Abstract
Insulin is a small but beautifully organized protein with a unique two-chain structure, the first protein to be sequenced. The mechanism of its biosynthesis invited much initial speculation but was finally clarified by the discovery of proinsulin, its single-chain precursor. The rich present-day field of protein precursor processing via post-translational proteolysis within the secretory pathway arose in the early 1970s as an offshoot of studies on insulin biosynthesis, which provided a novel paradigm for the generation of many other small neuroendocrine peptides. Before long, this mechanism was also found to play a role in the production of a much wider spectrum of proteins traversing the secretory pathway (receptors, growth factors, blood-clotting components, and even many viral envelope proteins) occurring in almost all eukaryotic cells. Indeed, yeast provided a key clue in the search for the proprotein convertases, the endoproteases that work along with carboxypeptidases and other modifying enzymes, such as the amidating enzyme complex (PAM), in converting inactive or less active precursor proteins into their fully active peptide products. In this “Reflections” article, I have tried to recount the people and events in my life that led to my involvement first in basic biochemical research and then on to insulin, proinsulin, and many relevant related areas that continue to fascinate and challenge my colleagues and me, as well as many other biomedical scientists today, as diabetes mellitus increasingly threatens human health throughout our contemporary world.
Keywords: Diabetes, Insulin, Insulin Synthesis, Insulin-like Growth Factor (IGF), Peptide Biosynthesis, Peptides
Early Scientific Developments Leading to Proinsulin
My biomedical career resulted from a chance encounter with a marvelous book. It was entitled Fearfully and Wonderfully Made: The Human Organism in the Light of Modern Science (1). I was then just twenty years old and a second-year student in a chemical engineering cooperative work/study program at the University of Cincinnati (my first “U of C”) working in the quality-control laboratory of a paper mill in West Carrollton, Ohio (a training rotation). One morning, while browsing in the nearby Dayton Public Library before reporting for the second shift at the mill, I chanced upon this illuminating volume. Having never before encountered biology or physiological chemistry in my high school studies, I was immediately swept off my feet by the concepts it conjured up of living cells carrying out thousands of highly coordinated chemical reactions simultaneously, such as the complete combustion of glucose at body temperature, through the catalytic action of myriad enzymes and with complete control of chirality, all coupled energetically to biosynthetic reactions that maintained the cells in a stable constant state, seemingly defying entropy. This book came as a wonderful revelation that could not be ignored, even though I barely understood much of it. Indeed, I was so enthralled by its revelations that, within just a few days, I decided that I must somehow gain access to this compelling new scientific field. I telephoned home to my mother (my father had died suddenly from a heart attack in 1939 at age 50 when I was nine years old) and tried to explain my sudden decision to transfer to a different program at the university, one that would allow me to apply for admission to a medical school, where I hoped to be able to realize the dream of doing research on living systems. My mother listened patiently to my plea, asked a few practical questions, and then graciously replied that, if I was sure that was what I wanted to do, she would support me as fully as possible. I might add that my mother had not completed high school herself because, as the eldest of five daughters, she was expected by her father to train as a secretary/stenographer and help support the family. This background proved a great advantage for her after the untimely death of my father because my mother had more business and legal experience than most of her peers. My parents had the highest regard for the pursuit of knowledge, which was very fortunate for me and my brother, Phares, who studied music, also in Cincinnati, and then went on to become an expert pipe organ builder and to earn a master's degree in organ performance at the University of Michigan. I owe my love of Bach to Phares!
In the autumn of 1950, when my new classes in the University of Cincinnati School of Arts and Sciences began, I quickly realized that at last I had found a goal that totally absorbed my interest, and my fascination with biology continued through the next two years until graduation in 1952. During that time, I happily studied general zoology, comparative anatomy, ecology, evolution, introductory genetics, and organic chemistry in addition to humanities courses, such as history, literature, psychology, and linguistics. I then applied for admission to the University of Chicago Medical School, as well as two other medical schools in Ohio. I wanted most to go to Chicago because of its well deserved reputation for excellence in research, and my very helpful professors of biology at Cincinnati also supported this ambitious plan. The day that my admission confirmation to Chicago (my second “U of C”) arrived in the mail from Dean of Students Joseph J. Ceithaml, himself a biochemist, was one of the happiest in my life!
The University of Chicago School of Medicine proved to be the academic haven I needed. I also had the good fortune to have two medical school classmates who shared my strong interest in research. They were Donald D. Brown and Phillip G. Nelson. Don was instrumental in helping me find my way into the biochemistry department, where both of our research interests flourished, although in different directions. He was greatly interested in genetics and aligned with the Chicago phage group created by Chairman Earl A. Evans, Jr., working with Lloyd Kozloff on the mechanism of bacteriophage infection (Fig. 1). Don became the pioneering molecular developmental biologist who transformed the excellent Carnegie Laboratory of Embryology in Baltimore and led it for many years. He also created the Life Sciences Research Foundation to provide better resources for postgraduate training for young scientists with funding from the pharmaceutical industry. Phil also was headed for a distinguished career in neural science; he signed on with the famous neurophysiologist Julian Tobias while at Chicago and went on to become head of the Section of Neurobiology in the National Institute of Child Health and Human Development at the National Institutes of Health. For several years as students, we shared an apartment and encouraged our mutual visions of research while enjoying a few diversions from the demands of the medical school curriculum. My old '36 Ford got us around and provided occasional bits of comic relief, including an ill fated canoeing/camping trip up to the Quetico country of Minnesota, which turned out to coincide with the monsoon season. Don affectionately called the old Ford the “gray beetle.” Despite its many shortcomings, it taught me a lot about cars. At age 6, I had accompanied my dad on the day he purchased it new in the midst of the Great Depression, but that is a story for another day.
FIGURE 1.
Shown is a photograph of Professor Earl A. Evans, Jr. (center) surrounded by several distinguished colleagues formerly associated with the University of Chicago Department of Biochemistry, including (from left) Eugene P. Kennedy, Konrad Bloch, Lloyd Kozloff, and Donald D. Brown, in 1975 at the time of a celebration honoring Evans, who was then nearing his 65th birthday and retirement, for his thirty years of service (1942–1972) as chairman. The setting was in front of a large outdoor sculpture by Arnaldo Pomodoro, just behind the Cummings Life Sciences Center (1973), which then housed biochemistry.
My first serious research efforts, during the final two years of medical school, were made under the guidance of Professor Herbert S. Anker (Fig. 2), in the biochemistry department, on the biosynthesis of antibodies in vitro. Anker, I later learned, had constructed by hand the mass spectrometer at Chicago (the second in a United States department of biochemistry, with the first being at Columbia) that had been used by Konrad Bloch in his Nobel Prize-winning work on cholesterol biosynthesis (see Fig. 1). Herbert also invented a simple windowless counter for gassing and counting radioactive planchets, which he gave to a chap named Lyle Packard, who worked in the nearby research institute machine shop, where it was built. Packard asked him if he might make copies for others and thereby began to build up a small manufacturing company, which grew into the Packard Instrument Company and, within a few years, developed the first commercial liquid scintillation counter.
FIGURE 2.
Professor Herbert S. Anker in the 1950s.
My research project was to develop a method for culturing spleen cells after an antigenic challenge to enable us to show that antibodies were made de novo rather than via transformation of pre-existing immunoglobulins, as postulated in Linus Pauling's “template hypothesis” (2). The success of this project won me a student research award and raised my interest in studying the biosynthesis of other proteins, such as insulin, the sequence and two-chain structure of which had just been reported that year (1955) by Fred Sanger. However, I was not able to find an insulin-producing tumor at that time, as they occur quite rarely.
The discovery of insulin in 1921 by Frederick Banting and Charles Best in Toronto was one of the great medical discoveries of the 20th century,1 but even after insulin was crystallized in 1926 by John Jacob Abel (3), the activity was considered to be possibly due to a smaller active molecule, analogous to thyroxine or adrenaline, that was adsorbed on protein impurities in the crystals. However, the work of Hans Jensen and Earl A. Evans, Jr., working in Abel's laboratories in the 1930s, established that the material extracted by acidic ethanol from the pancreas was indeed a small protein, and they demonstrated the presence of an N-terminal phenylalanine, the first “end group” to be identified in a protein (4). In the 1930s and 1940s, insulin's life-saving actions in the treatment of diabetes occupied center stage throughout the biomedical world, as many studies explored its role in normal physiology and diabetes therapy.
My interest in insulin developed much further as a result of my desire to extend my training in Seattle after completing my general medical internship at King County Hospital in 1957. Having grown up in Ohio and attended medical school at the University of Chicago, life on the West Coast strongly attracted me, despite the famously rainy weather of the Pacific Northwest. In fact, I had already begun to learn to sail with friends from Harborview Hospital on nice evenings on Lake Washington and to ski in the mountains on winter weekends. I also wanted to extend that training, too! However, by the time I decided to apply for residency, it was too late to be considered for the coming year. Instead, Robert H. Williams (Fig. 3), who was head of medicine at the very young University of Washington Medical School, offered me a fellowship in endocrinology to join his group studying the new oral hypoglycemic agents that were just emerging in the wake of the discovery of the hypoglycemic sulfonylureas, and I eagerly accepted. This worked out so well that I stayed on for an additional year and then continued my research while doing a year of medical residency at the University of Washington Hospital, with the aid of Vija Rouda, a very able technician in Williams' laboratory.
FIGURE 3.
Dr. Robert H. Williams, head of medicine and endocrinology at the University of Washington, in the 1960s.
Early Work on Insulin Action
In the summer of 1957, when I arrived in Williams' laboratory, work was focused on a new class of hypoglycemic agents, the biguanides, and Williams suggested that I look into their mechanism of action. Unlike the sulfonylureas, they did not require a functioning pancreas to lower blood glucose, but instead they appeared to stimulate the glycolytic pathway, resulting in increased glucose uptake but associated with a buildup of lactate in the blood of treated animals. This suggested a possible “Pasteur effect” on aerobic metabolism, and indeed, I was soon able to obtain evidence, using an available Warburg apparatus in the laboratory, of an inhibitory effect of phenformin (phenylethylbiguanide (PEBG)) on tissue respiration that we traced to mitochondrial oxidative systems, most likely at the level of cytochrome oxidase (5). This kind of action seemed to correlate with clinical findings of mild malaise, decreased appetite, and weight loss among patients treated for longer periods with PEBG in the clinic. Other work soon appeared supporting this concept, especially some findings of Arne N. Wick and colleagues in San Diego with adipose tissue in vitro, showing that PEBG treatment, in sharp contrast with insulin's actions, led to inhibition of the citric acid cycle and lacked any stimulatory effect on fatty acid biosynthesis (6). Williams and I authored an editorial on the biguanides for publication in the journal Diabetes warning clinicians and investigators that hypoglycemia per se was not necessarily indicative of a physiological insulin-like action (7). Of course, it is now well established that biguanides, such as the currently widely used metformin, activate AMP kinase, but it is not yet clear whether this is a direct effect or might result from some degree of inhibition of mitochondrial electron transport or oxidative phosphorylation.
This work stimulated my interest in insulin action, as I realized that essentially nothing was known at a molecular level about how insulin worked. It seemed obvious that we needed to know more about the biochemical basis of insulin's actions before trying to find substitutes for it. Of course, the effects of insulin in enhancing plasma membrane transport of glucose in peripheral tissues, such as muscle, had been established by the early beautiful studies of C. R. Park and co-workers (8), and later studies established its powerful stimulation of lipogenesis in adipose tissue, but nothing was known about its tissue receptors (the concept of specific hormone receptors did not yet exist) or the intracellular signaling pathways that might convey its signals to various target proteins or enzymes. Although insulin was known to directly act on muscle and fat cells, it was an open question as to whether it had any direct effects on the liver. And so, over the next decade, my students and I endeavored to find out more about insulin's biochemical action(s) in the liver. Our initial approach was to determine the time course of insulin's effects on key liver substrates, such as glucose 6-phosphate and glycogen, as well as on protein, RNA, and DNA synthesis, using the alloxan-induced severely diabetic rat as a model. I began this work in Seattle in Williams' laboratory (9) and then continued it after returning to the University of Chicago in 1960, when I joined the biochemistry faculty as an assistant professor.
My faculty appointment at the University of Chicago was a remarkably generous gift from friendly colleagues at my medical alma mater who responded to my inquiries in the spring of 1960 about several postdoctoral possibilities I was then considering. These included opportunities to work with Albert Lehninger at The Johns Hopkins University, Konrad Bloch at Harvard University, or Feodor Lynen in Munich. All three of these outstanding scientists had agreed to fellowship positions, but I had difficulties in deciding among such stellar laboratories. Earl A. Evans, Jr. was then chairman of the University of Chicago biochemistry department, and he telephoned me within just a few days after I brought the matter up with my friend and student mentor Herbert S. Anker at Chicago. I was very surprised when Evans immediately offered me a position as assistant professor! It was an offer I could not refuse, essentially because I could continue my insulin work, which was what I really preferred to do, and as Evans pointed out, the pay would be higher!
