Abstract
Sandalwood oil is one of the worlds most highly prized fragrances. To identify the genes and encoded enzymes responsible for santalene biosynthesis, we cloned and characterized three orthologous terpene synthase (TPS) genes SaSSy, SauSSy, and SspiSSy from three divergent sandalwood species; Santalum album, S. austrocaledonicum, and S. spicatum, respectively. The encoded enzymes catalyze the formation of α-, β-, epi-β-santalene, and α-exo-bergamotene from (E,E)-farnesyl diphosphate (E,E-FPP). Recombinant SaSSy was additionally tested with (Z,Z)-farnesyl diphosphate (Z,Z-FPP) and remarkably, found to produce a mixture of α-endo-bergamotene, α-santalene, (Z)-β-farnesene, epi-β-santalene, and β-santalene. Additional cDNAs that encode bisabolene/bisabolol synthases were also cloned and functionally characterized from these three species. Both the santalene synthases and the bisabolene/bisabolol synthases reside in the TPS-b phylogenetic clade, which is more commonly associated with angiosperm monoterpene synthases. An orthologous set of TPS-a synthases responsible for formation of macrocyclic and bicyclic sesquiterpenes were characterized. Strict functionality and limited sequence divergence in the santalene and bisabolene synthases are in contrast to the TPS-a synthases, suggesting these compounds have played a significant role in the evolution of the Santalum genus.
Keywords: Enzyme Kinetics, Enzyme Mechanisms, Evolution, Metabolism, Plant, Santalum, Gas Chromatography, Phylogenetics, Santalene, Terpene Synthase
Introduction
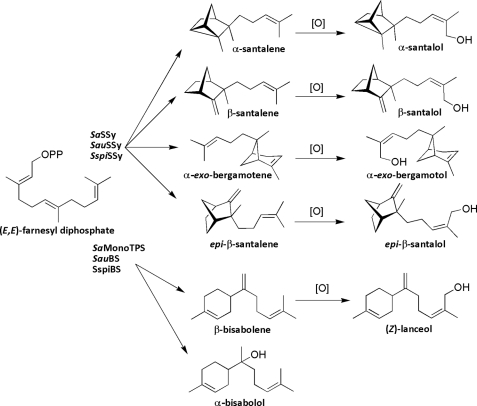
Santalum album L. is a slow-growing hemi-parasitic tree, which has long been exploited for its fragrant heartwood. Other species such as S. spicatum, native to the arid and semi-arid regions of Western Australia (WA) and S. austrocaledonicum from Vanuatu and New Caledonia, have also contributed substantially to the fragrance market (1). Unsustainable demand for sandalwood has led to the establishment of plantations to add supply. Knowledge of the molecular and physiological underpinnings of sandalwood oil biosynthesis will advance plantation development through improved management and selection. Sandalwood heartwood contains a complex mixture of sesquiterpene olefins and alcohols (Fig. 1) with (+)-α-santalene, (−)-β-santalene, (−)-α-exo-bergamotene, (+)-epi-β-santalene, and β-bisabolene frequently representing about 1–2% by weight of oil (2, 3). The more odor intensive compounds, α- and β-santalol, α-exo-bergamotol, and epi-β-santalol, as well as (Z)-lanceol and α-bisabolol make up the bulk of S. album oil, but are often lower and more variable in S. austrocaledonicum (4) and S. spicatum (5). The compounds found in sandalwood oil may play an important role in warding off pathogens, as sesquiterpenoids have been implicated in plant pathogen defense strategies (6). Santalol biosynthesis is proposed to proceed by multiple Wagner-Meerwein rearrangements of transoid farnesyl diphosphate, (E,E)-FPP,3 followed by oxidation at C12, probably via a cytochrome P450 (Fig. 1). Co-occurrence patterns of the santalenes and bergamotene, as well as other olefins and bisabolol, indicated that multi-product TPS enzymes may be responsible for the production of several sandalwood oil components (2). Multiple product formation from terpene synthase (TPS) enzymes in S. album was recently confirmed by characterization of two TPSs, however these were not responsible for santalene biosynthesis (7). The commercially exploited sandalwoods possess unique chemical phenotypes (3), whereas others such as S. accuminatum and S. murrayanum, endemic to southwestern WA, produce little or no oil (8). Despite this variation, the santalenes and their hydroxylated equivalents remain key components. To better understand essential oil biosynthesis as well as the molecular origins of chemical diversity in the Santalum genus, we cloned and functionally characterized several TPS cDNAs from three divergent oil-producing Santalum species, S. album, S. austrocaledonicum, and S. spicatum. We also compared genomic TPS sequences of the three commercial Santalum species to those of S. murranyanum to test if the absence of, or open reading frame mutations in the genes accounted for the oil-deficient phenotype of this species.
FIGURE 1.
Biosynthesis of sesquiterpenes in sandalwood commences with the TPS-catalyzed rearrangements of farnesyl diphosphate. Specific oxidation at C12 is proposed to occur via a cytochrome P450 enzyme.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All reagents, solvents, antibiotics, cloning kits, modifying enzymes, and precursor chemicals were purchased from commercial sources. (E,E)-FPP and geranyl diphosphate (GPP) were from Sigma. (Z,Z)-FPP was synthesized using standard methods (9, 10) and products verified by GC-MS, NMR, and IR spectroscopy. Monoterpene standards were from an in-house collection of commercially available standards. An olefin fraction of sandalwood oil yielded a santalene standard (see below under GC-MS analysis and product identification).
