Abstract
Reduced brain metabolism is an invariant feature of Alzheimer Disease (AD) that is highly correlated to the decline in brain functions. Decreased activities of key tricarboxylic acid cycle (TCA) cycle enzymes may underlie this abnormality and are highly correlated to the clinical state of the patient. The activity of the α-ketoglutarate dehydrogenase complex (KGDHC), an arguably rate-limiting enzyme of the TCA cycle, declines with AD, but the mechanism of inactivation and whether it can be reversed remains unknown. KGDHC consists of multiple copies of three subunits. KGDHC is sensitive to oxidative stress, which is pervasive in AD brain. The present studies tested the mechanism for the peroxynitrite-induced inactivation and subsequent reactivation of purified and cellular KGDHC. Peroxynitrite inhibited purified KGDHC activity in a dose-dependent manner and reduced subunit immunoreactivity and increased nitrotyrosine immunoreactivity. Nano-LC-MS/MS showed that the inactivation was related to nitration of specific tyrosine residues in the three subunits. GSH diminished the nitrotyrosine immunoreactivity of peroxynitrite-treated KGDHC, restored the activity and the immunoreactivity for KGDHC. Nano-LC-MS/MS showed this was related to de-nitration of specific tyrosine residues, suggesting KGDHC may have a denitrase activity. Treatment of N2a cells with peroxynitrite for 5 min followed by recovery of cells for 24 h reduced KGDHC activity and increased nitrotyrosine immunoreactivity. Increasing cellular GSH in peroxynitrite-treated cells rescued KGDHC activity to the control level. The results suggest that restoring KGDHC activity is possible and may be a useful therapeutic approach in neurodegenerative diseases.
Keywords: Alzheimer Disease, Glucose Metabolism, Mitochondria, Oxidative Stress, Protein Chemical Modification, Reactive Oxygen Species (ROS), Tricarboxylic Acid (TCA) Cycle
Introduction
Although part of classical biochemistry, the tricarboxylic acid (TCA)2 cycle has recently received considerable attention due to its significant roles in diminished glucose metabolism that accompanies neurodegenerative diseases including Alzheimer disease (AD). Oxidants that are increased in neurodegenerative diseases have been shown to regulate the TCA cycle enzymes at multiple levels. For example, oxidants modulate expression of genes in the TCA cycle (1–3) and alter enzyme activity (4–8). The experiments in this manuscript examine the post-translational modifications of one of these enzymes.
The current studies focus on the α-ketoglutarate dehydrogenase complex (KGDHC) because it is reduced in multiple neurodegenerative diseases and is sensitive to a variety of oxidants. KGDHC is arguably the rate controlling enzyme complex in the TCA cycle. KGDHC consists of multiple copies of three subunits: α-ketoglutarate dehydrogenase (E1k, EC 1.2.4.2), dihydrolipoyl succinyltransferase (E2k, EC 2.3.1.61), and dihydrolipoyl dehydrogenase (E3, EC 1.6.4.3). In Escherichia coli, multiple copies of E1k (12 dimers) and E3 (6 dimers) are arranged along the edges and faces, respectively, of an octahedral E2k core (24 subunits) to form a multienzyme complex with a molecular weight of around 5 × 106 dalton (9). The precise molecular weight and subunit composition of KGDHC and how it may vary with function in mammalian systems remains unknown.
The mechanism of the inactivation of KGDHC in disease is unknown. Diminished activity of KGDHC occurs in a number of neurodegenerative diseases including AD (10, 11), Parkinson disease (12) and progressive supranuclear palsy (13, 14). The relations are most extensively studied in AD in which impaired glucose metabolism and enhanced oxidative stress exist (15–19). In AD patients, the diminished activities of KGDHC and other TCA cycle enzymes are highly correlated to the clinical dementia rating score before the patients died (20). This suggests that these TCA cycle enzymes may play a role in the pathophysiology of AD. Thus, understanding the mechanism by which these enzymes are altered in these diseases would contribute to the knowledge of the disease pathology. The current studies focus on one of these enzymes, KGDHC. One subunit of the KGDH complex, E3, is shared with the pyruvate dehydrogenase complex (PDHC).