Of course I then had to inform all three of my projected postdoctoral mentors that I would not be accepting their offers. As it happened, four years later, in the autumn of 1964, I was traveling through Europe (my first time abroad) accompanied by my mother and one of her sisters who had previously visited Germany with her father some fifty years earlier, just before the outbreak of World War I. While we were in Munich, I decided to pay a visit to Professor Lynen and personally thank him for having considered me for a position in 1960. When I arrived at the Max Planck Institute for Cell Chemistry, which he then headed, I asked one of the students where Lynen's office might be, and he replied with something like “Ach, der hohe Adler!” (Oh, the high eagle!) “He is around the corner in that direction!” A bit mystified by such veneration, I arrived at his office to find his very excited secretary, who told me that he had just received word that morning from Stockholm that he had received the Nobel Prize and that she could not reach him, as he was away at a meeting. Just then, the phone rang, and she spoke excitedly for a few minutes and then turned to me and asked, “Do you know who is this fellow Konrad Bloch, who shares the Nobel Prize?” Professor Bloch (Fig. 1) had been one of my teachers in medical school in the early 1950s, so I informed her that he was an American scientist, now at Harvard University, who had also done outstanding work on cholesterol biosynthesis. She then passed this information on to a newspaper reporter. Having thus served as Konrad Bloch's pro tempore press agent, I left a brief congratulatory note for Lynen with her, and then my two companions and I continued our European explorations. A year or so later, I received a kind note of thanks attached to an autographed copy of Lynen's Nobel address.
When I arrived back at Chicago from Seattle in September 1960, Evans provided me with a well equipped laboratory in the basement of Abbott Hall that had been used originally by Fred C. Koch, followed years later by Frank Putnam and his graduate students, including the late Eugene Goldwasser of future erythropoietin fame, and more recently by another of my former mentors, Eugene P. Kennedy (Fig. 1), during his transition from the Ben May Laboratory for Cancer Research (now The Ben May Department for Cancer Research) at the University of Chicago to become the chairman of the Harvard Medical School Department of Biochemistry. I had gained quite a bit of practical laboratory knowhow from Gene during a brief rotation I spent in his laboratory as a third-year medical-cum-graduate student, and to now inherit his former laboratory was a remarkable privilege. Returning to Chicago also gave me an opportunity to learn more about this beautiful city. Somewhat later in my career, as I began to have the opportunity to visit many of the great cities of the world, I began to appreciate Chicago's breadth and depth of cultural merits much more than I had as a student!
Our research over time provided much evidence of a direct action of insulin on the livers of severely diabetic rats to rapidly stimulate dramatic increases in both glycogen content and RNA synthesis, followed by increased protein synthesis and, later, after 36–60 h of insulin stimulation, highly significant (∼65%) increases in total liver DNA (9). Later studies showed that this excess DNA content reflects hyperplasia but quickly reverts back to normal levels without histological signs of inflammation or necrosis of cells with a half-time of ∼14 days (10), indicating the existence of a much more rapid liver size-correcting process than normal hepatic DNA turnover. In retrospect, it is probably due to apoptosis, which at the time was thought to be mainly a developmental process. DNA had been measured in those experiments merely to serve as a presumably stable correction factor for rapid changes in cell volume and liver weight because of the massive increases in hepatocellular glycogen and water content that occur within the first 12 h after initiation of insulin treatment in such severely diabetic animals. Many further studies in my laboratory and others strongly indicated that insulin action in the liver controls the rates of expression of many genes encoding enzymes regulating metabolic pathways, especially those for glycolysis and gluconeogenesis, as well as stimulating many anabolic processes and even leading to dramatic temporary increases in total liver cell numbers in the period from 24 to 72 h after starting treatment (11). The direct action of insulin on the liver has been definitively established by recent experiments of C. Ronald Kahn and co-workers (12) using liver-specific insulin receptor knock-out mice.
Insulin Biosynthesis Beckons
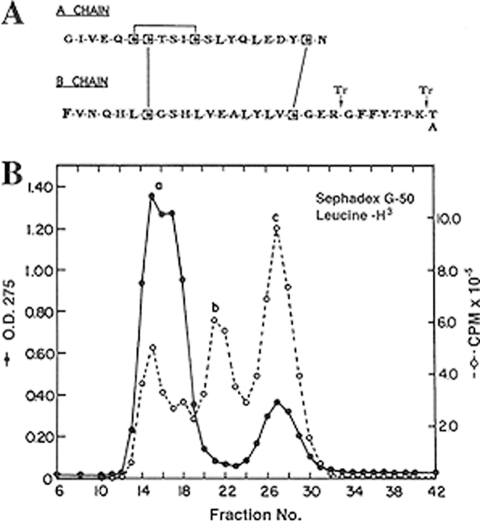
I received tenure at Chicago in 1965, although my work on the hepatic actions of insulin and glycogen synthase was incomplete. However, the truth was that methods to study the biosynthesis of specific messenger RNAs to directly assess the hypothesis that insulin altered gene readout were not yet available (13). This was still well before the molecular biological revolution that enabled such studies, but then, serendipity intervened. In the spring of 1965, I had casually mentioned, to one of the third-year medical students, my longstanding interest in insulin biosynthesis and asked him to alert me should he hear of any patients with insulin-producing adenoma of the pancreas being admitted for surgery at the university hospital. Remarkably, when such a case arrived on the wards in October of that year, the student, Nicholas Vick, remembered my request and informed me of it! However, when I checked with the endocrinologist in charge, I was told that the patient was to be operated on the very next morning! I was taken aback at the short lead time available to prepare a suitable experiment. Nonetheless, I could not resist giving it a try anyway. I had no preconceptions about the outcome, but I firmly believed that studying a pure beta cell tumor would surely provide the optimum approach to unraveling the problem of insulin biosynthesis, via either chain assembly or some sort of precursor. Such tumors are very rare, so, at the very least, it might serve as a feasibility study, if nothing more. We very fortunately already had several tritium-labeled amino acids in the laboratory, including [3H]leucine and [3H]phenylalanine, which we were using in attempts to label hepatic glycogen synthetase. I noticed that these two amino acids occupy very different positions within the insulin molecule (Fig. 4A) and therefore seemed likely to be strategically useful in characterizing any insulin-related biosynthetic products that might be obtained. Accordingly, incubation media containing each of these labeled amino acids were prepared, and I personally went to the operating room in scrubs to persuade the surgeon and the surgical pathologist to share a bit of the tumor with me in as fresh a state as possible. A small tumor in the pancreas was soon located, ∼1 cm in diameter, and I thankfully received about half of it (∼0.5 g) immediately after its excision. As all the ongoing work in my laboratory at that time was exclusively devoted to insulin action, I carried out the tumor incubation myself as an exploratory project, and as there was no grant support for such a project, I also performed the ensuing analyses in my spare time on weekends or in the evenings. A small portion of the tumor was also placed in cell culture with the hope that it might grow, but no growth occurred.
FIGURE 4.
A, structure of human insulin. Note the distribution of leucine (L) versus phenylalanine (F) within the A and B chains. Arrows indicate potential tryptic cleavage sites. B, elution profile, from a 1 × 50-cm column of Sephadex G-50 in 1 m acetic acid, of leucine-labeled acid/ethanol-soluble protein from incubated slices of a human insulinoma (from Ref. 15). Note the lack of A275 (O. D. 275) under peak b as opposed to peak c (insulin).
The ultimate outcome of this simple exploratory experiment greatly exceeded my wildest expectations, but progress was slow in the beginning, as I was exploring what for me was scientific terra incognita. Two batches of fresh tumor slices were incubated for 2.5 h, separately with each of the two available labeled amino acids, and then frozen awaiting further analysis. After due consideration of available literature, I chose an extraction method based on the widely used acid/ethanol procedure for insulin, coupled with the recently developed gel-filtration method to resolve the extracted components by size (14). Both labeled extracts contained a fair amount of almost pure insulin and also a higher molecular mass component, which proved to be of great interest. The main labeled components were designated a, b, and c, in order of decreasing size (Fig. 4B). Component a appeared to be a mixture of high molecular mass proteins eluting in the void volume of the column, whereas components b and c ranged in estimated size from 9 to 10 kDa for component b to ∼6 kDa for component c (identified as insulin). There were no peaks smaller than insulin in either extract, i.e. no evidence for free insulin chains. Peak b was of immediate interest because of its larger size and also because it lacked much associated A275, indicating that it was likely a protein of much higher specific activity than peak c, which represented mainly insulin stored in considerable amounts in the tumor. The three peaks, a, b, and c (labeled with either [3H]Leu or [3H]Phe); were all dried in vacuo; redissolved in dilute HCl; and stored for future use in countless studies probing their nature over the next year or so, mostly by myself and later with occasional help from several undergraduate students.
Using this material, we showed that component b was reactive with insulin antisera; could be converted to an insulin-like component by brief exposure to trypsin; and upon oxidative treatment to cleave disulfide bonds, did not dissociate into insulin chains. Because we had used phenylalanine to label one batch of slices, Philip Oyer, a first-year medical student who had joined the laboratory, was able to demonstrate that component b had a free N-terminal phenylalanine. Indeed, as predicted by these and other findings, tumor peak b consisted of a single-chain polypeptide that began with the B chain, ended with the A chain of insulin, and contained an additional connecting peptide sequence linking the chains together. Our first report on this work appeared in early 1967 (15). The role of peak b as a biosynthetic precursor of insulin was convincingly demonstrated later in 1967 by pulse-chase labeling studies with isolated rat islets of Langerhans.2 Once its precursor status was clearly established, we named it proinsulin (16), adopting a name that had been used by Su-Sun Wang and Frederick H. Carpenter in their earlier unsuccessful search for a “proinsulin” in bovine pancreas (17). Their inability to demonstrate proinsulin in pancreatic extracts only indicated that it was normally present in relatively small amounts (compared with insulin), which could not be detected by the strictly chemical approach they had used.
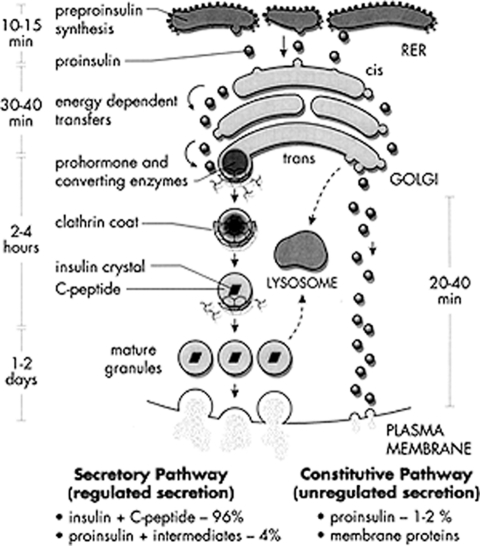
We were quite fascinated to observe in our earliest pulse-chase studies in rat islets that labeled proinsulin accumulated within the islets during an initial period of 20–25 min before beginning to undergo conversion to insulin (16), but once initiated, proteolytic conversion exhibited pseudo-first-order kinetics with a half-life of ∼30–60 min (18). What was even more remarkable was that the conversion process occurred entirely intracellularly, and the resultant labeled insulin was stored in the mature granules of the beta cells and then secreted in response to glucose stimulation, beginning ∼90 min after its biosynthesis. The cell biological implications of this time course led to the development of models predicting that newly synthesized proinsulin moved from the endoplasmic reticulum (ER) to the Golgi region within 15–20 min, a process requiring energy, and shortly thereafter was packaged into secretory granules and then converted to insulin (Fig. 5) (19). Our model was influenced, of course, by the elegant studies of Jim Jamieson and George Palade, who had delineated the intracellular progression of newly labeled exocrine pancreatic zymogens into nascent secretory granules (condensing vacuoles) via the Golgi apparatus (20). However, the pancreatic zymogens are not processed proteolytically within the cell but are stored and secreted as zymogens. On the other hand, our results clearly indicated that proinsulin conversion most likely was accomplished by specialized proteolytic enzymes within the newly formed secretory granules (19). This important postulate was subsequently supported by much additional evidence and finally was definitively demonstrated by Lelio Orci and co-workers, who showed an abundance of immunoreactive proinsulin within immature secretory granules in the trans-Golgi network of beta cells using monoclonal antibodies specific for proinsulin developed by Ole Madsen while a fellow in Chicago in the mid-1980s (21).
FIGURE 5.
Schematic representation of the beta cell secretory pathway showing the progression of newly synthesized proinsulin from the ER to insulin storage granules via the regulated arm (left). RER, rough endoplasmic reticulum.
Subsequent studies showed that small amounts of proinsulin and the connecting peptide derived from proinsulin by intracellular cleavage, which we later designated the C-peptide, also were secreted along with insulin in response to glucose stimulation (22). Moreover, the C-peptide and insulin were secreted in equivalent amounts, suggesting that the C-peptide is retained within the mature secretory granules along with insulin after the proteolytic cleavage of proinsulin within the maturing granules (Fig. 5).