Plant Material Collection and RNA Extraction
Several 25 mm holes were drilled into the lower stems of mature S. album, S. austrocaledonicum, S. spicatum, and S. murrayanum trees growing on land managed by the Forest Products Commission of Western Australia. Wood shavings from the heartwood-sapwood transition zone were collected and frozen immediately in liquid nitrogen. The samples were transported to the lab where RNA was extracted from 10 g of tissue using an established protocol (11). After precipitation by LiCl, RNA was stored at −80 °C until needed for reverse transcription and RACE. S. album RNA was transported to UBC Vancouver, Canada for cDNA library construction.
Santalum album cDNA Library Construction
Xylem total RNA from S. album (1.4 μg) was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and a cDNA library was constructed using the SMART-Creator kit with the pDNR-LIB vector (Clontech). The ligation mixture was transformed by electroporation into 25 μl of phage resistant electrocompetent Escherichia coli cells and Sangar-sequenced at the Michael Smith Genome Sciences Centre, Vancouver, Canada. Reads were assembled using the CAP3 program with default settings. The sequences (∼6000 unique reads) were compared with the GenBankTM database for key specialized metabolism genes, particularly prenyltransferases and TPS genes. The following sequences have been deposited into the GenBankTM database: SaSSy (HQ343276); SauSSy (HQ343277); SspiSSy (HQ343278); SauBS (HQ343279); SspiBS (HQ343280); SauSesquiTPS (HQ343281); SspiSesquiTPS (HQ343282), SaSTPS gDNA (JF746807); SauSTPS gDNA (JF746808); SspiSTPS gDNA (JF746809); SmSTPS gDNA (JF746810); SmSSy (JF746811); SaSSy (JF746812); SauBS gDNA (JF746813); SmBS (JF746814); SaMonoTPS1 gDNA (JF746815).
TPS Gene Discovery and RACE
cDNA was generated for S. austrocaledonicum, S. spicatum, and S. murrayanum in the same manner as before, except the cDNA was used directly as template for PCR. Primers based on the open reading frame (ORF) of previously identified S. album TPS genes were used for amplification (7) (supplemental Fig. S1). Where products could not be amplified, 5′- and 3′-RACE using the Clontech SMARTerTM kit was used to obtain the respective untranslated regions for more specific primer design. SaSesquiTPS1 gene orthologs were amplified in two rounds using a nested primer approach. All products were first cloned into a high-copy storage vector (TOPO Zero Blunt, Invitrogen, or pJET1.2, Fermentas) for sequencing before being subcloned into pET28b(+). Several individuals of S. album and S. spicatum were studied to examine if polymorphisms were present in the ORFs across populations of these two species. No TPS genes could be amplified from S. murrayanum cDNA, although this could be due to exceedingly low transcript abundance. Genomic DNA sequences for all three TPS genes were also cloned and sequenced for all three Santalum species. The same ORF primers used for successful cDNA amplifications were used on genomic DNA extracted from the same individuals from which RNA extractions were performed. These larger gDNA fragments (3–4 kb) were cloned into pJET1.2 vectors and sent for sequencing (Macrogen, Korea). Genomic DNA sequences of TPS genes from the three oil-bearing species were spliced in-silico with introns following the usual 3′ n/GT and 5′ AG/n pattern (12). Santalum murrayanum intron-exon patterns were determined by comparing the gDNA sequences to cDNAs of S. spicatum.
Phylogenetic Analysis and Alignment of TPS Genes
Sequences were aligned using ClustalX 2.1.0, trees were constructed using Phylip (13) and visualized with the Phylodraw 0.8 program. Sequences used in the phylogenetic analysis are listed in full under supplemental Fig. S2. Genomic DNA sequences of TPS genes from the three oil-bearing species were spliced in-silico with introns following the usual 3′ n/GT and 5′ AG/n pattern (12). S. murrayanum intron-exon patterns were determined by comparing the gDNA sequences to cDNA sequences of S. spicatum.
Bacterial Expression and Protein Isolation
TPS genes were cloned into the pET28b(+) expression vector (Novagen, San Diego CA) with a polyhistidine tag in-frame. Depending on the restriction sites available, the His6 tag was either N-terminal or C-terminal. Primers with appropriate restriction sites (supplemental Fig. S1) were used to amplify each gene and cloned into the pET28b(+) vector. Vectors containing the TPS genes were transformed into chemically competent C41 E. coli cells (Avidis, Saint-Beauzire, France) containing the pRARE 2 plasmid isolated from Rosetta 2 competent cells (Novagen). Colonies were grown on LB plates containing kanamycin and chloramphenicol (50 μg ml−1). Three independent colonies were picked and grown in a shaker overnight at 37 °C in 5 ml of LB with the same antibiotics and this culture was used to inoculate 400 ml of Terrific Broth. Cell suspensions were grown at 37 °C with shaking until the A600 = 0.8 and induced with isopropyl-β-d-thiogalacto-pyranoside (IPTG) to a final concentration of 0.2 mm, and shaken overnight at 16 °C. Cell suspensions were centrifuged at 4 °C and pellets (∼1 g) were frozen at −80 °C for future use. Cell pellets were resuspended in 5 ml of lysis buffer containing 1 mg ml−1 lysozyme, 1 mm MgCl2, 5 mm DTT, 0.01 mg ml−1 DNAse1, and RNAse1, 100 μl of protease inhibitor mixture (Sigma) and made in His-trap binding buffer (20 mm Na2HPO4, pH 7.4, 500 mm NaCl, 30 mm imidazole, pH 7.4)., Cells were stirred thoroughly on ice with a glass rod for 30 min followed by homogenized using a high pressure cell cruncher. The lysate was centrifuged at 12,000 × g at 4 °C for 1.25 h before being decanted. Cleared lysate (∼12 ml) was purified using Ni2+ affinity chromatography spin-columns (GE Healthcare) and eluted in 600 μl of elution buffer (20 mm Na2HPO4 pH 7.4, 500 mm NaCl, 500 mm imidazole, pH 7.4). The eluted protein was desalted on a PD-10 desalting column (GE Healthcare) using 25 mm 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) pH 7.4, 10% (v/v) glycerol and 100 mm KCl. Approximately 1-ml fractions from a 3.5 ml elution were collected. Protein concentrations were determined using a NanoDrop spectrophotometer with extinction coefficients calculated by amino acid composition (14). SDS-PAGE followed by Coomassie Blue staining visualized the purified recombinant proteins (supplemental Fig. S3).
