Abstract
C8 is one of five complement proteins that assemble on bacterial membranes to form the lethal pore-like “membrane attack complex” (MAC) of complement. The MAC consists of one C5b, C6, C7, and C8 and 12–18 molecules of C9. C8 is composed of three genetically distinct subunits, C8α, C8β, and C8γ. The C6, C7, C8α, C8β, and C9 proteins are homologous and together comprise the MAC family of proteins. All contain N- and C-terminal modules and a central 40-kDa membrane attack complex perforin (MACPF) domain that has a key role in forming the MAC pore. Here, we report the 2.5 Å resolution crystal structure of human C8 purified from blood. This is the first structure of a MAC family member and of a human MACPF-containing protein. The structure shows the modules in C8α and C8β are located on the periphery of C8 and not likely to interact with the target membrane. The C8γ subunit, a member of the lipocalin family of proteins that bind and transport small lipophilic molecules, shows no occupancy of its putative ligand-binding site. C8α and C8β are related by a rotation of ∼22° with only a small translational component along the rotation axis. Evolutionary arguments suggest the geometry of binding between these two subunits is similar to the arrangement of C9 molecules within the MAC pore. This leads to a model of the MAC that explains how C8-C9 and C9-C9 interactions could facilitate refolding and insertion of putative MACPF transmembrane β-hairpins to form a circular pore.
Keywords: Complement, Membrane, Post-translational Modification, Protein Self-assembly, Protein Structure, C8, MACPF, Cytolysins, Mannosyl Tryptophan, Membrane Attack Complex
Introduction
Assembly of the “membrane attack complex” (MAC)3 of complement on the surface of Gram-negative bacteria and other pathogenic organisms involves the sequential interaction of complement proteins C5b, C6, C7, C8, and C9 (1–3). Association of the first four components produces a membrane-bound tetrameric C5b-8 complex, which then initiates the recruitment and sequential binding of 12–18 C9 molecules to form a cylindrical transmembrane pore (supplemental Fig. S1). Pore formation leads to loss of membrane integrity and lysis of the cell under attack.
The sequence of interactions leading to MAC formation is well defined; however, the mechanism by which the MAC disrupts membrane organization is poorly understood. C6, C7, the C8α and C8β subunits, and C9 are homologous and together comprise the “MAC family” of proteins (4, 5). Until now, structures have not been determined for any of these proteins. All contain N- and C-terminal modules and a central 40-kDa “membrane attack complex/perforin” (MACPF) domain. The MACPF domain was named as such because of sequence similarity between the MAC family proteins and perforin. The modules range in number from three to eight; all are small domains of 40–60 amino acids that contain multiple disulfide bonds (supplemental Fig. S2). One type of module, thrombospondin type 1 (TSP1), contains several mannosylated tryptophans (6).
Several hundred MACPF-containing proteins have been identified; however, functions are known for only a few. MACPF proteins exhibit limited sequence similarity, but all contain the MACPF signature motif ((Y/W)G(T/S)H(F/Y)X6GG) (7). Recently published crystal structures of two bacterial MACPF-containing proteins (8, 9), the MACPF domain from human C8α (10, 11), and mouse perforin (12) revealed a fold similar to the bacterial pore-forming cholesterol-dependent cytolysins (CDCs). Thus, the MACPF domain may also be referred to as the “MACPF/CDC” domain.
The mechanism of pore formation by CDCs is well understood (13). 30–50 CDC monomers self-polymerize on target membrane surfaces forming a circular pre-pore, which upon completion inserts into the membrane as a functional pore. This process involves a concerted major conformational change whereby two regions of each CDC monomer, which are in an α-helical conformation in the pre-pore state (14), refold into an extended conformation and insert into the bilayer as two transmembrane β-hairpins (TMH). TMHs from neighboring molecules “share edges” of their β-hairpins to form a hydrogen-bonded β-barrel. Structural similarity between complement MACPF proteins and the CDCs suggests that complement uses a CDC-like mechanism for pore formation. Conservation of several key glycine residues known to be important for CDC refolding and pore formation supports this hypothesis (11, 13).
C8 is unique among the MAC components and other MACPF proteins in that it is a heterotrimer composed of three genetically distinct subunits (15). Strong noncovalent interactions mediate binding between the MACPF-containing C8α (NP_000553.1) and C8β (NP_000057.1) subunits, whereas the lipocalin-like C8γ (NP_000597.2) subunit is associated via a disulfide link to C8α. This arrangement differs from the recently characterized bacterial MACPF proteins and perforin which, like the CDCs, are monomers. In this report, we describe the structure of human C8 determined at 2.5 Å resolution. This is the first complete structure of a MAC family protein and of a protein containing two tightly associated MACPF domains. The unusual geometric arrangement of the C8α and C8β subunits provides new insight into how MAC components may interact to form a circular pore.
EXPERIMENTAL PROCEDURES
Protein Purification and Deglycosylation
C8 was purified from human plasma Cohn fraction III (Bayer Corp., Clayton, NC) (16). N-Linked oligosaccharides were removed using peptide:N-glycosidase F (New England Biolabs) in 50 mm sodium phosphate, 135 mm NaCl, pH 7.5. Released oligosaccharides and peptide:N-glycosidase F were removed by gel filtration. Hemolytic activity of deglycosylated C8 (DC8) was measured as described previously and is shown in supplemental Fig. S3 (17).
