative analyses of RNA transport in neuronal processes, the general methods provided here can be adopted for non-neuronal cells if the desired subcellular compartment can be isolated to purity. Mili et al. (2008) have used this general scheme for profiling mRNAs in pseudopodia of fibroblasts stimulated with fibronectin versus lysophosphatidic acid (23). Likewise, subcellular fractionation of synaptoneurosomes from post-mortem human brain was recently used to profile potential alterations in mRNA transport in Alzheimer disease (12). A key issue for success in these approaches is finding means to confirm the purity of the preparations; that is, validating the absence of cell body restricted RNAs. The universal expression of localizing β-actin and non-localizing γ-actin mRNAs is likely the best starting point with additional control transcripts based on those discussed and referenced above.
Acknowledgments
The methods presented here were developed using funds from the National Institutes of Health (R01-NS041596 and R01-NS049041 to JLT and K99-NR010797 to DEW).
Footnotes
Glass Pasteur pipettes are fire-polished to remove the sharp edge at the opening and to reduce the diameter of the opening. Once cooled, these fire-polished pipettes are used to mechanically dissociate the DRG tissue by gently pipetting up and down 15–20 times (i.e., triturate) until the DRG suspension is homogenous.
Real-time primers for use in comparative Ct experiments must first be tested in validation experiments to confirm their suitability for relative quantitation. In order to be used, the absolute value of the slope of the log input amount vs. ΔCt must be less than 0.1 over a minimum of 5 log units. In addition, the dissociation curves must show single melting peaks and there must be no non-specific product formation with the no template controls. For a more complete explanation on comparative Ct validation experiments, a good resource is the Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR, which can be found at appliedbiosystems.com.
All manipulations for the culture methods described here should be performed in a laminar flow hood (‘biosafety cabinet’) and all materials appropriately sterilized.
It takes a minimum of 2.5 ml of liquid to ensure that the bottom and top of the insert are fully covered. It is critical that the entire bottom surface of the insert be covered in order to facilitate axon growth. Also note that we typically use membranes with 8 μm diameter pores for the adult rat DRG neurons and the methods here describe that. Smaller pore diameters are preferred for longer cultures or cultures from adult mouse DRGs; we have used 3 μm diameter pores for both of these. Although our experience with embryonic preparations is limited, other groups have used membranes with 1 μm diameter pores for embryonic/early post-natal DRGs (21).
Higher concentrations of laminin will diminish the membrane pore size and this becomes very problematic for membranes with smaller diameter pores; the reader may consider a lower laminin concentration for membranes with 3 or 1 μm diameter pores.
Based on adult L4–5 DRG cultures, 2 ganglia per well produces optimal cell density for axons to penetrate the membrane pores and yields a robust preparation of axonal processes. Higher densities tend to decrease penetration and lower densities do not provide sufficient axon mass for subsequent manipulations.
This is the only antibiotic that the cells will be exposed to. Use a pair of micro-scissors to move each DRG from well to well during the rinse. This will ensure that the perineurium has been nicked to facilitate penetration of the collagenase in subsequent steps.
We have experienced variations in sources for cotton-tipped applicators that require re-optimizing this shearing approach. We have found that the most consistent source is the ‘cotton-tipped swabs’ available from local drug or department stores.
Leave an edge of about 3 mm of filter attached to the side to ensure that any cell body material remaining stays behind. In practice, using a #11 scalpel blade fitted to a surgical handle provides a 3 mm edge based on the thickness of the handle.
Note that all subsequent steps below require RNAse free solutions and RNAse free plasticware.
We prefer to use fluorometry to quantitate the small mass of RNA that this approach yields. The standard curve required for quantification by fluorometry gives confidence that the absorption values generated by the samples are within the dynamic range of the instrument. However, one can use other approaches that do not require the use of standard curve (e.g., nano-format UV spectrometry).
From our experience with adult rat DRG neurons, L4–5 DRGs from 3 animals (i.e., 12 ganglia) generate 25–100 ng of axonal RNA. With amplification, this is more than sufficient for generating probes for cDNA arrays (26) and quantitative RT-PCR approaches to gain a relative comparision of localized mRNA levels (see below) (25). Although analyses of axonal proteins requires more input since there are no means to exponentially amplify signals like there is for mRNA (i.e., with PCR), one can scale up and generate sufficient levels of protein for immunoblotting, immunoprecipitations, and even co-immunoprecipitation with the DRG cultures (20,25,28–30). Other methods for isolating axonal processes have similarly been used for such immunoprecipitations (31).