Enzyme Functional Characterization and Kinetic Assays
Enzyme assays for all recombinant proteins were done in triplicate using the GC vial method described by O'Maille et al. (15). For enzyme assays where only product identification was assessed, 10 μg of protein was used in a final volume of 500 μl of reaction buffer (25 mm HEPES, 10% (v/v) glycerol, 5 mm DTT, and 10 mm of either Mg2+ or Mn2+. Substrates (FPP and GPP) were added to a final reaction concentration of 100 μm. Vials were overlaid with 500 μl of hexane to trap volatile products and incubated at 30 °C for 2 h. Mixtures were vortexed for 1 min to extract all volatiles and the vials were centrifuged to separate the organic layer. For determination of steady-state enzyme kinetic constants, conditions were as described previously except the enzyme concentration was kept at 10 nm. Substrate concentrations ranged from 1 μm to 100 μm, and reactions were incubated at 30 °C for exactly 5 min. Critically, reactions were quenched at 5 min by the addition of 500 μl 0.5 m EDTA, pH 8.0 and vortexed, then stored at −80 °C immediately.
In Vivo Santalene Production in E. coli
An approach similar to the previously published method for diterpene production (16) was used for in vivo production of santalenes. The SaFPPS gene was identified in the xylem EST library (GenBankTM accession no. HQ343283) and was amplified from cDNA using primers with Nco1 and Not1 ends amenable to pCDFDuet-1 vector multiple cloning site 1. Likewise, SaSSy was amplified with Nde1 and Kpn1 sites adapted for use with the In-Fusion cloning system (Clontech) and cloned into the second multiple cloning site (supplemental Fig. S1). The dual expression vector construct, pCDFDuet-1:SaFPPS:SaSSy, as well as an empty pCDFDuet-1 vector control were transformed into chemically competent C41(DE3) cells and grown on 50 μg of ml−1 streptomycin selective media. Individual colonies were grown in 5 ml overnight cultures, and these were used to inoculate 200 ml shaking cultures of Terrific Broth. As cells were approaching the log-phase of growth (A600 = 0.6) at 37 °C the incubator was cooled to 20 °C for 1 h before inducing with IPTG at a final concentration of 0.5 mm. Cultures were shaken for a further 16 h before being centrifuged to pellet the cells. Cell pellets were lysed with 0.2 m NaOH and neutralized with acetic acid before being extracted twice with hexanes and reduced by evaporation. Media was extracted twice with 100 ml of hexane and these fractions were reduced by rotary evaporation to yield a thin oily residue. The residues were resuspended in 2 ml of hexane and analyzed by GC-MS using conditions described below.
GC-MS Analysis and Product Identification
Product mixtures were analyzed by GC-MS in scan mode for product identification. A standard containing the three santalenes and α-exo-bergamotene was prepared by flash chromatography of 2 ml of neat S. album oil over silica and eluted in hexane. A final yield of 25 mg was resuspended in EtOH, and purity was confirmed by GC. Monoterpene identification was aided through an in-house standard collection. All mass spectra were compared with the NIST 2005 library and the literature (17–23). The Kovat retention indices (KIs) were determined for all compounds (supplemental Figs. S4, S7, S10) using an n-alkane standard and compared with the literature wherever DB-WAX or similar phase column data were available (17–21, 23, 24). Where reliable retention data and pure standards were not available, a combination of mass spectral and retention properties was used to infer the candidate compound. GC-MS was performed on a Shimadzu GC2010 with a DB-WAX column and He as carrier gas. Conditions were as follows: Injector 200 °C, MS interface 240 °C, ion source 200 °C, Oven program: 40 °C for 3 min, then 8 °C min−1 to 180 °C, held 5 min, then 10 °C min−1 to 220 °C, held 10 min. Solvent cut time was set to 5 min. For product identification, total ion monitoring was used, scanning from m/z 41 to m/z 250. Incubations with (Z,Z)-FPP were analyzed on a HP5890 with a DB-WAX column and an initial oven temperature of 40 °C then ramped at 10 °C min−1 to 230 °C and held for 20 min. For kinetic assays, single ion monitoring (SIM) of the sesquiterpene base ions m/z 91, 93, and 94 were used. Likewise monoterpene base ions (m/z 69, 71, and 93) were monitored for GPP assays. An internal standard (isobutyl benzene, 30 μm) was added to the hexane used to overlay each reaction. Detector response factors were calculated based on the santalene standard which was prepared earlier. Product losses due to extraction inefficiency was accounted for by first adding the standard to assay buffer as an EtOH stock and extracting into the hexane layer as per sample assays.
