Abstract
Despite decades of speculation, the proton pumping mechanism of complex I (NADH-ubiquinone oxidoreductase) is unknown and continues to be controversial. Recent descriptions of the architecture of the hydrophobic region of complex I have resolved one vital issue: this region appears to have multiple proton transporters that are mechanically interlinked. Thus, transduction of conformational changes to drive the transmembrane transporters linked by a “connecting rod” during the reduction of ubiquinone (Q) can account for two or three of the four protons pumped per NADH oxidized. The remaining proton(s) must be pumped by direct coupling at the Q-binding site. Here, we present a mixed model based on a crucial constraint: the strong dependence on the pH gradient across the membrane (ΔpH) of superoxide generation at the Q-binding site of complex I. This model combines direct and indirect coupling mechanisms to account for the pumping of the four protons. It explains the observed properties of the semiquinone in the Q-binding site, the rapid superoxide production from this site during reverse electron transport, its low superoxide production during forward electron transport except in the presence of inhibitory Q-analogs and high protonmotive force, and the strong dependence of both modes of superoxide production on ΔpH.
Keywords: Bioenergetics, Membrane Energetics, NADH, Superoxide Ion, Ubiqinone, Complex I, NADH-Ubiquinone Oxidoreductase, Proton Translocation
Introduction
Complex I (NADH-ubiquinone oxidoreductase) is central to energy transformation in many prokaryotes and most eukaryotes (1, 2). It establishes a protonmotive force (Δp)2 by coupling the oxidation of NADH and reduction of ubiquinone (Q) or analogous quinones to the pumping of four protons across the membrane per two electrons transferred (4H+/2e) (3–6). The overall reaction is reversible and can be summarized by the following:
Despite the importance of complex I to energy transformation, the mechanism of proton translocation remains a notable unknown in bioenergetics and is currently under intense scrutiny.
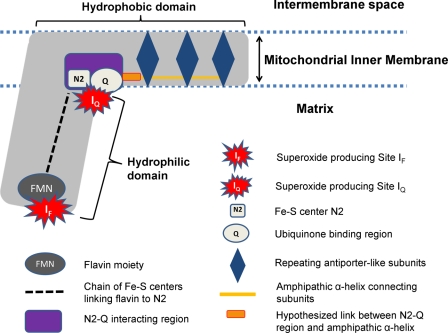
Complex I is L-shaped and comprises a hydrophilic peripheral arm and a hydrophobic membrane domain (Fig. 1). The structure of the hydrophilic arm from Thermus thermophilus shows that it contains the flavin mononucleotide, which accepts electrons from NADH. Electrons are channeled from the flavin along a chain of iron-sulfur centers to the terminal iron-sulfur center (designated N2) (7, 8). N2 subsequently reduces ubiquinone (or an analog), the hydrogen carrier in the lipid phase. The Q-binding region of complex I is located at the junction of the two domains (1).
FIGURE 1.
Mitochondrial complex I. Electrons enter the complex from NADH at the flavin and are channeled along a chain of Fe-S centers to the terminal Fe-S center, N2. Ubiquinone binds near center N2 and is reduced to ubiquinol by addition of two single electrons. The repeating antiporter-like structures in the hydrophobic domain are linked to redox changes in the N2-Q interacting region by an uncharacterized conformational change and to each other by a long amphipathic α-helix (shown in yellow), as proposed by Efremov et al. (13). A hypothetical link to transfer conformational changes in the N2-Q region to the amphipathic α-helix (shown in orange), which is required of all models of indirect proton pumping, is also included.
Advances in knowledge of the structure of complex I have led to crucial insight into its function. Despite general agreement (9) that four protons are translocated per pair of electrons (3–6), no simple consensus model of proton translocation has been constructed. Models that use only direct mechanisms have been proposed (10, 11) but involve complex interactions between multiple molecules of Q to fit the expected 4H+/2e stoichiometry. Friedrich (12) proposed a mixed model of proton translocation with a “black box” direct redox-driven proton translocation in combination with conformational changes that coupled additional indirect proton translocation in the hydrophobic domain. The recent structure of the hydrophobic domain of complex I (13) provides strong evidence that at least some of the protons are indeed pumped by an indirect mechanism.