Axonal preparations from adult DRGs show robust amplification of β-actin mRNA, but are devoid of γ-actin mRNA (which remains confined to the cell body) and the glial-specific mRNAs MAP2 and GFAP (25,26). Additional purity markers, such as mRNAs encoding histones and transcription factors, have proven useful in other culture methods using different neuronal cell types (22,24,27).
In this approach, we limit the neurotrophin exposure to the axonal compartment by immobilizing the ligands onto polystyrene microparticles that are of too large diameter to pass through the membrane pores (20 μm diameter particles versus 8 μm diameter pores).
Although the method described here is for NGF, a member of the neurotrophin family, it is easily adapted for other polypeptide ligands (e.g., semaphorins (26)) Care should be taken to ensure desired level of receptor occupancy. Proteoglycans can also be used, but this requires a passive adsorption process rather than covalent linkage used here. However, a more diffuse localized gradient of ligand in the media could be generated by use of passively adsorbed ligands as these will be more easily leached from the particles. Additionally, avidin-conjugated microparticles could effectively be used to increase affinity for any biotinylated agents.
These 2 spins, each concentrating the samples 20-fold and effectively exchanging 95% of the RNA lysis buffer for PBS, makes the flow-through sample compatible with NanoOrange reagent for protein quantitation.
Normalized RT reactions from pure axonal preparations treated with NGF or BSA are diluted 1:5 with RNase-free water and 1 μl assayed in quadruplicate from a minimum of 3 independent experiments. 96-well formats can also be used but they do not allow for the same level of throughput that is needed to simultaneously anlayze many samples and controls in quadruplicate.
Controlling for the variability in the number of axons that traverse through the pores of the tissue culture inserts (or into an isolated chamber as in other methods) presents a unique challenge in analyzing changes in axonal mRNA transport. We have controlled for axon number based on the total protein content of the isolated axons and have used this axonal protein amount to normalize the axonal load between the NGF- and BSA-treated samples (see section 3.2.4). We have also used a comparative threshold method to provide an additional control for axonal content as well as an internal control for reverse transcription efficiency. We prefer this to an absolute determination of localized RNA levels, since the 12S RNA that we use for normalization also gives an independent assessment of RT efficiency. When dealing with minute RNA quantities, such as here, efficiency of RT reaction can fall dramatically as the RNA template levels decrease. iScript has behaved well for us down to 15 ng of total RNA, but below this the efficiency quickly falls.
Since transport of some mRNAs that are classically considered as ‘housekeeping genes’ is robustly regulated by ligands (e.g., β-actin mRNA), care must be taken in choice of RNAs used for the the Ct calculation. We have used the 12S mitochondrial ribosomal RNA as mitochondria are frequent in axons and the mitochondrial genome is transcribed as a single polycistronic RNA (25).