RESULTS
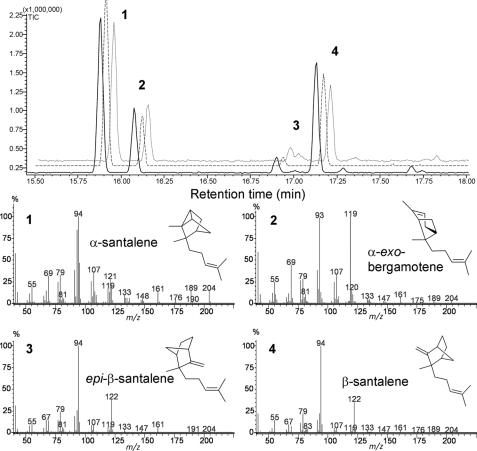
A full-length cDNA, SaSSy (S. album santalene synthase) homologous to previously reported angiosperm TPS-b group genes (25) was identified in the S. album xylem EST library and cloned. It encoded a 569 amino acid protein with 56% identity to SamonoTPS1 (7) and appeared to lack an N-terminal transit peptide. Upon heterologous expression in E. coli, the affinity-purified recombinant His6-tagged protein had a molecular mass of ∼66 kDa, similar to that of most monomeric TPS enzymes (supplemental Fig. S3). When incubated in the presence of 10 mm Mg2+ the enzyme converted (E,E)-FPP into α-, β-, and epi-β-santalene, α-exo-bergamotene, as well as traces of α- and β-farnesene (Fig. 2). Orthologous TPSs identified in two other oil-bearing species (SspiSSy from S. spicatum and SauSSy from S. austrocaledonicum) were also found to convert (E,E)-FPP into the santalenes in very similar proportions as SaSSy (Fig. 2). In all three orthologs, incubations with Mn2+ yielded mainly α-exo-bergamotene (supplemental S4). The larger metal ion likely distorts the active site, causing premature quenching of the bergamotyl carbocation intermediate. Each santalene synthase had an apparent Km of 1.4 (± 0.3) μm, indicating a biologically relevant, high affinity for (E,E)-FPP. Catalytic turnover rates were similar for all three enzymes, with a kcat for SaSSy of 0.34 s−1, 0.91 s−1 for SauSSy, and 2.6 s−1 for SpiSSy. All three santalene synthases produced linalool, geraniol, and terpineol along with traces of α-pinene and camphene when incubated with geranyl diphosphate (GPP) (supplemental Fig. S4). Although conversion of GPP did occur, a linear relationship existed between substrate concentration and V0, even at high (>100 μm) concentrations, rather than an asymptotic curve indicative of active site saturation. Monoterpenes have only been reported in sandalwood oil at very low concentrations (23, 26).
FIGURE 2.
GC-MS chromatogram of in vitro assays with recombinant santalene synthases; SaSSy (solid trace), SauSSy (dashed trace), and SspiSSy (dotted trace) using (E,E)-FPP as substrate. Peaks: 1) α-santalene, 2) α-exo-bergamotene, 3) epi-β-santalene, 4) β-santalene. Traces of α- and β-farnesene isomers are also found in the mixture. Mass spectral data of the main compounds are shown.
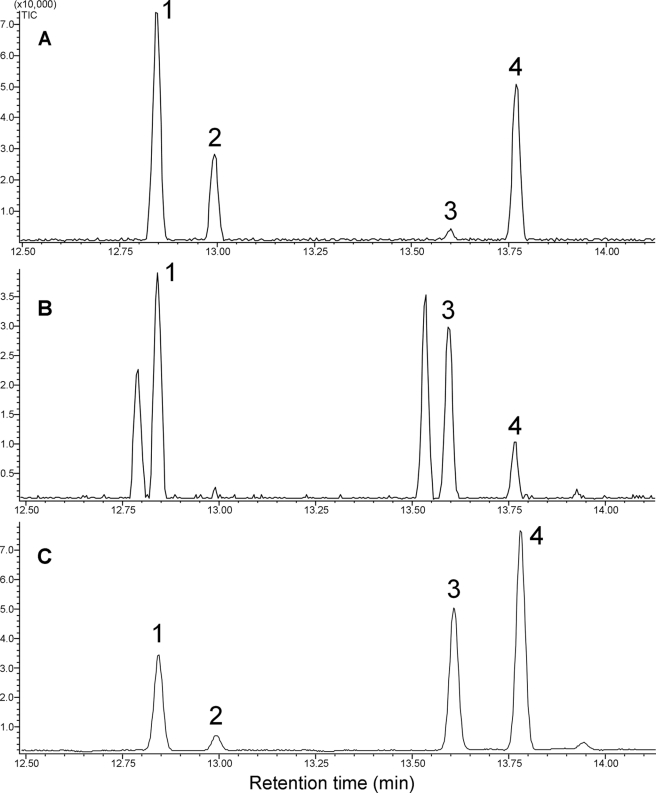
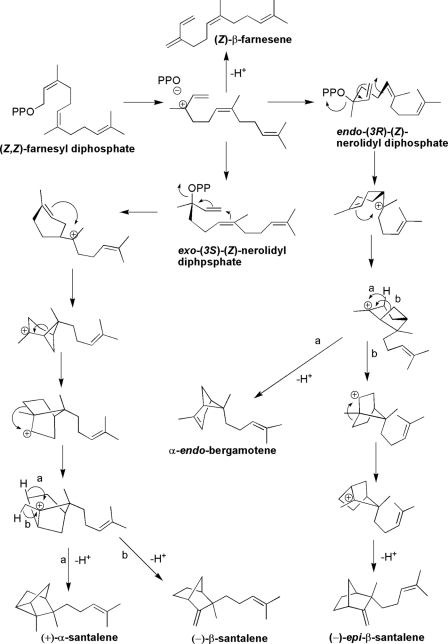
Incubations of SaSSy with (Z,Z)-FPP indicated the enzyme was also catalytically active on this isomer. In the presence of 10 mm Mg2+ (Z,Z)-FPP was converted into α-endo-bergamotene, α-santalene, (Z)-β-farnesene, epi-β-santalene, and β-santalene (Fig. 3A). The product profile resulting from incubations of SaSSy with (E,E)-FPP (Fig. 3B) resemble the authentic essential oil of S. album (Fig. 3C) more closely than that of the (Z,Z)-FPP incubations.