Protein Crystallization
Initial crystallization conditions were determined by surveying results from the high throughput screening facility at Hauptman-Woodward Medical Research Institute, Inc. (Buffalo, NY) (18). DC8 was concentrated to 4–6 mg/ml in 25 mm imidazole, 135 mm NaCl, pH 7.4, and crystallized at 23 °C by hanging-drop vapor diffusion using 14–16% polyethylene glycol 4000, 0.10 m TAPS, 400 mm NaCl, 10 mm SrCl2, pH 9.0, as the precipitant solution. Crystals were flash-frozen at 100 K in a mixture of 16% (v/v) ethylene glycol and 84% (v/v) precipitant solution.
Structure Determination and Refinement
Data were collected at 100 K on beamline 22-ID of the SER CAT at the APS, Argonne National Laboratory, and processed with HKL-2000 (19). Crystals were prone to decay, and a strategy to cover the asymmetric part of the reciprocal space with minimum exposure was used. The initial model was obtained by molecular replacement using Phaser software (20) from the CCP4 suite of programs (21). The following fragments were fitted in this sequence: 1) the MACPF domain of C8α; 2) C8γ; and 3) homology model of the upper subdomain of C8β. These fragments were derived from the αMACPF-γ structure (Protein Data Bank entry 2RD7) (11). This model constituted about 60% of the scattering power. An extensive search for heavy atom derivatives was not successful. Therefore, the remaining residues were built to electron density maps phased with partial structures in an iterative process using Turbo (22) and Coot (23) for molecular graphics and CNS (24) and Refmac5 (25) for crystallographic refinements. Previous high resolution analysis of C8γ showed internal water molecules in the lipocalin cavity (26). These molecules were observed in C8 electron density maps but only at 0.6 σ level. Thus, it appears that the noise level in the final maps is quite high, and therefore no solvent structure was included. The final model reports 1149 residues out of 1274. Relevant statistics are in Table 1. The initial homology model for the upper part of C8β and the homology model of C9 were constructed using Turbo (22). Superpositions were calculated using Lsqkab software (27). Accessible surface calculations were carried out with PISA website (28). Ribbon representations were generated using Molscript (29) and Raster3D (30). Fig. 3B and supplemental Figs. S4–S6 were prepared using Turbo (22). Figs. 2 and 5C were prepared using PyMOL (31).
TABLE 1.
Crystallographic data and refinement statistics for C8
Data were collected on a single crystal. Values in parentheses correspond to the highest resolution shell.
| Data collection | |
| Space group | P63 |
| Cell dimensions | |
| a, b, c and α, β, γ | 139.58 Å, 139.58 Å, 127.16 Å and 90°, 90°, 120° |
| Resolution | 50.0 to 2.5 Å (2.54 to 2.50 Å) |
| Rmerge | 10.0% (41.7%) |
| I/σ(I) | 15.8 (1.4) |
| Completeness | 93.3% (63.7%) |
| Redundancy | 3.2 (1.8) |
| Refinement | |
| Resolution | 2.5 Å |
| No. of reflections/unique reflections | 144,790/44,803 |
| R/Rfree, Refmac5 | 24.9%/33.7% |
| No. of atoms | |
| Protein | 9235 |
| Ligand/ion | 2 |
| Water molecules | 0 |
| Mean B-factor Overall | 52.0 Å2 |
| Protein chain A | 52.5 |
| Protein chain B | 50.6 |
| Protein chain C | 55.2 |
| Wilson plot statistics | 62.4 |
| Root mean square deviations | |
| Bond length | 0.015 Å |
| Angles | 1.8° |
| Ramachandran | |
| Residues in most favored regions | 740 (76.4%) |
| Residues in additional allowed regions | 206 (21.3%) |
| Residues in generously allowed regions | 22 (2.3%) |
| Residues in disallowed regions | 0 (0.00%) |
FIGURE 3.
Superposition of C8α (red) on C8β (blue) based on their upper MACPF subdomains. MACPF domains are in dark colors, and modules are in light colors. This superposition yields a 24° angle between strands of the β-sheets in the lower subdomain. The region with conserved glycine pairs 318–319, 395–396 and 297–298, 375–376 (numbering for C8α and C8β, respectively) is located at the bends of the sheets (circled). Two orthogonal views are shown.
FIGURE 2.
Features of the C8α and C8β MACPF domains. Surface of the C8 molecule is shown. β-sheets of the C8α (red) and C8β (blue) MACPF domains are shown as stick models. This view shows “fanning out” of the MACPF subdomains. The upper part likely forms the outer or extracellular part of the pore. The lower part spreads out making the base wider. A stereo version is available online (supplemental Fig. S4).
FIGURE 5.
