Abstract
Monoglyceride lipase (MGL) influences energy metabolism by at least two mechanisms. First, it hydrolyzes monoacylglycerols (MG) into fatty acids and glycerol. These products can be used for energy production or synthetic reactions. Second, MGL degrades 2-arachidonoyl glycerol (2-AG), the most abundant endogenous ligand of cannabinoid receptors (CBR). Activation of CBR affects energy homeostasis by central orexigenic stimuli, by promoting lipid storage, and by reducing energy expenditure. To characterize the metabolic role of MGL in vivo, we generated an MGL-deficient mouse model (MGL-ko). These mice exhibit a reduction in MG hydrolase activity and a concomitant increase in MG levels in adipose tissue, brain, and liver. In adipose tissue, the lack of MGL activity is partially compensated by hormone-sensitive lipase. Nonetheless, fasted MGL-ko mice exhibit reduced plasma glycerol and triacylglycerol, as well as liver triacylglycerol levels indicative for impaired lipolysis. Despite a strong elevation of 2-AG levels, MGL-ko mice exhibit normal food intake, fat mass, and energy expenditure. Yet mice lacking MGL show a pharmacological tolerance to the CBR agonist CP 55,940 suggesting that the elevated 2-AG levels are functionally antagonized by desensitization of CBR. Interestingly, however, MGL-ko mice receiving a high fat diet exhibit significantly improved glucose tolerance and insulin sensitivity in comparison with wild-type controls despite equal weight gain. In conclusion, our observations implicate that MGL deficiency impairs lipolysis and attenuates diet-induced insulin resistance. Defective degradation of 2-AG does not provoke cannabinoid-like effects on feeding behavior, lipid storage, and energy expenditure, which may be explained by desensitization of CBR.
Keywords: Adipose Tissue, Lipase, Lipolysis, Liver, Monoacylglycerol, 2-Arachidonoyl Glycerol, Endocannabinoids
Introduction
Monoacylglycerols (MG)3 are short lived intermediates of lipid catabolism derived from extracellular or intracellular sources. Pancreatic lipase and lipoprotein lipase generate MG by the hydrolysis of dietary triacylglycerols (TG) and circulating lipoproteins, respectively (1, 2). The lipolytic products, MG and free fatty acids (FFA), are subsequently taken up by cells, and MG are hydrolyzed into FFA and glycerol or re-esterified by the monoacylglycerol acyltransferase reaction (3). Within cells, MG are derived from the hydrolysis of glycerophospholipids or TG. Glycerophospholipids may be degraded by phospholipase C generating sn-1,2-diacylglycerols (DG), which are further hydrolyzed by sn-1-specific DG lipase resulting in the formation of 2-MG (4). The breakdown of TG is initiated by adipose triglyceride lipase (ATGL), and the produced DG is hydrolyzed by hormone-sensitive lipase (HSL) (5). The stereospecificity of ATGL has not been studied so far. Yet, similar to DG lipase, HSL hydrolyzes DG preferentially in sn-1(3) position generating 2-MG (6). Monoglyceride lipase (MGL) degrades sn-1(3) and 2-MG at identical specific rates (7). The enzyme is expressed in most cell types and is considered the rate-limiting enzyme in the degradation of MG (8, 9).
In addition to its metabolic role, MGL was identified as an important component of the endocannabinoid system. The enzyme degrades 2-arachidonoyl glycerol (2-AG), the most abundant endogenous ligand of cannabinoid receptors (CBR1 and CBR2) (10). 2-AG predominantly derives from the degradation of glycerophospholipids by the consecutive action of phospholipase C and DG lipase (4). Endogenous CBR-activating compounds are collectively designated as endocannabinoids (EC). The best characterized ECs are N-arachidonoylethanolamine (AEA, anandamide) and 2-AG. Their biological effect is mimicked by Δ9-tetrahydrocannabinol, the major psychoactive component of marijuana (11). In the brain, EC are released from postsynaptic neurons and cause retrograde suppression of synaptic transmission. In peripheral tissues, the EC system is active in the autonomic nervous system, in immune cells, and several other cell types, including hepatocytes and adipocytes. EC affect numerous neuronal processes, including learning, motor control, cognition, and pain. Moreover, the EC system is involved in the regulation of food intake and lipid metabolism. This has been demonstrated by the effects of treating obese patients with the CBR1 antagonist rimonabant and in animal models lacking CBR1 (12–14). In general, CBR1 deficiency or antagonist-mediated blockade of CBR1 reduces food intake and inhibits peripheral lipogenesis. Conversely, an overactive EC system affects energy homeostasis via a central orexigenic effect by promoting lipogenesis in peripheral tissues, including liver and WAT, and by reducing energy expenditure. Because of their capability of modulating energy homeostasis, ECs have been linked to the pathogenesis of metabolic diseases such as obesity and type 2 diabetes (15).
To characterize the role of MGL in energy metabolism, we generated a mouse model systemically lacking the enzyme (MGL-ko). MGL deficiency results in accumulation of 2-AG and other MG species in various tissues, reduces adipose tissue lipolysis, liver VLDL secretion, and attenuates diet-induced insulin resistance. Yet defective degradation of 2-AG does not produce cannabinoid-like effects such as stimulation of food intake and peripheral lipogenesis, which may be explained by desensitization of CBRs.
EXPERIMENTAL PROCEDURES
Animals
Mice totally lacking MGL were bred by crossing mice heterozygous for the mutant MGL allele, which were backcrossed 4–5 times on the C57Bl/6 background. In all experiments, we used wild-type littermates as controls. Mice were maintained on a regular light-dark cycle (12 h light, 12 h dark) with a room temperature of 22 ± 1 °C and kept on a standard laboratory chow diet (4.5% w/w fat). For diet studies, mice were kept on a high fat diet for 12 weeks (54% w/w fat; Sniff, Soest, Germany). Animals were anesthetized with IsoFlo®/isoflurane (Abbott) and euthanized by cervical dislocation. The study was approved by the Austrian ethics committee and is in accordance with the council of Europe Convention (ETS 123).