FIGURE 3.
GC-MS chromatogram of incubations of (Z,Z)-FPP with SaSSy (A), (E,E)-FPP with SaSSy (B), and the olefin fraction of authentic sandalwood oil for comparison (C). Peaks: 1) α-endo-bergamotene, 2) α-santalene, 3) α-exo-bergamotene, 4) (Z)-β-farnesene, 5) epi-β-santalene, 6) β-santalene, 7) (E)-β-farnesene.
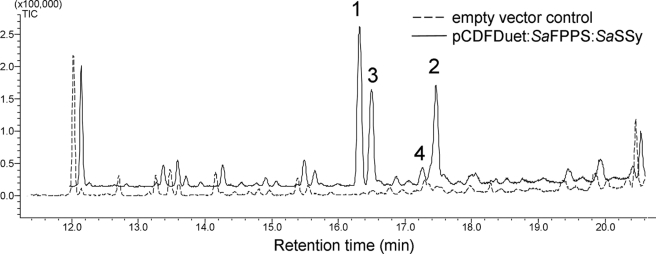
Continuity of santalene biosynthesis from FPP produced by a S. album FPP synthase (SaFPPS) was confirmed using an in vivo E. coli expression system. SaFPPS and SaSSy were cloned into the dual expression vector pCDFDuet-1 (Novagen) and transformed into C41 chemically competent cells. No detectable levels of the santalenes or bergamotene were found in the cell pellet extraction, or in the empty vector control extracts, but all four compounds were detected in hexane extracts of media from overnight cultures (Fig. 4). These results validated the in vivo activity of SaSSy and demonstrate the feasibility of metabolically engineering a santalene synthase into an appropriate host microorganism for in vivo production of santalenes using established methods (16, 27).
FIGURE 4.
In vivo production of santalenes in overnight E. coli cultures. Peaks: 1) α-santalene, 2) α-exo-bergamotene, 3) epi-β-santalene, 4) β-santalene.
To further explore the origins of chemical diversity in the genus Santalum, the orthologous TPS gene pair SauBS (S. austrocaledonicum β-bisabolene synthase) and SspiBS (S. spicatum α-bisabolol synthase) were cloned and characterized using cDNA as PCR template with primers originally developed from S. album (7). Recombinant SauBS produced almost exclusively β-bisabolene and only traces of α-bisabolol with (E,E)-FPP, while only limonene and terpineol were produced when incubated with GPP (supplemental Figs. S5 and S6). SspiBS produced a mixture of β-bisabolene and α-bisabolol, along with traces of α-bisabolene and farnesene isomers. The functions of these two enzymes were very similar to the previously identified SaMonoTPS1 from S. album which produced mostly β-bisabolene and traces of α-bisabolol from (E,E)-FPP, but could also convert GPP into monoterpenes analogous in structure to the bisabolenes (7). Extending our investigation of Santalum sesquiterpene synthases into the TPS-a subfamily, two orthologous cDNAs; SspiSesquiTPS and SauSesquiTPS, from S. spicatum and S. austrocaledonicum, respectively, were cloned and characterized. The translated amino acid sequences of these genes were similar to the previously characterized SaSesquiTPS1 from S. album (7) but the recombinant TPSs of these species yielded markedly different sesquiterpene profiles. SauSesquiTPS produced α-humulene and δ-cadinene, along with β-elemene, which is the thermal rearrangement product of germacrene A (28) and several other bicyclic sesquiterpenes when incubated with (E,E)-FPP (supplemental Fig. S7). The cadinenes may also be the result of heat-induced dehydration rearrangements (7). In contrast, SspiSesquiTPS produced only three main compounds; β-elemol (the thermal rearrangement product of hedycaryol (29)), guaiol and bulnesol (supplemental Fig. S8). Both enzymes produced only traces of linalool with GPP (supplemental Fig. S9).
Each set of orthologous TPS across the Santalum genus showed sequence homology to previously reported TPS with key domains being well conserved (supplemental Fig. S10). As with all angiosperm TPSs, the R(R/P)X8W motif implicated in prenyl diphosphate ionization (30) and the aspartate-rich divalent metal ion binding domain (31) (DDXXD) are present. Residues likely to be responsible for product specificity and substrate preference were identified in the α14 helix, and the α18-α19 helix turn respectively, based on those identified by Kamparinas et al. (32).
