Abstract
Sinorhizobium meliloti forms a symbiosis with the legume alfalfa, whereby it differentiates into a nitrogen-fixing bacteroid. The lipid A species of S. meliloti are modified with very long-chain fatty acids (VLCFAs), which play a central role in bacteroid development. A six-gene cluster was hypothesized to be essential for the biosynthesis of VLCFA-modified lipid A. Previously, two cluster gene products, AcpXL and LpxXL, were found to be essential for S. meliloti lipid A VLCFA biosynthesis. In this paper, we show that the remaining four cluster genes are all involved in lipid A VLCFA biosynthesis. Therefore, we have identified novel gene products involved in the biosynthesis of these unusual lipid modifications. By physiological characterization of the cluster mutant strains, we demonstrate the importance of this gene cluster in the legume symbiosis and for growth in the absence of salt. Bacterial LPS species modified with VLCFAs are substantially less immunogenic than Escherichia coli LPS species, which lack VLCFAs. However, we show that the VLCFA modifications do not suppress the immunogenicity of S. meliloti LPS or affect the ability of S. meliloti to induce fluorescent plant defense molecules within the legume. Because VLCFA-modified lipids are produced by other rhizobia and mammalian pathogens, these findings will also be important in understanding the function and biosynthesis of these unusual fatty acids in diverse bacterial species.
Keywords: Bacteria, Fatty Acid, Lipopolysaccharide (LPS), Plant, Toll-like Receptors (TLR), Legume, Sinorhizobium meliloti, Symbiosis, Very Long-chain Fatty Acid
Introduction
The α-proteobacterium Sinorhizobium meliloti forms a symbiosis with the agriculturally important legume Medicago sativa (alfalfa) (for recent reviews, see Refs. 1 and 2). The symbiosis is initiated by the legume, which secretes flavonoids in the absence of a suitable nitrogen source within the soil. S. meliloti then produce nodulation factors and colonize the root hairs, where they induce the formation of plant-derived structures called infection threads. The bacteria then traverse down these infection threads and are eventually endocytosed into cortical root cells and encompassed in plant-derived, acidic membrane compartments called symbiosomes. Within the symbiosome compartment, S. meliloti differentiates into a morphologically distinct cell type known as a bacteroid (3), which fixes nitrogen gas into ammonia for the legume. The lipopolysaccharide (LPS) structure of S. meliloti has been found to play a key role in bacteroid development within legumes (4, 5).
LPS forms the outermost leaflet of the outer membrane of Gram-negative bacteria, such as S. meliloti, and is composed of lipid A, a sugar core, and O-antigen. The biosynthesis of LPS has been most comprehensively studied in Escherichia coli (6). However, unlike E. coli LPS, the LPS species of S. meliloti, other rhizobia, and Brucella species differ, because they are modified with an unusual very long-chain fatty acid (VLCFA)4 of either 27-OHC28:0, 27-O (βOme C4:0) C28:0, or 29-OHC30:0 (7, 8). The S. meliloti acpXL and lpxXL genes, which encode a VLCFA acyl carrier protein and a VLCFA acyl transferase, respectively, are essential for the biosynthesis of VLCFA-modified lipid A in cultured S. meliloti (9, 10). The LpxXL VLCFA acyl transferase was also found to be essential for the biosynthesis of VLCFA-modified lipid A in S. meliloti bacteroids within alfalfa (5). Both S. meliloti AcpXL and LpxXL proteins were demonstrated to confer a competitive advantage to S. meliloti during the alfalfa symbiosis and play central but distinct roles in bacteroid development (5, 9, 10). S. meliloti acpXL and lpxXL mutants also displayed an increased sensitivity to environmental stresses in their free living state relative to the parent strain (9, 10). Therefore, S. meliloti VLCFA-modified lipid A is important for the legume symbiosis and for survival of S. meliloti within the rhizosphere. It was also recently determined that the lipid A VLCFA modification is essential for the symbiosis of Sinorhizobium NGR234 with different types of legume hosts (11), further confirming the importance of identifying the other gene products involved in VLCFA biosynthesis.
The S. meliloti acpXL and lpxXL genes form a six-gene cluster (Fig. 1A), where the other four genes are also hypothesized to encode proteins involved in the biosynthesis of the lipid A VLCFA modification (Fig. 1A) (9, 10, 12). This six-gene cluster is also conserved in Rhizobium leguminosarum, where it was shown that the fabZXL, fabF2XL, and fabF1XL genes are part of the same operon and that the FabF2XL gene product is involved in the biosynthesis of the VLCFA-modified lipid A (12). The adhA2XL gene is also predicted to form a two-gene operon with the lpxXL gene (10). However, roles for the other cluster gene products in the biosynthesis of the lipid A VLCFA remained to be determined. Due to the importance of the VLCFA-modified lipid A in both the free living and the symbiotic life of rhizobia, we constructed insertional mutants in the fabZXL, fabF2XL, fabF1XL, and adhA2XL genes in S. meliloti. Through biochemical analysis of the lipid A species produced by these mutant strains, we found that all of the genes in the acpXL-lpxXL cluster encode proteins involved in the biosynthesis of the VLCFA-modified lipid A.
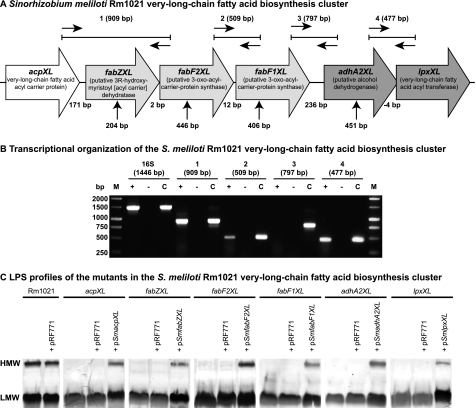
FIGURE 1.