Generation of MGL-deficient Mice
To generate the MGL gene targeting vector, a synthetic oligonucleotide encoding a loxP sequence flanked by the EcoRV restriction site at the 5′ end and KpnI and HindIII restriction site at the 3′ end, respectively, was digested with EcoRV and HindIII restriction enzymes and ligated into pBluescript II KS(−) phagemid vector (Stratagene, La Jolla, CA). The 5′ part of the long arm of the gene targeting vector, a 2.0-kb MGL genomic sequence encompassing MGL exon 3 and 4, was amplified by PCR from genomic murine HM-1 embryonic stem (ES) cell DNA using PhusionTM High Fidelity DNA polymerase (Finnzymes, Espoo, Finland). The following PCR primers were used in the reaction: forward, 5′-GGA CTA GTA TGT GTG ATC TCC TAA TGC TGC-3′; reverse, 5′-TTG ATA TCT GGC CAG TTC AAC ATG CAC-3′. The PCR primers introduced a synthetic SpeI and EcoRV restriction site (underlined). The PCR product was digested with SpeI and EcoRV restriction enzymes and ligated into the plasmid harboring the loxP sequence. In the following step, a 2.0-kb MGL intron 2 sequence was amplified by PCR generating the short arm of the targeting vector. The following PCR primers were used in the reaction: forward, 5′-GAG CGG CCG CCA CCA GGA GCT AAG AGC GAT G-3′; reverse, 5′-GGA CTA GTC TGC AGC ATT AGG AGA TCA CAC-3′. The PCR product contained a synthetic NotI and SpeI restriction site (underlined) introduced by the amplification reaction. After a NotI-SpeI digest, the PCR product was ligated into the NotI-XbaI restriction site of pBK-CMV phagemid vector (Stratagene) harboring a loxP-flanked neomycin resistance cassette. The resulting plasmid was digested with NotI and SpeI restriction enzymes, and the 3.2-kb DNA fragment encompassing the short arm of the targeting vector and the selection cassette was cloned into the plasmid containing the insertion of the 5′ part of the long arm and the single loxP site. To generate the 3′ part of the long arm of the targeting vector, a 5.0-kb MGL intron 4 sequence was amplified from genomic HM-1 ES cell DNA by PCR. The PCR was performed with the following primer pair: forward, 5′-TTG ATA TCG TTT GTG CAT GTT GAA CTG GC-3′; reverse, 5′-CCA TCG ATT GGC CCG AGT CAC AGA CAT AG-3′. The PCR primers introduced a synthetic EcoRV and ClaI restriction site (underlined). After EcoRV-ClaI digest, the DNA fragment was cloned into the HindIII filled in ClaI restriction site of the plasmid harboring the insertion of the short arm and 5′ part of the long arm of the targeting vector. A diphtheria toxin fragment A cassette was introduced 5′ to the short arm to generate the final targeting vector. The linearized targeting vector was transferred into HM-1 ES cells by electroporation. G418-resistant clones were picked and propagated. Clones that underwent homologous recombination were transfected with a Cre-expressing plasmid to generate ES cell clones harboring an MGL null allele. Two independent clones were injected into 3.5-day-old C57Bl/6 blastocysts and transferred into pseudo-pregnant recipient mice. Chimeric animals with high degree of coat color chimerism were bred with C57Bl/6 mice. Germ line transmission of the MGL manipulated allele was observed by coat color and verified by Southern blotting.
Southern Blotting Analysis
Genomic DNA was prepared with the DNeasy® blood and tissue kit (Qiagen Inc., Hilden, Germany) according to the manufacturer's protocol. For restriction fragment analysis, 10 μg of genomic DNA were digested with KpnI. DNA fragments were separated by agarose gel electrophoresis followed by depurination with 0.2 n HCl and denaturation with 0.5 n NaOH. After neutralization with Tris-HCl, pH 7.5, DNA was transferred onto a Hybond-N+ membrane (GE Healthcare) and hybridized with a 32P-labeled probe. The external probe for DNA analysis was generated by PCR from HM-1 genomic ES cell DNA using the following primer pair: forward, 5′-CGG AAT TCC ACA GCC TGG TCC TCA GAA AG-3′; reverse, 5′-CGG AAT TCC GTA ACC ACC ATG CTC AAT CG-3′. The PCR primers introduced EcoRI restriction enzyme sites (underlined). The PCR product was ligated into respective restriction enzyme site of pBluescript II KS(−) phagemid vector. Gene-specific probe for Southern blot analysis was labeled with [α-32P]dCTP (GE Healthcare) using the Prime-a-Gene® DNA labeling kit according to the manufacturer's protocol (Promega, Madison, WI). After hybridization and washing, signals were visualized by exposure to PhosphorImager Screen (GE Healthcare).
Western Blotting Analysis
Proteins of tissue homogenates were separated by 10% SDS-PAGE according to standard protocols, blotted onto polyvinylidene fluoride membrane (Carl Roth GmbH, Karlsruhe, Germany), and hybridized with a rabbit polyclonal antibody raised against murine MGL or SREBP-1c (Abcam, Cambridge, UK). Specifically bound immunoglobulins were detected in a second reaction using horseradish peroxidase-conjugated anti-rabbit IgG antibody and visualized by enhanced chemiluminescence detection (ECL, GE Healthcare). For detection of nuclear SREBP-1, liver nuclear extracts were prepared with NE-PER® nuclear and cytoplasmic extraction reagents obtained from Pierce.
Quantitative Real Time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse-transcribed using high capacity cDNA reverse transcription kit (Applied Biosystems, CA). Quantitative RT-PCR (20 μl) contained 8 ng of cDNA, 10 pmol of each primer, and 10 μl of SYBR Green master mix (Fermentas) and was performed using StepOne Plus real time PCR system (Applied Biosystems). Relative mRNA levels were quantified using the ΔΔCt method with β-actin as reference gene. The following primer pairs were used for quantitative RT-PCR: β-actin forward, 5′-AGC CAT GTA CGT AGC CAT CCA-3′, and reverse, 5′-TCT CCG GAG TCC ATC ACA ATG-3′; fatty-acid synthase forward, 5′-TGC TCC CAG CTG CAG GC-3′, and reverse, 5′-GCC CGG TAG CTC TGG GTG TA-3′; HSL forward, 5′-GCT GGG CTG TCA AGC ACT GT-3′, and reverse, 5′-GTA ACT GGG TAG GCT GCC AT-3′; ABHD12 forward, 5′-TTG ACT GGT TCT TCC TCG ACC CC-3′, and reverse, 5′-CCG AGA TGG TGC AGC AAT GTT G-3′; and ABHD6 forward, 5′-AAG ACC AGG TGC TTG ATG TGT CC-3′, and reverse, 5′-TCC ATC ACT ACC GAA TGG CCA CAG-3′.
Determination of MG Hydrolase (MGH) Activity
Tissues were washed with phosphate-buffered saline (PBS) and homogenized on ice in lysis buffer A (0.25 m sucrose, 1 mm EDTA, 20 μm dithiothreitol, 0.1% Triton X-100, 20 μg/ml leupeptin, 2 μg/ml antipain, 1 μg/ml pepstatin, pH 7.0) using an Ultra Turrax (IKA, Staufen, Germany). Lysates were centrifuged at 20,000 × g for 1 h at 4 °C, and the lipid-poor infranatant was used for measuring MGH activity. COS-7 cells expressing HSL were disrupted by sonication and centrifuged at 1,000 × g. The post-nuclear fraction was used for the determination of MGH activity. Substrates for determination of MGH activity contained 1 mm rac-1(3)-oleoyl glycerol (OG), 2-OG, or 2-AG (Sigma) bound to defatted BSA in an equimolar ratio in 50 mm potassium phosphate buffer, pH 7.0, as described previously (16). Protein concentrations of tissue and cell lysates were determined using the Bio-Rad protein assay according to the manufacturer's instructions (Bio-Rad) and bovine serum albumin (BSA) as standard. Depending on the tissue, 1–20 μg of protein was incubated with 100 μl of MG substrate for 30 min at 37 °C. Thereafter, the reaction was stopped by the addition of 100 μl of chloroform and vortexing. Phases were separated by centrifugation at 10,000 × g for 5 min at 4 °C. Glycerol concentrations in the aqueous phase were determined using a commercial kit (free glycerol reagent, Sigma).