Proinsulin Circles the Globe: First Presentations, 1967
In mid-1967, I embarked on an around-the-world trip beginning in Aachen, Germany, in response to an invitation from Helmut Zahn, the organic chemist who had put together one of the three pioneering groups that had accomplished the first complete chemical synthesis of insulin via chain combination in 1963. He had sent an emissary (one of his students) to visit me in my laboratory while attending a peptide meeting in Chicago in April 1967, and he extended the invitation. I agreed to go and talk about the proinsulin work in Aachen just before attending the International Diabetes Federation meetings in Stockholm in mid-July of that year, at which time I planned to make the first public presentation on the topic. To get to Aachen, I chose to arrive in Paris by air and spend a day or two there to get over the jet lag. While in Paris, I toured around and found the Collège de France. In front of it was a beautiful large statue of Claude Bernard. As I stood there admiring it, a young Japanese gentleman stepped up to me and asked if I would mind taking a photo of him standing by the sculpture. I obliged and then asked if he would mind returning the favor with my camera (Fig. 6). I then surprised him by asking whether he might be on his way to Stockholm, and he replied that indeed he was. I told him that I had guessed as much when he wanted to be photographed with Claude Bernard and admitted that I was also going to the same meeting. His name was Goto, he said, and he was a diabetologist. I believe he later gained fame as the developer of the Goto-Kakizaki rat, a widely used model for diabetes studies.
FIGURE 6.
The author paying homage to Claude Bernard in Paris in July 1967.
In Aachen, I was very cordially received by Helmut Zahn and his large group of young chemists. Another American also was visiting, Arnold Marglin from R. Bruce Merrifield's laboratory at Rockefeller University, and we each presented a talk. Arnie had succeeded in making synthetic A and B chains by solid-phase methods and combined them to reconstitute insulin. I presented a preview of the biosynthetic studies with islets, not yet published, that I intended to present in Stockholm. Clearly, Zahn had realized immediately the significance of proinsulin after our first publication appeared in February, and he was among the first to welcome me to the insulin fold, as a friend, rather than a threat. Both Arnie and I were very warmly received and, after our presentations, were invited to a centuries-old and very picturesque pub in the main square of Charlemagne's famous city for beers and supper, followed by one of Helmut's famous (as I later learned) pub crawls, which lasted until quite late in the night!
From Aachen, I boarded a train to Copenhagen, where I stopped to visit scientists at the Novo Company, the Danish insulin producer that had helped my work by supplying insulin samples for isolation of proinsulin and other minor components. We had a wonderful meeting: they were very interested and enthusiastic about proinsulin and also very willing to help in any way possible. During that meeting, I met Jørgen Schlichtkrull, the developer of their Lente insulin preparations and who was then the director of research; Lisa Heding, who later helped to develop excellent immunoassays for measuring serum proinsulin; and many others with whom we collaborated. They quickly agreed to gel-filter large amounts of partially purified insulin lots and provide the higher molecular mass component b-like material, which became our main source for the purification of bovine proinsulin in Chicago, and I subsequently freely provided information on methods we developed for purification of proinsulin and intermediate forms by column chromatography, which they adapted for industrial usage, resulting in great improvements in the purity of their therapeutic insulin, e.g. the development of highly purified “monocomponent” insulin.
Shortly afterward, in Stockholm, I made the first public presentation on proinsulin. It was well attended and well received. If any in the audience had doubts about proinsulin beforehand, they must have been dispelled by the pulse-chase studies with rat islets that I presented and that were then just about to appear in Science (16). As I recall, there were few if any questions asked, but then one person in the audience rose, walked to a microphone, and quietly stated, “What we have just heard is a quantum leap!” That ended the session, to my great relief. Then, as I returned to my seat, a young man came over and sat down. He smiled and held up his name tag to introduce himself. It said “René Humbel!” He then whispered to me, “Can we meet after this session?” Although I had not met René before, I knew that he had published a paper in 1965 (35) that claimed to show that the A and B chains were made in parallel rather than in series, rendering the single-chain concept unlikely.3 I nodded yes and then heard little of the next presentation, as I worried that René would soon tell me that he had found something terribly wrong with my findings. When we finally got outside the hall and exchanged friendly greetings, René said, “That is very nice work, but do you think it is possible that your proinsulin might have two chains?” I immediately sensed his pain but could tell him only that all our evidence indicated that it was indeed a single chain. I felt badly for René, but, fortunately he did not give up on insulin. He went on to spend the next fifteen to twenty years extending his work with Rudi Froesch in Zurich purifying and sequencing the peptides comprising “nonsupressible insulin-like activity” (also known as serum sulfation factor or somatomedin), which circulates in the blood. Rather ironically, perhaps, he was the first to show that these proteins are single-chain relatives of insulin (proinsulin, actually) that contain an integral C domain that is in part necessary for their binding to a receptor that is closely related to the insulin receptor, i.e. the two insulin-like growth factors, IGF-1 and IGF-2, which bind and strongly activate the IGF-1 receptor. This was, indeed, a major achievement that had enormous implications for our understanding of the relationship of insulin to the regulation of both growth and metabolism through gene duplication and diversification in the evolution of vertebrates. What a wonderful scientific comeback!
After Stockholm, I spent an interval of about 10 days traveling on around the world, stopping off in Greece, Israel, Istanbul, Thailand, Hong Kong, and Kyoto as a tourist to finally reach Tokyo, where I presented a second report on proinsulin in early August at the International Congress of Biochemistry to a different but equally interested audience. I returned home via Honolulu and San Francisco to a lot of work awaiting me in the lab!
Race to Isolate and Sequence Proinsulin
The isolation, characterization, and sequencing of proinsulin required relatively large amounts of starting material and were possible only with material provided by both the Novo Company in Denmark and Eli Lilly Research Laboratories in Indianapolis. As our biosynthetic studies with isolated islets proceeded, we began to examine small amounts (100–200 mg) of commercial insulin samples from these generous sources and soon found that a small but significant amount of proinsulin-like material (component b) was present as a contaminant (0.5–2.0% of the total insulin) and could be separated by gel filtration even from insulin that had been recrystallized 10 times, a product of Novo for research that was essentially free of glucagon, a common contaminant of the animal insulin preparations of that era. The reason for this is that proinsulin self-associates just like insulin to form hexamers in the presence of zinc, as first shown by Bruce Frank at Lilly, and such mixed proinsulin/insulin hexamers can be efficiently incorporated at low levels into insulin crystals. To facilitate our structural studies on bovine proinsulin, colleagues at the Novo Research Laboratories in the summer of 1967 began to carry out gel filtration of large amounts of crystalline bovine insulin from their production lots, dried down the 1–2% of b component it contained, and sent it to us in gram amounts in Chicago, where it served as starting material for further purification (23). This “b component” fraction proved to consist of multiple components when examined by polyacrylamide gel electrophoresis, a relatively new method I was encouraged to try by Simon Pilkis, who at that time was working on his Ph.D. thesis on glucokinase with Professor Mike Krahl in the nearby department of physiology. Simon became a great friend and always had a wonderful sense of humor, despite his personal battle with juvenile-onset diabetes, which unfortunately greatly shortened his productive life. Several years later, in 1973, Simon married Jo Borg, a fine technician who had been working for several years in my laboratory, and they moved shortly thereafter to Vanderbilt University, where Simon distinguished himself scientifically through his path-breaking work on fructose 2,6-bisphosphate, an important regulator of gluconeogenesis, and by the discovery and characterization of the unusual bifunctional enzyme that synthesizes and regulates its level in liver.
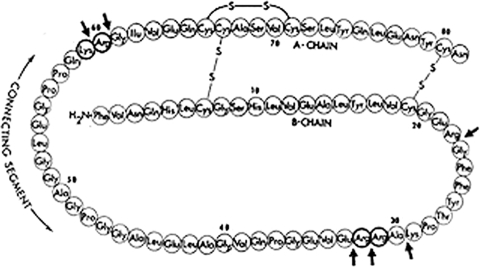
The major protein constituents of the component b-like fraction from commercial insulin preparations turned out to be intact single-chain proinsulin, along with several intermediate cleavage products and ∼30% of an unrelated covalently linked insulin dimer, a transpeptidation artifact generated by the strongly acidic (acid/ethanol) extraction procedures then used, which also caused the formation of small amounts of both ethylated and desamidated products. We developed gradient-elution ion-exchange chromatographic methods using urea-containing buffers to separate all of the above-mentioned components (23). These procedures later were adapted and modified at Novo for the large-scale production of “monocomponent insulin,” which proved to be much less immunogenic than earlier less pure insulin formulations and was a generally superior product. This work was overseen by Jørgen Schlichtkrull, who earlier had developed the Lente insulin-zinc formulations that provided more prolonged action profiles. Jørgen also showed that the higher molecular mass component, the a fraction from crystalline insulin, contained insulin bound to larger proteins, which enhanced its immunogenicity. Similar production improvements were also adopted rapidly at Eli Lilly and Company. Free C-peptide was absent from commercial insulin preparations, but the C-peptide could be purified from fresh animal pancreas in amounts that were equimolar with insulin, as would be expected if both insulin and the C-peptide were retained together in the beta cells of the pancreas after cleavage of proinsulin (24), as mentioned earlier. The sequence analysis of bovine proinsulin was carried out in collaboration with Chris Nolan and Emanuel Margoliash at the Abbott Research Laboratories in North Chicago, beginning in 1967. Emanuel Margoliash was renowned for his comparative sequence studies on cytochrome c from many species, and this seemed to bode well for getting a proinsulin sequence quickly. However, well over 500 mg of the bovine prohormone purified in my laboratory were expended in their efforts, and the tools that made short work of cytochrome c seemed to falter when applied to proinsulin. Well before our sequence analysis could be completed, Ronald Chance and his colleagues at Lilly succeeded in isolating and sequencing porcine proinsulin (25). Their analysis verified our proposed structural organization for human and bovine proinsulin and clearly revealed the paired basic residue sequences at the processing sites that soon became recognized as the hallmark sites for recognition and cleavage by the prohormone convertases (Fig. 7).
FIGURE 7.
Amino acid sequence of bovine proinsulin (81 residues), with arrows illustrating sites of potential tryptic hydrolysis. Basic residues in boldface circles are removed on conversion to insulin, releasing insulin and C-peptide (modified from Ref. 26).
Identification and Isolation of the Proinsulin C-peptide
In mid-1967, a graduate student, Jeffrey Clark, joined the laboratory and carried out an extensive series of biochemical and biosynthetic studies using isolated rat islets (22). He first prepared a good supply of rat insulin (rats and mice both have two non-allelic insulin genes) from a kilogram of rat pancreas, purified the two rat insulins, isolated and identified both proinsulins, and, in doing so, corrected the structure of rat insulin-2. In the course of his biosynthetic studies, he also identified a fast-moving labeled band on PAGE, which he identified as the rat C-peptide by labeling it with various radioactive amino acids. These studies revealed the absence of Phe and the presence of both Leu and Pro, as expected (22). On the basis of these findings, we then undertook isolation of the bovine C-peptide from pancreas via a purification procedure that began with modified acid/ethanol extraction, followed by gel-exclusion chromatography (24). On gel filtration in acetic acid, the C-peptide co-migrates with insulin, which explains why, in earlier experiments with the leucine-labeled human proinsulin fraction from the tumor studies, treatment with low amounts of trypsin resulted in an apparently seamless conversion of proinsulin to insulin (15, 18). We then purified the bovine C-peptide from the insulin by paper electrophoresis in 30% formic acid, which (after drying) gave a sharp band with ninhydrin staining but was negative to two other protein stains, Sakaguchi and Pauly, which recognize Arg and His/Tyr, respectively (24). These amino acids do not occur in the bovine C-peptide but are present in insulin. Having pure preparations of bovine pancreatic C-peptide available, we undertook its amino acid sequence determination ourselves to demonstrate unequivocally that it was identical to the connecting peptide segment of bovine proinsulin then still being sequenced by our collaborators in North Chicago. This work was performed mainly by Philip Oyer and another graduate student, Jim Peterson, while I was writing up a manuscript, but to our great surprise, our C-peptide sequence differed slightly from that region in bovine proinsulin. We checked our data, and Chris Nolan re-examined his and, in fact, found that he had misplaced an Ala residue in a chain of several glycines, a difficult sequence to sort out (Fig. 7). It was fortunate that we had gone the extra mile of sequencing in addition to simply peptide mapping the bovine C-peptide, as we could be confident that both sequences were correct, and their identical structures also strongly supported the origin of the pancreatic C-peptide from proinsulin (24, 26). Biosynthetic studies carried out by Jeffrey Clark and Arthur H. Rubenstein, who had joined my laboratory group in 1968, demonstrated that the molar ratio of insulin to C-peptide both within rat islets and in the medium after secretion was very close to unity, confirming that the C-peptide and insulin are stored together (in the secretory granules) after conversion and then co-secreted upon stimulation by glucose (27).