Organization of the S. meliloti acpXL-lpxXL gene cluster and analysis of cluster mutant LPS. A, the S. meliloti acpXL-lpxXL gene cluster is encoded on the chromosome of strain Rm1021. All gene products are predicted to be involved in the biosynthesis of the lipid A VLCFA. The vertical arrows below indicate the position of the plasmid insertions in the mutant strains. The expected sizes of the RT-PCR/PCR products (1–4) using the different primer pairs are indicated (see “Experimental Procedures” for primers used). B, RT-PCR was performed using the primer pairs shown in A with S. meliloti Rm1021 RNA. 16 S rRNA primers were used as a positive control for the integrity of the RNA. Reactions with (+) and without reverse transcriptase (−) added are shown. For control reactions (c), the RNA was replaced with a colony of Rm1021. C, SDS-PAGE analysis of the LPS from the indicated strains (pRF771 is a broad host range control plasmid; pSmacpXL, pSmfabZXL, pSmfabF2XL, pSmfabF1XL, pSmadhA2XL, and pSmlpxXL are wild-type versions of the disrupted genes cloned into pRF771). HMW and LMW LPS species are shown.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
The bacterial strains and plasmids used in this study are shown in Table 1. E. coli strains were grown in Luria-Bertani (LB) broth (10 g/liter NaCl) (13) or on LB agar (1.5%, w/v) supplemented with the appropriate antibiotics at 37 °C. S. meliloti strains were grown in either LB broth or on LB agar (1.5%, w/v) supplemented with 2.5 mm MgSO4 and 2.5 mm CaCl2 (LB/MC) and the appropriate antibiotics, where required, at 30 °C. For the salt gradient plates, a modified LB medium without NaCl (LB−NaCl) was used such that NaCl could be added to the specified concentration.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. meliloti strains | ||
| Rm1021 | Smr derivative of SU47 | Ref. 36 |
| acpXL | Rm1021 acpXL::pk18mobGII Nmr | Ref. 9 |
| lpxXL | Rm1021 lpxXL::pJH104 Nmr Hmr | Ref. 9 |
| adhA2XL | Rm1021 adhA2XL::pk18mobGII Nmr | This study |
| fabF1XL | Rm1021 fabF1XL::pk18mobGII Nmr | This study |
| fabF2XL | Rm1021 fabF2XL::pk18mobGII Nmr | This study |
| fabZXL | Rm1021 fabZXL::pk18mobGII Nmr | This study |
| msbA2 | Rm1021 msbA2::pJH104 Nmr | Ref. 25 |
| E. coli strains | ||
| DH5α | supE44ΔlacU169 (w80lacZDM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| MT616 | MM294A recA56 (pRK600) Cmr | Ref. 17 |
| Plasmids | ||
| pRK600 | pRK2013 npt::Tn9 Cmr | Ref. 17 |
| pk18mobGII | mob lacZα gusA Kmr | Ref. 16 |
| pRF771 | pTE3 with an alternative polylinker TcR | Ref. 18 |
| pSmfabZXL | pRF771 carrying fabZXL with a 31 bp upstream of the start codon of the gene TcR | This study |
| pSmfabF1XL | pRF771 carrying fabF2XL with a 54 bp upstream of the start codon of the gene TcR | This study |
| pSmfabF2XL | pRF771 carrying fabF1XL and 31 bp upstream of the start codon of the gene TcR | This study |
| pSmadhA2XL | pRF771 carrying adhA2XL and 31 bp upstream of the start codon of the gene TcR | This study |
| pSmacpXL | pRF771 carrying acpXL and 40 bp upstream of the start codon of the gene TcR | This study |
| pSmlpxXL | pRF771 carrying IpxXL and 33 bp upstream of the start codon of the gene TcR | This study |
RNA Isolation and RT-PCR
All primers used in this study are listed in Table 2. RNA was extracted from 20 ml of exponential phase Rm1021 culture (A600 ∼0.1) using a previously published protocol (14). In brief, cultures were rapidly microcentrifuged, and pellets were frozen for 10 min in liquid nitrogen. Total RNA was extracted using the RNeasy minikit (Qiagen). Cells were lysed in RLT buffer (supplied with the kit) using Fast Protein tubes (Qbiogen) and the FastPrep FP120 cell disrupter (30 s at 6.5 m/s2). The RNA was then purified using the RNeasy minikit protocol. RT-PCR was performed using the SuperScript One-Step RT-PCR kit (Invitrogen) according to the manufacturer's protocol. Co-transcription of acpXL, fabZXL, and fabF2XL was analyzed using primers smc04277_−241_F and smc04275_+188_R, co-transcription of fabF2XL and fabF1XL using primers smc04273_−155_F and smc04273_+352_R, co-transcription of fabF1XL and adhA2XL using primers smc04273_+960_F and smc04270_+235_R, and co-transcription of adhA2XL and lpxXL using primers smc04270_+698_F and smc4268_+149_R (Table 2). To assess the quality of the extracted RNA, the universal ribosomal RNA primers 8F and 1492R were also used (15). For all primer pairs, PCRs were also set up using the extracted RNA or an Rm1021 colony as a template using GoTaq DNA polymerase (Promega). To confirm the correct identity of the RT-PCR products, and the DNA fragments were purified (illustra GFXTM PCR DNA and Gel Band Purification Kit) and sequenced.
TABLE 2.
Primers
Restriction enzyme recognition sites are underlined.
| Primer name | Sequence (5′–3′) |
|---|---|
| pkX_DS_R | CGAAAGGGGGATGTGCTGC |
| smc04278_−33_PstI_F | TCTTAGCTGCAGATAGGGTTCGCCAAGACTC |
| smc04278_stop_XbaI_R | CGTACGTCTAGACTCAACCCGCTTTTGC |
| smc04270_+78_EcoRI_F | GATTACGAATTCCGAGGTGACGCTGCGGG |
| smc04270_+451_HindIII_R | AGTGCCAAGCTTCGAAGGTGACCGGCGCCA |
| smc04270_−270_F | CGCCAGCCTTGTCATGAC |
| smc04270_−35_PstI_F | GCTTGGCTGCAGCTCGCGCGACAACGACATATG |
| smc04270_stop_XbaI_R | CGTACGTCTAGATCAATCCATCCGCAACACGATC |
| smc04273_+33_PstI_F | GTCGACCTGCAGTCCGATCGTTGCCGTC |
| smc04273_+406_HindIII_R | AGTGCCAAGCTTGGAACCGATTATAGGCGTCG |
| smc04273_−155_F | CGCTCTCGCTCGGACAC |
| smc04273_−31_NsiI_F | GTGATCATGCATCGTGCTTGCCGCGGAAT |
| smc04273_+1307_XbaI_R | CGTACGTCTAGACAGAGGGGGCCTTTTTCTTCCAC |
| smc04275_+36_PstI_F | GTCGACCTGCAGGGTAGGCATCGTGACGAGC |
| smc04275_+446_HindIII_R | AGTGCCAAGCTTGCCAGAAACAGCGTCGG |
| smc04275_−126_F | GATATGCGATGCTCAGCTG |
| smc04275_−54_PstI_F | GCTTGGCTGCAGGCGTGCGGAAGAACTGGG |
| smc04275_stop_XbaI_R | CGTACGTCTAGATTATTCCGCGGCAAGCA |
| smc04277_+36_PstI_F | ATCTACCTGCAGCGAAACCGTCGATCTG |
| smc04277_+204_HindIII_R | AGTGCCAAGCTTGAATTTCGTCGCGGCC |
| smc04277_−241_F | CGAGGAGTACTTCGTGCTGA |
| smc04277_−31_PstI_F | GCTTGGCTGCAGCTTGAGCGCGAAACGAATG |
| smc04277_stop_XbaI_R | CGTACGTCTAGATCACTTCTCCGAACCGGC |
| smc04268_−40_PstI_F | TCTTAGCTGCAGAGTCGCGCCAGATCTTC |
| smc04268_stop_XbaI_R | CGTACGTCTAGATCAAAGAGACCGCTTGATG |
| smc04273_+352_R | AAGCTCGAAACGGGCATT |
| smc04273_+960_F | CACGCCCGAAAACGAC |
| smc04270_+235_R | GGGAGCACATTGGCGA |
| smc04270_+698_F | AGGGCGTCGATGTCGTC |
| smc4268_+149_R | GCAACGCGATCCATGAAG |
| smc04275_+188_R | CGGAATCTGCTGCGACC |
| 8_F | AGAGTTTGATCCTGGCTCAG |
| 1492_R | GGTTACCTTGTTACGACTT |
Constructions of Insertional Mutants
All primers used in this study are listed in Table 2. The insertional mutant in adhA2XL was obtained by amplifying a 374-bp internal fragment of the gene using primers smc04270_+78_EcoRI_F and smc04270_+451_HindIII_R. For the mutant in fabF1XL a 374-bp internal fragment using primers smc04273_+33_PstI_F and smc04273_+406_HindIII_R, for the mutant in fabF2XL a 411-bp internal fragment using primers smc04275_+36_PstI_F and smc04275_+446_HindIII_R, and for the fabZXL mutant a 169-bp internal fragment using primers smc04277_+36_PstI_F and smc04277_+204_HindIII_R was amplified by PCR. The PCR fragments were then purified, digested with the respective restriction enzymes according to the manufacturer's protocols, ligated into the respective restriction sites of pk18mobGII (16), and transformed into E. coli DH5α. The plasmids were conjugated into the S. meliloti Rm1021 parent strain by triparental mating using the E. coli strain MM294A carrying the helper plasmid pRK600 (17). Transconjugates were selected on LB/MC containing Sm (500 μg/ml) and Nm (200 μg/ml) for 4 days. The correct insertions were confirmed by PCR using the plasmid-specific primer pkX_DS_R and the chromosome-specific primers smc04270_−270_F for the adhA2XL mutant, smc04273_−155_F for the fabF1XL mutant, smc04275_−126_F for the fabF2XL mutant, and smc04277_−241_F for the fabZXL mutant. The disrupted genes were then transduced into Rm1021 using φM12, and the mutants were selected on LB Sm Nm agar plates (500 and 200 μg/ml, respectively).