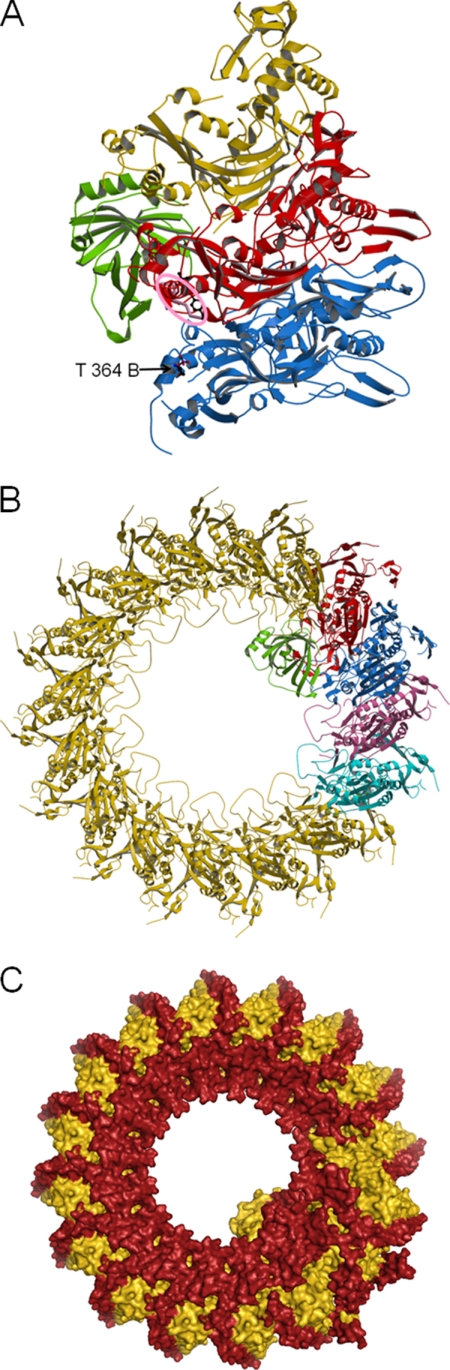
Atomic model of the MAC. The model was generated assuming that the binding mode between C8α and C8β MACPF domains is conserved for C6, C7, and C9. A, view of the modeled C8-C9 complex. Colors correspond to C8α (red), C8β (blue), C8γ (green), and C9 (gold). The putative CD59-binding site in C8α is circled in pink. The nearby position of phosphorylated C8β Thr364 is also shown. A more detailed view of these features is available in supplemental Fig. S7. B, partial model of the MAC. View is of the side facing the membrane. Proteins are colored C6 (cyan), C7 (pink), C8β (blue), C8α (red), C8γ (green), and C9 (gold). Absent are C5b, the CCP and FIM modules of C6 and C7, and the additional N-terminal TSP1 module in C6, the locations of which could not be predicted. Current evidence suggests C5b binds to the C7 FIMs (41); it is assumed the other missing components do not interfere with circular packing of the MACPF domains. The inner diameter of the ring is 110 Å with 80 Å between the protruding loops. The outer diameter is ∼220 Å and the height is 90 Å. This agrees well with EM images of pores formed by poly-C9, which generally have dimensions of ∼100 and 210 Å for the inner and outer diameters, respectively (42). C, molecular envelope of the model shown in B with modules in red and MACPF domains in gold. View is of the side facing away from the membrane. The modules account for a substantial fraction of the surface. It can be speculated that they may partially protect the MACPF domains from proteolytic factors at sites of inflammation as they contain multiple disulfides and carbohydrates.
RESULTS
Structure of C8
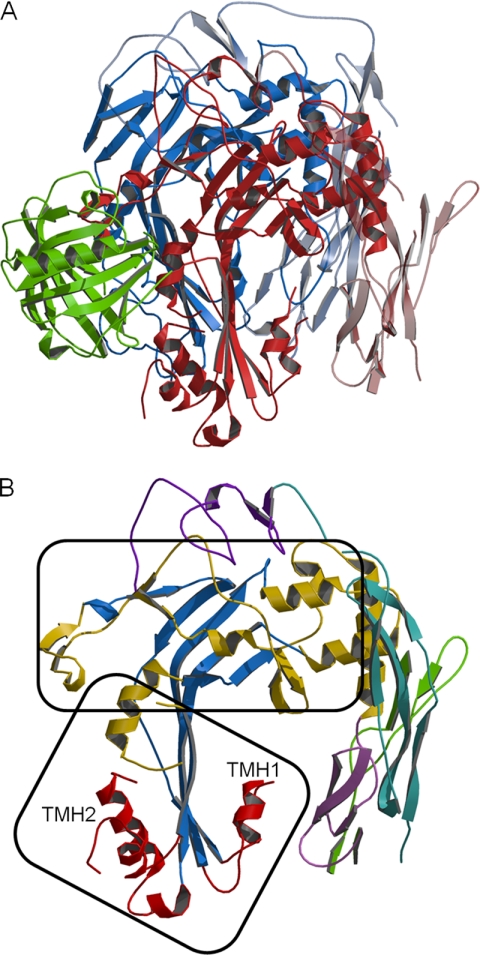
The C8 structure is shown in Fig. 1A. C8α and C8β have similar folds as predicted by 32% amino acid sequence identity. The sequence of C9 is 27 and 26% identical to the corresponding regions of C8α and C8β, respectively; thus, C9 likely has a similar fold. The central parts of the C8α and C8β MACPF domains are large, four-stranded, antiparallel β-sheets with a bend and twist in the middle, which gives them a “Γ” shape (Fig. 1B). Their lower part is flanked by two α-helical bundles that are predicted to refold and form two TMHs during MAC formation. Together with another helix, they form the “lower” MACPF subdomain. The upper part of the sheet and helices surrounding it forms the “upper” subdomain, which makes contacts with the modules, and most likely with other MAC proteins.
FIGURE 1.
Structure of C8. A, ribbon model showing C8α (red), C8β (blue), and C8γ (green). MACPF domains are in dark colors and modules in light colors. B, view of C8α oriented as in A. The central MACPF β-sheet is blue; α-helices in TMH1 and TMH2 are red, and the remainder of the MACPF domain is gold. The LDLRA domain is violet; EGF-like domain is lavender, N-terminal TSP1 is malachite, and C-terminal TSP1 is green. The upper and lower MACPF subdomains are identified with frames.