To obtain the human C-peptide sequence, Sooja Cho in our group collected pancreatic material from autopsies in the hospital on a weekly basis for more than a year and extracted it. It had usually undergone some autolysis, which lowered yields. However, Philip Oyer was able to secure enough pure material to determine its sequence, and we could thus predict the structure of human proinsulin (28). Arthur H. Rubenstein spearheaded studies showing the presence of secreted proinsulin in both the blood and urine of human subjects (29). The C-peptide radioimmunoassay was first devised in 1970 (30) to demonstrate its presence in human serum. Arthur and Kenneth Polonsky then accomplished the ensuing feat of translational medicine by further developing, refining, and calibrating the C-peptide assay, which is now widely utilized to measure endogenous insulin production under a wide variety of conditions (31, 32).
Emergence of a New Field
The discovery of proinsulin seemed to come somewhat as a bombshell to the scientific community and was greeted both with excitement and enthusiasm by some and with great skepticism by many others. Among the more enthusiastic were Christian B. Anfinsen and co-workers, whose hypothesis that protein primary sequence is the major determinant of protein folding was supported by our single-chain precursor findings (33). The need for such a precursor form actually had been posited by them in 1965 (34) but was followed shortly later in the literature by evidence supporting the parallel synthesis of the insulin A and B chains in islets from a teleost fish (35). Also, by 1964, peptide chemists, e.g. Panayotis Katsoyannis in New York, Helmut Zahn and co-workers in Aachen, and Chinese chemists in Shanghai, all had succeeded in the complete synthesis of insulin, with impressive yields ranging up to 50% or more (reviewed in Ref. 18). Their success seemed to support the view that a precursor was unnecessary. However, the Anfinsen hypothesis was basically correct, and Anfinsen later received the Nobel Prize in Chemistry for his elegant work on the spontaneous refolding of denatured ribonuclease, chymotrypsinogen, and other small proteins, sharing the 1972 award with Stanford Moore and William Stein at Rockefeller University, who had pioneered the field of protein analysis and invented the amino acid analyzer.
In 1968, Ron Chance and I were invited to make presentations on our proinsulin studies at the annual Federation of American Societies for Experimental Biology meetings in a biochemical symposium on proteins. Ron had just completed the sequence analysis of porcine proinsulin (25), and I had completed a series of studies on the refolding of bovine proinsulin, demonstrating its strong propensity to refold spontaneously and correctly with good yields in vitro. Both of our presentations were well received and created great interest. As I recall, the room was quite crowded, and after our talks, there was a break. As the audience exited the room, an elderly gentleman came over and congratulated me on the proinsulin work. It was Vincent du Vigneaud, and I was greatly honored by his kind words. Also, Chris Anfinsen very graciously communicated our findings on proinsulin refolding to the Proceedings of the National Academy of Sciences (33).
Many scientists were initially skeptical of our findings because tumor material had been used in the original study. Actually, human pancreatic insulinomas are usually well differentiated and generally retain many normal basic biochemical mechanisms but may exhibit poorly regulated proteolytic processing or secretory behavior. Since our initial study, insulinoma cells have come to be extensively used in laboratories and, through the work of Douglas Hanahan and co-workers, are now often induced experimentally in various species to provide an invaluable source of material for studies on the insulin-producing beta cell (36).
Although the initial interpretation of our discovery that proinsulin was required for the facile biosynthesis and assembly of insulin had much to recommend it, it proved to be far too narrow a concept. In 1964, Howard Sachs had produced suggestive evidence for a precursor of vasopressin, but it proved difficult to isolate and identify (37). Meanwhile, precursors for other bioactive peptides completely lacking the exacting assembly requirements of the insulin molecule began to be identified in the early 1970s, including proparathyroid hormone (John Potts and co-workers), proglucagon (Howard Tager), and promelanocyte-stimulating hormone/ACTH/endorphin or proopiomelanocortin (POMC) (Elizabeth Eipper and Richard Mains) (38). In 1967, Michel Chrétien and C. H. Li had shown that α- and γ-lipotropins, later shown to be fragments derived from POMC, contained β-promelanocyte-stimulating hormone bracketed by basic residues as in proinsulin (see discussion in Ref. 18, page 218). Most neuroendocrine peptides are small and devoid of secondary or tertiary structure. Their larger precursors presumably satisfy a proposed minimum size requirement for their efficient synthesis by the ribosomal mechanism of protein synthesis, about which more was constantly being learned throughout this period. However, another kind of proprotein was also discovered around 1970, exemplified by proalbumin, which had only a simple hexapeptide extension at its N terminus. A similar propeptide was found a year or so later in proparathyroid hormone and still later in the N-terminal propeptides of many coagulation factor zymogens synthesized in the liver. All of these precursors shared the common feature of closely similar processing sites made up usually of paired basic amino acids such as Lys-Arg or Arg-Arg, suggesting the existence of specialized proteolytic enzymes within the secretory pathway in many cells. With the advent of molecular cloning in the late 1970s, the structural elucidation of precursor proteins rapidly increased with many interesting and exciting revelations of larger polyprotein precursors containing multiple copies of structurally or functionally related biologically active peptides (38). In fact, proinsulin was just the tip of the iceberg, indicating the existence of an extensive and widespread system of precursor-processing proteolytic enzymes in the secretory pathway of all eukaryotic cells, which eventually emerged as the kexin/subtilisin-like prohormone convertase family of proprotein-processing endoproteases with its associated exopeptidases and amidating components (39).
We are also now well aware that the vast majority of secreted and membrane-bound proteins in both prokaryotic and eukaryotic cells have a more cryptic initial precursor form, or preprotein, bearing a signal peptide that is the initial translated product and that places new constraints on the N-terminal sequence of prosecretory proteins. Signal peptides are usually cotranslationally cleaved by signal peptidases during their translocation into the ER lumen, as was first beautifully demonstrated by Günther Blobel and Bernhard Dobberstein (40, 41). This additional layer of precursor processing has added another brilliant chapter to the cell biology of the secretory pathway. Space does not permit me to cover this aspect of precursor processing in any detail except to say that, when such an extension was first found in a nascent immunoglobulin light chain and then in the in vitro translation product of parathyroid hormone mRNA in the early 1970s, we were all very excited and quickly looked into this possibility. My colleague Shu Jin Chan, then a graduate student in my laboratory, became one of the first to translate islet mRNA in cell-free systems and succeeded in identifying and characterizing rat preproinsulin (42). Shortly afterward, Dennis Shields, working with Blobel, identified several preproinsulins in teleost fishes (43). During the late 1970s, preproteins became a major focus of research throughout the world, resulting in rapid progress in elucidating the cellular machinery underlying the translocation and processing of preproteins in both prokaryotes and eukaryotes, an area that is also actively thriving today.
Some Research Diversions
Although after 1967 insulin biosynthesis and processing were dominant themes in my laboratory, they were by no means the only work going on. We soon became interested in the evolution of insulin and the islets of Langerhans as work on proinsulin progressed, and this interest was facilitated greatly by Sture Falkmer, a Swedish pathologist/biologist who was actively exploring this topic and who provided excellent opportunities in the early 1970s to work with early vertebrates like hagfishes (an extant jawless vertebrate) at the Royal Swedish Marine Biology Station at Kristineberg on the western coast of Sweden, north of Gothenburg (Fig. 8). A bit later, Åke Lernmark joined my laboratory, from Umeå, Sweden, in 1974 and initiated studies on beta cell plasma membrane proteins, which led to his future discoveries of beta cell autoimmunity and autoantigens in type 1 diabetes, a field that he continues to explore vigorously today.
FIGURE 8.
Research at the Kristineberg Marine Biology Station in 1971. Upper panel, view of the Gullmar Fjord from above the station's wharf. Lower panel, Sture Falkmer (left) and Ingve Östberg (right) isolating islet organs from hagfish (Myxine glutinosa) for insulin studies. (I also helped!)
We also retained an interest in insulin action, and in 1969, when a new medical student, Susan Terris, appeared on the scene, we discussed various possibilities and decided that it might be good if she learned to isolate hepatic cell plasma membranes to look for putative insulin-binding sites or receptors, a field that then appeared ripe for study. Susan worked all summer on the membrane isolation technique and helped with some other experiments before entering her first year of medical studies. She now had plasma membranes but not enough time to look for binding of insulin until she rejoined the laboratory after completing the second year. In the meantime, of course, Pedro Cuatrecasas, Jesse Roth, and co-workers identified and characterized insulin-binding sites in plasma membranes and made considerable progress. When Susan returned to the laboratory in 1971, she was very disappointed at having been scooped and shed a few bitter tears in my office. Not quite knowing what to say, I tried to comfort her a bit by telling her that it surely was not all over in that field. For example, I said, no one had studied the binding of insulin to intact liver cells. Such preparations already were available in our laboratory; in fact, as Shu Jin Chan had developed a very effective method for isolating intact hepatocytes before he shifted his thesis project from insulin action in the liver to the more pressing problem of characterizing preproinsulin, as described above. I suggested that Susan should try using such intact cell preparations to look at insulin binding and look for any differences that might be of interest: I was convinced that there was much more to be learned about insulin binding, even though I was not sure what that might be!
Susan went to work and soon ran into the problem of insulin degradation, which had been an especially difficult problem with isolated plasma membranes, where contaminating enzymes released during their preparation tended to complicate binding studies if not controlled with inhibitors. There was no evidence that such degradation was connected in any way with insulin binding. In contrast with that situation, however, she found that very clean hepatocyte preparations showed very low levels of contaminating proteases capable of degrading labeled insulin in the medium, but when the cells were incubated with insulin, it was rapidly degraded. She then went on to show that it was not the free insulin that was being degraded but rather the receptor-bound hormone. This was certainly a very different behavior than expected, but she soon amassed an impressive body of evidence that strongly indicated that the liver cells not only preferentially degraded the bound insulin but much more of it than could be accounted for by the estimated number of receptors on the surface at any one time. In short, it seemed that the receptors catalyzed the degradation in a way that led to its complete breakdown into free iodotyrosine (44). Similar results were then obtained using perfused rat liver preparations (45). Over a several-year period, as Susan progressed in her work, we began to realize that insulin receptors were being taken up, probably via endocytosis, and the receptor-bound insulin was then degraded within the cells while the receptors were rapidly returned to the plasma membrane to actively repeat the uptake cycle. Susan had, in fact, discovered receptor-mediated uptake and intracellular degradation of receptor-bound insulin, a very important mechanism in insulin turnover in vivo. Her findings were further corroborated by mechanistic studies carried out by Cecilia Hofmann, who followed her in the laboratory (46). What Susan and Cecilia found was, in fact, quite analogous to the findings of Michael Brown and Joseph Goldstein with the seemingly unrelated system of LDL receptor binding, uptake of LDL, and its degradation to import cholesterol. Because hormone receptors were not yet known to exhibit such behavior, Susan was initially very worried that veterans in the field would not believe her results, but that turned out to be a groundless fear because she had done her work so thoroughly that its validity was never questioned when she first presented it publicly in 1976.
One good discovery often leads to others! In the late 1970s, Howard Tager and Arthur Rubenstein discovered the first patients with mild diabetes due to insulin gene mutations. Several mutations at proinsulin cleavage sites causing hyperproinsulinemia were already known, but these new mutations were different, as they were located at specific sites within the insulin molecule (FB24S and FB25L), which explained their lower biological activity. One of the puzzling features of these patients was the very high insulin levels they all shared in their blood, hence, the syndrome name of “hyperinsulinemic diabetes.” Kishio Nanjo at Wakayama Medical University soon discovered a third insulin mutation of this type in several Japanese families (VA3L) that lowered the biological potency by ∼1000-fold. Thus, when Tager and Steven Shoelson used HPLC to separate the plasma insulin components, they were amazed to find that the levels of the mutant insulin were always manyfold higher than the normal insulin level, as though the mutant insulins were secreted in excessive amounts (47). This seemed unlikely because the patients were all heterozygous for the mutations and therefore should have been producing equal amounts of the mutant and normal hormones, but then we realized what must be happening: because mutant insulins do not bind well to the insulin receptor, they would be poorly cleared from the blood and accumulate there, a direct consequence of the receptor-mediated pathway of insulin metabolism that Susan Terris had discovered! This supposition was subsequently confirmed to be the case by Howard Tager and colleagues.
Interestingly, the much more recent discovery that other mutations in the human insulin gene account for a considerable proportion of cases of permanent neonatal diabetes, in my colleague Graeme Bell's laboratory (48), has led to the identification of many new point mutations in all four domains of the preproinsulin protein, underscoring the unique functional importance of both the pre- and proregions of the precursor for the successful biosynthesis, translocation, folding, and processing of proinsulin in the biosynthesis of insulin. Most of these mutations impair primarily the folding and structural organization of the proinsulin molecule, leading to increased degradation of misfolded material, associated with activation of the unfolded protein response and increased ER stress, resulting eventually in beta cell death via apoptosis (49). This is currently an area of great interest because increased levels of beta cell ER stress also have been found to be associated with the more common type 2 diabetes as well, but for reasons that remain unclear.