References
- 1.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–30. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–91. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–89. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 4.Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo S, van Niekerk EA, Merianda TT, Twiss JL. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–6. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willis DE, Twiss JL. Regulation of protein levels in subcellular domains through mRNA transport and localized translation. Mol Cell Proteomics. doi: 10.1074/mcp.R900005-MCP200. in press [PMID: 20167945] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis L, Katz F, Pfenninger KH. Nerve growth cones isolated from fetal rat brain. J Neurosci. 1985;5:1393–1401. doi: 10.1523/JNEUROSCI.05-06-01393.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witzmann FA, Arnold RJ, Bai F, Hrncirova P, Kimpel MW, Mechref YS, McBride WJ, Novotny MV, Pedrick NM, Ringham HN, Simon JR. A proteomic survey of rat cerebral cortical synaptosomes. Proteomics. 2005;5:2177–201. doi: 10.1002/pmic.200401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jimenez CR, Eyman M, Lavina ZS, Gioio A, Li KW, van der Schors RC, Geraerts WP, Giuditta A, Kaplan BB, van Minnen J. Protein synthesis in synaptosomes: a proteomics analysis. J Neurochem. 2002;81:735–44. doi: 10.1046/j.1471-4159.2002.00873.x. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto M, Setou M, Inokuchi K. Transcriptome analysis reveals the population of dendritic RNAs and their redistribution by neural activity. Neurosci Res. 2007;57:411–23. doi: 10.1016/j.neures.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Williams C, Mehrian Shai R, Wu Y, Hsu YH, Sitzer T, Spann B, McCleary C, Mo Y, Miller CA. Transcriptome analysis of synaptoneurosomes identifies neuroplasticity genes overexpressed in incipient Alzheimer’s disease. PLoS One. 2009;4:e4936. doi: 10.1371/journal.pone.0004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiler IJ, Greenough WT. Metabotropic glutamate receptors trigger postsynaptic protein synthesis. Proc Natl Acad Sci, USA. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemmer P, Smit AB, Li KW. Proteomics analysis of immuno-precipitated synaptic protein complexes. J Proteomics. 2009;72:82–90. doi: 10.1016/j.jprot.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Schrimpf SP, Meskenaite V, Brunner E, Rutishauser D, Walther P, Eng J, Aebersold R, Sonderegger P. Proteomic analysis of synaptosomes using isotope-coded affinity tags and mass spectrometry. Proteomics. 2005;5:2531–41. doi: 10.1002/pmic.200401198. [DOI] [PubMed] [Google Scholar]
- 16.Cotrina ML, Lin JH, Lopez-Garcia JC, Naus CC, Nedergaard M. ATP-mediated glia signaling. J Neurosci. 2000;20:2835–44. doi: 10.1523/JNEUROSCI.20-08-02835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slezak M, Pfrieger FW. New roles for astrocytes: regulation of CNS synaptogenesis. Trends Neurosci. 2003;26:531–5. doi: 10.1016/j.tins.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Shea TB. Toxic and trophic effects of glial-derived factors on neuronal cultures. Neuroreport. 1994;5:797–800. doi: 10.1097/00001756-199403000-00015. [DOI] [PubMed] [Google Scholar]
- 19.Torre ER, Steward O. Protein synthesis within dendrites: glycosylation of newly synthesized proteins in dendrites of hippocampal neurons in culture. J Neurosci. 1996;16:5967–78. doi: 10.1523/JNEUROSCI.16-19-05967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng JQ, Kelly T, Chang B, Ryazantsev S, Rajasekaran A, Martin K, Twiss J. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–4. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–9. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–9. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis D, Li KW, Zheng JQ, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–91. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–80. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–59. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 29.van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci U S A. 2007;104:12913–8. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, van Niekerk E, Willis DE, Twiss JL. RNA transport and localized protein synthesis in neurological disorders and neural repair. Develop Neurobiol. 2007;67:1166–82. doi: 10.1002/dneu.20511. [DOI] [PubMed] [Google Scholar]
- 31.Eng H, Lund K, Campenot RB. Synthesis of beta-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knowles RB, Kosik KS. Neurotrophin-3 signals redistribute RNA in neurons. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14804–8. doi: 10.1073/pnas.94.26.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 35.Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Segal-Ruder Y, Vuppalanchi D, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss J, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–52. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olink-Coux M, Hollenbeck PJ. Localization and Active Transport of mRNA in Axons of Sympathetic Neurons in Culture. Journal of Neuroscience. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moccia R, Chen D, Lyles V, Kapuya E, Kalachikov YES, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–17. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha-Chowdhury R, Leung KM, Holt CE, Fawcett JW. Axonal mRNAs: characterisation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42:102–15. doi: 10.1016/j.mcn.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillefors M, Gioio A, Mameza M, Kaplan B. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007 doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu HI, Cheng GH, Wong YY, Lin CM, Fang W, Chow WY, Chang YC. A lab-on-a-chip platform for studying the subcellular functional proteome of neuronal axons. Lab Chip. 10:647–53. doi: 10.1039/b918217a. [DOI] [PubMed] [Google Scholar]
- 42.Rishal I, Michaelevski I, Rozenbaum M, Shinder V, Medzihradszky KF, Burlingame AL, Fainzilber M. Axoplasm isolation from peripheral nerve. Dev Neurobiol. 2009 doi: 10.1002/dneu.20755. [DOI] [PMC free article] [PubMed] [Google Scholar]