Genomic sequences of a set of Santalum TPS genes were compared with determine whether differences at the genomic level might be related to the chemotypic differences observed among oil producing (S. album, S. spicatum, S. austrocaledonicum) and oil-deficient (S. murrayanum) species. All four species have genomic copies of the target TPS genes (supplemental Fig. S11) although cDNAs could not be amplified from a xylem-derived RNA pool of S. murrayanum. The deduced amino acid sequences of the santalene synthase, bisabolene/bisabolol synthase, and TPS-a sesquiterpene synthase gDNAs in S. murrayanum are highly homologous to those of the functionally characterized TPS cDNAs of the other species (Fig. 7) and possibly encode functional TPS enzymes. Intron-exon structure of sandalwood TPS genes were typical of angiosperm TPS-a and TPS-b genes; each bearing 6 introns and 7 exons (12) (supplemental Fig. S11). Intron boundaries observed a 3′ ∼N▾GT, 5′ AG▾N∼ pattern. An ortholog of SauBS in S. murrayanum, labeled SmBS, showed a mutation encoding a stop codon in exon 3. We also cloned a variant SaSSy gDNA sequence from S. album with a 10-nucleotide deletion in exon 3, producing a frameshift and subsequent stop codon. No frame shifts or premature stop codons were present in the other TPS gDNAs sequenced.
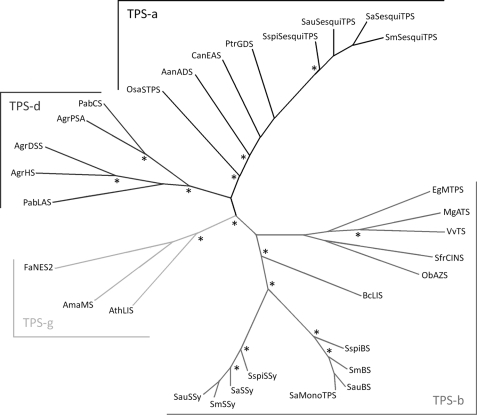
FIGURE 7.
Neighbor-joining phylogenetic tree of the TPSs compared in this study. The santalene synthase and bisabolene/bisabolol synthase orthologs group with the TPS-b clade. Abbreviations and GenBankTM accession numbers are listed under “Experimental Procedures.” The asterisk indicates a bootstrap value greater than 95%.
DISCUSSION
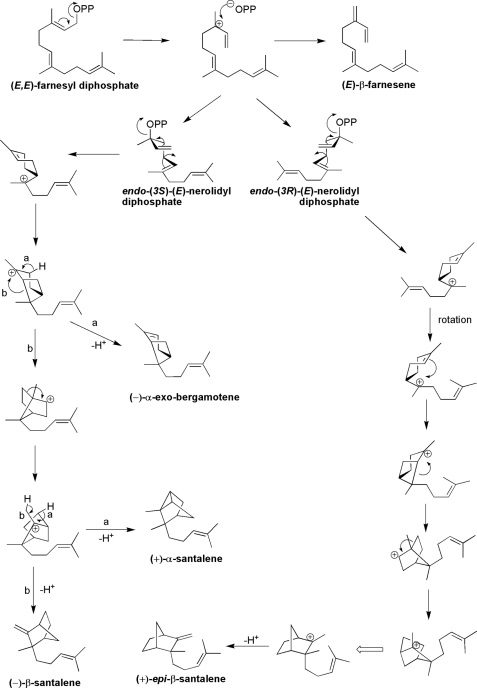
The genes and encoded enzymes for santalene biosynthesis have been isolated from three divergent sandalwood species. SaSSy, SauSSy, and SpiSSy are bona fide sesquiterpene synthases for santalene formation, despite being phylogenetically aligned with the TPS-b subfamily, which comprises mainly of angiosperm monoterpene synthases. Low Km values for each santalene synthase indicated a high affinity for (E,E)-FPP, while no active site saturation was apparent with GPP, indicating they are genuine sesquiterpene synthases. Vestigial activity with GPP suggests that these santalene synthases may have evolved from a monoterpene synthase ancestor through loss of the plastid target peptide and subsequent specialization of the active site for (E,E)-FPP. The putative ancestral enzyme which gave rise to SaSSy, SauSSy, and SspiSSy likely had latent plasticity in the active site, enabling it to accommodate both GPP and FPP. Similarly, it has been shown in the case of fruit flavor biosynthesis in wild and cultivated strawberry (33) that molecular evolution of the corresponding TPSs involved plasticity of the active site to accommodate alternative substrates when sub-cellular localization of the enzymes is changed through loss of a plastid target peptide. Most intriguing, SaSSy was able to convert (Z,Z)-FPP into α-, β-, and epi-β-santalenes and α- endo-bergamotene (Fig. 3). Sallaud et al. (34) discovered a santalene/bergamotene synthase in wild tomato, which was able to accommodate (Z,Z)-FPP but was unreactive toward (E,E)-FPP (Fig. 5). SaSSy is uniquely able to accommodate both transoid and cisoid isomers of FPP and surprisingly, produce a similar suite of compounds. These rearrangements are likely to proceed via the initial ionization of (Z,Z)-FPP into either (3S) or (3R)-nerolidyl diphosphate (Fig. 6). Cyclization may occur through an endo- conformation, resulting in α-endo-bergamotene and (−)-epi-β-santalene, while the exo-cyclization would lead to α- and β-santalene. Sandalwood oil contains traces of α-endo-bergamotene (26) thus it is plausible that the native enzyme is able to access both isomers of FPP in the cell. To our knowledge this level of plasticity in the active site of a TPS is unprecedented.
FIGURE 5.