Construction of Complementing Plasmids
For complementation of the insertional mutants, the complete genes, including their ribosomal binding site, were cloned into pRF771 (18) at the appropriate restriction sites, under the control of the trp promoter. The acpXL gene, including 40 bp upstream of the start codon, was amplified using primers smc04268_−40_PstI_F and smc04268_stop_XbaI_R; the fabZXL gene and a 31-bp region upstream of its start were amplified using primers smc04277_−31_PstI_F and smc04277_stop_XbaI_R; the fabF2XL gene and a region 54 bp upstream of it were amplified using primers smc04275_−54_PstI_F and smc04275_stop_XbaI_R; the fabF1XL gene was amplified, including 31 bp upstream of the start codon, using primers smc04273_−31_NsiI_F and smc04273_+1307_XbaI_R; the adhA2XL gene and 35 bp upstream of the start codon were amplified using primers smc04270_−35_PstI_F and smc04270_stop_XbaI_R; and the lpxXL gene was amplified, including 33 bp upstream of the start codon, using primers smc04278_−33_PstI_F and smc04278_stop_XbaI_R. All genes were then cloned into the PstI and XbaI sites of pRF771 to generate pSmacpXL, pSmfabZXL, pSmfabF2XL, pSmfabF1XL, pSmadhA2XL, and pSmlpxXL, respectively.
LPS Isolation
The LPS species were isolated using either SDS lysis followed by SDS-PAGE or hot water/phenol extraction as described previously (10, 19). For SDS lysis, the strains were grown for 48 h (72 h for the S. meliloti lpxXL mutant) to mid-exponential phase (A600 ∼2.0), and then 1 ml of culture was used for the extraction procedure. The hot water/phenol extraction was performed from 4 liters of the strains grown in LB/MC supplemented with the relevant antibiotics until mid-exponential phase as earlier. After extraction, the lyophilized water phase fractions were dissolved in deionized water at a concentration of 10 μg/ml and then purified by ultracentrifugation (100,000 × g, 8 °C) for 3.5 h. The purified LPS pellets were then resuspended in deionized water and lyophilized.
Lipid A Analysis by MALDI-TOF MS
The LPS species isolated into the aqueous phase were washed with 95% (v/v) ethanol at 4 °C to remove phospholipids, resuspended in water and freeze-dried. Lipid A was released from 2 mg of LPS by mild acid hydrolysis using 1% (v/v) HOAc. Briefly, samples (5 mg/ml in 1% (v/v) HOAc) were hydrolyzed at 100 °C for 1 h with constant stirring, freeze-dried, and extracted with Bligh-Dyer solvent (2 CHCl3, 2 MeOH, 1.8 H2O; v/v/v). The organic phase was collected, and extraction was repeated with chloroform only. Combined organic phases were extracted back with H2O and dried with a stream of N2. Samples were dissolved in a CHCl3/IPA/H2O mixture (5/3/0.25; v/v/v) and were then mixed with 1:1 (v/v) of 0.5 m 2′,4′,6′-trihydroxyacetophenone monohydrate matrix in methanol, and 0.5 μl (1 μg) was spotted onto the stainless steel target. Finally, the spectra were recorded in negative reflectron ion mode using the AB SCIEX TOF/TOF 5800 system (Applied Biosystems). All acquisitions were calibrated with previously described lipid A preparations (20, 21), and observed ions represent an average of the most abundant [M − H]− ions.
Quantification of the Lipid A Fatty Acids and Sugars by GC-MS
The identity and quantity of the lipid A fatty acid components as their trimethylsilyl fatty acid methyl esters was determined using Tri-Sil reagent (Pierce) after methanolysis with 2 m methanolic HCl at 80 °C for 16 h and analyzed by combined gas chromatography-mass spectrometry (GC-MS) (22, 23). The sugar residues were quantified by treating the purified LPS samples with 2 m methanolic HCl at 80 °C for 16 h in the presence of an internal standard of inositol, followed by N-acetylation with pyridine and acetic anhydride in methanol. Treatment with Tri-Sil reagent at 80 °C for 30 min resulted in the trimethyl methyl glycoside derivatives. Products were dissolved in hexane and analyzed on a 30-m capillary HP-1 column (Hewlett-Packard) in a 5890 Series-Trio-1 GC-MS (Hewlett-Packard).
S. meliloti-Alfalfa Interaction Experiments
Alfalfa seedlings were inoculated with the defined S. meliloti strains, exactly as described previously (24). Competition assays were also performed as described before (8, 25). In brief, 3-day-old alfalfa seedlings were inoculated with a 1:1 ratio of Rm1021 and the mutant strain. Four weeks after the inoculation, the bacteria were extracted and recovered on LB/MC Sm and LB/MC SmNm agar. Because both the parent and mutant strains grow on LB/MC Sm, whereas only the mutant strains will grow on LB/MC SmNm agar, the percentage occupancy of each strain could be determined.
Fluorescence Microscopy of Nodule Sections
To detect the production of plant defense fluorescent polyphenolics in the root nodules, the nodules were fixed and analyzed as described previously (25) using a Zeiss Axioskop microscope with UV excitation and the DAPI fluorescence filter set (Filter Set 01, BP 365/12).
Salt Gradient Experiments
The ability of the different strains to grow in the presence of a salt gradient was performed as described previously (9), with slight modifications. Gradients were prepared in large square culture dishes (22.5 × 22.5 cm). A bottom layer was poured using 200 ml of LB−NaCl agar and a top layer of 200 ml of LB agar supplemented with 0.5 m NaCl. Late exponential phase cultures were washed three times in LB−NaCl and diluted to an A600 of ∼0.2, and then 60 μl was streaked across the plate. After 72 h at 30 °C, the minimum salt requirement for growth was determined assuming a linear gradient of NaCl across the plate.
Preparation of Monocytes and LPS Stimulation Studies
All studies were preapproved by the institutional ethics committee. Whole venous blood was collected from two healthy individuals who were not undergoing any drug therapy. The blood was diluted 1:1 in RPMI 1640 (Sigma-Aldrich), layered onto Histopaque-1077 (Sigma-Aldrich), and centrifuged at 800 × g for 30 min. Peripheral blood mononuclear cells were removed from the interface and washed twice (10 min, 300 × g) in RPMI before being counted and resuspended in growth media comprising RPMI 1640 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Invitrogen), and 10% human AB serum (Sigma-Aldrich). Monocytes were plated at a density of 4 × 105 cells/well in 96-well plates and incubated for 90 min at 37 °C/5% CO2. Non-adherent cells were removed with two rinses of PBS. The effect of different S. meliloti LPS preparations were assessed by stimulating the monocytes for 6 h, and then the supernatants were collected. An E. coli commercial LPS preparation (0111:B4; Sigma catalog no. L4391) was used as a positive control. E. coli and S. meliloti LPS preparations used were 1 μg/ml in the monocyte stimulation experiments.
Assay of Human Cytokines in Culture Supernatants
The concentrations of TNF-α and IL-10 in culture supernatant were measured simultaneously using a human inflammatory cytokine cytometric bead array (BD Biosciences). Samples were analyzed on a multifluorescence BD FACSCaliburTM flow cytometer using BD CellQuestTM software and BDTM cytometric bead array software. The assay sensitivities for both cytokines were 10 pg/ml. The coefficients of variation of all cytokine assays were less than 10%.
Statistical Analysis
The statistical significance of the data was analyzed using Student's t test in Microsoft Excel.