Thin Layer Chromatography (TLC) Analysis of WAT MG
Total lipids were extracted using the Folch extraction procedure. Aliquots of the lipid extract were applied onto silica gel plates (Merck) and separated with chloroform/acetone/acetic acid (90:8:1) as the solvent system. rac-1(3)-OG and 2-OG were used as standards. Lipids were visualized by carbonization after spraying the plates with 10% CuSO4 (w/v), 10% H3PO4 (v/v) and heating them to 120 °C for 30 min.
Enzymatic Determination of Tissue MG and TG Content
For determination of tissue TG concentrations, lipid extracts were dried and resuspended by sonication in 0.1% Triton X-100 (two times for 10 s, 10% output, Sonicator 4000, Misonix). The TG content of lipid suspensions was determined using a commercial kit (InfinityTM triglycerides, Thermo Scientific).
For the determination of total tissue MG concentrations, lipid extracts corresponding to ∼0.3 g of tissue were dried under nitrogen and solubilized in 300 μl of buffer B (150 mm NaCl, 50 mm potassium phosphate buffer, pH 7.0, 0.1% Triton X-100, 5% fatty acid-free BSA) by sonication (two times for 10 s, 10% output, Sonicator 4000, Misonix). Subsequently, emulsified lipids were incubated for 30 min at 37 °C in the presence of 1 μg of recombinant His-tagged mouse MGL (mMGL) purified from Escherichia coli. mMGL exhibited a specific activity of 4 mmol of MG/h·mg protein. Subsequently, the mixture was washed two times with a 4-fold volume of water-saturated butanol and centrifuged. Aliquots of the aqueous phase were used for glycerol determination using a commercial kit (free glycerol reagent, Sigma).
HPLC-MS Analysis of MG
For the determination of specific MG species, we used HPLC-MS. Lipids were extracted according to Bligh and Dyer and resuspended in 200 μl of chloroform/methanol (1:1). MG 17:0 was added as internal standard prior to lipid extraction. For liver (∼0.2 g) and plasma samples (50 μl), the amount of internal standard added was 2 nmol. For brain samples (∼0.2 g), the amount was 20 nmol. For separation of lipids by HPLC, we used the Hypersil GOLD C18 column (100 × 1 mm, 1.9 μm particle size, Thermo Scientific). Water (solvent A) and methanol/2-propanol (5:2, solvent B) both containing 1% ammonium acetate and 0.1% formic acid were used as solvent system. The gradient started at 2% of solvent B, increased to 70% within 7 min and then to 98% within 4 min, and remained at this level for 3 min. The flow rate was 250 μl/min. 2-AG was determined by a TSQ triple quadrupole instrument in positive ESI mode (Thermo Scientific). The spray voltage was set to 4500 V, capillary voltage to 35 V, and vaporizer temperature to 370 °C. 2-AG and MG 17:0 (internal standard) were detected by single reaction monitoring. The transitions were m/z 379 to m/z 287 at 11 eV collision energy for 2-AG and m/z 345 to m/z 253 at collision energy of 14 eV. Peak areas were calculated by QuanBrowser for 2-AG and the internal standard. Quantitation was done by calculating 2-AG/MG 17:0 ratios and using a calibration curve for each batch of samples.
Plasma Parameters
Blood samples of fed or fasted mice were collected by retro-orbital puncture. Plasma levels of TG, glycerol, cholesterol, ketone bodies, and FFA were determined using commercial kits (Thermo Electron Corp., Victoria, Australia; Sigma; Roche Diagnostics). Blood glucose was determined using Accu-Check glucometer and Accutrend® Plus (Roche Diagnostics). Plasma insulin was determined by rat insulin ELISA kit (Chrystal Chem Inc., Downers Grove, IL).
Organ Cultures of WAT
Gonadal fat pads were surgically removed and washed extensively with PBS. Tissue pieces (∼20 mg) were incubated in Dulbecco's modified Eagle's medium (Invitrogen) containing 2% fatty acid-free bovine serum albumin (Sigma) either in the presence or absence of 10 μm forskolin at 37 °C for 1 h. Thereafter, fat pads were transferred into identical fresh medium and incubated for another 60 min at 37 °C. Thereafter, aliquots of the medium were removed and analyzed for FFA and glycerol content using commercial kits (Wako Chemicals; Sigma). For protein determinations, fat pads were washed extensively with PBS and lysed in 0.3 n NaOH, 0.1% SDS. Protein measurements were performed using the BCA reagent (Pierce).
Animal Experiments
To measure spontaneous physical activity, O2 consumption, CO2 production, and food intake, mice were housed in metabolic cages allowing continuous measurement of these parameters (LabMaster, TSE Systems GmbH, Bad Homburg, Germany). Before monitoring, animals were familiarized with these cages for at least 3 days.
Insulin and Glucose Tolerance Tests
For insulin and glucose tolerance tests (ITT and GTT, respectively), mice were fasted for 4 and 6 h, respectively. Mice were injected intraperitoneally with bovine insulin (Sigma, 0.75 units/kg) or with 20% glucose (1.5 g/kg). Blood samples were taken at 0, 15, 30, 120, and 180 min.
Statistical Analyses
Data are shown as mean ± S.D. Areas under the curve were calculated by manual integration. Statistical significance was determined by Student's unpaired t test (two-tailed). Group differences were considered different for p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
RESULTS
Generation of MGL-deficient Mice
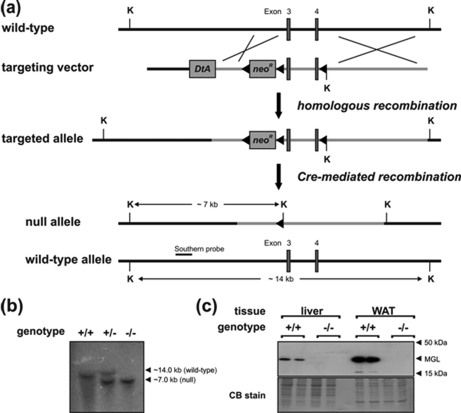
MGL-ko mice were generated using the Cre/loxP recombination system (Fig. 1a). For that purpose, we designed a targeting vector harboring a loxP-flanked neomycin resistance gene cassette upstream to exon 3 and an additional loxP recombination site within intron 4 of the murine MGL gene. This vector was introduced into ES cells by electroporation, and cells that underwent homologous recombination were subsequently transfected with a plasmid encoding Cre-recombinase. ES cell clones harboring an MGL null allele were used for blastocyst injection. Southern blotting analysis confirmed disruption of the MGL gene (Fig. 1b). MGL protein expression was not detectable in tissues of mice homozygous for the targeted allele (Fig. 1c). MGL-ko mice were born within the Mendelian frequency. Male as well as female MGL-ko mice were fertile and exhibited no obvious phenotype.
FIGURE 1.
Targeting strategy and generation of MGL-ko mice. a, homologous recombination of the targeting vector with the wild-type allele resulted in the introduction of a loxP site (◀) into intron 4 and a neomycin resistance gene cassette flanked by loxP sites into intron 2 of the MGL gene. Subsequent Cre-recombinase-mediated recombination among the distal loxP sites resulted in the deletion of exon 3 and 4 of the MGL gene. The targeted allele was identified by KpnI (K) restriction digest and hybridization with an external Southern probe (solid bar) revealing a 7.0-kb DNA fragment. b, genomic DNA from mice was digested with KpnI and analyzed by Southern blotting using an external probe specific for intron 2 of the MGL gene. Autoradiography signals obtained from DNA fragments of 14.0 and 7.0 kb corresponded to wild-type (+) and targeted MGL (−) alleles, respectively. c, Western blotting analysis of MGL protein expression levels was performed with lysates of WAT and liver using a rabbit polyclonal MGL antiserum. The polyvinylidene fluoride membrane was stained with Coomassie Blue (CB) as loading control.