Detailed mechanism explaining the formation of products resulting from incubations of SaSSy with (E,E)-farnesyl diphosphate in the presence of Mg2+.
FIGURE 6.
Detailed mechanism explaining the formation of products resulting from incubations of SaSSy with (Z,Z)-farnesyl diphosphate in the presence of Mg2+.
In addition to characterizing the santalene synthases, continuity of the biosynthetic pathway from FPP to the santalenes (Fig. 4) was demonstrated using an in vivo production system, highlighting the potential for metabolic engineering of microorganisms to produce precursors of valuable fragrance compounds of limited availability from plants. This system will also serve as a useful platform for exploring further downstream metabolic processes such as hydroxylation to the santalols.
Like the santalene synthases, the active sites of the bisabolene/bisabolol synthases are sufficiently plastic to accommodate the same series of carbocation rearrangements for both C10 and C15 substrates, as has been found with a cineole synthase mutant from Salvia fruticosa (32). Santalum spicatum oil contains variable amounts of α-bisabolol relative to other sesquiterpenoids (5) and allelic variation (35) in the ORF of SspiBS, particularly in the α14 helix may be partly responsible for the diversity of phenotypes observed across the distribution of S. spicatum. Within-species variation in the santalene synthase ORFs were not evident for the species studied here.
Phylogenetically, SauBS and SspiBS also cluster with the santalene synthases in the TPS-b group (Fig. 7). Thus, TPSs of two phylogenetic clades, TPS-a and TPS-b, contribute to the sesquiterpene profiles of sandalwood oils.
The TPS-a sesquiterpene synthases from the three oil-bearing Santalum species all produced markedly different combinations of sesquiterpenes (supplemental Figs. S7–S9). This was true to a lesser extent for the bisabolene/bisabolol synthases (supplemental Figs. S5 and S6) yet in both cases the diversity of function is in contrast to the highly conserved product profiles of the three orthologous santalene synthases (Fig. 2).
Genomic sequences revealed that all four species of Santalum studied here had copies of TPS genes, and that no frame shifts or mutations in the open reading frames were apparent (supplemental Fig. S11). Even the genome of S. murrayanum contains a complete ORF of the santalene synthase, despite possessing an oil-deficient phenotype. This suggests factors controlling the spatial or temporal patterns of TPS expression, rather than the absence of, or mutations in the ORFs of the genes themselves, are likely to be responsible for the low- or no-oil phenotype.
All oil producing species of Santalum, and even ancestral genera within the Santalaceae family (36), contain detectable levels of the santalenes, α-bergamotene, bisabolene, and bisabolol, and all species studied here contain complete genomic copies of the santalene synthase ORF. The uniformity of santalene product profiles of SaSSy, SauSSy, and SspiSSy is in contrast to the variety of compounds produced by the bisabolene/bisabolol synthases, and more so by the TPS-a group sesquiterpene synthases in the Santalum genus. The higher amino acid sequence identity of the santalene synthase orthologs (94–98%) compared with the TPS-a genes (89–93%) further support a genetic basis of functional conservation of santalene biosynthesis (supplemental Fig. S10). Function of SauBS and SaMonoTPS (7) is also well conserved between S. album and S. austrocaledonicum (99% identity) however SspiBS differs in both sequence identity (92%) and catalytic function (supplemental Fig. S2). These findings may suggest positive selection for santalene and to a lesser extent bisabolene biosynthesis in the Santalum genus, and possibly in the Santalaceae family more generally. As sesquiterpenes have been implicated in defense against pathogens (6) it is possible that selection pressure in the form of disease has allowed for the survival of populations with the santalene and bisabolene phenotype.
In conclusion, we have identified several terpene synthases responsible for the production of key fragrant compounds in three commercially exploited species of sandalwood. These findings are of great significance for the flavor and fragrance industry, as well as the growing plantation sandalwood industry.
Supplementary Material
Acknowledgments
We thank Len Norris, Karen Reid, Lina Madilao, Maria Li, Hanna Henderson, and Harpreet Kaur for technical support. We thank Drs. Christopher Keeling and Dawn Hall for critical comments and discussion.
This work was funded by the Australian Research Council (ARC) and the Forest Products Commission of Western Australia (FPC) as part of linkage project LP0882690 (to J. A. P. and E. L. G.) and a Discovery Grant of the Natural Sciences and Engineering Council of Canada (NSERC) (to J. B.).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) HQ343276, HQ343277, HQ343278, HQ343279, HQ343280, HQ343281, HQ343282, JF746807, JF746808, JF746809, JF746810, JF746811, JF746812, JF746813, JF746814, JF746815.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11.
- FPP
- farnesyl diphosphate
- TPS
- terpene synthase.