RESULTS
The acpXL-lpxXL Gene Cluster Is Organized into Two Operons
The acpXL-lpxXL gene cluster of rhizobia, including S. meliloti Rm1021 (Fig. 1A), is predicted to consist of three distinct transcriptional units (10, 12). To confirm the transcriptional organization of this gene cluster, RT-PCR was performed using RNA extracted from S. meliloti Rm1021. We obtained 909- and 509-bp RT-PCR fragments using primer pairs internal to the acpXL and fabF2XL genes and fabF2XL and fabF1XL genes, respectively (Fig. 1, A and B), whereas in the absence of reverse transcriptase, no PCR products were observed with any primer pairs (Fig. 1B). In addition, no RT-PCR fragment was observed using S. meliloti RNA in combination with primers internal to the fabF1XL and adhA2XL genes, indicating that these two genes are not co-transcribed (Fig. 1, A and B). Therefore, these findings show that in S. meliloti Rm1021, the acpXL, fabZXL, fabF2XL, and fabF1XL genes form part of the same transcriptional unit. Because the RT-PCR using primers internal to the adhA2XL and lpxXL genes yielded a fragment of 477 bp, this confirmed that the adhA2XL and lpxXL genes constitute the second transcriptional unit in the proposed lipid A VLCFA biosynthesis gene cluster (Fig. 1, A and B).
The Cluster Mutants Lack High Molecular Weight LPS
The six genes in the S. meliloti acpXL-lpxXL gene cluster are hypothesized to encode proteins involved in the biosynthesis of the VLCFA-modified lipid A (10). To investigate whether the other four genes in the acpXL-lpxXL cluster are also involved in LPS biosynthesis, we constructed S. meliloti insertional mutants in the fabZXL, fabF2XL, fabF1XL, and adhA2XL genes (Fig. 1A). Previously, an S. meliloti acpXL mutant was found to lack high molecular weight (HMW) LPS species (10). SDS lysis followed by SDS-PAGE/periodate-silver staining also determined that, unlike the parent strain, which produced both HMW and low molecular weight (LMW) LPS species, the cluster mutant strains only produced LMW LPS species (Fig. 1C). Hence, these analyses showed that all of the mutants in the acpXL-lpxXL gene cluster have LPS structural alterations relative to the parent strain and lack the ability to synthesize HMW LPS.
To confirm that the disrupted genes are responsible for the absence of HMW LPS in the mutant strains, the wild-type versions of these genes were cloned into the broad host range vector pRF771 under control of the constitutively active trp promoter (18) and then mated into the appropriate mutant strain. The LPSs of these strains were then analyzed by SDS-lysis followed by SDS-PAGE/periodate-silver staining (Fig. 1C). We found that the complementing plasmids, but not the control plasmid (pRF771), restored the ability of the mutant strains to produce HMW LPS (Fig. 1C), confirming that the disrupted genes alone were responsible for the LPS structural alterations in the mutant strains. Therefore, the fabZXL, fabF2XL, fabF1XL, and adhA2XL gene products, like the acpXL and lpxXL gene products, are involved in S. meliloti LPS biosynthesis.
The Cluster Mutants Have Defects in Lipid A VLCFA Biosynthesis
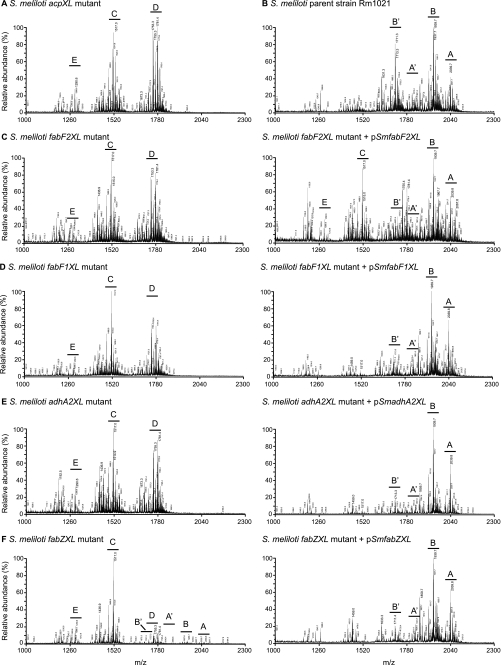
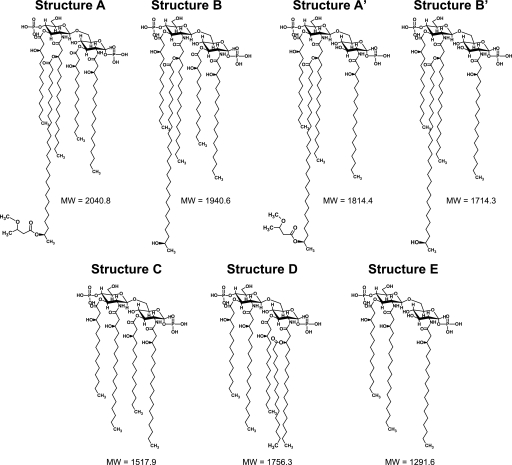
To determine whether all the acpXL-lpxXL cluster gene products (Fig. 1A) are involved in the biosynthesis of VLCFA-modified lipid A, the LPS species from the four cluster mutants were isolated by hot water/phenol. The lipid A species in the aqueous phases were then purified from the rest of the LPS molecule by mild acid hydrolysis and analyzed by MALDI-TOF MS (Fig. 2). As controls, the lipid A species produced by the S. meliloti acpXL insertional mutant and parent strain were also analyzed (Fig. 2, A and B, respectively). We found that the S. meliloti parent strain produced four major VLCFA-modified lipid A species, two of which are penta-acylated (Figs. 2B and 3, structures A and B) and two of which are tetra-acylated (Figs. 2B and 3, structures A′ and B′). As observed previously (9), the S. meliloti acpXL mutant completely lacked VLCFA-modified lipid A species and produced a mixture of penta-, tetra-, and triacylated lipid A species (Figs. 2A and 3, structures D, C, and E, respectively). We found that the S. meliloti fabF2XL, fabF1XL, and adhA2XL insertional mutants also completely lacked VLCFA-modified lipid A species (Fig. 2, C, D, and E, respectively) and produced the same three lipid A species (Fig. 3, structures D, C, and E) as the S. meliloti acpXL insertional mutant (Fig. 2A). In contrast, MALDI-TOF MS analysis of the S. meliloti fabZXL mutant lipid A sample showed that it contained a mixture of lipid A species with (Figs. 2F and 3, structures A, A′, B, and B′) and without the VLCFA modification (Figs. 2F and 3, structures D, C, and E). Therefore, this analysis showed that all of the mutants in the S. meliloti acpXL-lpxXL gene cluster have defects in the biosynthesis of VLCFA-modified lipid A.
FIGURE 2.
The S. meliloti acpXL-lpxXL cluster mutants have alterations in their lipid A biosynthesis. Shown is MALDI-TOF MS of the lipid A species produced by each strain (acpXL (A), Rm1021 (B), fabF2XL without and with pSmfabF2XL (C), fabF1XL without and with pSmfabF1XL (D), adhA2XL without and with pSmadhA2XL (E), and fabZXL without and with pSmfabZXL (F)). The capital letters indicate the major lipid A species (see Fig. 3 for structures) in each sample (pSmacpXL, pSmfabZXL, pSmfabF2XL, pSmfabF1XL, pSmadhA2XL, and pSmlpxXL are wild-type versions of the disrupted genes in the mutant strains cloned into pRF771).
FIGURE 3.
Proposed lipid A structures produced by the S. meliloti parent strain and acpXL-lpxXL cluster mutants. Structures A, A′, B, and B′ contain a VLCFA modification, whereas structures C–E lack VLCFAs. Structure D lacks the VLCFA modification but is still penta-acylated due to the addition of a non-hydroxylated fatty acid. The example shown contains C16:0, but based on our GC-MS analysis (Table 3), this could also be replaced by a C18:0 or C18:1.