The hydrophobic domain from Escherichia coli and T. thermophilus contains a long amphipathic α-helix that spans much of the domain and lies parallel to the membrane surface. This helix is proposed to act as a “connecting rod,” linking three putative membrane-domain proton-translocating subunits (13). A similar linking element has also been reported for the eukaryotic complex I (14). If conformational changes in the N2-Q interacting region of the hydrophilic arm are linked to this connecting rod, then the redox chemistry in the N2-Q region can be coupled to proton pumping in the hydrophobic domain (13). However, having a maximum of three conformationally driven proton pumps creates a significant problem for models that rely solely on indirect mechanisms for proton translocation because there is no simple way to explain the 4H+/2e stoichiometry by indirect pumping alone.
Because models that use only direct or indirect mechanisms do not readily fit the observed 4H+/2e stoichiometry for complex I, mixed models are becoming more attractive (12, 13, 15). These combine direct proton translocation via Q-redox reactions coupled to conformational changes that extend into the hydrophobic domain to move additional protons through the interconnected proton pumps. However, none of these models specifies the nature of the direct proton pump beyond a generic black-box mechanism. We propose such a mixed model here that, unlike previous mixed models, has specific constraints for the direct redox-driven pump. These constraints are primarily extensions of the characteristics of mitochondrial complex I superoxide production integrated with the redox chemistry of complex I.
Integrating Superoxide Production, Q-Redox Reactions, and Δp
Along with its role in energy transformation, complex I (and upstream dehydrogenases) also produces superoxide (16–21). Single electrons can reduce molecular oxygen to superoxide at the active site flavin (site IF) during NADH oxidation. Superoxide production from site IF occurs at a maximum rate when the flavin is fully reduced (22, 23). However, the rate of superoxide production by complex I is much higher under conditions that drive reverse electron transport (16–20, 24), with no greater reduction of the flavin (25). The higher rate, along with differential responses to inhibitors and substrates, strongly implicates a second site in superoxide production by complex I (16, 17), although this remains contentious (19, 20, 26, 27). It is thought that this second site (site IQ) is a semiquinone in the region where the iron-sulfur center N2 interacts with Q (N2-Q). It is important to note that N2 is a single electron carrier; therefore, a semiquinone must be formed in the conversion of Q to QH2, or vice versa, by complex I.
Initially observed and reviewed by Liu (28), the superoxide produced by mitochondria or well coupled submitochondrial particles during reverse electron transport is sensitive to dissipation of Δp (16, 17, 29–31). In mitochondria, site IQ is especially sensitive to ΔpH; superoxide production is strongly inhibited by addition of nigericin (in a high K+ medium) or phosphate (16, 17), both of which lead to dissipation of ΔpH and a compensatory increase in membrane potential (Δψ) with no change in overall Δp.
The Δp-dependence of superoxide production at site IQ during reverse electron transport indicates that the superoxide producer is involved, at least indirectly, in the translocation of protons by complex I. However, the key feature widely overlooked is that site IQ superoxide production is much more sensitive to ΔpH than to Δψ or Δp (17). The effect of ΔpH is not simply a result of the matrix acidification that occurs when ΔpH is dissipated, as it is not replicated by acidification of the whole incubation (16, 17). This important observation indicates that the superoxide-producing species must be closely linked to a largely electroneutral movement of protons across the membrane (and is less strongly linked to the electrogenic steps of the mechanism). We propose that this superoxide is generated when an unstable semiquinone is formed in the N2-Q interacting region of complex I.
Superoxide production by the flavin of complex I in intact mitochondria during forward electron transport is relatively low, but this rate increases when complex I Q-site inhibitors (rotenone, myxothiazol, or piericidin) are added. The addition of NADH-generating substrates in concert with inhibition of Q reduction by complex I results in a highly reduced mitochondrial NADH pool, maximizing superoxide generation by the flavin of complex I (17, 32). However, even under these conditions, generation of Δp by addition of ATP markedly increases superoxide production by activating site IQ (17). Importantly, this additional superoxide production is also ΔpH-sensitive and quantitatively equivalent to the rate at the same Δp and ΔpH during reverse electron transport (17). The equivalence of these rates suggests that the site IQ superoxide producer can become accessible when electrons enter at the flavin and move into the complex in the physiological forward direction. However, special conditions of 1) a highly reduced NADH pool, 2) a high Δp with sufficiently high ΔpH component, and 3) blockage of electron escape to the bulk Q pool are required to create significant amounts of superoxide from this site during forward electron flow.