REFERENCES
- 1. Jones C. G., Plummer J. A., Barbour E. L., Byrne M. (2009) Silvae Genet. 58, 279–286 [Google Scholar]
- 2. Jones C. G., Ghisalberti E. L., Plummer J. A., Barbour E. L. (2006) Phytochemistry 67, 2463–2468 [DOI] [PubMed] [Google Scholar]
- 3. Howes M. J., Simmonds M. S., Kite G. C. (2004) J. Chromatogr. A 1028, 307–312 [DOI] [PubMed] [Google Scholar]
- 4. Page T., Southwell I. A., Russell M., Tate H., Tungon J., Sam C., Dickinson G., Robson K., Leakey R. R. (2010) Chemistry Biodiversity 7, 1990–2006 [DOI] [PubMed] [Google Scholar]
- 5. Piggot M. J., Ghisalberti E. L., Trengove R. D. (1997) Flavour. Fragr. J. 12, 43–46 [Google Scholar]
- 6. Gershenzon J., Dudareva N. (2007) Nat. Chem. Biol. 3, 408–414 [DOI] [PubMed] [Google Scholar]
- 7. Jones C. G., Keeling C. I., Ghisalberti E. L., Barbour E. L., Plummer J. A., Bohlmann J. (2008) Arch. Biochem. Biophys. 477, 121–130 [DOI] [PubMed] [Google Scholar]
- 8. Applegate G. B., Chamberlain J., Daruhi G., Feigelson J. L., Hamilton L., McKinnell F. H., Neil P. E., Rai S. N., Rodehn P., Statham P., Stemmermann L. (1990) in Proceedings of the Symposium on Sandalwood in the Pacific April 9–11, (Hamilton L., Conrad C. E. eds), pp 1–11, US Forest Service, Honolulu, Hawaii [Google Scholar]
- 9. Xie H., Shao Y., Becker J. M., Naider F., Gibbs R. A. (2000) J. Org. Chem. 65, 8552–8563 [DOI] [PubMed] [Google Scholar]
- 10. Shao Y., Eummer J. T., Gibbs R. A. (1999) Org. Lett. 1, 627–630 [DOI] [PubMed] [Google Scholar]
- 11. Kolosova N., Miller B., Ralph S., Ellis B. E., Douglas C., Ritland K., Bohlmann J. (2004) BioTechniques 36, 821–824 [DOI] [PubMed] [Google Scholar]
- 12. Trapp S. C., Croteau R. B. (2001) Genetics 158, 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Felsenstien J. (1989) Cladistics 5, 164–166 [Google Scholar]
- 14. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., D A. R., A B. (2005) in The Proteomics Protocols Handbook (Walker J. M. ed), pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 15. O'Maille P. E., Chappell J., Noel J. P. (2004) Anal. Biochem. 335, 210–217 [DOI] [PubMed] [Google Scholar]
- 16. Cyr A., Wilderman P. R., Determan M., Peters R. J. (2007) J. Am. Chem. Soc. 129, 6684–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paolini J., Muselli A., Bernardini A.-F., Bighelli A., Casanova J., Costa J. (2007) Flavour. Fragr. J. 22, 479–487 [Google Scholar]
- 18. Skaltsa H. D., Demetzos C., Lazari D., Sokovic M. (2003) Phytochemistry 64, 743–752 [DOI] [PubMed] [Google Scholar]
- 19. Chung T. Y., Eiserich J. P., Shibamoto T. (1993) J. Agric. Food Chem. 41, 1693–1697 [Google Scholar]
- 20. Choi H. S. (2003) J. Agric. Food Chem. 51, 2687–2692 [DOI] [PubMed] [Google Scholar]
- 21. Adams R. P. (1995) Identification of essential oil components by gas chromatography/mass spectrometry, 2 Ed., Allured Publishing Corporation, Carol Stream, Illinois [Google Scholar]
- 22. Palá-Paúl J., Pérez-Alonsoa M. J., Velasco-Negueruela A., Vadaré J., Villa A. M., Sanz J., Brophy J. J. (2005) J. Chromatogr. A 1074, 235–239 [DOI] [PubMed] [Google Scholar]
- 23. Valder C., Neugebauer M. (2003) J. Essent. Oil. Res. 15, 178–186 [Google Scholar]
- 24. Davies N. W. (1990) J. Chromatogr. A 503, 1–24 [Google Scholar]
- 25. Bohlmann J., Meyer-Gauen G., Croteau R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Braun N. A., Meier M., Hammerschmidt F.-J. (2005) J. Essent. Oil. Res. 17, 477–480 [Google Scholar]
- 27. Huang Q., Roessner C. A., Croteau R., Scott A. I. (2001) Bioorg. Med. Chem. 9, 2237–2242 [DOI] [PubMed] [Google Scholar]
- 28. de Kraker J. W., Franssen M. C., de Groot A., Konig W. A., Bouwmeester H. J. (1998) Plant Physiol. 117, 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Southwell I. A. (1970) Phytochemistry 9, 2243–2245 [Google Scholar]
- 30. Williams D. C., McGarvey D. J., Katahira E. J., Croteau R. (1998) Biochemistry 37, 12213–12220 [DOI] [PubMed] [Google Scholar]
- 31. Lesburg C. A., Zhai G., Cane D. E., Christianson D. W. (1997) Science 277, 1820–1824 [DOI] [PubMed] [Google Scholar]
- 32. Kampranis S. C., Ioannidis D., Purvis A., Mahrez W., Ninga E., Katerelos N. A., Anssour S., Dunwell J. M., Degenhardt J., Makris A. M., Goodenough P. W., Johnson C. B. (2007) Plant Cell 19, 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aharoni A., Giri A. P., Verstappen F. W., Bertea C. M., Sevenier R., Sun Z., Jongsma M. A., Schwab W., Bouwmeester H. J. (2004) Plant Cell 16, 3110–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sallaud C., Rontein D., Onillon S., Jabes F., Duffe' P., Giacalone C., Thoraval S., Escoffier C., Herbette G., Leonhardt N., Causse M., Tissiera A. (2009) Plant Cell 21, 301–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Köllner T. G., Schnee C., Gershenzon J., Degenhardt J. (2004) Plant Cell 16, 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krieipl A. T., König W. A. (2004) Phytochemistry 65, 2045–2049 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.