Our fascination with the insulin receptor has continued in more recent studies directed toward learning more about how insulin binds to and activates the insulin receptor, an important problem that remains unsolved to this day. This work is being carried out by Shu Jin Chan in Chicago collaboratively with Michael Lawrence and Colin Ward at the Walter and Eliza Hall Institute in Parkville, Australia, and Michael Weiss and Jonathan Whittaker at Case Western Reserve University in Cleveland (50) and is focused on the role of the C-terminal segment of the α-subunit in insulin binding, an unexpected discovery made in the laboratory in 1994 by Takeshi Kurose, a postdoctoral fellow, using a novel photo-cross-linking approach (51).
Unraveling the Prohormone-processing Mechanism
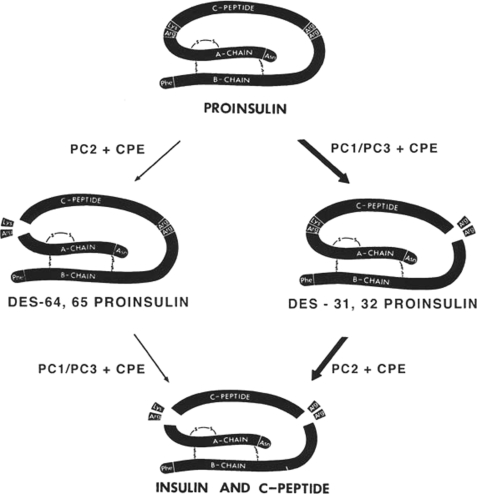
Our earliest studies on proinsulin had demonstrated its great sensitivity to trypsin, which was later explained by identification of the paired basic residues (Arg-Arg and Lys-Arg) that link the C-peptide to the insulin chains in proinsulin (25, 26). However, before this structural feature was known, I had noticed that the presence of proinsulin could be detected easily in partially purified higher molecular mass “b component-like” fractions separated from commercial crystalline insulin preparations. During incubation of aliquots of this material with small amounts of trypsin at pH ∼7.6, I carefully observed the tube contents. If proinsulin was abundant, the clear solution became very turbid within the first minute or so of incubation and then slowly cleared. I soon noticed that this phenomenon did not happen when buffers in the range of pH 8.5 were used. When Ron Chance published his classic study on the structure of porcine proinsulin in 1968, he reported that trypsin treatment yielded desalanyl insulin, a biologically active form of insulin that lacked the last amino acid of the B chain, along with the tripeptide Ala-Arg-Arg (25). It thus seemed likely that the initial cleavage of proinsulin by trypsin was because of the rapid excision of the C-peptide by cleavage after the two pairs of basic residues, giving rise to diarginyl insulin, which might precipitate due to its higher pI (near 7.5), followed by its slower cleavage between Lys-B29 and Ala-B30 to release the tripeptide and restore the normal pI of ∼5.3 to the desalanyl insulin product, which would then redissolve. Larger amounts of trypsin were required to cleave at Arg-B22 to release desoctapeptide insulin and a larger heptapeptide (Fig. 7). However, these findings posed somewhat of a paradox because Wang and Carpenter had reported in the literature that trypsin cleaves insulin preferentially after Arg-B22 to produce only desoctapeptide insulin and an octapeptide (B23–B30) and that subsequent cleavage of this peptide at Lys-B29 then generates the heptapeptide and free alanine, i.e. that it is impossible to generate desalanyl insulin from insulin using trypsin (52)!
A possible explanation for this paradox was that the proximity of the Arg-Arg sequence upstream of Lys-B29 in proinsulin might attract trypsin and thus enhance its susceptibility to trypsin. To test this idea, I tried adding an excess of carboxypeptidase B during the tryptic incubation to see if it might quickly remove the C-terminal basic residues as they were generated by trypsin and thus protect the C-terminal alanine from removal by the modest amount of trypsin required to release the C-peptide. This approach worked remarkably well! There was no longer any visible clouding during incubation. We then tried various combinations of these two proteases and soon found a suitable ratio that quantitatively converted the proinsulin to intact insulin and C-peptide without generating any detectable desalanyl insulin (53). This combination of enzymes thus provided a model system for the in vivo system, as it produced the same natural products of processing that are normally found in vivo in greatest abundance in the pancreas: native insulin and intact C-peptide lacking any basic residues. In earlier in vitro studies with isolated actively processing islet secretory granule preparations, we were also unable to find any evidence for the release of basic dipeptides from [3H]arginine-labeled proinsulin during its conversion (54).
All these observations provided us early on with a compelling model for conversion of prohormones in vivo by a combination of trypsin-like and carboxypeptidase B-like converting enzymes. All we had to do was find them! However, to do this took almost twenty years and involved the efforts of a large number of talented investigators (as described below). Despite the fact that Mother Nature uses her own more elaborate version of this simple enzyme combination to produce a huge variety of biologically active natural products, the trypsin/carboxypeptidase B combination has proved to be highly useful as a simple, inexpensive, and convenient (as well as previously unpatented) method for the conversion of biosynthetic human proinsulin to insulin in a commercial process that has been used for many years at Lilly in the production of high-quality recombinant human insulin for diabetes therapy (55). As it turns out, the natural convertases, despite their precision and elegance, are not sufficiently active and stable under industrial conditions to be useful thus far in biotechnology.
Search for the Convertases
Although our earlier studies had demonstrated that isolated intact secretory granules from islets were a major site of conversion of proinsulin to insulin in vivo, we were unable to recover any activity after lysis of the granules (54). We used a variety of approaches, including synthetic substrates, various proteinase inhibitors, and active site-labeling techniques without any success. Even though our model system predicted that a trypsin-related enzyme(s) might be involved and despite the close developmental relationship of the islets with the pancreas, the putative enzymes proved elusive and possibly unstable. However, a glimmer of hope arose in 1975 when Dr. Hartmut Zühlke, a visiting scientist from a diabetes center in East Germany working with islet extracts, found evidence for the existence of an acidic carboxypeptidase B-like activity (56). Seeing as how we believed that the processing reactions occurred in the mildly acidic environment within maturing secretory granules, this finding was encouraging. However, because of our preoccupation with the endoprotease activity (first things first!), we did not pursue it further. A few years later, Lloyd Fricker at Johns Hopkins found a similar activity in brain extracts and designated it carboxypeptidase E (CPE). When cloned and sequenced, it turned out to be a homolog of carboxypeptidase B with an acidic pH optimum (57). A subsequent discovery by Jürgen Naggert, Ed Leiter, and co-workers at The Jackson Laboratory of a mouse with severe obesity and mild diabetes associated with CPE deficiency confirmed its importance for proinsulin processing (58). Our joint studies of these mutant mice demonstrated elevated circulating proinsulin levels and accumulation of C-terminally extended diarginyl insulin in islets, clearly confirming the role of CPE in proinsulin processing. Disruption of normal neuropeptide processing also occurs in the brains of CPE−/− mice and may account for the hyperphagia and obesity associated with frank diabetes, which occurs more prominently in males (58). The accumulation of high concentrations of C-terminally extended conversion intermediates in the secretory granules very likely also inhibits the converting endoproteases (59).
Where was the all-important endoprotease that had become our holy grail? We were diverted from the search for a trypsin-related serine protease in the early 1980s by some new findings in our laboratory that seemed to implicate a cathepsin-like enzyme in this role (60). This proved to be a false lead and was soon set aside. However, more recent evidence indicates that some procathepsin B normally passes into immature beta cell secretory granules after their formation in vivo but is subsequently sorted into lysosomes, where it is activated (61). A possible role for cathepsin L in neuropeptide processing in the brain has been proposed recently by Vivian Hook and co-workers. One possibility might be that it plays a role in activating or stabilizing some of the proprotein convertases.
Yeast Genetics to the Rescue!
Sometime in the mid-1970s, Jeremy Thorner visited the University of Chicago as a biochemistry job candidate (Fig. 9). At that time, he mentioned to me that yeast produces some small bioactive peptides, such as α-mating factor, that were likely processed from larger precursors similarly to prohormones. I encouraged him to look into the possibility that yeast might provide a novel means to identify the enzymes involved. A few years later, in May 1979, I received a letter from Jeremy, then at the University of California at Berkeley, requesting some reprints and including some of his recent publications reporting progress in this direction. Some five years later, in 1984, he informed me that the search for a convertase had been successful and that he and his co-workers had cloned several novel enzymes, including a calcium-dependent endoprotease, which they called Kex2p, that had high specificity for paired basic residues, as well as a carboxypeptidase and an amino dipeptidase that were all required for conversion of pro-α-factor to its active form (62). David Julius' beautiful work with Jeremy on the identification and cloning of Kex2p (kexin), the yeast endoprotease responsible for α-factor cleavage, provided at long last the key to unlocking the mystery of the nature of the mammalian proprotein convertases. However, we were not there yet! Kex2p at first exhibited some confusing properties: it was inhibited by some thiol protease inhibitors and did not seem to be related to trypsin. This puzzle soon was resolved by the discovery that it was homologous to the subtilases, a structurally different family of serine proteases (63).
FIGURE 9.
Jeremy Thorner, Ph.D., in the mid 1980s. This photograph was kindly provided by Thorner.
Enter the Mammalian Prohormone Convertase Family
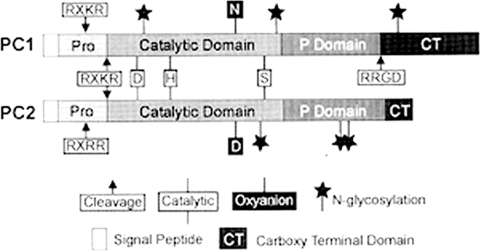
Building on this latter discovery, Steven Smeekens, who had joined my laboratory in 1986 and spent two years in fruitless attempts to use a mating-defective Kex2p null yeast strain in complementation experiments with cDNAs from islets or human islet cell tumors, turned to PCR, exploiting likely conserved sequences near the catalytic residues to design suitable probes for mammalian homologs of Kex2p. He announced his exciting new findings to me on a memorable date, April 1, 1989! He showed me a cDNA sequence from a human insulinoma that clearly encoded a Kex2p-like protein, which he called PC2 (for protease candidate 2). Its predicted size was only ∼68 kDa. There were a few other problems, i.e. the catalytic oxyanion residue was Asp rather than Asn, as in all other known subtilisin-like proteases, and it lacked both a Ser/Thr-rich segment and a transmembrane domain (64), which accounted for its much smaller size than Kex2p (∼97.5 kDa). However, all these differences could be rationalized as being adaptations to its localization within the more acidic secretory granules, where prohormones or proneuropeptides are processed proteolytically to their bioactive forms and then stored or secreted. Moreover, Steve also had found another similar partial cDNA in the insulinoma library that differed in sequence from PC2. In a short time, a full-length clone for this enzyme was obtained from a cDNA library derived from a pituitary corticotropic AtT20 cell line, and it was designated PC3 (now known as PC1/3). It proved to be PC2's companion convertase in the beta cells, and it also lacked a transmembrane anchor (65). Nabil Seidah and Michel Chrétien in Montreal quickly reported similar discoveries using PCR in endocrine cell cDNA libraries, and then many other reports followed! Together, these two convertases have proved to be the major prohormone convertase family members involved in processing a large number of prohormones and neuropeptides and are highly expressed throughout the vertebrate neuroendocrine system (Fig. 10) (66). The presence of two calcium-dependent processing enzymes in the beta cells was first proposed by Howard Davidson, Chris Rhodes, and John Hutton in 1988 (67), and these activities were soon shown to be identical to PC2 and PC1/3. Also, workers at Chiron, in collaboration with Robert Fuller and Jeremy Thorner at Berkeley, published a report in Science in 1989 (68) describing yet another putative mammalian convertase, PACE1 or furin, a protein identified originally as a possible receptor tyrosine kinase by a group in The Netherlands. It was a closer homolog of kexin than PC1/3 or PC2, as it had a transmembrane domain and a Cys-rich domain in the N-terminal juxtamembrane region, where Kex2p has a Ser/Thr-rich domain (66). Furin has proved to be the most ubiquitous of the mammalian convertases, and unlike PC1/3 and PC2, it functions mainly in the constitutive arm of the secretory pathway, participating in the cleavage of proteins, such as the insulin and IGF-1 receptor precursors, viral glycoprotein precursors, blood-clotting factor, serum albumin proproteins, and many others. Today, the total count of related subtilisin-like proprotein convertases stands at 10: furin, PC1/3, PC2, PC4, PACE4, PC5/6A, PC5/6B, PC7, SKI1/S1P, and PCSK9 (69).
FIGURE 10.
Comparative structures of PC1/3 and PC2 with various features and domains indicated. Autoactivation occurs via dual cleavages of the prodomain at the polybasic sites illustrated. The conserved P domain has been shown by modeling and x-ray studies to be an eight-strand β-barrel. It appears to add stability to the catalytic domain and is present in all the convertases having multibasic residue specificity. See Ref. 80 for details (reproduced with permission from Ref. 80).