RESULTS
General Model
Because N2 reduces Q by two sequential single electron additions, the overall reaction for complex I must contain a semiquinone intermediate. We propose a model of complex I proton translocation in which there is a semiquinone intermediate that is also a superoxide producer, and the steady-state concentration of the semiquinone (and the rate of superoxide production) depends strongly on ΔpH and more weakly on Δψ and Δp. The unique aspect of our model is that we incorporate this ΔpH sensitivity with the redox reactions forming the semiquinone as part of the direct proton translocation mechanism. Our model also proposes where the energetics of the redox reactions could be favorable for coupling conformational changes in the N2-Q region to the indirect translocation of protons by pumps in the hydrophobic domain.
Requirements of the Model
The mechanism must result in the movement of four protons across the membrane per pair of electrons. At least one of these protons must move via redox reactions in the N2-Q site, with the remaining protons pumped by indirect mechanisms in the hydrophobic domain. The superoxide producer QH• or the anionic Q̇̄ must be formed as an intermediate, with its steady-state concentration strongly dependent on ΔpH and, more weakly, on Δψ and Δp. Complex I generally runs close to equilibrium, with the strongly exergonic oxidation of NADH by Q balanced by the strongly endergonic reaction of proton translocation against Δp. Reverse electron transport is readily observed when Δp is maintained by oxidation of other substrates or hydrolysis of ATP and the membrane pool of Q is kept reduced, resulting in demonstrable reduction of NAD+. Thus, the overall proton pumping reaction must be reversible while remaining consistent with the observed characteristics of complex I superoxide production.
Characteristics Important to the Model
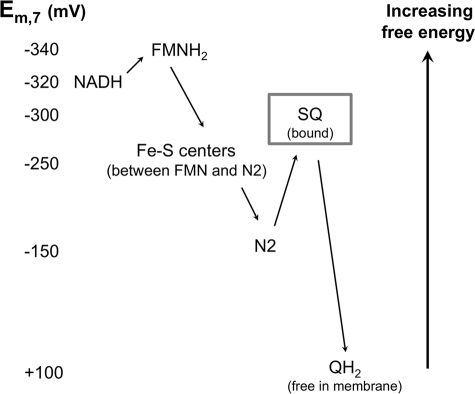
At least two semiquinone signals associated with complex I have been observed by EPR (33), one of which, designated SQNf, is uncoupler-sensitive and can only be observed in very well coupled submitochondrial particles at high Δp (33). The Δp sensitivity of this semiquinone strongly suggests that it is intimately involved in the generation of Δp. However, it is not clear if these signals come from two separate Q molecules or whether a single semiquinone can exist in different states (1). Rapid kinetic measurements on preparations of purified complex I with a 1:1 stoichiometrically bound Q molecule indicate that the terminal iron-sulfur center, N2, is very rapidly reduced with no appearance of an EPR signal consistent with a semiquinone (34). Verkhovskaya et al. (34) conclude that the Em,7 for the Q/semiquinone couple in complex I is quite low, and estimate −300 mV as an upper limit. In contrast, the Em,7 of N2 is the highest among complex I iron-sulfur centers (reported values range from −50 to −200 mV (35, 36)) and is much higher than NADH (-320 mV) or the flavin (-340 mV) (36). Thus, the first electron from NADH is held on N2, and whenever it reduces Q to the semiquinone, a second electron from NADH and N2 rapidly completes the reduction to QH2, keeping the steady-state concentration of semiquinone very low. Because the Em,7 for the Q/semiquinone couple approaches the Em,7 of the NAD+/NADH couple, whereas the Q-pool has an Em,7 of +100 mV (Fig. 2), it seems likely that it is not the reduction of Q to the semiquinone but the reduction of the semiquinone to QH2 that is associated with a large energy drop that could be coupled to proton translocation (34). The instability of the semiquinone and the coupling to proton pumping of its reduction to QH2 are central to our model.
FIGURE 2.
Reduction potentials of complex I redox chemistry. Values for NADH, FMNH2, iron-sulfur centers, and the membrane Q-pool are taken from Refs. 11 and 36 and references therein. Note that values for N2 range as high as −50 mV to as low as −200 mV. The potential of the bound semiquinone (SQ, highlighted by a gray box) is unknown, but an upper limit of −300 mV is estimated by Verkhovskaya et al. (34). This would indicate that the bound semiquinone is unstable and its formation is a barrier to electron flow between N2 and the Q-pool.