To confirm that the disrupted genes are responsible for the absence/decrease of the VLCFA-modified lipid A in the cluster mutants, the lipid A species of the mutants with the respective wild-type copy of the disrupted gene cloned into pRF771 were also analyzed by MALDI-TOF MS (Fig. 2, C–F). We found that, with the exception of the S. meliloti fabF2XL mutant (Fig. 2C), the lipid A alterations of the cluster mutants were restored by the presence of the respective complementing plasmids (Fig. 2, D–F). This showed that disruption of fabF1XL, fabZXL, or adhA2XL in the mutant strains was responsible for their lipid A phenotypes. In the case of the S. meliloti fabF2XL mutant with pSmfabF2XL, we only found partial complementation of its lipid A alteration relative to the parent strain (Fig. 2, compare C and B). However, because partial complementation was observed, this finding confirmed that FabF2XL is involved in the biosynthesis of VLCFA-modified lipid A.
The S. meliloti fabZXL Mutant Is Substantially Reduced in Its Ability to Produce VLCFA-modified Lipid A
To determine the precise changes in the lipid A fatty acid content of the mutant strains relative to the parent strain, we used GC-MS (Table 3). Consistent with the MALDI-TOF MS findings, GC-MS analysis of the lipid A fatty acids showed that the S. meliloti fabF2XL, fabF1XL, and adhA2XL insertional mutants completely lack 27OH-C28:0 and 29OH-C30:0. Therefore, the fabF2XL, fabF1XL, and adhA2XL gene products play essential roles in the biosynthesis of the VLCFA-modified lipid A in S. meliloti. The VLCFA content of the S. meliloti fabF1XL and adhA2XL mutant lipid A was also fully restored by the respective complementing plasmid (Table 3). In contrast, although the 29OH-C30:0 level was fully restored in the S. meliloti fabF2XL mutant by pSmfabF2XL, the 27OH-C28:0 level amount was only partially restored by this plasmid, and 25OH-C26:0 was detected in this sample (Table 3). This finding confirms that we were only partially able to complement the lipid A alteration in this mutant strain. We also found that the S. meliloti fabZXL insertional mutant lipid A contained substantially reduced amounts of 27OH-C28:0 and 29OH-C30:0 fatty acids relative to the parent strain (Table 3). Interestingly, the lipid A of this mutant strain contained increased amounts of 29OHC30:0 compared with 27OHC28:0 and also contained trace amounts of 25OH-C26:0, which was absent in the parent strain lipid A preparation (Table 3). Because the plasmid, pSmfabZXL, restored the lipid A VLCFA content of the S. meliloti fabZXL mutant to that of the parent strain, these data showed that the fabZXL gene product, although not essential, plays an important role in the biosynthesis of VLCFA-modified lipid A in S. meliloti.
TABLE 3.
Fatty acid composition of the mutants in the acpXL-lpxXL cluster
| Fatty acyl residue | EI/MS detector response (percentage of area)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rm1021 | acpXL | fabZXL | fabZXL + pSmfabZXL | fabF2XL | fabF2XL + pSmfabF2XL | fabF1XL | fabF1XL + pSmfabF1XL | adhA2XL | adhA2XL + pSmadhA2XL | |
| VLCFA | ||||||||||
| 27OH-C28:0 | 25.6 | ND | TR | 25.2 | ND | 8.5 | ND | 29.9 | ND | 37.8 |
| 29OH-C30:0 | 5.4 | ND | 1.7 | 7.3 | ND | 9.7 | ND | 6.0 | ND | 4.8 |
| 25OH-C26:0 | ND | ND | TR | ND | ND | 1.5 | ND | ND | ND | ND |
| Non-hydroxylated fatty acids | ||||||||||
| C18:1 | TR | 17.5 | 13.3 | 3.1 | 19.9 | 7.1 | 17.0 | 3.3 | 30.2 | 10.3 |
| C16:0 | ND | 4.0 | 2.0 | ND | 3.7 | TR | 2.2 | ND | 3.7 | TR |
| C18:0 | 2.0 | 4.8 | 6.7 | TR | 7.5 | 3.4 | 9.1 | 2.2 | 10.0 | 2.7 |
| Hydroxylated fatty acids | ||||||||||
| 3OH-C14:0 | 21.7 | 34.7 | 32.9 | 22.2 | 29.5 | 25.6 | 34.7 | 23.2 | 14.9 | 17.9 |
| 3OH-C16:0 | 11.3 | 6.8 | 6.6 | 5.5 | 6.4 | 7.6 | 7.3 | 5.1 | 5.7 | 3.8 |
| 3OH-C18:0 | 28.8 | 28.8 | 33.2 | 30.9 | 30.0 | 31.0 | 27.7 | 27.1 | 31.7 | 19.2 |
| 3OH-C18:1 | 5.2 | 3.4 | 3.6 | 5.9 | 3.0 | 5.5 | 2.2 | 3.3 | 3.8 | 3.6 |
a ND, not detected; TR, trace amount detected (not included in the area percentage calculation); 3OH-C17:0 and 3OH-C19:1 were found in trace amounts or not detected in all samples.
The lipid A preparation of the S. meliloti parent strain contains mainly hydroxylated fatty acids, with only trace or minor amounts of non-hydroxylated fatty acids (Table 3). We showed previously that the lipid A of the S. meliloti acpXL mutant has elevated amounts of non-hydroxylated fatty acids because it can transfer a non-hydroxylated shorter chain fatty acid onto the lipid A in the absence of the VLCFA to create a penta-acylated lipid A species (9) (Fig. 3, structure D). Our analysis found that the lipid A samples from the S. meliloti fabF2XL, fabF1XL, fabZXL, and adhA2XL mutant strains contained elevated amounts of non-hydroxylated lipid A fatty acids relative to the parent strain (Table 3). In all cases, the increase in non-hydroxylated fatty acids in the mutant strains was reduced by the respective complementing plasmid (Table 3). Consequently, these findings confirmed our MALDI-TOF MS results and showed that in the absence of either FabF2XL, FabF1XL, FabZXL, or AdhA2XL, S. meliloti mutants can transfer a non-hydroxylated shorter chain fatty acid onto the lipid A to create a penta-acylated lipid A species (see Fig. 3, structure D). In contrast to the non-hydroxylated fatty acids, we did not observe any substantial differences in the amount of 3-hydroxylated lipid A fatty acids in the cluster mutant strains relative to the parent strain (Table 3).
Cluster Mutants Do Not Have Alterations in Their LPS Sugar Composition
In bacterial species lacking VLCFA-modified LPS, the LMW and HMW LPS bands observed by SDS-PAGE are defined as rough and smooth LPS, respectively (26). Smooth LPS is the complete LPS consisting of O-antigen, core, and lipid A, whereas rough LPS lacks the O-antigen. It was shown previously that the S. meliloti acpXL insertional mutant, which lacks HMW LPS species, did not have a significant difference in its sugar composition relative to the parent strain to account for loss of the O-antigen subunit (10). Therefore, it was proposed that in S. meliloti, HMW LPS species are polymerized LPS molecules containing the VLCFA modification. To confirm that this was also the case with other cluster mutants, we analyzed the sugar composition of the purified aqueous phase LPS extracted from the S. meliloti lpxXL and adhA2XL insertional mutants by GC-MS. However, we found no significant difference in the sugar composition of the S. meliloti lpxXL and adhA2XL insertional mutant LPS relative to the parent strain LPS (data not shown). This provides further support that the absence of HMW LPS in the S. meliloti cluster mutants is not due to the absence of the O-antigen subunit.