Convertase Knock-outs Prove Their Function
The identification of the genes encoding the mammalian subtilisin-like calcium-dependent family of proprotein convertases finally enabled the fulfillment of our earlier postulate that the only definitive proof of the role of any proposed processing enzyme could be achieved through the deletion or mutation of its gene (70). This postulate was first tested with both PC2 and PC4, a testis-specific convertase, in 1997 when gene disruptions in mice preventing the formation of these two respective active enzymes produced specific proteolytic processing defects in each case that fit well with their known patterns of tissue-specific expression (39). Thus, PC2 nulls fail completely to process proglucagon to glucagon in the pancreatic islet alpha cells, as anticipated from previous studies of Yves Rouille in our laboratory, and as a result have lower than normal blood glucose levels at all times. They also show many other processing defects affecting insulin and a major proportion of many other hormones or CNS neuropeptides. Similarly, knock-out of the neuroendocrine protein 7B2, which is required for the conversion of pro-PC2 to its active form, produces a closely similar phenotype to that of PC2 nulls when tested on the same genetic background in mice (39, 72).
On the other hand, mice null for PC1/3 displayed alterations in a different subset of neuroendocrine peptides, reflecting subtle differences in the tissue specificity of its expression compared with that of PC2 within neuroendocrine tissues (73). In contrast, disruptions of the genes encoding furin, PACE4, and PC5/6 (74), as well as PAM (75), the peptide-amidating monooxygenase that accomplishes the C-terminal amidation of many neuroendocrine peptides, all result in developmental defects that prevent the normal development of the fetus, usually through defective activation of various growth factors required for specific developmental processes. Therefore, targeted deletions have been undertaken for some of these genes, e.g. a targeted deletion of furin only in the pancreatic beta cells results in a prohormone-processing defect due to defective activation of the vacuolar ATPase that acidifies the maturing secretory granules to create the proper conditions for processing by PC1/3 and PC2 of their complement of precursors, which consists mainly of proinsulin and proislet amyloid polypeptide (pro-IAPP) (76).
Because both PC1/3 and PC2 are required for efficient conversion of proinsulin to insulin, the proinsulin-processing defects in PC1/3 versus PC2 nulls differ in characteristic ways, reflecting their individual preferences for certain sites and intermediate forms as substrates. Recent studies have verified that PC2 prefers many substrates having the sequence motif Lys-Arg within cleavage sites, whereas PC1/3 tends to prefer those with Arg-Arg (77, 78). This preference is consistent with the current view that, in the islet beta cells, PC1/3 cleaves proinsulin first at the B chain/C-peptide (Arg-Arg) cleavage site, followed by CPE to generate the des-31,32-proinsulin intermediate, a preferred substrate for cleavage by PC2 at the C-peptide/A chain site (Lys-Arg) (Fig. 11) (79). Similarly, proglucagon, which contains three glucagon-related peptides, e.g. glucagon, GLP-1 and GLP-2, each having differing biological actions, is processed in islet alpha cells so as to release only (or mainly) glucagon. This is due to cleavage exclusively by PC2 acting at Lys-Arg sites. On the other hand, the same proglucagon molecule is processed differently in intestinal L cells by PC1/3, acting alone to release only GLP-1 and GLP-2 via cleavage at sites containing only arginine (Arg-Arg or X-Arg). This differential processing of multifunctional precursors is not unusual. However, both PC2 and PC1/3 often cleave at both types of basic pairs, as well as others, such as Lys-Lys and Arg-Lys, which occur more rarely (80). Precursor conformation seems to be a major determinant, but little is known about this for most peptide precursors.
FIGURE 11.
Pathways of conversion of proinsulin to insulin and C-peptide in the beta cell. The pathway shown on the right is predominant normally because PC1/3 acts somewhat earlier than PC2, forming a preferred intermediate for PC2 cleavage, the des-31,32-proinsulin intermediate. In the absence of PC1/3, only ∼10–15% of the proinsulin is processed to insulin. The B chain/C-peptide junction is a better convertase substrate because it is more accessible and has an upstream basic residue (lysine) at the P4 position (B29). The C-peptide/A chain site lacks a P4 basic residue. (See Refs. 79 and 84 for further discussion.)
The discovery of IAPP (amylin) in 1986 by Per Westermark introduced another minor secretor product of the beta cell. IAPP is normally produced at levels well below 5% of those of insulin, mainly in the islet beta cells (81). Human IAPP is especially fascinating due to its unique tendency to self-associate to form amyloid fibrils and to form deposits in the islets of elderly persons but to a much greater extent in patients with type 2 diabetes of many years duration. Its possible role in the pathogenesis of diabetes remains unresolved and is a topic of continued research and pharmaceutical exploration. The IAPP precursor (81) requires proteolytic processing at two sites, sharing the same cleavage enzymes, PC1/3, PC2, and CPE, with proinsulin, as shown recently by Bruce Verchere and co-workers in Vancouver (82). However, PC2 is the more major endoprotease involved in processing pro-IAPP, in contrast with proinsulin, where PC1/3 is more important.
As mentioned earlier, defects in the expression of CPE can have far-reaching effects on neuroendocrine peptide processing, also affecting insulin biosynthesis and a broad spectrum of other peptides and (in mice) resulting in severe obesity (58). The absence of CPE or its companion homolog, CPD, which appears to act primarily in the constitutive arm of the secretory pathway in concert with convertases such as furin, PACE4, and PC5/6B, allows the accumulation of intermediate peptides with C-terminal basic residue extensions, such as diarginyl insulin and C-peptide-Lys-Arg, in the beta cell (58). Such extended peptides appear to be potent inhibitors of the convertases, especially at the millimolar concentrations that can easily occur in actively processing secretory granules in the absence of CPE (59). However, because CPE is normally present in relative excess to the PCs, reduced enzyme levels due to hemizygosity for CPE have no detectable effects on processing, whereas hemizygosity for PC2 or especially PC1/3 is reflected in minor processing defects, as manifested in partially impaired processing of proinsulin in beta cells (79). Like CPE deficiency, decreased PC1/3 activity also leads to obesity, presumably mainly due to imbalances in the hypothalamic processing of orexic versus anorexic peptides involved in regulating feeding behavior in both man and experimental animals.
Postlude
As time has passed, the dimensions of the field of precursor processing have continually enlarged far beyond the simple secretory pathway paradigm of proinsulin and its role in insulin biosynthesis. It has been a great pleasure, perhaps like that of a parent (which I do not happen to be, other than as a surrogate parent to the many wonderful students, postdoctoral fellows, and younger colleagues with whom I have been privileged to work), to observe its steady growth, development, and expansion over the past forty-five years. Since 1992, there has been a biennial Gordon Research Conference held every other July entitled “Proprotein Processing, Trafficking and Secretion,” which has been well attended. Like many other discoveries, e.g. protein phosphorylation, which seemed at first to apply only to one particular special enzyme system (glycogen breakdown4), precursor processing as a cellular biosynthetic and regulatory mechanism participates in the production of a great variety of proteins involved in many diverse regulatory processes, well beyond insulin and neuropeptides. One of the richest rewards a scientist can enjoy is to have this experience.
Another very enjoyable aspect of our research activities over the years has been our efforts to explore the evolutionary origins, first of proinsulin, insulin, and C-peptide and then of the insulin gene family, with the help of Sture Falkmer and his graduate student Stefan Emdin. Interestingly, our initial studies on hagfish at the Kristineberg Marine Research Station required the isolation of islet organs from ∼10,000 hagfish to obtain enough of the insulin to sequence and characterize (Fig. 8, lower panel). Fortunately, Stefan was able to crystallize it just before a visit in 1972 from Dorothy Hodgkin to the University of Chicago, where she was to receive an honorary degree for her post-Nobel Prize work focused on elucidating the structure of porcine insulin in 1969. The beautiful big tetragonal crystals of hagfish insulin that Stefan grew, in the absence of zinc, were all it took to convince Dorothy that the hagfish x-ray structure should be done. This was carried out with her associates, John and Susan Cutfield, and their structure showed that, despite substitutions at ∼38% of positions, the overall structure of hagfish insulin was closely similar to that of the mammalian hormones. Later on, with the availability of cloning technology, Shu Jin Chan was able to determine the primary structure of hagfish preproinsulin using just a few islet organs to make a cDNA library. He was thus able to extend our evolutionary search to the insulin/IGF gene family, as well as their receptor molecules. This work is still in progress but already has revealed the likelihood that the IGF subfamily is a vertebrate invention, first arising via gene duplication, probably after the protochordates, as exemplified by amphioxus, which appears to have a single insulin/IGF-like protein (termed insulin-like protein) (83). The mainline insulin gene per se, however, is much more ancient and is present in recognizable form in most metazoans, although more work needs to be done in this area. Multiple insulin-like peptides are present in Drosophila and other insects, as well as in nematodes. Likewise, the insulin-like receptor protein is remarkably well conserved both in structure and in function, including metabolic regulation, growth, and control of longevity. However, molecules similar to IGFs and their receptors first appeared in jawless vertebrates like the hagfishes, which have a single IGF-like molecule and one typical IGF receptor, in addition to the insulin/insulin receptor system. Then, in the teleosts and higher vertebrates, two IGFs are minimally present with a single (IGF-1-type) receptor. Many of the teleosts are tetraploid, however, so these numbers may be doubled. Thus, the vertebrates have likely benefitted in evolution from the separation of metabolic and growth regulatory functions via successive insulin gene duplications.
The cloning of the convertases has provided another gene family for evolutionary exploration that has turned out to be highly conserved throughout the metazoans but does not appear to be present in plants, protozoans, or bacteria. PC2 appears to be the most highly conserved convertase of the family, especially with its unique aspartyl oxyanion residue, which worried us so much when we first identified this enzyme. Its conservation perhaps is related also to its unique requirement for activation via interaction with neuroendocrine protein 7B2, as shown so convincingly by Iris Lindberg and her colleagues. Other convertase family members, e.g. PC1/3, PC5/6 and furin, also are very well conserved, indicating the likelihood that proteolytic processing of precursor proteins is an evolutionarily important mechanism for making and/or activating biologically active peptides and creating greater diversity, as well as consolidating related bioactivities through the evolution of large multifunctional polyprotein precursors like POMC, proglucagon, and many others. This topic thus presents fertile ground for further exploration in contemplating the ever-widening development of our incredibly rich natural world. It is remarkable to consider how vastly our knowledge in the area of regulatory bioscience has expanded within the last hundred years and how important this knowledge has become already for the survival and enrichment of human life.
A final thought: I and others of my generation have been especially fortunate to develop our careers in an era when resources were initially small, but there was always enough to get by as long as one was willing to use a little ingenuity and make the special reagents that he or she might need, and positions were so much more available in the tenure track system than is now the case. The steady erosion of opportunities for young investigators over the past thirty years greatly troubles me. It is clearly imperative to make more openings available at the entry level so that individuals do not have to wait for positions and resources while their most creative years evaporate. Early placement of promising young investigators in responsible positions must become one of our highest priorities if we wish to preserve the unparalleled scientific productivity that we have achieved. This is one of the greatest challenges we face today in science. Unfortunately, I have no easy solutions to propose, other than the observation that, clearly, we older scientists must reduce our consumption of resources to make room for the young to flourish.
Acknowledgments
I am indebted to many students, postdoctoral fellows, and younger collaborating colleagues who have contributed to the work from my laboratory discussed in this “Reflections” article. In particular, I would like to mention Robert Roskoski, Judy King, Lee Younger, Dennis Cunningham, Philip Oyer, Jeffrey Clark, Wolfgang Kemmler, Hiroyuki Sando, Henrik Ege, Ole Hallund, Jørgen Schlichtkrull, Lisa Heding, Jørgen Gliemann, Franco Melani, Arthur Rubenstein, Jo Borg, Simon Pilkis, Howard Tager, Ron Chance, Bruce Frank, James Peterson, Shu Jin Chan, Rosa Choy, Edward Schlenk, Sooja Cho Nehrlich, Susan Terris, Ray Carroll, Ole Madsen, Jens Hoiriis Nielsen, Sture Falkmer, Stefan Emdin, Åke Lernmark, Takahiro Kanatsuna, Cecilia Hofmann, Jon Marsh, Hisako Ohgawara, Christoph Patzelt, Dorothy Hodgkin, Guy and Eleanor Dodson, Simon Kwok, Masakazu Haneda, Kevin Docherty, John Hutton, Chris Rhodes, Philippe Halban, David Nielsen, Michael Welsh, Kishio Nanjo, Susumu Seino, Craig Blackstone, Steve Smeekens, Graeme Bell, Kenneth Polonsky, Erika Plisetskaya, Steve Duguay, Cun Ming Duan, Motoshige Miyano, Masahiro Nishi, Shinya Ohagi, Machi and Hiroto Furuta, Shinya Nagamatsu, Yasunao Yoshimasa, Jonathan Whittaker, Tadashi Hanabusa, Yves Rouillé, Jeremy Paul, Mohammad Pashmforoush, Isaiah Pitman IV, Satoe Nakagawa, Lelio Orci, Louis Philipson, Gregory Lipkind, Sean Martin, Tony Oliva, An Zhou, Gene Webb, Joe Bass, Xiaorong Zhu, Takeshi Kurose, Arunangsu Dey, Jie Wang, Margaret Milewski, Jeff Stein, Iris Lindberg, Gary Thomas, Lloyd Fricker, Jens Rehfeld, Per and Gunilla Westermark, Michael Weiss, Colin Ward, Michael Lawrence, and Soo Young Park. My early mentors were Herbert S. Anker, Ph.D. (deceased), Earl A. Evans, Jr., Ph.D. (deceased), Eugene P. Kennedy, Ph.D., and Robert H. Williams, M.D. (deceased). To them all, I am greatly indebted. This article is dedicated to the memory of two special colleagues (both now deceased), Philip C. Hoffmann (pharmacology) and Eugene Goldwasser (biochemistry), who were my steadfast friends/advisers and luncheon companions for many years. Work in my laboratory has been supported by National Institutes of Health Grants DK13914 and DK20595 and by the Howard Hughes Medical Institute. My thanks to Steve Roess and Gladys Paz for assistance with the production of this manuscript and figures.