The pKa of N2 depends on its reduction state: the pKa of the reduced form is >8.5, and the pKa of the oxidized form is <6 (10). A redox-Bohr proton pumping mechanism has been suggested to play a role at the N2-Q interacting region (10), but mutation studies on Yarrowia lipolytica indicate that the loss of this redox-Bohr effect has no impact on the stoichiometry or rate of proton pumping by the complex (37). Nevertheless, these traits mean that at physiological pH, wild-type N2 will be protonated when reduced and deprotonated when oxidized. We incorporated this characteristic into our model, but it is not an essential feature of the proton pumping mechanism (in the mutant, the precise sites of protonation and deprotonation will be altered with no net effect on proton pumping).
Forward Electron Transport
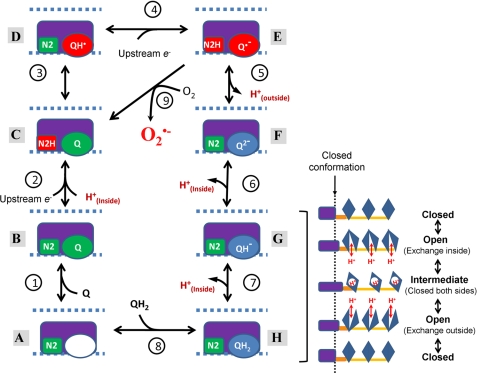
The model is illustrated in Fig. 3. The sequential steps are numbered, starting from the complex with no Q or QH2 bound in the N2-Q interacting region (state A, bottom). Inside refers to the mitochondrial matrix or prokaryotic cytoplasm, and outside refers to the intermembrane or periplasmic space. The model assumes the simplest pathway for electron flow. Electrons enter at N2 and pass sequentially to Q to form QH2. Conformational coupling to the membrane domain occurs when the unstable semiquinone relaxes to the more stable quinol. N2 protonates when reduced and deprotonates when oxidized. We then added protonations and deprotonations of Q at appropriate points to generate the observed ΔpH dependence of superoxide formation from the semiquinone. States A–C and H are stable, and states D–G are less stable.
FIGURE 3.
Model of electron transfer and proton translocation by complex I. The orientations of the membrane, Q-binding site, Fe-S center N2, and their interacting region in relation to the mechanical linkage to the three indirect proton pumps in the membrane domain are the same as Fig. 1. Oxidized redox centers are shown in green, those that have been reduced by a single electron are shown in red, and two electron reduction states are shown in blue. The transitions from state G to state H result in conformational changes in the N2-Q interacting region (represented by a purple box) that are coupled to the indirect movement of three protons in the membrane domain (as hypothesized in Ref. 13). See text for details.
1. Q Binding
The N2-Q pocket is initially empty, with proton access from the inside (state A), and binds oxidized Q to form state B.
2. First Reduction Step
N2 accepts an electron from upstream FeS centers (originally from NADH) and is immediately protonated from the inside to form state C. Note that steps 1 and 2 may occur in either order.
3. Internal Electron and Proton Transfer
N2 passes its electron (and, optionally, its proton) to Q, producing QH• and the very unstable state D. The large difference in Em,7 between N2 and the bound Q/semiquinone couple (34) suggests that this reaction is not favored. Moreover, there is little drop in free energy from the NAD+/NADH couple to this state, making this reaction unlikely to be coupled to proton translocation.
4. Second Reduction Step
When state D is formed, a second electron from the other upstream Fe-S centers rapidly reduces N2, forming state E. This transition to state E alters the gating of proton channels from the inside to the outside. We assume the anionic form of the semiquinone, Q̇̄, because the Δp-dependent SQNF is thought to be anionic (33) (see below for an alternative two-proton pumping formulation where a second proton from inside forms QH·). If a proton is released to the inside at step 3, it is taken up again here from the inside. The distribution between steps 4 and 5 of net charge transfer across the membrane depends on the location of the proton from N2 in the membrane once the gating is to the outside. To explain the strong ΔpH sensitivity of superoxide formation by state E, we assume that that proton from N2 moves to the outer surface of the membrane so that most of the charge translocation occurs in step 4.