The relative positions of the C8α and C8β MACPF domains differ as a result of a different bend and twist in their central β-sheets. The upper MACPF subdomains form very thin tabular structures tightly packed together with the β-sheets facing one another (Fig. 2 and supplemental Fig. S4). These interactions are likely responsible for the strong binding between C8α and C8β. The lower subdomains differ in that the sheets are not facing one another but are nearly coplanar; a small gap between them shows no edge sharing. Separation of the lower subdomains could facilitate refolding of the TMHs during insertion into membranes. The tightly packed upper subdomains are unlikely to change their relative position and may have a role in directing and maintaining the circular shape of the pore.
In CDCs, during transition from pre-pore to pore, the bend in the central β-sheet is reduced, and the sheet changes from the “Γ” shape to a “(” shape, and the TMHs refold from an α-helical to an extended conformation, thus elongating the central β-sheet and allowing insertion of THMs into the membrane (13). This transition requires a large movement between the upper and lower subdomains and is thought to be facilitated by four glycines located at the bend of the central β-sheets (Fig. 3). These residues are conserved in C8α and C8β and likely have a similar role in refolding of the MACPF domains.
N- and C-terminal Modules
C8α and C8β each contain four modules. These are not essential for C8 activity as the C8α MACPF domain alone can bind C8β, C8γ, and C9 to form a MAC that has reduced but significant lytic activity (32). The C8β MACPF domain likewise can bind C8α-γ and express C8 activity (33). The modules constitute about a third of the length of C8α and C8β and are located on the periphery of the structure.
The N-terminal TSP1 modules of C8α and C8β form three-stranded antiparallel domains that are similar to one another and to homologous modules found in other proteins. In C8α, the central part of the module forms a ladder-like array of side chains Leu11–Arg31–Trp14–Arg29–Trp17–Lys27 clamped by two disulfides. In C8β, the array is Leu13–Ala33–Trp16–Arg31–Trp19–Arg29. Replacement of the first arginine with an alanine creates a fairly large hydrophobic cavity enclosed by Leu13, Trp16, and Phe48. An electron density feature within the cavity indicates the presence of a bound ligand (supplemental Fig. S5). The pattern of disulfide bridges is the same in C8α and C8β and is analogous to that reported for F-spondin (34). Surprisingly, this pattern (9–44, 20–54, and 28–60 in C8α; 11–46, 22–56, and 25–62 in C8β) is different from that reported for C9 (35). The four cysteines with altered disulfide bond connectivity are conserved between C8α, C8β, and C9; however, they are fairly close to one another, and the connectivity proposed for C9 may not require an unlikely change of the overall TSP1 fold.
The low density lipoprotein receptor (LDLRA) domains are located on the top of C8α and C8β (Fig. 1). They make contact with each other but not with the other modules. Each has a tightly wound Ca2+-binding site with the metal ion in a regular octahedral coordination, as found in homologous domains in other proteins. In the structure of LDL-receptor-associated protein complex (Protein Data Bank code 2FCW), there are also two LDLRA domains in contact with each other, but their mode of interaction is entirely different.
Similarity of the C8α and C8β EGF domains to the EGF protein is the lowest among the modules. The differences are greatest near the first disulfide where the position of the cysteine residues is not conserved, and the loops are quite different (supplemental Fig. S6). The EGF domains appear to be more compact and less flexible in C8 than in EGF, as judged by a greater similarity between C8α and C8β EGF domains (root mean square displacement of 1.7 Å, based on Cα atoms of 30 residues) than between two copies of EGF proteins in the unit cell in their crystal structure (root mean square displacement of 3.3 Å; based on Cα atoms of 24 residues) (36). The pattern of disulfide bonds agrees with those in C9 with an additional bond present between Cys467 and Cys514 in C8α and Cys449 and Cys498 in C8β.
The C-terminal TSP1 domains, which are on the periphery of C8, have relatively poor density, especially in C8α. There are only two pairs of intradomain disulfide bonds; their pattern could not be positively established due to disorder, but in C8β the connection Cys503–Cys536 (and by default Cys514–Cys526) appears likely because of the proximity of the residues.
Post-translational Modifications
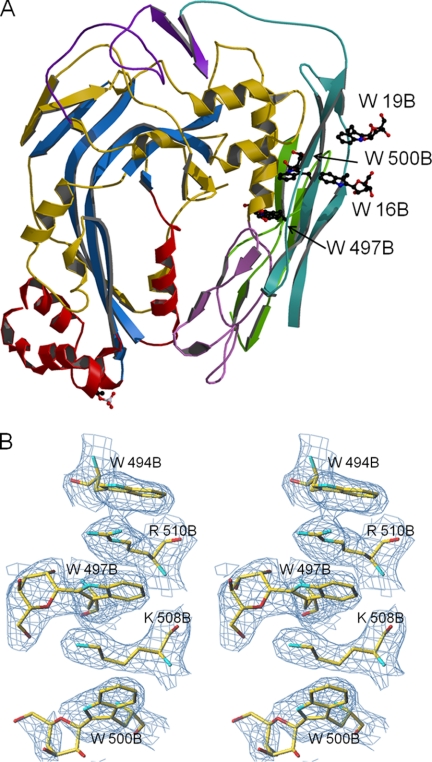
An infrequent modification of proteins is the covalent attachment of a mannopyranosyl moiety to Cδ1 of tryptophan via a C–C bond. NMR studies of a peptide derived from mannosylated human RNase indicated that an α-mannopyranose is attached (37). However, thus far there is no reported crystal structure of a peptide or protein containing a mannosyl tryptophan. All TSP1 domains present in C6, C7, C8, and C9 have some tryptophan residues mannosylated (6). In C8α, the N- and C-terminal TSP1 domains have 1 and 3 tryptophans modified, respectively, whereas the C8β domains each have 2. Electron density maps confirmed the presence of mannopyranosyl moieties at the expected positions. The mannosyl tryptophan residues from both modules cluster together (Fig. 4A). Interestingly, one of the corresponding densities is excellent and shows that a better fit can be obtained using a β- rather than α-mannosyl substituent (Fig. 4B). A previously identified single phosphorylation site on C8β Thr364 is located at the end of the TMH2 region (38). The N-linked carbohydrates in C8 used for crystallization were enzymatically removed. Potential glycosylation sites are all on the surface, and electron density maps suggest that removal was complete.