Biochemical Characterization
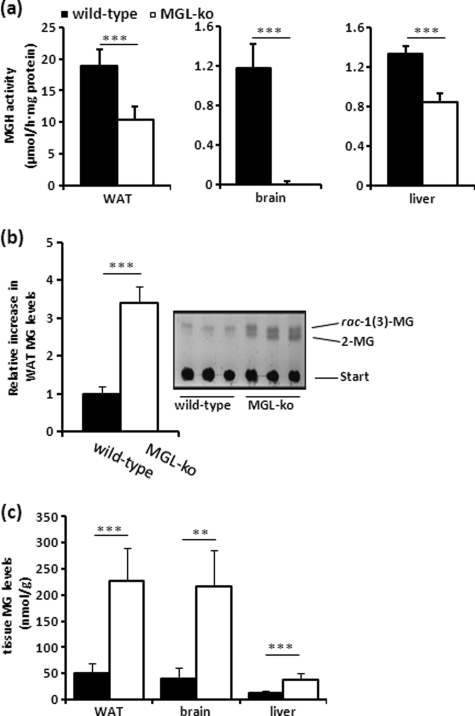
MGH activity assays indicated a decrease in neutral MGH activity in WAT (−45%) and liver (−35%) and an almost undetectable activity in brain (Fig. 2a). RT-PCR analysis indicated that the lack of MGL was not associated with a compensatory up-regulation of mRNA levels of other MG hydrolases in these tissues such as ABHD6, ABHD12, and HSL (supplemental Fig. S1) (17, 18). TLC analysis of wild-type WAT lipids revealed a single band co-migrating with the sn-1(3)-MG standard (Fig. 2b). In MGL-ko WAT extracts, we observed the accumulation of two bands co-migrating with sn-1(3)- and 2-MG standards. These signals completely disappeared when tissue lipids were treated with recombinant purified mMGL confirming the accumulation of MG (data not shown). Densitometric quantitation of TLC signals (Fig. 2b) revealed that MG levels were increased ∼3.5-fold. Similar results were obtained when we determined the amounts of glycerol released by digestion of lipid extracts with mMGL (Fig. 2c). These determinations revealed concentrations of ∼50 and 225 nmol MG/g WAT obtained from wild-type and MGL-ko mice, respectively. Total MG levels in brain and liver of MGL-ko mice were increased ∼5- and 3-fold, respectively (Fig. 2c).
FIGURE 2.
MGL-ko mice exhibit reduced MGH activity and accumulate MG in WAT, brain, and liver. a, MGH activities were determined in 20,000 × g infranatants of WAT, brain, and liver using rac-1(3)-oleoylglycerol (OG) as substrate. b, lipid extracts of WAT were separated by TLC and visualized by charring at 120 °C. Quantitation of TLC signals was performed using ImageQuant software. c, lipid extracts of WAT, brain, and liver were digested in a buffer containing purified recombinant mMGL exhibiting a specific activity of 4 mmol of MG/h··mg protein. The release of glycerol from lipid extracts was determined using a commercial kit (Sigma). Data are presented as mean ± S.D. (n = 5–6 for each genotype). **, p < 0.01; ***, p < 0.001.
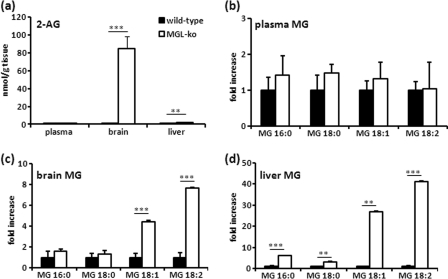
To detect specific MG species, we analyzed lipid extracts using HPLC-MS. The concentrations of 2-AG and other MG species were not significantly changed in plasma (Fig. 3, a and b). In brain, we detected an enormous increase in 2-AG (58-fold, Fig. 3a) and elevated levels of other unsaturated MG species (Fig. 3c). In liver, saturated as well as unsaturated MG species were increased between 2- and 40-fold (Fig. 3, a and d). Together, our measurements confirm a major role for MGL in the hydrolysis of MG in WAT, brain, and liver.
FIGURE 3.
MGL-ko mice accumulate 2-AG and other MG species in brain and liver. Tissue lipids were extracted with chloroform/methanol (1:1), and MG species were quantified by HPLC-MS using an internal standard (17:0 MG) and a standard curve for 2-AG. a, 2-AG concentrations in plasma, brain, and liver of wild-type and MGL-ko mice. b–d, relative changes in saturated and unsaturated MG species in plasma, brain, and liver. Data are presented as mean ± S.D. (n = 5 for each genotype). **, p < 0.01; ***, p < 0.001.
Role of MGL in WAT Lipolysis
To investigate the role of MGL in WAT lipolysis, we determined fatty acid and glycerol release in organ cultures of WAT (Fig. 4, a and b). In comparison with wild-type controls, forskolin-stimulated FA and glycerol release of MGL-ko fat pads were decreased by 24 and 43%, respectively. Nevertheless, hormonal stimulation enhanced basal glycerol release of MGL-ko fat pads 8-fold indicating that the lack of MGL activity is partially compensated by other enzymes.
FIGURE 4.
MGL and HSL are involved in MG degradation in WAT. a and b, basal and forskolin-stimulated release of FFA and glycerol from gonadal WAT. Fat pads (∼20 mg) were preincubated in Dulbecco's modified Eagle's medium, containing 2% defatted BSA in the absence or presence of 10 μm forskolin for 1 h at 37 °C. Thereafter, WAT pieces were transferred into identical fresh medium, and FFA and glycerol release were determined after incubation for another hour (n = 5 for each genotype). c, MGH activity in WAT was determined in 20,000 × g infranatants using various concentrations of rac-1(3)-OG as substrate. Experiments were performed in the absence or in the presence of the HSL inhibitor 76-0079 (n = 5 for each genotype). d, MGH activities were inhibited in 20,000 × g infranatants of WAT of wild-type mice using various concentrations of specific inhibitors for MGL (JZL 184), HSL (76-0079), or both inhibitors in combination (JZL184/76-0079). e, activity of HSL was determined in post-nuclear lysates (1000 × g supernatant) of COS-7 cells expressing murine HSL using 2-AG, rac-1(3)-OG, or 2-OG as substrate. The MGH activity detected in cells expressing β-galactosidase was set as blank. Data are presented as mean ± S.D. and are representative for at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
HSL is the rate-limiting lipase in DG hydrolysis in WAT and is also known to possess MGH activity (18). To test the involvement of HSL in MG hydrolysis, we performed MGH assays in WAT lysates in the presence of a specific HSL inhibitor (19). As shown in Fig. 4c, under saturating substrate concentrations MGH activity in MGL-ko lysates was reduced by ∼50% in comparison with wild-type lysates. In the presence of the HSL inhibitor (76-0079), MGH activity in MGL-ko lysates was strongly reduced. In wild-type WAT lysates, 76-0079 inhibited MGH activity up to 35% (Fig. 4d) indicating that the lack of MGL results in a moderate up-regulation of HSL activity, which is not accompanied by increased mRNA levels (supplemental Fig. S1). The MGL-specific inhibitor JZL184 (20) reduced MGH activity by 68% and the combined use of MGL and HSL inhibitors led to almost complete loss of MGH activity in wild-type adipose tissue (Fig. 4d). We also investigated MG hydrolase activity of HSL overexpressed in COS-7 cells. This enzyme is capable of hydrolyzing sn-1(3)-OG, 2-OG, and 2-AG with similar specific activity (Fig. 4e). The apparent Km values of the reactions of 0.27 ± 0.06, 0.25 ± 0.01, and 0.26 ± 0.05 mm for 2-AG, sn-1(3)-, and 2-OG, respectively, are close to the values reported for MGL (9). Together, these observations suggest that HSL accounts for most of the MGH activity detected in MGL-deficient WAT and partially compensates for the lack of MGL activity.