5. Internal Electron Transfer and Deprotonation
The second internal electron reduction of Q̇̄ to state F is coupled to the release of a proton to the outside, completing the directly coupled proton transport step (in the alternative two-proton formulation below, two protons are released from N2H and QH· to the outside).
6. First Scalar Proton Uptake
Q2− forms QH− by taking up a proton from the inside to generate state G. The pKa for this protonation is likely very high (>11 in 80% ethanol (38)). Steps 5 and 6 involve deprotonation to the outside and protonation from the inside but only partial net charge transfer across the membrane, so, crucially, the relative occupancies of states E and G are strongly affected by ΔpH but less so by Δψ.
7. Second Scalar Proton Uptake
QH− relaxes to QH2 by taking up a second proton from the inside and completing the catalytic cycle at state H. The pKa for this protonation is similarly expected to be >11 (38). We propose that the unstable QH− interacts with the local protein environment, so relaxation of G to H in step 7 initiates conformational changes that drive the proton pumps in the hydrophobic domain (shown on the right of Fig. 3). These conformational changes in the N2-Q region work, by mechanical analogy, as a piston driving the proximal proton pump. The long amphipathic α-helix reported by Efremov et al. (13) acts as a connecting rod, linking the other indirect proton pumps in the membrane domain to the proximal pump. We use a schematic demonstration of a mechanism similar to Ref. 13 in that three proton pumps, each with high similarity to known ion antiporters, are forced through a proton pumping cycle by the conformational changes that occur in the N2-Q region. This is the power stroke for the conformationally driven proton pumps in the hydrophobic domain, driven by the unstable G relaxing to the stable H.
8. QH2 Release
The reduced QH2 is released to the membrane lipid environment, leaving the N2-Q interacting region empty (state A) and ready for another cycle.
Reverse Electron Transport
At sufficiently high Δp, complex I can reverse and transfer electrons from QH2 to NAD+. Our model feasibly works in both directions and accounts for the known reversibility of the overall reaction. Also central to our proposed model, superoxide production is highest under these conditions of reverse electron transport.
Under conditions of high Δp, the indirect proton pumps will provide the energetic drive to reverse reactions 8–5, producing state E. Once the NAD+ pool and the upstream flavin mononucleotide and FeS centers are fully reduced by several turnovers of reverse electron transport, step 4 will become irreversible because of the lack of an electron acceptor. State E will have its maximum occupancy, and superoxide production from site IQ by reaction 9 will also be maximal, as observed. In this condition, the relative population of states E, F, and G will be dependent on ΔpH. Dissipation of ΔpH by addition of nigericin or phosphate will shift the equilibrium from state E to states F and G, in which Q is fully reduced and does not generate superoxide. Superoxide production by reaction 9 will fall dramatically, as observed. In addition, the total combined population of states E, F, and G will depend on the reversal of proton pumping at step 7, driven by Δp, so superoxide production at state E will also depend on Δp, as observed.
Our model predicts that the formation of the relatively unstable Q̇̄ will be a barrier to the rate of reverse electron transport to NAD+ from QH2. Collapsing ΔpH with nigericin inhibits the rate of reverse electron transport but not the maximal steady state NAD+ reduction when driven by succinate oxidation in isolated mitochondria (17). Thus, it is Δp that sets the final reduction state of the intramitochondrial NADH pool (and the ΔpH-insensitive superoxide production from site IF) during reverse electron transport, but the loss of ΔpH substantially inhibits the rate of this process by lowering the steady-state concentration of state E, the transition state intermediate.
Superoxide Production from Site IQ during Forward Electron Transport
Mitochondrial superoxide production by complex I in the presence of NADH-generating substrates is increased by complex I Q-site inhibitors such as rotenone. These inhibitors maximize NADH/NAD+ while abolishing Δp (16, 27, 30, 32). We assert that this increase is predominantly from the flavin of complex I (site IF), which becomes more reduced in concert with the intramitochondrial cofactor pool. However, under these same conditions of high NADH/NAD+ and the presence of complex I Q-site inhibitors, establishing Δp by addition of ATP markedly increases superoxide production. This increase is diminished by collapsing ΔpH with nigericin (16). How can our model explain these results, given that these inhibitors block the transfer of electrons from complex I to the membrane Q-pool?