FIGURE 4.
Post-translationally modified residues in C8β. A, central β-sheet is shown in blue, TMHs in red, and the remainder of the MACPF domain in gold. The LDLRA domain is in violet, EGF in lavender, N-terminal TSP1 in malachite, and C-terminal TSP1 in green. Mannosyl tryptophans are clustered together on the periphery of the molecule; phosphothreonine 364 is at the bottom of the structure. B, electron density for mannosylated Trp497 in C8β. The 2FoFc composite omit map is contoured at a 1σ level.
Structure of C8γ
The structure of C8γ within C8 is very similar to that determined for C8γ alone (26). In C8α, the binding site for C8γ includes a 19-residue insertion (indel) within the MACPF domain. This segment also contains C8α Cys164 that covalently links to C8γ Cys40 (39). C8γ is a member of the lipocalin family of proteins that display a β-barrel fold that forms a binding pocket for a small, generally hydrophobic ligand (40). The existence of a natural small molecule ligand for C8γ has not been established, and inspection of the binding cavity in C8 does not show electron density for a ligand. Also, the entrance to the cavity is covered by the tightly bound “indel” portion of C8α making it unlikely that a ligand was removed during the purification process. Because C8 was purified from blood, it appears that C8γ does not normally carry a physiologically relevant small molecule.
Subunit Interfaces
Analysis of contacts between subunits indicates that residues buried upon C8α-C8β dimer formation constitute 10% of the surface of each subunit. Trimer formation buries an additional 4% of C8α surface and 12% of C8γ surface. There is no contact between C8β and C8γ. Almost all contacts between C8γ and C8α are through the indel. When contacts are defined as distances <4.0 Å for hydrophobic side chains and <3.5 Å for hydrophilic side chains, 30 residues from C8α form contacts with 28 residues from C8β. The interface is quite hydrophilic with 27 residues charged, 21 polar and 10 hydrophobic. The most evolutionary distant sequence is for C8α from the nurse shark. This protein is 44% identical to human C8α with 50% of the contact residues conserved. Three contact residues are identical in human C8α, C8β, and C9; an additional 14 are conserved in two of the three proteins.
Model of the MAC Pore
Although the structure of C8 is of a pair of MACPF proteins in a “non-pore” state, the unusual geometrical relationship between the C8α and C8β subunits has important functional implications regarding MAC assembly. Most frequently, homodimeric proteins are related by a 180° rotation. In contrast, C8α and C8β are related by a rotation of 22°, and the translation between their centers is almost perpendicular to the rotation axis, resulting in only a small (1.6 Å) translational component parallel to the rotation axis. It is well established that C8 mediates the binding and incorporation of C9 into the MAC and that this involves a specific C9-binding site located within the C8α MACPF domain (32). Assuming the modes of binding between C9 and C8α and between C9 and C9 are similar to that between C8α and C8β, with no translational component, we constructed a model for the C8-C9 complex. To do this, a homology model of C9 was generated based on the structure of C8α and rotated (∼22°) according to the geometrical relationship between C8α and C8β. Rotations of the C9 model by multiplicities of 22° yielded the positions of further C9 molecules and resulted in a model for the circular MAC pore (Fig. 5). This pore model contains 16 molecules per ring, which is in excellent agreement with low resolution EM studies of pores formed by poly-C9 and the MAC (3, 42).
Comparison of C8 and Perforin Structures
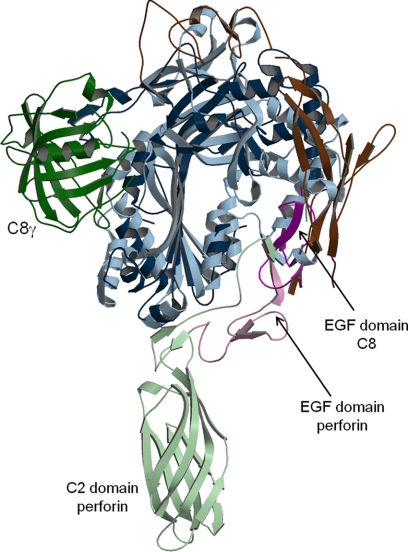
A comparison of C8 to the recently reported mouse perforin structure (Protein Data Bank code 3NSJ) (12) reveals that their similarity is limited to the MACPF/CDC domains (Fig. 6). The location of the EGF domain, which is the only module present in both proteins, is very different and suggests different roles for this module. Also, the position of C8γ does not correspond to that of the membrane-interacting C2 domain of perforin. The amino acid sequence identity of the combined MACPF + EGF domains of C8α and perforin is 21%.
FIGURE 6.
Comparison of C8 and perforin structures. C8 is in dark colors and perforin in light colors. C8γ is in green, and C8β is omitted. The MACPF/CDC domains are in blue, EGF domains in purple, and remaining domains in brown. The putative membrane location would be horizontal and below the models.