Plasma Lipid and Carbohydrate Analysis
To investigate how MGL deficiency affects energy metabolism in vivo, we analyzed plasma carbohydrate and lipid parameters in fed and fasted mice (Table 1). In the fed state, we observed a decrease in plasma glycerol levels by 33% in comparison with wild-type animals. All other blood parameters, including FFA, TG, cholesterol, and glucose concentrations, were unchanged. Fasted MGL-ko mice exhibited reduced ketone bodies (−16%), glycerol (−25%), and TG levels (−60%) indicative for impaired lipolysis. However, circulating FFA concentrations remained unchanged suggesting a rather moderate effect on lipolysis.
TABLE 1.
Plasma metabolites of fed and fasted mice
Values were obtained from male mice at the age of 12 weeks receiving a normal chow diet. Data are presented as mean ± S.D. (n = 6 for each genotype).
| Parameter | Fed |
Fasted (16 h) |
||
|---|---|---|---|---|
| Wild type | MGL-ko | Wild type | MGL-ko | |
| FFA (mm) | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.8 ± 0.2 | 0.8 ± 0.1 |
| Glycerol (mm) | 0.3 ± 0.1 | 0.2 ± 0.1a | 0.4 ± 0.1 | 0.3 ± 0.1b |
| TG (mm) | 0.9 ± 0.3 | 0.7 ± 0.5 | 0.7 ± 0.3 | 0.3 ± 0.1b |
| Cholesterol (mg/dl) | 118 ± 25 | 127 ± 26 | 112 ± 25 | 105 ± 23 |
| Glucose (mg/dl) | 164 ± 24 | 160 ± 19 | 87 ± 17 | 104 ± 22 |
| Ketone bodies (mm) | NAc | NA | 1.1 ± 0.2 | 0.92 ± 0.11a |
a p < 0.05.
b p < 0.001.
c NA means not analyzed.
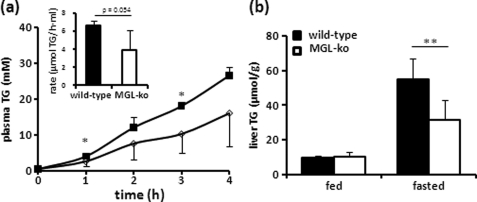
VLDL Secretion and Liver TG levels
To test whether the pronounced reduction in plasma TG levels is caused by reduced VLDL secretion, fasted wild-type and MGL-ko mice were treated with poloxamer 407 (P407), and plasma TG levels were monitored over a period of 4 h. P407 inhibits lipoprotein lipase activity and prevents the degradation of TG-rich lipoproteins (21). As shown in Fig. 5a, TG secretion in P407-treated MGL-ko mice was decreased by 40% compared with wild-type controls. Liver TG levels were unchanged in the fed state and reduced by 40% in the fasted state (Fig. 5b) suggesting that the reduction in VLDL secretion in fasted mice may be caused by a reduced transport of WAT-derived FFA to the liver. Activation of CBR1 was shown to promote liver steatosis through the activation of SREBP-1c (14). However, despite a substantial increase in liver 2-AG levels (Fig. 4a), the level of nuclear SREBP-1 and mRNA levels of its target gene fatty-acid synthase remained unchanged (supplemental Fig. S2).
FIGURE 5.
Fasted MGL-ko mice exhibit reduced VLDL secretion and liver TG levels. a, VLDL secretion was determined in mice after a 16-h fasting period. To inhibit degradation of TG-rich lipoproteins, mice were treated with poloxamer 407 by intraperitoneal injection (1 mg/kg). TG secretion rate was calculated by linear regression from the increase in plasma TG over a period of 4 h (n = 4 for wild-type and n = 5 for MGL-ko mice, respectively). b, liver TG content of mice was determined in the fed state and after a 16-h fasting period (n = 6 for each genotype). Data are presented as mean ± S.D. *, p < 0.05; **, p < 0.01.
Metabolic Characterization and Food Intake
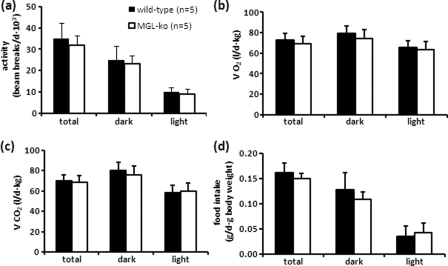
To investigate whether overactive EC signaling affects energy expenditure and produces orexigenic effects in MGL-ko mice, animals were housed in metabolic cages allowing the continuous measurement of metabolic parameters and food intake. We could not find significant changes in locomotor activity, daily O2 consumption, CO2 production, and food consumption in comparison with wild-type controls (Fig. 6, a–d) indicating that the lack of MGL activity does not provoke changes in energy expenditure and feeding behavior. Among other effects, CBR agonists have been shown to stimulate food intake in mice at doses that do not significantly compromise locomotor activity. At high doses, cannabinoids cause hypomotility associated with decreased food intake (22). To test whether MGL-ko animals respond normally to CBR activation, mice fasted for 12 h received the CBR agonist CP 55,940 by intraperitoneal injection. Subsequently, locomotor activity, O2 consumption, and food intake were monitored over a period of 2 h. We could not find significant differences between wild-type and mutant mice treated with the carrier solution (Fig. 7, a–c) or treated with 0.05 mg/kg CP 55,940 (Fig. 7, d–f). Yet we observed a significant increase in food intake in wild-type as well as in MGL-ko animals 26 and 39 min after agonist treatment in comparison with carrier-treated mice (indicated by #; compare Fig. 7, c and f). This suggests that MGL-ko mice respond to the orexigenic stimuli produced by low doses of the CBR agonist. At a dose of 0.15 mg of CP 55,940/kg, we could see clear differences between animal groups. In comparison with MGL-ko mice, locomotor activity and O2 consumption were significantly decreased in wild-type mice within the 1st h after injection (Fig. 7, g and h), which was also associated with a reduction in food intake (Fig. 7i). Thus, MGL-ko mice exhibit an increased tolerance to the hypometabolic activity produced by high doses of CP 55,940. This might represent an adaptation to the increased 2-AG levels in the brain of MGL-ko mice and functionally antagonize the elevated 2-AG concentrations.
FIGURE 6.