Footnotes
Two excellent sources for further historical details are the authoritative book by Michael Bliss entitled The Discovery of Insulin, published by the University of Chicago Press in 1982, and a more recent book by Thea Cooper and Arthur Ainsberg entitled Breakthrough: Elizabeth Hughes, the Discovery of Insulin, and the Making of a Medical Miracle, published by St. Martin's Press in 2010.
We had tried a number of approaches to studying insulin biosynthesis, such as fetal or newborn rat pancreas, without any success. Just about that time, an enzymatic method for isolating islets became available. Still, we made many trials before being able to isolate even a few islets. Once we cleared that hurdle, we had inconsistent results in demonstrating proinsulin by gel filtration after labeling the islets. Soon I realized that, although we added unlabeled insulin as a carrier during extraction to prevent losses, it did not effectively prevent adsorption of the tiny amounts of labeled proinsulin to the glass tubes in our fraction collectors. I discovered that large proinsulin peaks could be eluted from the empty tubes with an ammonia/BSA wash. We then tried coating them before use with BSA dissolved in 50% acetic acid, which was then baked dry in a glassware oven. Only then could we consistently obtain reproducible proinsulin peaks, as shown in Ref. 16. When we finally obtained such results, it was late one evening, and I was alone in the laboratory, watching the counts appear in the scintillation counter from one of the first successful labeling experiments. As beautiful symmetrical peaks of labeled proinsulin began to emerge just as hoped, I heard myself exclaiming “wow” and “gee whiz.” Suddenly, I remembered that Gene Kennedy had once remarked, “There are only two kinds of scientists: those who say ‘gee whiz’ and those who say ‘so what’ when something new and exciting appears.” I am clearly one of the former!
I well remember the night in late 1966 that I stopped by Ira Wool's office in Abbott Hall. (We both often worked late into the evenings). I had decided to divulge my new findings on insulin biosynthesis to my knowledgeable elder colleague. When I told him about the outcome of the tumor study, he shocked me by saying, “Donald, don't you know that's impossible!?” I was dumbfounded and asked him why he said that. He told me he had just attended a small meeting on insulin at Brookhaven at which a young scientist from Rockefeller named René Humbel had presented a very convincing report on the biosynthesis of insulin, showing that the insulin chains were made separately and then combined. I was stunned by this news and asked where it was to be published. He said he thought it was to be in the Proceedings of the National Academy of Sciences of the United States of America. The next morning, I rushed to the library and tried to find it. It apparently was not yet published, but, as I searched through recent copies of the journal, I found an article by Givol et al. (34) that presented strong arguments in favor of a single-chain precursor model for insulin biosynthesis. I decided that the algebraic sum of our knowledge about insulin biosynthesis at that time was zero and resolved not to worry any more about the matter. My own data alone had to be paramount. A month or so later, when I had prepared my first manuscript on proinsulin (15), Ira read it for me and made a number of helpful suggestions; he especially urged me to state my conclusions more explicitly, that the data clearly demonstrated the existence of a single polypeptide chain larger than insulin that began with the B chain, extended through a connecting segment of approximately 3 kDa, and terminated with the A chain. For this sage advice, I am eternally grateful to Ira.
In early 1960, when I was trying to find a postdoctoral position that might allow me to stay in Seattle, I spoke to several members of Hans Neurath's excellent biochemistry department at the University of Washington. None of them, including Neurath himself, had any positions open. I especially recall my conversation with Edwin Krebs because, at that time, his laboratory was fully concentrated on the mechanism of phosphorylase activation, whereas my work in the department of medicine was on glycogen biosynthesis via the new Leloir pathway involving UDP-glucose and the possible effects of insulin on synthase activity in the liver. I told Ed what I had been doing, and he said, in effect, “Sorry, but we are only working on glycogen breakdown!” Interestingly, in the early 1960s, Chris Nolan, who later assisted us in sequencing bovine proinsulin around 1967–1969, was working on the structure of the phosphopeptides from glycogen phosphorylase with Krebs in Seattle (71). Of course, within about ten or fifteen years, Ed's interest in insulin's effects on protein phosphorylation greatly increased, but despite occasional temptations, I avoided ever reminding him of his parochial former view. We always had a pleasant relationship, and Ed was certainly among the most gracious colleagues I have known. I later enjoyed many pleasant visits with Neurath, Krebs, and Ed Fischer on trips to Seattle. In fact, I skied with Hans and his wife, Suzi, along with several other protein chemists on several occasions at meetings in western settings, when they were probably in their early 90s. Hans greatly enjoyed skiing!
REFERENCES
- 1. Eulenburg-Wiener R. V. (1938) Fearfully and Wonderfully Made: The Human Organism in the Light of Modern Science, Macmillan, New York [Google Scholar]
- 2. Steiner D. F., Anker H. S. (1956) On the synthesis of antibody protein in vitro. Proc. Natl. Acad. Sci. U.S.A. 42, 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murnaghan J. H., Talalay P. (1967) John Jacob Abel and the crystallization of insulin. Perspect. Biol. Med. 10, 334–380 [DOI] [PubMed] [Google Scholar]
- 4. Jensen H., Evans E. A., Jr. (1935) Studies on crystalline insulin. XVIII. The nature of the free amino groups in insulin and the isolation of phenylalanine and proline from crystalline insulin. J. Biol. Chem. 108, 1–9 [Google Scholar]
- 5. Steiner D. F., Williams R. H. (1958) Respiratory inhibition and hypoglycemia by biguanides and decamethylene diguanidine. Biochim. Biophys. Acta 30, 329–340 [DOI] [PubMed] [Google Scholar]
- 6. Wick A. N., Larson E. R., Serif G. S. (1958) A site of action of phenethylbiguanide, a hypoglycemic compound. J. Biol. Chem. 233, 296–298 [PubMed] [Google Scholar]
- 7. Steiner D. F., Williams R. H. (1959) Actions of phenethylbiguanide and related compounds (editorial). Diabetes 8, 154–157 [DOI] [PubMed] [Google Scholar]
- 8. Morgan H. E., Cadenas E., Regen D. M., Park C. R. (1961) Regulation of glucose uptake in muscle. II. Rate-limiting steps and effects of insulin and anoxia in heart muscle from diabetic rats. J. Biol. Chem. 236, 262–268 [PubMed] [Google Scholar]
- 9. Steiner D. F., Williams R. H. (1959) Some observations concerning hepatic glucose 6-phosphate content in normal and diabetic rats. J. Biol. Chem. 234, 1342–1346 [PubMed] [Google Scholar]
- 10. Steiner D. F. (1978) Insulin-induced liver hyperplasia. Evidence for a negative liver-size-correcting process. CIBA Found. Symp. 55, 229–246 [DOI] [PubMed] [Google Scholar]
- 11. Younger L. R., King J., Steiner D. F. (1966) Hepatic proliferative response to insulin in severe alloxan diabetes. Cancer Res. 26, 1408–1414 [PubMed] [Google Scholar]
- 12. Michael M. D., Kulkarni R. N., Postic C., Previs S. F., Shulman G. I., Magnuson M. A., Kahn C. R. (2000) Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87–97 [PubMed] [Google Scholar]
- 13. Steiner D. F. (1966) Insulin and the regulation of hepatic biosynthetic activity. Vitam. Horm. 24, 1–61 [DOI] [PubMed] [Google Scholar]
- 14. Davoren P. R. (1962) The isolation of insulin from a single cat pancreas. Biochim. Biophys. Acta 63, 150–153 [DOI] [PubMed] [Google Scholar]
- 15. Steiner D. F., Oyer P. C. (1967) The biosynthesis of insulin and a probable precursor of insulin by a human islet cell adenoma. Proc. Natl. Acad. Sci. U.S.A. 57, 473–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steiner D. F., Cunningham D., Spigelman L., Aten B. (1967) Insulin biosynthesis evidence for a precursor. Science 157, 697–700 [DOI] [PubMed] [Google Scholar]
- 17. Wang S. S., Carpenter F. H. (1965) A compositional assay for insulin applied to a search for “proinsulin.” J. Biol. Chem. 240, 1619–1625 [PubMed] [Google Scholar]
- 18. Steiner D. F., Clark J. L., Nolan C., Rubenstein A. H., Margoliash E., Aten B., Oyer P. E. (1969) Proinsulin and the biosynthesis of insulin. Recent Prog. Horm. Res. 25, 207–282 [DOI] [PubMed] [Google Scholar]
- 19. Steiner D. F., Clark J. L., Nolan C., Rubenstein A. H., Margoliash E., Melani F., Oyer P. E. (1970) The biosynthesis of insulin and some speculations regarding the pathogenesis of human diabetes. In: The Pathogenesis of Diabetes Mellitus. Nobel Symposium 13 (Cerasi E., Luft R. eds) pp. 57–80, Almqvist and Wiksell International, Stockholm [Google Scholar]
- 20. Jamieson J. D., Palade G. E. (1967) Intracellular transport of secretory proteins in the pancreatic exocrine cell. I. Role of the peripheral elements of the Golgi complex. II. Transport to condensing vacuoles and zymogen granules. J. Cell Biol. 34, 577–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Orci L., Ravazzola M., Amherdt M., Madsen O., Vassalli J. D., Perrelet A. (1985) Direct identification of prohormone conversion site in insulin-secreting cells. Cell 42, 671–681 [DOI] [PubMed] [Google Scholar]
- 22. Clark J. L., Steiner D. F. (1969) Insulin biosynthesis in the rat. Demonstration of two proinsulins. Proc. Natl. Acad. Sci. U.S.A. 62, 278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Steiner D. F., Hallund O., Rubenstein A., Cho S., Bayliss C. (1968) Isolation and properties of proinsulin, intermediate forms, and other minor components from crystalline bovine insulin. Diabetes 17, 725–736 [DOI] [PubMed] [Google Scholar]
- 24. Steiner D. F., Cho S., Oyer P. E., Terris S., Peterson J. D., Rubenstein A. H. (1971) Isolation and characterization of proinsulin C-peptide from bovine pancreas. J. Biol. Chem. 246, 1365–1374 [PubMed] [Google Scholar]
- 25. Chance R. E., Ellis R. M., Bromer W. W. (1968) Porcine proinsulin. Characterization and amino acid sequence. Science 161, 165. [DOI] [PubMed] [Google Scholar]
- 26. Nolan C., Margoliash E., Peterson J. D., Steiner D. F. (1971) Structure of bovine proinsulin. J. Biol. Chem. 246, 2780–2795 [PubMed] [Google Scholar]
- 27. Rubenstein A. H., Clark J. L., Melani F., Steiner D. F. (1969) Secretion of proinsulin C-peptide by pancreatic B cells and its circulation in blood. Nature 224, 697–699 [Google Scholar]
- 28. Oyer P. E., Cho S., Peterson J. D., Steiner D. F. (1971) Studies on human proinsulin. Isolation and amino acid sequence of the human pancreatic C-peptide. J. Biol. Chem. 246, 1375–1386 [PubMed] [Google Scholar]
- 29. Rubenstein A. H., Cho S., Steiner D. F. (1968) Evidence for proinsulin in human urine and serum. Lancet 291, 1353–1355 [DOI] [PubMed] [Google Scholar]
- 30. Melani F., Rubenstein A. H., Oyer P. E., Steiner D. F. (1970) Identification of proinsulin and C-peptide in human serum by a specific immunoassay. Proc. Natl. Acad. Sci. U.S.A. 67, 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polonsky K. S., Rubenstein A. H. (1986) Current approaches to measurement of insulin secretion. Diabetes Metab. Rev. 2, 315–329 [DOI] [PubMed] [Google Scholar]
- 32. Palmer J. P., Fleming G. A., Greenbaum C. J., Herold K. C., Jansa L. D, Kolb H., Lachin J. M., Polonsky K. S., Pozzilli P., Skyler J. S., Steffes M. W. (2004) C-peptide is the appropriate outcome measure for type 1 diabetes clinical trials to preserve beta-cell function. Report of an ADA workshop, 21–22 October 2001. Diabetes 53, 250–264 [DOI] [PubMed] [Google Scholar]