If these complex I Q-site inhibitors allow Q binding at step 1 but preferentially block the release of bound QH2 at reaction 8, then our model explains these observations. NADH will reduce complex I, and in the presence of Q-site inhibitors and the absence of Δp, the primary species formed will be H. This condition will result in negligible site IQ superoxide production. However, establishing Δp by hydrolysis of ATP will reverse the membrane domain proton pumps and drive electrons back to E, where they will stall because of reduction of the other Fe-S centers by NADH. This will result in significant site IQ superoxide production by reaction 9, which will be particularly dependent on ΔpH, as discussed above. In the absence of Q-site inhibitors, the QH2 will be lost to the Q pool, and the reaction will not reverse when Δp is generated. In this way, the model explains the strict requirements that are observed for superoxide production from site IQ during forward electron transport: a highly reduced NADH pool, a high Δp with a high enough ΔpH component, and blockage of electron escape to the bulk Q pool (16).
DISCUSSION
A recent model of complex I proton pumping suggests that only two of the hydrophobic domain proton pumps are active (15, 39). The argument that only two protons are pumped in the membrane domain is based on the finding that mutation of conserved acidic residues in the NuoN subunit (one of the indirect pumps in the model by Efremov et al. (13)) of E. coli complex I does not affect proton pumping (40). In phylogenetically “higher” metazoans, the subunit equivalent to NuoN, subunit ND2 by mammalian nomenclature, has been truncated, losing three of the 14 highly conserved α-helices. These helices could be important to the structure required for proton translocation (41). Therefore, in these species, indirect proton pumping may also be reduced to 2H+ per indirect cycle, although this has not been directly demonstrated. Our model can accommodate this possibility with only minor alterations to pump two protons per pair of electrons in the N2-Q site instead of one. This can be done by adding an extra protonation from the inside at step 4 and an extra deprotonation to the outside at step 5 (with the proviso that the semiquinone intermediate in state E will now be QH• instead of the anionic Q̇̄). Similarly, if this modification is allowed and three hydrophobic domain pumps operate, the model can generate a proton pumping stoichiometry of 5H+/2e.
In conclusion, we present a simple general model of the proton pumping mechanism of complex I that is consistent with current understanding of the structure, reversibility, and proton pumping stoichiometry of the complex. Crucially, it explains the otherwise puzzling characteristics of superoxide production by complex I, which provide strong constraints that allow us to produce a model with unprecedented empirical support. No other model has attempted to explain this important side reaction of the complex, but any alternative model, or modification of this model, should be able to do so if it is to be taken seriously.
This study was supported, in whole or in part, by National Institutes of Health Grants P01 AG025901, PL1 AG032118, and R01 AG033542. This work was also supported by The Ellison Medical Foundation Grant AG-SS-2288–09.
- ΔpH
- pH gradient
- Q
- ubiquinone
- N2
- terminal iron-sulfur center
- Δp
- protonmotive force
- site IF
- active site flavin
- site IQ
- second site
- Δψ
- membrane potential.
REFERENCES
- 1. Brandt U. (2006) Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 2. Friedrich T., Scheide D. (2000) FEBS Lett. 479, 1–5 [DOI] [PubMed] [Google Scholar]
- 3. Brown G. C., Brand M. D. (1988) Biochem. J. 252, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galkin A. S., Grivennikova V. G., Vinogradov A. D. (1999) FEBS Lett. 451, 157–161 [DOI] [PubMed] [Google Scholar]
- 5. Pozzan T., Miconi V., Di Virgilio F., Azzone G. F. (1979) J. Biol. Chem. 254, 12000–12005 [PubMed] [Google Scholar]