DISCUSSION
Our proposed model for MAC formation assumes conservation of binding geometry between MAC components. It is generally accepted that the MAC family proteins evolved from an ancestral pre-perforin cytolysin (43). This interpretation is consistent with amino acid sequence analyses and suggests that the first components in the contemporary MAC assembly pathway (i.e. C6 and C7) appeared later in complement evolution (44). This concept is in agreement with the hypothesis put forward by Horowitz for metabolic pathways. Horowitz (45) proposed a retrograde model, with the last enzyme in a pathway (equivalent to C9) appearing first. Horowitz (46) later extended this hypothesis by proposing that consecutive enzymes within a pathway are evolutionarily related. His rationale was that enzymes, especially those catalyzing reversible reactions, must bind both the substrate and product; thus an emerging enzyme would already have substrate binding function, and only new catalytic machinery would need to evolve. It became apparent that this later hypothesis is only occasionally true because during pathway development recruitment of enzymes with appropriate catalytic function, rather than substrate binding function, was much more frequent (47). For instance, in the glycolytic pathway pyruvate kinase and enolase, which both bind pyruvate, are the only consecutive enzymes that are evolutionarily related (48). The concepts by Horowitz (45, 46) appear to be entirely applicable to the evolution of proteins involved in MAC formation because regulatory rather than novel catalytic functions were added as complement evolved. The functions of the ancestral lysin, from which complement evolved, included self-oligomerization. The oligomerization function is conserved in contemporary terminal complement proteins, as C9 binds to C8α, C8α to C8β, etc. Thus, the mode of binding observed between C8α and C8β should reflect that between the ancestral complement/lysin molecules and consequently that between contemporary molecules of the MAC.
There is currently no experimental evidence for formation of a pre-pore or insertion of putative TMHs by the MAC proteins. The latter is assumed to occur because of structural similarity between the complement MACPF proteins and CDCs. Efficient membrane lysis by complement requires incorporation of multiple C9 molecules into the MAC in the final step. Although no organized pore or ring-like structure has been observed for the intermediate C5b-8 complex, C5b-8 at high concentrations can partially disrupt the membrane of simple target cells such as heterologous erythrocytes (49). Photolabeling studies using membrane-restricted probes identified C8α as the major C5b-8 component inserted in the membrane under these conditions (50). This finding is consistent with sequences of the putative TMH regions of the MAC proteins, which show that the most hydrophobic TMHs are those of C8α (11). Upon formation of C5b-8, these C8α regions would likely be the first to form transmembrane hairpins because interactions between their hydrophobic side chains and membrane lipid could overcome the energy deficit of unsatisfied hydrogen bonding at the open edges of the hairpins. The hydrogen bonding capability at the edges of the C8α hairpins could then become the driving force for membrane insertion of the more hydrophilic TMHs from C9. For effective pore formation, hairpins must have a partially hydrophilic character to enable migration of water along the inner wall of the pore and to repel membrane lipid. Divergent evolution of C9 and C8α may have enabled their functions to be separated. Initial membrane penetration by the very hydrophobic TMHs of C8α could facilitate insertion of the less hydrophobic pore-forming TMHs of C9, thereby creating a highly controlled yet efficient system for pore formation.
There is considerable similarity between the structures of perforin and the C8α and C8β subunits; however, there is a significant difference between the intermolecular arrangement proposed for the perforin pore and that suggested by our model of the MAC. Based on EM studies, it was proposed the MACPF/CDC domains within perforin pores (12) have an opposite orientation than in CDC pores (51). A longitudinal cut through the pore cylinder can be schematically represented by “( )” for the perforin pore and “) (” for CDC pores, where the curvature refers to the central β-sheets and the space between them the pore opening. The model for the MAC pore based on C8α/C8β orientation within C8 is “) (,” and it agrees with the model of the CDC pore rather than the perforin pore. The evolutionary distance between perforin and C8 is significantly smaller than between perforin and CDCs. Thus, the concept that during evolution the function (cell lysis) and structure (MACPF/CDC domain) were conserved, whereas the intermolecular binding mode, crucial for pore self-assembly, was changed is quite revolutionary.
The potential harm to “self” cells by activated complement is under the control of an array of proteins. MAC formation itself is regulated through interaction with CD59, a widely distributed, membrane-anchored glycoprotein that binds to C8α and C9 and thereby restricts MAC assembly and function. Earlier studies showed the CD59-binding site in C8α is within a segment defined by residues 334–385 in TMH2 (52). More recently, it was suggested the binding site lies within residues 350–355 (53). In the C8 structure, these residues are in an extended conformation and link the C terminus of the C8α Ser339–Leu350 α-helix to disordered loop Glu355–His368 (Fig. 5A and supplemental Fig. S7). The residues are in the vicinity of C8β and are not accessible. This is consistent with the observation that in solution C8 and C9 do not bind CD59 unless they are partially unfolded (54). Apparently, partial refolding of C8α TMH2 occurs as C8 binds to C5b-7, and this allows CD59 to bind. This interaction would restrict further refolding and insertion into the membrane. If so, membrane insertion of the TMHs must be significantly slower than CD59 binding, otherwise complement inhibition by this process would not work. It can be further speculated that inhibition of MAC formation would be most efficient if CD59 binding interferes with C9 recruitment. For this to occur, the C9-binding site must fully form only if C8α TMH refolding progresses beyond the state in which CD59 binding takes place.