Locomotor activity, energy expenditure, and food consumption of MGL-ko mice are unchanged. Mice were housed in a laboratory animal monitoring system (LabMaster, TSE Systems), which allows the simultaneous measurement of locomotor activity (a), O2 consumption (VO2) (b), CO2 production (VCO2) (c), and food consumption (d). Mean values were calculated from a 72-h monitoring period of mice that have been familiarized with metabolic cages for at least 3 days (n = 5 for each genotype). Data are presented as mean ± S.D.
FIGURE 7.
MGL-ko mice are more tolerant to the hypometabolic effect of the CBR agonist CP 55,940. Mice were fasted for 12 h and then treated with carrier solution alone (control, a–c), with 0.05 mg/kg (d–f), and with 0.15 mg/kg (g–i) of CP 55,940 (solubilized in PBS containing 5% ethanol and 5% Emulphor®) by intraperitoneal injection. Subsequently, locomotor activity (a, d, and g), O2 consumption (b, e, and h), and food intake (c, f, and i) were monitored for 2 h. Data are presented as mean ± S.D. (n = 6 for each genotype). *, comparison of wild-type and MGL-ko mice; #, comparison of agonist- and carrier-treated mice; *, #, p < 0.05; **, ##, p < 0.01; ***, p < 0.001.
Role of MGL in High Fat Diet-induced Obesity and Insulin Resistance
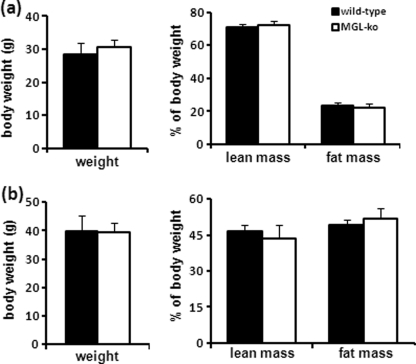
To investigate whether MGL deficiency affects the development of obesity, we determined fat and lean mass of mice set on a normal chow diet and on a high fat diet using NMR. In comparison with wild-type controls, body weight, fat, and lean mass of MGL-ko mice were unchanged on the chow and on the high fat diet (Fig. 8, a and b). Glucose tolerance tests (GTT) and ITT revealed no differences between animal groups when fed a chow diet (Fig. 9a). Importantly, however, MGL-ko mice fed a high fat diet showed a significant improvement in insulin sensitivity and glucose intolerance in comparison with wild-type mice (Fig. 9b). Analysis of plasma parameters of mice receiving high fat diet revealed reduced glycerol, TG, and insulin levels in MGL-ko mice (Table 2). These observations imply that the lack of MGL does not influence WAT mass but attenuates diet-induced insulin resistance.
FIGURE 8.
MGL-ko exhibit normal weight, lean, and fat mass. Lean and fat mass of mice were analyzed by NMR (the minispec, NMR Analyzer, Bruker, Ettlingen, Germany). a, weight, lean, and fat mass of male mice on a normal chow diet at the age of 3 months (n = 6 per genotype); b, male mice at the age of 18 weeks fed a high fat diet or for 12 weeks (n = 10 and 11 for wild-type and MGL-ko mice, respectively). Data are presented as mean ± S.D.
FIGURE 9.
MGL-ko mice receiving a high fat diet exhibit improved insulin sensitivity and glucose tolerance. For insulin and glucose tolerance tests (ITT and GTT), male mice were fasted for 4 and 6 h, respectively. Mice received an intraperitoneal injection of 0.75 IU of insulin or 1.5 mg of glucose per kg of body weight. Insets show the area under the curve (AUC). a, ITT and GTT were performed with male mice at the age of 12 weeks receiving a normal chow diet (n = 6 per genotype). b, ITT and GTT were performed with mice set on a high fat diet for 12 weeks (n = 10 for each genotype). Data are presented as mean ± S.D. *, p < 0.05; **, p < 0.01.
TABLE 2.
Plasma metabolites and insulin levels in mice set on an HFD
Values were obtained from male mice receiving a high fat diet for 12 weeks. Data are presented as mean ± S.D. (n = 6 for each genotype).
| Parameter | Wild type | MGL-ko |
|---|---|---|
| FFA (mm) | 0.5 ± 0.1 | 0.6 ± 0.1 |
| Glycerol (mm) | 0.5 ± 0.1 | 0.4 ± 0.1a |
| TG (mm) | 1.1 ± 0.4 | 0.5 ± 0.1a |
| Cholesterol (mg/dl) | 237 ± 27 | 235 ± 40 |
| Glucose (mg/dl) | 230 ± 59 | 247 ± 35 |
| Plasma insulin (μg/liter) | 3.2 ± 0.4 | 2.2 ± 0.3a |
a p < 0.01.
DISCUSSION
This study focused on the biochemical and metabolic characterization of a mouse model systemically lacking MGL. MGL-ko mice exhibit reduced MGH activity in brain, liver, and adipose tissue accompanied by the accumulation of 2-AG and other MG species. These observations confirm a major role of the enzyme in MG catabolism. Recently, two other enzymes of the α/β-hydrolase superfamily, ABHD6 and ABHD12, have been shown to hydrolyze MG esterified with long chain FFA (17). Interestingly, mutations in ABHD12 are causative for an inherited neurodegenerative disease (PHARC) (23), and ABHD6 was shown to modulate the efficacy of 2-AG in activating CBR (24). In comparison with these lipases, MGL exhibits a much higher specific activity against MG (17). However, the relative contribution of MGL and other lipases to MG hydrolysis may depend on their subcellular localization and on their cell type-specific expression level. Therefore, ABHD6 or ABHD12 may be important in cell types where MGL is not expressed such as specific regions of the brain (24).
Enzymes catalyzing the hydrolysis of long chain MG in WAT have already been described in the 1970s (7, 25). These studies already suggested a major role of MGL in this process, and it was estimated that HSL accounts for ∼20% of the total activity against MG. Our data demonstrate that MGL-deficient WAT is still able to degrade MG in response to lipolytic stimulation leading to an increased release of glycerol. Using a specific inhibitor, we show that HSL accounts for most of this remaining MGH activity in WAT of MGL-ko animals implying that this enzyme is involved in MG hydrolysis and partially compensates for the lack of MGL activity. However, our data also provide evidence for a role of MGL in lipolysis. We show that MGL-ko mice exhibit reduced plasma glycerol levels suggesting that glycerol partially remains trapped in MG, which are generated by the degradation of lipid stores. In ex vivo studies, the lack of MGL activity resulted in reduced FFA and glycerol release from cultured fats pads. Moreover, liver TG levels, VLDL secretion, and circulating ketone bodies are decreased in fasted MGL-ko mice. Together, these observations suggest a reduced flux of FFA from WAT into the liver. However, we could not find significant changes in circulating FFA levels implying that MGL deficiency is associated with a more moderate defect in lipolysis in comparison with animal models lacking upstream lipases ATGL or HSL. ATGL is the rate-limiting enzyme in lipolysis, and mice lacking this enzyme exhibit reduced plasma FFA levels even in the fed state. Under fasting conditions or in response to exercise, ATGL-deficient animals are unable to mobilize sufficient energy to maintain normal energy metabolism and develop hypoglycemia (26, 27). Prolonged fasting induces a torpor-like state in these mice characterized by strongly reduced oxygen consumption and hypothermia (28). The lipolytic defect in HSL-deficient mice is less severe and primarily caused by defective degradation of DG in WAT. Yet HSL-deficient mice exhibit strongly reduced plasma ketone bodies, TG, and FFA levels in the fasted state (29, 30).