- 6. Wikström M. (1984) FEBS Lett. 169, 300–304 [DOI] [PubMed] [Google Scholar]
- 7. Hinchliffe P., Sazanov L. A. (2005) Science 309, 771–774 [DOI] [PubMed] [Google Scholar]
- 8. Sazanov L. A., Hinchliffe P. (2006) Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 9. Brand M. D. (2005) Biochem. Soc. Trans. 33, 897–904 [DOI] [PubMed] [Google Scholar]
- 10. Brandt U. (1997) Biochim. Biophys. Acta 1318, 79–91 [DOI] [PubMed] [Google Scholar]
- 11. Dutton P. L., Moser C. C., Sled V. D., Daldal F., Ohnishi T. (1998) Biochim. Biophys. Acta 1364, 245–257 [DOI] [PubMed] [Google Scholar]
- 12. Friedrich T. (2001) J. Bioenerg. Biomembr. 33, 169–177 [DOI] [PubMed] [Google Scholar]
- 13. Efremov R. G., Baradaran R., Sazanov L. A. (2010) Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 14. Hunte C., Zickermann V., Brandt U. (2010) Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 15. Ohnishi S. T., Salerno J. C., Ohnishi T. (2010) Biochim. Biophys. Acta 1797, 1891–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambert A. J., Brand M. D. (2004) J. Biol. Chem. 279, 39414–39420 [DOI] [PubMed] [Google Scholar]
- 17. Lambert A. J., Brand M. D. (2004) Biochem. J. 382, 511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adam-Vizi V., Chinopoulos C. (2006) Trends Pharmacol. Sci. 27, 639–645 [DOI] [PubMed] [Google Scholar]
- 19. Brand M. D. (2010) Exp. Gerontol. 45, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hirst J., King M. S., Pryde K. R. (2008) Biochem. Soc. Trans. 36, 976–980 [DOI] [PubMed] [Google Scholar]
- 21. Murphy M. P. (2009) Biochem. J. 417, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Esterházy D., King M. S., Yakovlev G., Hirst J. (2008) Biochemistry 47, 3964–3971 [DOI] [PubMed] [Google Scholar]
- 23. Kussmaul L., Hirst J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jastroch M., Divakaruni A. S., Mookerjee S., Treberg J. R., Brand M. D. (2010) Essays Biochem. 47, 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lambert A. J., Buckingham J. A., Brand M. D. (2008) FEBS Lett. 582, 1711–1714 [DOI] [PubMed] [Google Scholar]
- 26. Ohnishi S. T., Shinzawa-Itoh K., Ohta K., Yoshikawa S., Ohnishi T. (2010) Biochim. Biophys. Acta 1797, 1901–1909 [DOI] [PubMed] [Google Scholar]
- 27. Ohnishi S. T., Ohnishi T., Muranaka S., Fujita H., Kimura H., Uemura K., Yoshida K., Utsumi K. (2005) J. Bioenerg. Biomembr. 37, 1–15 [DOI] [PubMed] [Google Scholar]
- 28. Liu S. S. (1997) Biosci. Rep. 17, 259–272 [DOI] [PubMed] [Google Scholar]
- 29. Votyakova T. V., Reynolds I. J. (2001) J. Neurochem. 79, 266–277 [DOI] [PubMed] [Google Scholar]
- 30. Liu Y., Fiskum G., Schubert D. (2002) J. Neurochem. 80, 780–787 [DOI] [PubMed] [Google Scholar]
- 31. Korshunov S. S., Skulachev V. P., Starkov A. A. (1997) FEBS Lett. 416, 15–18 [DOI] [PubMed] [Google Scholar]
- 32. Treberg J. R., Quinlan C. L., Brand M. D. (2010) FEBS J. 277, 2766–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Magnitsky S., Toulokhonova L., Yano T., Sled V. D., Hägerhäll C., Grivennikova V. G., Burbaev D. S., Vinogradov A. D., Ohnishi T. (2002) J. Bioenerg. Biomembr. 34, 193–208 [DOI] [PubMed] [Google Scholar]
- 34. Verkhovskaya M. L., Belevich N., Euro L., Wikström M., Verkhovsky M. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3763–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Medvedev E. S., Couch V. A., Stuchebrukhov A. A. (2010) Biochim. Biophys. Acta 1797, 1665–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohnishi T. (1998) Biochim. Biophys. Acta 1364, 186–206 [DOI] [PubMed] [Google Scholar]
- 37. Zwicker K., Galkin A., Dröse S., Grgic L., Kerscher S., Brandt U. (2006) J. Biol. Chem. 281, 23013–23017 [DOI] [PubMed] [Google Scholar]
- 38. Rich P. R. (1984) Biochim. Biophys. Acta 768, 53–79 [DOI] [PubMed] [Google Scholar]
- 39. Ohnishi T., Nakamaru-Ogiso E., Ohnishi S. T. (2010) FEBS Lett. 584, 4131–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amarneh B., Vik S. B. (2003) Biochemistry 42, 4800–4808 [DOI] [PubMed] [Google Scholar]
- 41. Birrell J. A., Hirst J. (2010) FEBS Lett. 584, 4247–4252 [DOI] [PubMed] [Google Scholar]