The structure of C8 shows that the location of C8γ with respect to C8α is very different from that observed in the αMACPF-γ structure (11). Apparently, in the absence of C8β, a hydrophobic surface of αMACPF is exposed, and C8γ being linked by the flexible indel is able to migrate and bind to that surface. This differs from the C8 structure in that binding between C8α and C8γ is mediated primarily by the indel (supplemental Fig. S8). In our MAC model (Fig. 5), C8γ extends out into the pore, partially interacting not only with C8α but also C9. It does not appear to extend down to the membrane surface, in agreement with previous data suggesting it is located on the periphery of the MAC (50, 55). Also, C8γ does not bind Ca2+, and although the surface directed toward the membrane is mostly hydrophilic, it does not contain arginine or lysine residues that typically interact with bilayer phosphates. In our model, C9 appears to make contact with C8γ. This may lead to a more efficient recruitment of the first C9 molecule, which could explain why C8γ significantly enhances hemolytic and bactericidal activity when added to a noncovalent C8α-C8β complex (17, 56). The presence in shark C8α of an indel segment with a conserved Cys164 suggests C8γ appeared early in complement evolution (44).
In summary, the oligomeric nature of C8 allowed us to extend its crystal structure into an atomic model of the MAC. Structure-function analysis of the model yielded new insights into the final steps of pore formation by complement and the mechanism of its inhibition by CD59.
Supplementary Material
Acknowledgments
We thank W. Minor and M. Chruszcz (University of Virginia) and D. J. Slade for their help in the early stages of this project. We are grateful for the assistance of the high throughput crystallization laboratory at Hauptman-Woodward Medical Research Institute, Inc., Buffalo, NY, and the staff at SER-CAT beamlines at Advanced Photon Source.
This work was supported, in whole or in part, by National Institutes of Health Grant GM042898 (to J. M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
The atomic coordinates and structure factors (code 3OJY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MAC
- membrane attack complex
- MACPF
- membrane attack complex perforin
- CDC
- cholesterol dependent cytolysin
- TMH
- transmembrane β-hairpin
- TSP1
- thrombospondin type 1
- LDLRA
- low density lipoprotein receptor class A
- TAPS
- 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid.
REFERENCES
- 1. Müller-Eberhard H. J. (1988) Annu. Rev. Biochem. 57, 321–347 [DOI] [PubMed] [Google Scholar]
- 2. Esser A. F. (1994) Toxicology 87, 229–247 [DOI] [PubMed] [Google Scholar]
- 3. DiScipio R. G. (1991) J. Immunol. 147, 4239–4247 [PubMed] [Google Scholar]
- 4. Plumb M. E., Sodetz J. M. (1998) in The Human Complement System in Health and Disease (Volanakis J. E., Frank M. M. eds) pp. 119–148, Marcel Dekker, Inc., New York [Google Scholar]
- 5. Lebioda L., Sodetz J. M. (2005) in Structural Biology of the Complement System (Morikis D., Lambris J. D. eds) pp. 233–250, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 6. Hofsteenge J., Blommers M., Hess D., Furmanek A., Miroshnichenko O. (1999) J. Biol. Chem. 274, 32786–32794 [DOI] [PubMed] [Google Scholar]
- 7. Ponting C. P. (1999) Curr. Biol. 9, R911–R913 [DOI] [PubMed] [Google Scholar]
- 8. Rosado C. J., Buckle A. M., Law R. H., Butcher R. E., Kan W. T., Bird C. H., Ung K., Browne K. A., Baran K., Bashtannyk-Puhalovich T. A., Faux N. G., Wong W., Porter C. J., Pike R. N., Ellisdon A. M., Pearce M. C., Bottomley S. P., Emsley J., Smith A. I., Rossjohn J., Hartland E. L., Voskoboinik I., Trapani J. A., Bird P. I., Dunstone M. A., Whisstock J. C. (2007) Science 317, 1548–1551 [DOI] [PubMed] [Google Scholar]
- 9. Xu Q., Abdubek P., Astakhova T., Axelrod H. L., Bakolitsa C., Cai X., Carlton D., Chen C., Chiu H. J., Clayton T., Das D., Deller M. C., Duan L., Ellrott K., Farr C. L., Feuerhelm J., Grant J. C., Grzechnik A., Han G. W., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kozbial P., Krishna S. S., Kumar A., Lam W. W., Marciano D., Miller M. D., Morse A. T., Nigoghossian E., Nopakun A., Okach L., Puckett C., Reyes R., Tien H. J., Trame C. B., van den Bedem H., Weekes D., Wooten T., Yeh A., Zhou J., Hodgson K. O., Wooley J., Elsliger M. A., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2010) Acta Crystallogr. F Struct. Biol. Cryst. Comm. 66, 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hadders M. A., Beringer D. X., Gros P. (2007) Science 317, 1552–1554 [DOI] [PubMed] [Google Scholar]
- 11. Slade D. J., Lovelace L. L., Chruszcz M., Minor W., Lebioda L., Sodetz J. M. (2008) J. Mol. Biol. 379, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Law R. H., Lukoyanova N., Voskoboinik I., Caradoc-Davies T. T., Baran K., Dunstone M. A., D'Angelo M. E., Orlova E. V., Coulibaly F., Verschoor S., Browne K. A., Ciccone A., Kuiper M. J., Bird P. I., Trapani J. A., Saibil H. R., Whisstock J. C. (2010) Nature 468, 447–451 [DOI] [PubMed] [Google Scholar]