The Cluster Mutants Have Reduced Alfalfa Competitiveness and a Minimum Salt Requirement for Growth
S. meliloti acpXL and lpxXL insertional mutants are symbiotically competent with alfalfa but have a reduced competitiveness in infecting legume nodules relative to the parent strain (9, 10). We also found that all of the other S. meliloti acpXL-lpxXL cluster mutants were able to nodulate alfalfa and support plant growth to a similar extent as the parent strain after a 4-week growth period (data not shown), showing that these gene products are not essential for the S. meliloti-legume symbiosis. To investigate whether the other cluster mutants were also reduced in their alfalfa competitiveness relative to the parent strain, alfalfa seedlings were co-inoculated with the parent and individual mutant strains in a 1:1 ratio. Because the mutant strains contain a neomycin resistance cassette, the percentage of each mutant strain relative to the total number of bacteria within each nodule was determined (Table 4). We found, consistent with previous observations for the S. meliloti lpxXL mutant (9), that the nodules could be subdivided into two distinct classes, Class I and Class II, which were primarily infected by either the parent or the mutant strains, respectively (Table 4). Because the majority of nodules were Class I and the mutant bacteria only represented a small percentage of the total bacterial population within these nodules (Table 4, Class I nodules), these findings showed that the mutant strains were all reduced in their alfalfa competitiveness relative to the parent strain. Within the Class I nodules, we observed no significant difference in the ability of the different mutant strains to infect these nodules relative to each other. In contrast, small differences were observed in the number of Class II nodules (Table 4) found in the competition experiments with different mutant strains. However, because the number of Class II nodules is so low, no significant conclusions could be drawn on whether the acpXL-lpxXL cluster mutant strains have slight differences in their alfalfa competitiveness.
TABLE 4.
Competitiveness of the mutants in the acpXL-lpxXL cluster
| S. meliloti mutant strain used in the competition assaya | Amount of each mutant strain within the nodules (percentage of total bacteria within the nodules)b |
|
|---|---|---|
| Class I nodules | Class II nodules | |
| % | % | |
| acpXL | 3.3 ± 8.5 (n = 16) | 53.3 (n = 1) |
| fabZXL | 5.9 ± 9.6 (n = 22) | 92.9 ± 10.1 (n = 2) |
| fabF2XL | 3.9 ± 9.0 (n = 18) | 85.2 ± 15.5 (n = 6) |
| fabF1XL | 6.1 ± 11.4 (n = 21) | 94.1 ± 10.3 (n = 3) |
| adhA2XL | 3.1 ± 9.1 (n = 21) | 90.3 ± 13.1 (n = 3) |
a The mutant and parent strains were inoculated onto alfalfa seedlings in a 1:1 ratio.
b The nodules analyzed fell into two classes, Class I and Class II nodules, which were infected primarily by either the parent or mutant strains, respectively. The data sets shown are average values from the defined number of nodules (n).
S. meliloti mutants lacking the lipid A VLCFA are known to be sensitive to high osmolarity relative to the parent strain (9). We also found that the S. meliloti fabF2XL, fabF1XL, fabZXL, and adhA2XL mutants have slightly increased sensitivities to hyperosmotic salt stress relative to the parent strain (data not shown). However, more significantly, we found that unlike the parent strain, all of the cluster mutants have a minimum salt requirement for growth and that this, with the exception of the S. meliloti fabF2XL mutant with pSmfabF2XL, is overcome by the presence of the respective complementing plasmid (Fig. 4A). Consistent with our biochemical characterization of the lipid A, we found that pSmfabF2XL only partially complemented the minimum salt requirement growth phenotype of the S. meliloti fabF2XL mutant (Fig. 4A). We also determined that the S. meliloti lpxXL mutant has the highest salt requirement for growth among the cluster mutants (Fig. 4A).
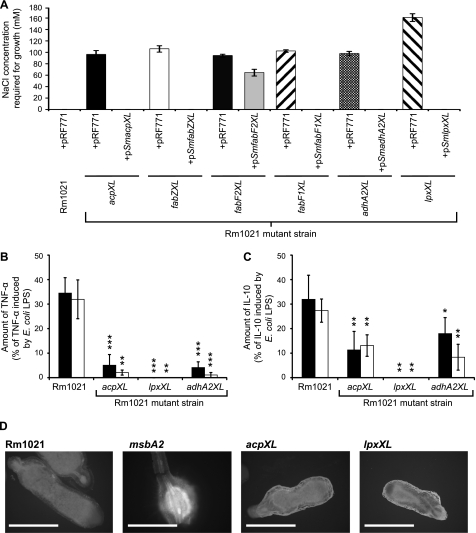
FIGURE 4.
The S. meliloti acpXL-lpxXL mutants have a minimum salt requirement for growth, and their LPS is less immunogenic than the parent strain LPS. A, the S. meliloti parent strain with the control plasmid (pRF771) or the mutant strains with either pRF771 or the respective complementing plasmid were streaked onto a gradient of LB−NaCl supplemented with NaCl ranging from 0 to 500 mm. The minimum salt concentration for growth was determined assuming a linear salt gradient across the agar. The absence of a bar indicates that the strain grows in the absence of sodium chloride. B and C, human monocytes from two individuals were exposed to 1 μg/ml purified LPS from the defined strains, and then the amount of TNF-α (B) and IL-10 (C) in the supernatant was determined after 6 h of incubation. The cytokine amounts shown are expressed as a percentage of the cytokine induction by E. coli LPS, which induced TNF-α and IL-10 levels from human monocytes of 973.2 ± 73.0 and 228.1 ± 34.3 pg/ml for individual 1 (filled bars) and 2241.3 ± 216.4 and 291.6 ± 27.5 pg/ml for individual 2 (open bars), respectively. The significance values (***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05) represent comparisons of the individual LPS responses induced by the mutant strains relative to the S. meliloti parent strain. D, the autofluorescence of plant defense polyphenolics in alfalfa nodules infected with the designated strains was determined using fluorescent microscopy. The nodules from five different alfalfa plants for each strain were analyzed, and representative nodules images are shown. Scale bars, Rm1021, acpXL, and lpxXL (2 mm) and msbA2 (1 mm). Error bars, S.D.
The VLCFA Modification Does Not Suppress the Immunogenicity of S. meliloti LPS or Affect the Production of Fluorescent Plant Defense Polyphenolics
The LPS of rhizobia, including S. meliloti, are substantially reduced in their ability to induce cytokine production from human monocytes relative to E. coli LPS (27). Because E. coli LPS lacks VLCFA modifications (6), one possibility is that the VLCFA modification is important to prevent S. meliloti LPS-induced activation of human monocytes. Likewise, it was previously hypothesized that the VLCFA modification is important to suppress the induction of the plant defense response during S. meliloti infection of the legume root nodules (9, 10). To investigate the effect of the VLCFA modification on the immunogenicity of S. meliloti LPS, we analyzed the ability of purified LPS from the S. meliloti acpXL, adhA2XL, and lpxXL mutant strains to induce human monocytes to produce cytokines relative to purified LPS from the S. meliloti parent strain and an E. coli commercial LPS preparation (Fig. 4, B and C). We found that the S. meliloti parent strain LPS was substantially reduced in its ability to induce both TNF-α and IL-10 cytokines from human monocytes relative to E. coli LPS (Fig. 4, B and C, respectively). However, we determined that the LPS species produced by the VLCFA biosynthesis cluster mutants, which completely lack the VLCFA modification, were even more reduced in their ability to induce cytokine production from human monocytes relative to the parent strain (Fig. 4, B and C). In fact, no cytokine induction was observed with LPS from the lpxXL mutant. Consequently, these findings show that the lipid A VLCFA modification is not suppressing the activation of human monocytes by S. meliloti LPS but is required for the activation process.