Numerous studies demonstrate that CBR1 agonists as well as the EC anandamide and 2-AG stimulate food intake (31). 2-AG concentrations in the brain of MGL-ko mice were enormously elevated suggesting that these mice exhibit increased EC signaling that could produce orexigenic effects. However, MGL-ko mice did not show significant changes in food intake and fat mass implying that defective 2-AG degradation is not sufficient to provoke changes in feeding behavior. Recently, two studies were published, which provide an explanation for our observations. Both studies were focused on the role of 2-AG in the regulation of pain sensitivity. Schlosburg et al. (32) showed that the MGL-specific small molecule inhibitor JZL 184 produces CBR-dependent analgesia. Chronic treatment of mice with JZL 184 led to the loss of the analgesic activity of this compound. This was explained by impaired synaptic plasticity and desensitized brain CBR1. MGL-ko mice did not exhibit sustained analgesia but were less sensitive to CBR1 agonists suggesting that chronically elevated 2-AG levels in these mice lead to pharmacological tolerance. Similarly, Chanda et al. (33) demonstrated that MGL-ko mice do not show alterations in neuropathic and inflammatory pain sensitivity. Mice were less sensitive to CBR1 agonists due to down-regulation of CBR1 density and signaling in the brain. Our studies confirm the concept of CBR desensitization showing that MGL-ko mice are less sensitive to the hypometabolic effects of the CBR agonist CP 55,940. Yet MGL-ko mice respond to the orexigenic stimulus produced by doses of this compound, which do not compromise locomotor activity. Apparently, the EC system is still functional but less sensitive to hyperactivation. An analogous pharmacological tolerance is observed in laboratory animals chronically treated with plant-derived or synthetic cannabinoids (34).
The EC system was also shown to affect liver TG accumulation and lipoprotein metabolism. Activation of CBR1 in hepatocytes promotes liver steatosis through the activation of the lipogenic transcription factor SREBP-1c resulting in increased de novo fatty acid synthesis (14). Despite a severalfold increase in liver 2-AG levels, MGL deficiency was not associated with increased liver TG levels, and we could not detect changes in the expression of SREBP-1 and its target gene fatty-acid synthase. Ruby et al. (35) showed that acute pharmacological inhibition of EC-degrading enzymes increases plasma TG levels. Ruby et al. (35) proposed that an overactive EC system leads to an impaired apoE-mediated clearance of TG-rich lipoproteins. The unchanged and reduced plasma TG levels in fed and fasted MGL-ko mice, respectively, do not support this conclusion. However, we cannot exclude that circulating lipid concentrations as well as liver TG levels are influenced by desensitization of CBR.
Interestingly, our studies suggest that the lack of MGL activity has no effect on high fat diet-induced obesity but counteracts insulin resistance. EC are considered as regulators of energy homeostasis. It has been postulated that aberrant EC signaling contributes to the development of metabolic diseases by modulating behavioral and metabolic processes, including appetite, lipid and energy metabolism, as well as insulin sensitivity. In general, it is assumed that EC signaling is overactive in obese patients, which promotes food intake and lipid storage, and reduces energy expenditure (15, 36). However, it is also known that the availability of FFA substantially influences insulin sensitivity (37). Excess fatty acids can cause cellular dysfunction in non-adipose tissues, a phenomenon known as lipotoxicity (38), and MGL is clearly involved in the lipolytic release of FFA in WAT and non-adipose tissues. Moreover, reduced lipolysis can strongly improve systemic insulin sensitivity as evident from observations in ATGL-ko mice (28, 39). Thus, MGL deficiency could influence the pathogenesis of metabolic diseases by at least two mechanisms. First, defective degradation of 2-AG could stimulate anabolic signaling processes promoting lipid storage and consequently the development obesity-related diseases. Second, the lack of MGL activity could impair lipolysis in adipose tissue and non-adipose tissues counteracting the development of insulin resistance. The observed improvement of insulin sensitivity and glucose tolerance suggests that protective effects prevail in the MGL-ko animal model. However, considering the observations that MGL deficiency is associated with impaired lipolysis, increased 2-AG levels, and desensitized CBRs, the role of MGL in the pathogenesis of metabolic diseases may be more complex and requires further investigations.
In conclusion, our study demonstrates that MGL deficiency in mice reduces lipolysis and VLDL synthesis and attenuates high fat diet-induced insulin resistance. Despite an enormous increase of 2-AG in brain and peripheral tissues, MGL deficiency does not affect food consumption, fat mass, energy expenditure, and locomotor activity. The absence of a more severe phenotype may be explained by desensitization of CBR resulting in increased tolerance to 2-AG-mediated signaling processes and by partial compensation of the lipolytic defect through other enzymes capable of hydrolyzing MG such as HSL, ABHD6, and ABHD12.
Supplementary Material
Acknowledgment
We thank F. Spener for reviewing the manuscript.
This work was supported by Austrian Science Funds, Project P21296, the DK Molecular Enzymology Grant W901-B05 DK, and by the Austrian Federal Ministry for Science and Research, Project “GOLD-Genomics of Lipid-associated Disorders” in the framework of the Austrian Genome Project “GEN-AU Genome Research in Austria.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- MG
- monoacylglycerol
- 2-AG
- 2-arachidonoyl glycerol
- ATGL
- adipose triglyceride lipase
- CBR
- cannabinoid receptor
- DG
- diacylglycerol
- EC
- endocannabinoid
- FFA
- free fatty acids
- HSL
- hormone-sensitive lipase
- MGL
- monoglyceride lipase
- MGH
- monoacylglycerol hydrolase
- OG
- oleoyl glycerol
- TG
- triacylglycerol
- WAT
- white adipose tissue
- mMGL
- mouse MGL
- ITT
- insulin tolerance test
- GTT
- glucose tolerance test.