- 13. Tweten R. K. (2005) Infect. Immun. 73, 6199–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rossjohn J., Polekhina G., Feil S. C., Morton C. J., Tweten R. K., Parker M. W. (2007) J. Mol. Biol. 367, 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ng S. C., Rao A. G., Howard O. M., Sodetz J. M. (1987) Biochemistry 26, 5229–5233 [DOI] [PubMed] [Google Scholar]
- 16. Steckel E. W., York R. G., Monahan J. B., Sodetz J. M. (1980) J. Biol. Chem. 255, 11997–12005 [PubMed] [Google Scholar]
- 17. Schreck S. F., Plumb M. E., Platteborze P. L., Kaufman K. M., Michelotti G. A., Letson C. S., Sodetz J. M. (1998) J. Immunol. 161, 311–318 [PubMed] [Google Scholar]
- 18. Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., DeTitta G. T. (2003) J. Struct. Biol. 142, 170–179 [DOI] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collaborative Computational Project No 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 22. Roussel A., Cambillau C. (1991) Turbo Frodo, Silicon Graphics Geometry, Silicon Graphics, Mountain View, CA [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Acta Crystallog. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Ortlund E., Parker C. L., Schreck S. F., Ginell S., Minor W., Sodetz J. M., Lebioda L. (2002) Biochemistry 41, 7030–7037 [DOI] [PubMed] [Google Scholar]
- 27. Kabsch W. (1976) Acta Crystallogr. A: Found. Crystallogr. 32, 922–923 [Google Scholar]
- 28. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 29. Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 30. Merritt E. A., Bacon D. J. (1997) Methods Enzymol. 277, 505–524 [DOI] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC, New York [Google Scholar]
- 32. Slade D. J., Chiswell B., Sodetz J. M. (2006) Biochemistry 45, 5290–5296 [DOI] [PubMed] [Google Scholar]
- 33. Brannen C. L., Sodetz J. M. (2007) Mol. Immunol. 44, 960–965 [DOI] [PubMed] [Google Scholar]
- 34. Paakkonen K., Tossavainen H., Permi P., Rakkolainen H., Rauvala H., Raulo E., Kilpelainen I., Guntert P. (2006) Protein Struct. Funct. Bioinformatics 64, 1268–1277 [DOI] [PubMed] [Google Scholar]
- 35. Lengweiler S., Schaller J., Rickli E. E. (1996) FEBS Lett. 380, 8–12 [DOI] [PubMed] [Google Scholar]
- 36. Lu H. S., Chai J. J., Li M., Huang B. R., He C. H., Bi R. C. (2001) J. Biol. Chem. 276, 34913–34917 [DOI] [PubMed] [Google Scholar]
- 37. de Beer T., Vliegenthart J. F., Löffler A., Hofsteenge J. (1995) Biochemistry 34, 11785–11789 [DOI] [PubMed] [Google Scholar]
- 38. Tang L. Y., Deng N., Wang L. S., Dai J., Wang Z. L., Jiang X. S., Li S. J., Li L., Sheng Q. H., Wu D. Q., Li L., Zeng R. (2007) Mol. Cell. Proteomics 6, 1952–1967 [DOI] [PubMed] [Google Scholar]
- 39. Plumb M. E., Sodetz J. M. (2000) Biochemistry 39, 13078–13083 [DOI] [PubMed] [Google Scholar]
- 40. Flower D. R., North A. C., Sansom C. E. (2000) Biochim. Biophys. Acta 1482, 9–24 [DOI] [PubMed] [Google Scholar]
- 41. Thai C. T., Ogata R. T. (2004) J. Immunol. 173, 4547–4552 [DOI] [PubMed] [Google Scholar]
- 42. DiScipio R. G., Berlin C. (1999) Mol. Immunol. 36, 575–585 [DOI] [PubMed] [Google Scholar]
- 43. Podack E. R., Hengartner H., Lichtenheld M. G. (1991) Annu. Rev. Immunol. 9, 129–157 [DOI] [PubMed] [Google Scholar]
- 44. Aybar L., Shin D. H., Smith S. L. (2009) Fish Shellfish Immunol. 27, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horowitz N. H. (1945) Proc. Natl. Acad. Sci. U.S.A. 31, 152–157 [Google Scholar]
- 46. Horowitz N. H. (1965) in Evolving Genes and Proteins (Bryson V., Vogel H. J. eds) pp. 15–23, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 47. Teichmann S. A., Rison S. C., Thornton J. M., Riley M., Gough J., Chothia C. (2001) J. Mol. Biol. 311, 693–708 [DOI] [PubMed] [Google Scholar]
- 48. Lebioda L., Stec B. (1988) Nature 333, 683–686 [DOI] [PubMed] [Google Scholar]
- 49. Zalman L. S., Müller-Eberhard H. J. (1990) Mol. Immunol. 27, 533–537 [DOI] [PubMed] [Google Scholar]
- 50. Steckel E. W., Welbaum B. E., Sodetz J. M. (1983) J. Biol. Chem. 258, 4318–4324 [PubMed] [Google Scholar]
- 51. Tilley S. J., Orlova E. V., Gilbert R. J., Andrew P. W., Saibil H. R. (2005) Cell 121, 247–256 [DOI] [PubMed] [Google Scholar]
- 52. Lockert D. H., Kaufman K. M., Chang C. P., Hüsler T., Sodetz J. M., Sims P. J. (1995) J. Biol. Chem. 270, 19723–19728 [DOI] [PubMed] [Google Scholar]
- 53. Huang Y., Qiao F., Abagyan R., Hazard S., Tomlinson S. (2006) J. Biol. Chem. 281, 27398–27404 [DOI] [PubMed] [Google Scholar]
- 54. Ninomiya H., Sims P. J. (1992) J. Biol. Chem. 267, 13675–13680 [PubMed] [Google Scholar]
- 55. Brickner A., Sodetz J. M. (1985) Biochemistry 24, 4603–4607 [DOI] [PubMed] [Google Scholar]
- 56. Parker C. L., Sodetz J. M. (2002) Mol. Immunol. 39, 453–458 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.