As an initial means to investigate the effect of the lipid A VLCFA on the plant defense response, we analyzed the production of autofluorescent plant defense polyphenolics produced in the root nodules after infection with different S. meliloti strains using microscopy (25). Using this methodology, we observed a clear induction of root nodule autofluorescent defense molecules with an S. meliloti msbA2 insertional mutant (Fig. 4D), which was previously shown to induce this response (25). In contrast, we did not observe any significant reproducible difference in root nodule autofluorescence after infection with either the S. meliloti acpXL and lpxXL insertional mutants relative to the parent strain (Fig. 4D). Therefore, the S. meliloti acpXL and lpxXL genes are not required to suppress the induction of root nodule autofluorescent defense molecules in alfalfa.
DISCUSSION
Why Is the Cluster FabZXL Gene Not Essential for Lipid A VLCFA Biosynthesis?
In this study, we identified three novel genes, fabF1XL, fabZXL, and adhA2XL, which are involved in the biosynthesis of VLCFA-modified lipid A in S. meliloti. In addition, we determined that that the fabZXL gene product is important but not essential for this process. Another putative fabZ gene is annotated in the S. meliloti Rm1021 genome (28). We determined that this putative fabZ encodes a protein with 47% similarity (26% identity) to the cluster FabZXL protein. The fabZ gene is located on the S. meliloti chromosome in a separate gene region predicted to be involved in lipid A biosynthesis (6, 28). This region also contains the gene encoding LpxA, which is one of the most conserved genes in lipid A biosynthesis (6, 28). Therefore, the possibility exists that this second putative FabZ protein is able to partially compensate for the absence of the cluster FabZXL protein in the biosynthesis of VLCFA-modified lipid A. E. coli FabZ is involved in the dehydration of short-chain hydroxyacyl-acyl carrier proteins (hydroxyacyl-ACPs) (29). However, the VLCFA-containing lipid A in the fabZXL mutant contains greater levels of 29OH-C30:0 than 27OH-C28:0 and also detectable levels of 25OH-C26:0. Therefore, this suggests that the second potential FabZ dehydratase may have broader chain length specificity and dehydrate both short- and long-chain lipid A fatty acids. The appearance of trace amounts of 25OH-C26:0 in the S. meliloti fabZXL mutant but not in the parent strain also suggests that the second potential FabZ protein is less effective at dehydrating longer chain hydroxyacyl-ACPs than the cluster FabZXL protein, leading to the accumulation of this slightly shorter VLCFA.
Why Did We Only Observe Partial Complementation of the S. meliloti fabF2XL Mutant?
We found that, as in R. leguminosarum (12), the S. meliloti FabF2XL protein is essential for the biosynthesis of VLCFA-modified lipid A in S. meliloti. However, we were unable to fully complement the lipid A alteration of this mutant strain. Because the fabF2XL gene is the third gene in a four-gene operon (Fig. 1A), the plasmid insertion may be exerting a polar effect on the downstream fabF1XL gene. Therefore, full complementation of this mutant strain might require both the plasmid-encoded fabF2XL and fabF1XL genes. However, because we did not experience complementation problems with any of the other cluster insertional mutants, this suggests that a promoter on the plasmid used for the insertional mutagenesis can drive transcription of the downstream genes. Another possibility to account for our inability to fully complement the lipid A alteration of the S. meliloti fabF2XL might be that the level of FabF2XL is critical for the proper biosynthesis of the VLCFA-modified lipid A and that the plasmid-encoded fabF2XL gene did not enable the correct expression level of this protein. Both the FabF1XL and FabF2XL proteins are predicted to encode 3-oxo-ACP synthases, which catalyze the reaction of the acyl-ACP with malonyl-ACP to produce a 3-oxoacyl-ACP (10). Therefore, if both of these proteins have different preferences for the chain length of the acyl-ACP, changing the levels of FabF2XL might affect the biosynthesis of VLCFA-modified lipid A. However, we found that the plasmid-encoded fabF1XL gene fully complemented the lipid A alteration of the S. meliloti fabF1XL mutant, suggesting this was not the case with FabF1XL. As observed in the S. meliloti fabZXL mutant, we also detected small amounts of 25OH-C26:0 in the S. meliloti fabF2XL mutant with pSmfabF2XL. Because this fatty acid is not normally found in the parent strain LPS, this suggests that too much FabF2XL might also lead to an imbalance in synthesis, leading to the accumulation of 25OH-C26:0.
Why Is the VLCFA Biosynthesis Cluster Important for Nodule Colonization?
We determined that the fabF2XL, fabF1XL, fabZXL, and adhA2XL gene products enhanced the competitiveness of S. meliloti within alfalfa nodules. Although we did observe a small number of nodules where the mutant strains out-competed the parent strain, it is likely that this is due to variability in the plant response because the alfalfa seedlings are not clonal. Upon entry into the root nodules, S. meliloti encounters a number of different stresses within the symbiosome compartment, including low oxygen, acidic pH, osmotic changes, and nodule-specific cysteine-rich peptides (1). Based on our salt growth assay, we discovered that the VLCFA-modified lipid A is critical for growth of S. meliloti in the absence of sodium chloride. Because low levels of cations can help stabilize interactions between different LPS species (6), these findings provide further evidence that the VLCFA modification plays a key role in outer membrane stability, enabling S. meliloti to grow in the absence of salt-LPS stabilizing conditions. We found previously that, unlike the other cluster mutants, the S. meliloti lpxXL mutant is unable to synthesize penta-acylated lipid A (9). Because the S. meliloti lpxXL mutant has the greatest salt requirement for growth of the cluster mutants, this suggests that the inability of this mutant to synthesize penta-acylated lipid A dramatically decreases the stability of its outer membrane relative to the other mutants. Therefore, the reduced competitiveness of the cluster mutants in legume colonization might just arise through an increased sensitivity to symbiosome-encountered stresses relative to the parent strain due to weakened outer membranes.
It was hypothesized previously that the VLCFA might affect the ability of rhizobia to suppress the plant defense response upon entry into the plant cell (9, 10, 30, 31). However, we observed no substantial increase in the production of autofluorescent plant defense polyphenolics in the root nodules of alfalfa after infection with either the S. meliloti acpXL or lpxXL mutants. Because we showed previously that LpxXL is essential for the biosynthesis of S. meliloti bacteroid VLCFA-modified lipid A (5), these findings provide evidence that the lipid A VLCFA is not essential to suppress the plant defense response upon infection of S. meliloti. This finding is consistent with a previous study whereby the purified LPS from an S. meliloti acpXL mutant was as effective as the parent strain LPS in suppressing an artificially induced oxidative burst in Medicago truncatula and alfalfa tissue culture systems (32). However, we cannot rule out at this stage the possibility that the altered lipid A structures in our VLCFA biosynthesis mutants have more subtle effects on the legume defense response relative to the parent strain, which could not be detected using this experimental approach.
Why Is the Cluster Mutant LPS Less Immunogenic than the Parent Strain LPS?
It is well established that the VLCFA-modified LPS species produced by rhizobia are less immunogenic than E. coli LPS, which lack these modifications (27, 33). The VLCFA-modified LPS of Rhizobium sin-1 also antagonizes the ability of E. coli LPS to induce monocytes to produce cytokines, and using a chemical synthesis approach, the VLCFA modification was shown to be important for the antagonist activity of synthetic lipid A (33, 34). In this study, we found that purified S. meliloti LPS from the cluster mutants, which completely lacks the VLCFA, was reduced or unable to induce human monocytes to produce cytokines relative to the VLCFA-modified LPS of the parent strain. Therefore, this finding demonstrates that the low immunogenicity of S. meliloti LPS relative to E. coli LPS is not due to the presence of VLCFAs. The bacterial lipid A component of the LPS is recognized by monocytes via Toll-like receptor 4 (TLR4) (35). E. coli lipid A is hexa-acylated whereas S. meliloti LPS is penta- and tetra-acylated (6). Consequently, the differences in the ability of S. meliloti and E. coli LPS to induce cytokine production from monocytes might be due to differences in the degree of acylation of their LPS affecting their interaction with TLR4. This could also account for why the LPS species of the cluster mutants are less immunogenic than the S. meliloti parent strain LPS because a proportion of their LPS species are only triacylated. This could also explain the results obtained with LPS from the lpxXL mutant because, unlike the LPS from the other mutant strains, it does not contain any penta-acylated lipid A species (9). In addition, the LPS from the cluster mutants might be less effective than the parent strain LPS because the VLCFA modification may aid the interaction with the Toll-like receptors. Because we found that the cluster mutant LPS lacks HMW species, HMW LPS might also be better at inducing monocyte cytokine production relative to LMW LPS. However, further studies would need to be performed to determine how the different lipid A species are recognized by the human monocyte receptors.