- 33. Steiner D. F., Clark J. L. (1968) The spontaneous reoxidation of reduced beef and rat proinsulins. Proc. Natl. Acad. Sci. U.S.A. 60, 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Givol D., Delorenzo F., Goldberger R. F., Anfinsen C. B. (1965) Disulfide interchange and the three-dimensional structure of proteins. Proc. Natl. Acad. Sci. U.S.A. 53, 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Humbel R. E. (1965) Biosynthesis of the two chains of insulin. Proc. Natl. Acad. Sci. U.S.A. 53, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Efrat S., Linde S., Kofod H., Spector D., Delannoy M., Grant S., Hanahan D., Baekkeskov S. (1988) Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc. Natl. Acad. Sci. U.S.A. 85, 9037–9041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sachs H., Takabatake Y. (1964) Evidence for a precursor in vasopressin biosynthesis. Endocrinology 75, 943–948 [DOI] [PubMed] [Google Scholar]
- 38. Douglass J., Civelli O., Herbert E. (1984) Polyprotein gene expression. Generation of diversity of neuroendocrine peptides. Annu. Rev. Biochem. 53, 665–715 [DOI] [PubMed] [Google Scholar]
- 39. Zhou A., Webb G., Zhu X., Steiner D. F. (1999) Proteolytic processing in the secretory pathway. J. Biol. Chem. 274, 20745–20748 [DOI] [PubMed] [Google Scholar]
- 40. Blobel G., Dobberstein B. (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67, 835–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blobel G., Dobberstein B. (1975) Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67, 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan S. J., Keim P., Steiner D. F. (1976) Cell-free synthesis of rat preproinsulins. Characterization and partial amino acid sequence determination. Proc. Natl. Acad. Sci. U.S.A. 73, 1964–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shields D., Blobel G. (1977) Cell-free synthesis of fish preproinsulin and processing by heterologous mammalian microsomal membranes. Proc. Natl. Acad. Sci. U.S.A. 74, 2059–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Terris S., Steiner D. F. (1975) Binding and degradation of 125I-insulin by rat hepatocytes. J. Biol. Chem. 250, 8389–8398 [PubMed] [Google Scholar]
- 45. Terris S., Steiner D. F. (1976) Retention and degradation of 125I-Insulin by perfused rat livers. J. Clin. Invest. 57, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terris S., Hofmann C., Steiner D. F. (1979) Mode of uptake and degradation of 125I-labelled insulin by isolated hepatocytes and H4 hepatoma cells. Can. J. Biochem. 57, 459–468 [DOI] [PubMed] [Google Scholar]
- 47. Shoelson S., Haneda M., Blix P., Nanjo A., Sanke T., Inouye K., Steiner D., Rubenstein A., Tager H. (1983) Three mutant insulins in man. Nature 302, 540–543 [DOI] [PubMed] [Google Scholar]
- 48. Støy J., Edghill E. L., Flanagan S. E., Ye H., Paz V. P., Pluzhnikov A., Below J. E., Hayes M. G., Cox N. J., Lipkind G. M., Lipton R. B., Greeley S. A., Patch A. M., Ellard S., Steiner D. F., Hattersley A. T., Philipson L. H., Bell G. I. (2007) Insulin gene mutations as a cause of permanent neonatal diabetes. Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steiner D. F., Park S. Y., Støy J., Philipson L. H, Bell G. I. (2009) A brief perspective on insulin production. Diabetes Obes. Metab. 11, 189–196 [DOI] [PubMed] [Google Scholar]
- 50. Smith B. J., Huang K., Kong G., Chan S. J., Nakagawa S., Menting J. G., Hu S. Q., Whittaker J., Steiner D. F., Katsoyannis P. G., Ward C. W., Weiss M. A., Lawrence M. C. (2010) Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc. Natl. Acad. Sci. U.S.A. 107, 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurose T., Pashmforoush M., Yoshimasa Y., Carroll R., Schwartz G. P., Burke G. T., Katsoyannis P. G., Steiner D. F. (1994) Cross-linking of a B25 azidophenylalanine insulin derivative to the carboxyl-terminal region of the alpha-subunit of the insulin receptor. Identification of a new insulin-binding domain in the insulin receptor. J. Biol. Chem. 269, 29190–29197 [PubMed] [Google Scholar]
- 52. Wang S. S., Carpenter F. H. (1969) Kinetics of the tryptic hydrolysis of zinc-free bovine insulin. J. Biol. Chem. 244, 5537–5543 [PubMed] [Google Scholar]
- 53. Kemmler W., Peterson J. D., Steiner D. F. (1971) Studies on the conversion of proinsulin to insulin. I. Conversion in vitro with trypsin and carboxypeptidase B. J. Biol. Chem. 246, 6786–6791 [PubMed] [Google Scholar]
- 54. Kemmler W., Steiner D. F., Borg J. (1973) Studies on the conversion of proinsulin to insulin. III. Studies in vitro with a crude secretion granule fraction isolated from rat islets of Langerhans. J. Biol. Chem. 248, 4544–4551 [PubMed] [Google Scholar]
- 55. Chance R. E., Frank B. H. (1993) Research, development, production, and safety of biosynthetic human insulin. Diabetes Care 16, Suppl. 3, 133–142 [DOI] [PubMed] [Google Scholar]
- 56. Zühlke H., Steiner D. F., Lernmark A., Lipsey C. (1976) Carboxypeptidase B-like and trypsin-like activities in isolated rat pancreatic islets. CIBA Found. Symp. 41, 183–195 [DOI] [PubMed] [Google Scholar]
- 57. Fricker L. D., Evans C. J., Esch F. S., Herbert E. (1986) Cloning and sequence analysis of cDNA for bovine carboxypeptidase E. Nature 323, 461–464 [DOI] [PubMed] [Google Scholar]
- 58. Naggert J. K., Fricker L. D., Varlamov O., Nishina P. M., Rouille Y., Steiner D. F., Carroll R. J., Paigen B. J., Leiter E. H. (1995) Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase E mutation which reduces enzyme activity. Nat. Genet. 10, 135–142 [DOI] [PubMed] [Google Scholar]
- 59. Day R., Lazure C., Basak A., Boudreault A., Limperis P., Dong W., Lindberg I. (1998) Prodynorphin processing by proprotein convertase 2. Cleavage at single basic residues and enhanced processing in the presence of carboxypeptidase activity. J. Biol. Chem. 273, 829–836 [DOI] [PubMed] [Google Scholar]
- 60. Docherty K., Carroll R., Steiner D. F. (1983) Identification of a 31,500 molecular weight islet cell protease as cathepsin B. Proc. Natl. Acad. Sci. U.S.A. 80, 3245–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuliawat R., Klumperman J., Ludwig T., Arvan P. (1997) Differential sorting of lysosomal enzymes out of the regulated secretory pathway in pancreatic beta-cells. J. Cell Biol. 137, 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Julius D., Brake A., Blair L., Kunisawa R., Thorner J. (1984) Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha factor. Cell 37, 1075–1089 [DOI] [PubMed] [Google Scholar]
- 63. Mizuno K., Nakamura T., Ohshima T., Tanaka S., Matsuo H. (1988) Yeast Kex2 gene encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem. Biophys. Res. Commun. 156, 246–254 [DOI] [PubMed] [Google Scholar]
- 64. Smeekens S. P., Steiner D. F. (1990) Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J. Biol. Chem. 265, 2997–3000 [PubMed] [Google Scholar]
- 65. Smeekens S. P., Avruch A. S., LaMendola J., Chan S. J., Steiner D. F. (1991) Identification of a cDNA encoding a second putative prohormone convertase related to PC2 in AtT20 cells and islets of Langerhans. Proc. Natl. Acad. Sci. U.S.A. 88, 340–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Steiner D. F., Smeekens S. P., Ohagi S., Chan S. J. (1992) The new enzymology of precursor processing endoproteases. J. Biol. Chem. 267, 23435–23438 [PubMed] [Google Scholar]
- 67. Davidson H. W., Rhodes C. J., Hutton J. C. (1988) Intraorganellar calcium and pH control proinsulin cleavage in the pancreatic beta cell via two distinct site-specific endopeptidases. Nature 333, 93–96 [DOI] [PubMed] [Google Scholar]
- 68. Fuller R. S., Brake A. J., Thorner J. (1989) Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science 246, 482–486 [DOI] [PubMed] [Google Scholar]
- 69. Seidah N. G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008) The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol. 40, 1111–1125 [DOI] [PubMed] [Google Scholar]
- 70. Docherty K., Steiner D. F. (1982) Post-translational proteolysis in polypeptide hormone biosynthesis. Annu. Rev. Physiol. 44, 625–638 [DOI] [PubMed] [Google Scholar]
- 71. Nolan C., Novoa W. B., Krebs E. G., Fischer E. H. (1964) Further studies on the site phosphorylated in the phosphorylase B to A reaction. Biochemistry 3, 542–551 [DOI] [PubMed] [Google Scholar]
- 72. Peinado J. R., Laurent V., Lee S. N., Peng B. W., Pintar J. E., Steiner D. F., Lindberg I. (2005) Strain-dependent influences on the hypothalamo-pituitary-adrenal axis profoundly affect the 7B2 and PC2 null phenotypes. Endocrinology 146, 3438–3444 [DOI] [PubMed] [Google Scholar]
- 73. Zhu X., Zhou A., Dey A., Norrbom C., Carroll R., Zhang C., Laurent V., Lindberg I., Ugleholdt R., Holst J. J., Steiner D. F. (2002) Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc. Natl. Acad. Sci. U.S.A. 99, 10293–10298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Scamuffa N., Calvo F., Chrétien M., Seidah N. G., Khatib A. M. (2006) Proprotein convertases. Lessons from knockouts. FASEB J. 20, 1954–1963 [DOI] [PubMed] [Google Scholar]
- 75. Czyzyk T. A., Ning Y., Hsu M. S., Peng B., Mains R. E., Eipper B. A., Pintar J. E. (2005) Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev. Biol. 287, 301–313 [DOI] [PubMed] [Google Scholar]
- 76. Louagie E., Taylor N. A., Flamez D., Roebroek A. J., Bright N. A., Meulemans S., Quintens R., Herrera P. L., Schuit F., Van de Ven W. J., Creemers J. W. (2008) Role of furin in granular acidification in the endocrine pancreas. Identification of the V-ATPase subunit Ac45 as a candidate substrate. Proc. Natl. Acad. Sci. U.S.A. 105, 12319–12324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang X., Pan H., Peng B., Steiner D. F., Pintar J. E., Fricker L. D. (2010) Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J. Neurochem. 112, 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Remacle A. G., Shiryaev S. A., Oh E. S., Cieplak P., Srinivasan A., Wei G., Liddington R. C., Ratnikov B. I., Parent A., Desjardins R., Day R., Smith J. W., Lebl M., Strongin A. Y. (2008) Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J. Biol. Chem. 283, 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhu X., Orci L., Carroll R., Norrbom C., Ravazzola M., Steiner D. F. (2002) Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc. Natl. Acad. Sci. U.S.A. 99, 10299–10304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cameron A., Apletalina E. V., Lindberg I. (2001) The enzymology of PC1 and PC2. In: The Enzymes, Vol. XXII, Academic Press, New York [Google Scholar]
- 81. Nishi M., Sanke T., Nagamatsu S., Bell G. I., Steiner D. F. (1990) Islet amyloid polypeptide. A new beta 61476 cell secretory product related to islet amyloid deposits. J. Biol. Chem. 265, 4173–4176 [PubMed] [Google Scholar]
- 82. Marzban L., Rhodes C. J., Steiner D. F., Haataja L., Halban P. A., Verchere C. B. (2006) Impaired NH2-terminal processing of human proislet amyloid polypeptide by the prohormone convertase PC2 leads to amyloid formation and cell death. Diabetes 55, 2192–2201 [DOI] [PubMed] [Google Scholar]
- 83. Chan S. J., Cao Q. P., Steiner D. F. (1990) Evolution of the insulin superfamily. Cloning of a hybrid insulin/insulin-like growth factor cDNA from amphioxus. Proc. Natl. Acad. Sci. U.S.A. 87, 9319–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Furuta M., Carroll R., Martin S., Swift H. H., Ravazzola M., Orci L., Steiner D. F. (1998) Incomplete processing of proinsulin to insulin accompanied by elevation of des-31, 32 proinsulin intermediates in islets of mice lacking active PC2. (1998) J. Biol. Chem. 273, 3431–3437 [DOI] [PubMed] [Google Scholar]