REFERENCES
- 1. Lowe M. E. (1997) Annu. Rev. Nutr. 17, 141–158 [DOI] [PubMed] [Google Scholar]
- 2. Goldberg I. J., Merkel M. (2001) Front. Biosci. 6, D388–405 [DOI] [PubMed] [Google Scholar]
- 3. Yen C. L., Cheong M. L., Grueter C., Zhou P., Moriwaki J., Wong J. S., Hubbard B., Marmor S., Farese R. V., Jr. (2009) Nat. Med. 15, 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao Y., Vasilyev D. V., Goncalves M. B., Howell F. V., Hobbs C., Reisenberg M., Shen R., Zhang M. Y., Strassle B. W., Lu P., Mark L., Piesla M. J., Deng K., Kouranova E. V., Ring R. H., Whiteside G. T., Bates B., Walsh F. S., Williams G., Pangalos M. N., Samad T. A., Doherty P. (2010) J. Neurosci. 30, 2017–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. (2004) Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 6. Rodriguez J. A., Ben Ali Y., Abdelkafi S., Mendoza L. D., Leclaire J., Fotiadu F., Buono G., Carrière F., Abousalham A. (2010) Biochim. Biophys. Acta 1801, 77–83 [DOI] [PubMed] [Google Scholar]
- 7. Tornqvist H., Belfrage P. (1976) J. Biol. Chem. 251, 813–819 [PubMed] [Google Scholar]
- 8. Karlsson M., Contreras J. A., Hellman U., Tornqvist H., Holm C. (1997) J. Biol. Chem. 272, 27218–27223 [DOI] [PubMed] [Google Scholar]
- 9. Karlsson M., Tornqvist H., Holm C. (2000) Protein Expr. Purif. 18, 286–292 [DOI] [PubMed] [Google Scholar]
- 10. Sugiura T., Kishimoto S., Oka S., Gokoh M. (2006) Prog. Lipid Res. 45, 405–446 [DOI] [PubMed] [Google Scholar]
- 11. Di Marzo V. (2009) Pharmacol. Res. 60, 77–84 [DOI] [PubMed] [Google Scholar]
- 12. Leite C. E., Mocelin C. A., Petersen G. O., Leal M. B., Thiesen F. V. (2009) Pharmacol. Rep. 61, 217–224 [DOI] [PubMed] [Google Scholar]
- 13. Cota D., Marsicano G., Tschöp M., Grübler Y., Flachskamm C., Schubert M., Auer D., Yassouridis A., Thöne-Reineke C., Ortmann S., Tomassoni F., Cervino C., Nisoli E., Linthorst A. C., Pasquali R., Lutz B., Stalla G. K., Pagotto U. (2003) J. Clin. Invest. 112, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osei-Hyiaman D., DePetrillo M., Pacher P., Liu J., Radaeva S., Bátkai S., Harvey-White J., Mackie K., Offertáler L., Wang L., Kunos G. (2005) J. Clin. Invest. 115, 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunos G., Osei-Hyiaman D., Liu J., Godlewski G., Bátkai S. (2008) J. Biol. Chem. 283, 33021–33025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heier C., Taschler U., Rengachari S., Oberer M., Wolinski H., Natter K., Kohlwein S. D., Leber R., Zimmermann R. (2010) Biochim. Biophys. Acta 1801, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blankman J. L., Simon G. M., Cravatt B. F. (2007) Chem. Biol. 14, 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fredrikson G., Tornqvist H., Belfrage P. (1986) Biochim. Biophys. Acta 876, 288–293 [DOI] [PubMed] [Google Scholar]
- 19. Schweiger M., Schreiber R., Haemmerle G., Lass A., Fledelius C., Jacobsen P., Tornqvist H., Zechner R., Zimmermann R. (2006) J. Biol. Chem. 281, 40236–40241 [DOI] [PubMed] [Google Scholar]
- 20. Long J. Z., Li W., Booker L., Burston J. J., Kinsey S. G., Schlosburg J. E., Pavón F. J., Serrano A. M., Selley D. E., Parsons L. H., Lichtman A. H., Cravatt B. F. (2009) Nat. Chem. Biol. 5, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Millar J. S., Cromley D. A., McCoy M. G., Rader D. J., Billheimer J. T. (2005) J. Lipid Res. 46, 2023–2028 [DOI] [PubMed] [Google Scholar]
- 22. Wiley J. L., Burston J. J., Leggett D. C., Alekseeva O. O., Razdan R. K., Mahadevan A., Martin B. R. (2005) Br. J. Pharmacol. 145, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiskerstrand T., H'mida-Ben Brahim D., Johansson S., M'zahem A., Haukanes B. I., Drouot N., Zimmermann J., Cole A. J., Vedeler C., Bredrup C., Assoum M., Tazir M., Klockgether T., Hamri A., Steen V. M., Boman H., Bindoff L. A., Koenig M., Knappskog P. M. (2010) Am. J. Hum. Genet. 87, 410–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marrs W. R., Blankman J. L., Horne E. A., Thomazeau A., Lin Y. H., Coy J., Bodor A. L., Muccioli G. G., Hu S. S., Woodruff G., Fung S., Lafourcade M., Alexander J. P., Long J. Z., Li W., Xu C., Möller T., Mackie K., Manzoni O. J., Cravatt B. F., Stella N. (2010) Nat. Neurosci. 13, 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tornqvist H., Krabisch L., Belfrage P. (1974) J. Lipid Res. 15, 291–294 [PubMed] [Google Scholar]
- 26. Schoiswohl G., Schweiger M., Schreiber R., Gorkiewicz G., Preiss-Landl K., Taschler U., Zierler K. A., Radner F. P., Eichmann T. O., Kienesberger P. C., Eder S., Lass A., Haemmerle G., Alsted T. J., Kiens B., Hoefler G., Zechner R., Zimmermann R. (2010) J. Lipid Res. 51, 490–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huijsman E., van de Par C., Economou C., van der Poel C., Lynch G. S., Schoiswohl G., Haemmerle G., Zechner R., Watt M. J. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E505–E513 [DOI] [PubMed] [Google Scholar]
- 28. Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 29. Haemmerle G., Zimmermann R., Strauss J. G., Kratky D., Riederer M., Knipping G., Zechner R. (2002) J. Biol. Chem. 277, 12946–12952 [DOI] [PubMed] [Google Scholar]
- 30. Osuga J., Ishibashi S., Oka T., Yagyu H., Tozawa R., Fujimoto A., Shionoiri F., Yahagi N., Kraemer F. B., Tsutsumi O., Yamada N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maccarrone M., Gasperi V., Catani M. V., Diep T. A., Dainese E., Hansen H. S., Avigliano L. (2010) Annu. Rev. Nutr. 30, 423–440 [DOI] [PubMed] [Google Scholar]
- 32. Schlosburg J. E., Blankman J. L., Long J. Z., Nomura D. K., Pan B., Kinsey S. G., Nguyen P. T., Ramesh D., Booker L., Burston J. J., Thomas E. A., Selley D. E., Sim-Selley L. J., Liu Q. S., Lichtman A. H., Cravatt B. F. (2010) Nat. Neurosci. 13, 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chanda P. K., Gao Y., Mark L., Btesh J., Strassle B. W., Lu P., Piesla M. J., Zhang M. Y., Bingham B., Uveges A., Kowal D., Garbe D., Kouranova E. V., Ring R. H., Bates B., Pangalos M. N., Kennedy J. D., Whiteside G. T., Samad T. A. (2010) Mol. Pharmacol. 78, 996–1003 [DOI] [PubMed] [Google Scholar]
- 34. Lichtman A. H., Martin B. R. (2005) Handb. Exp. Pharmacol. 168, 691–717 [DOI] [PubMed] [Google Scholar]
- 35. Ruby M. A., Nomura D. K., Hudak C. S., Mangravite L. M., Chiu S., Casida J. E., Krauss R. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14561–14566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Di Marzo V. (2008) Diabetologia 51, 1356–1367 [DOI] [PubMed] [Google Scholar]
- 37. Hue L., Taegtmeyer H. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E578–E591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaffer J. E. (2003) Curr. Opin. Lipidol. 14, 281–287 [DOI] [PubMed] [Google Scholar]
- 39. Kienesberger P. C., Lee D., Pulinilkunnil T., Brenner D. S., Cai L., Magnes C., Koefeler H. C., Streith I. E., Rechberger G. N., Haemmerle G., Flier J. S., Zechner R., Kim Y. B., Kershaw E. E. (2009) J. Biol. Chem. 284, 30218–30229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.