In conclusion, this study, combined with previous work (9, 10), has found that all of the genes in the S. meliloti acpXL-lpxXL cluster are involved in the biosynthesis of VLCFA-modified LPS. Other rhizobia and Brucella species, which form prolonged mammalian infections, also produce VLCFA-modified LPS and have an acpXL-lpxXL gene cluster (7). Therefore, these studies will provide important insights into the role of this unusual bacterial lipid modification in both plant and mammalian infections.
This work was supported in part by Biotechnology and Biological Sciences Research Council Grant BB/D000564/1 and Medical Research Council New Investigator Grant G0501107 (to G. P. F.). Structural analysis was supported in part by Department of Energy Grant DE-FG02-09ER20097 (to the Complex Carbohydrate Research Center).
- VLCFA
- very long-chain fatty acid
- HMW
- high molecular weight
- LMW
- low molecular weight
- ACP
- acyl carrier protein.
REFERENCES
- 1. Gibson K. E., Kobayashi H., Walker G. C. (2008) Annu. Rev. Genet. 42, 413–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones K. M., Kobayashi H., Davies B. W., Taga M. E., Walker G. C. (2007) Nat. Rev. Microbiol. 5, 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mergaert P., Uchiumi T., Alunni B., Evanno G., Cheron A., Catrice O., Mausset A. E., Barloy-Hubler F., Galibert F., Kondorosi A., Kondorosi E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campbell G. R., Reuhs B. L., Walker G. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3938–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haag A. F., Wehmeier S., Beck S., Marlow V. L., Fletcher V., James E. K., Ferguson G. P. (2009) J. Bacteriol. 191, 4681–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhat U. R., Carlson R. W., Busch M., Mayer H. (1991) Int. J. Syst. Bacteriol. 41, 213–217 [DOI] [PubMed] [Google Scholar]
- 8. Ferguson G. P., Datta A., Baumgartner J., Roop R. M., 2nd, Carlson R. W., Walker G. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferguson G. P., Datta A., Carlson R. W., Walker G. C. (2005) Mol. Microbiol. 56, 68–80 [DOI] [PubMed] [Google Scholar]
- 10. Sharypova L. A., Niehaus K., Scheidle H., Holst O., Becker A. (2003) J. Biol. Chem. 278, 12946–12954 [DOI] [PubMed] [Google Scholar]
- 11. Ardissone S., Kobayashi H., Kambara K., Rummel C., Noel K. D., Walker G. C., Broughton W. J., Deakin W. J. (2011) J. Bacteriol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vanderlinde E. M., Muszynski A., Harrison J. J., Koval S. F., Foreman D. L., Ceri H., Kannenberg E. L., Carlson R. W., Yost C. K. (2009) Microbiology 155, 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., p. A.1, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 14. Serrania J., Vorhölter F. J., Niehaus K., Pühler A., Becker A. (2008) J. Biotechnol. 135, 309–317 [DOI] [PubMed] [Google Scholar]
- 15. Galkiewicz J. P., Kellogg C. A. (2008) Appl. Environ. Microbiol. 74, 7828–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katzen F., Becker A., Ielmini M. V., Oddo C. G., Ielpi L. (1999) Appl. Environ. Microbiol. 65, 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finan T. M., Kunkel B., De Vos G. F., Signer E. R. (1986) J. Bacteriol. 167, 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wells D. H., Long S. R. (2002) Mol. Microbiol. 43, 1115–1127 [DOI] [PubMed] [Google Scholar]
- 19. Reuhs B. L., Kim J. S., Badgett A., Carlson R. W. (1994) MPMI 7, 240–247 [DOI] [PubMed] [Google Scholar]
- 20. Gudlavalleti S. K., Forsberg L. S. (2003) J. Biol. Chem. 278, 3957–3968 [DOI] [PubMed] [Google Scholar]
- 21. Muszynski A., Laus M., Kijne J. W., Carlson R. W. (2011) Glycobiology 21, 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhat U. R., Forsberg L. S., Carlson R. W. (1994) J. Biol. Chem. 269, 14402–14410 [PubMed] [Google Scholar]
- 23. York W. S., Darvill A. G., Mcneil M., Stevenson T. T., Albersheim P. (1986) Methods Enzymol. 118, 3–40 [Google Scholar]
- 24. Leigh J. A., Signer E. R., Walker G. C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beck S., Marlow V. L., Woodall K., Doerrler W. T., James E. K., Ferguson G. P. (2008) Microbiology 154, 1258–1270 [DOI] [PubMed] [Google Scholar]
- 26. Schild S., Lamprecht A. K., Reidl J. (2005) J. Biol. Chem. 280, 25936–25947 [DOI] [PubMed] [Google Scholar]
- 27. Tsukushi Y., Kido N., Saeki K., Sugiyama T., Koide N., Mori I., Yoshida T., Yokochi T. (2004) J. Endotoxin Res. 10, 25–31 [DOI] [PubMed] [Google Scholar]
- 28. Galibert F., Finan T. M., Long S. R., Puhler A., Abola P., Ampe F., Barloy-Hubler F., Barnett M. J., Becker A., Boistard P., Bothe G., Boutry M., Bowser L., Buhrmester J., Cadieu E., Capela D., Chain P., Cowie A., Davis R. W., Dreano S., Federspiel N. A., Fisher R. F., Gloux S., Godrie T., Goffeau A., Golding B., Gouzy J., Gurjal M., Hernandez-Lucas I., Hong A., Huizar L., Hyman R. W., Jones T., Kahn D., Kahn M. L., Kalman S., Keating D. H., Kiss E., Komp C., Lelaure V., Masuy D., Palm C., Peck M. C., Pohl T. M., Portetelle D., Purnelle B., Ramsperger U., Surzycki R., Thebault P., Vandenbol M., Vorholter F. J., Weidner S., Wells D. H., Wong K., Yeh K. C., Batut J. (2001) Science 293, 668–672 [DOI] [PubMed] [Google Scholar]
- 29. Mohan S., Kelly T. M., Eveland S. S., Raetz C. R., Anderson M. S. (1994) J. Biol. Chem. 269, 32896–32903 [PubMed] [Google Scholar]
- 30. Vedam V., Haynes J. G., Kannenberg E. L., Carlson R. W., Sherrier D. J. (2004) MPMI 17, 283–291 [DOI] [PubMed] [Google Scholar]
- 31. Vedam V., Kannenberg E., Datta A., Brown D., Haynes-Gann J. G., Sherrier D. J., Carlson R. W. (2006) J. Bacteriol. 188, 2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheidle H., Gross A., Niehaus K. (2005) New Phytol. 165, 559–565 [DOI] [PubMed] [Google Scholar]
- 33. Vandenplas M. L., Carlson R. W., Jeyaretnam B. S., McNeill B., Barton M. H., Norton N., Murray T. F., Moore J. N. (2002) J. Biol. Chem. 277, 41811–41816 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y., Wolfert M. A., Boons G. J. (2007) Bioorg. Med. Chem. 15, 4800–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hold G. L., Bryant C. E. (2011) in Bacterial Lipopolysaccharides (Knirel Y. A., Valvano M. A. eds) 1st Ed., Springer, Berlin [Google Scholar]
- 36. Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. (1982) J. Bacteriol. 149, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]