Post-translational modification (PTM) of proteins has been proposed to be involved in the pathogenesis of many diseases including neurodegenerative diseases. A commonly found oxidative modification in neurodegenerative diseases is the nitration of tyrosines, which is mainly induced by peroxynitrite (21). Excessive nitration occurs in brains of patients that died with AD. Peroxynitrite is a reactive nitrogen species (RNS) formed by the near diffusion-limited reaction of superoxide with nitric oxide (22, 23). Superoxide can be generated from the mitochondrial electron chain and KGDHC. Nitric oxide is produced by the three isoforms of the nitric-oxide synthase. KGDHC is inactivated by reactive oxygen species (ROS) (6). For example, hydrogen peroxide inactivates KGDHC activity in intact cardiac mitochondria, fibroblasts, and N2a cells (4, 5, 7, 8). Peroxynitrite reduces KGDHC activity and selectively alters immunoreactivity of E1k and E2k (24). However, the inhibitory mechanism of KGDHC by peroxynitrite including possible post-translational modification has not been reported. This could be due to the complexity and low abundance of KGDHC in brain, which has made identification of exact modification sites of KGDHC by mass spectrometry very difficult.
We focused these studies on nitration because it is stable over a long time of period compared with other labile modifications such as S-nitrosation or glutathionylation. Nitration is likely to be reversed as growing evidence supports the denitration of proteins in many biological systems including tissue homogenates, activated macrophages and crude extracts from different organs (25–30). However, the molecular identity of the denitrase activity is still unknown.
The large molecular weight and complex structure of KGDHC makes investigation of its oxidative modifications difficult. Thus, the current experiments employed multiple approaches. Clear- or Blue-Native gel has the advantage of allowing the study of protein complexes with molecular weight up to 10 MDa and preserving enzyme activity after electrophoresis (31). By combining proteomic tools including denaturing and native gel electrophoresis and a nano-LC-MS/MS system, the current studies tested whether distinct nitration sites underlie the oxidant induced deficits in KGDHC and whether the resulting nitration could be reversed.
EXPERIMENTAL PROCEDURES
Purification and Treatment of KGDHC
KGDHC (200 μl; Cat No. K1502; Sigma) was further purified by loading the protein onto a 2% ABT agarose column (0.7 × 50 cm) (Agarose Bead Technologies, Tampa, FL) to remove bovine serum albumin. The five fractions with the highest KGDHC activity from four column purifications were pooled and treated with 0, 1, or 10 μm peroxynitrite (Cayman, Ann Arbor, MI) at 37 °C for 15 min. Samples were then concentrated using an Amicon Ultra-15 Centrifugal Filter Unit 100 (Millipore; Billerica, MA). The concentrated samples were immediately processed for KGDHC activity measurement, SDS gel, Clear- or Blue-native gel electrophoresis and Western blotting detection.
In experiments testing the reversal of nitration by GSH, purified KGDHC was treated with 0 and 10 μm peroxynitrite at 37 °C for 15 min (two identical reactions were included for the 10 μm peroxynitrite treatment). Then, 5 mm GSH was added to one of the peroxynitrite-treated samples. All samples including 0, 10 μm peroxynitrite, and 10 μm peroxynitrite + 5 mm GSH were incubated for another 15 min at 37 °C. All samples were then processed as described in the previous paragraph.
Cell Culture and Treatment
N2a neuroblastoma cells were purchased from American Type Culture Collection (Manassas; VA). The cells in 100-mm dish were maintained at 37 °C in a humidified incubator under 5% CO2 and 95% air in the complete medium (DMEM supplemented with 10% fetal bovine serum). Cells were treated with peroxynitrite as described previously with slight modification (32) Cells were treated with different concentrations of peroxynitrite (0, 0.0625, 0.125, 0.25, 0.5, and 1 mm) for 5 min and allowed to recover in normal medium or medium containing 1 mm NAC for 24 h. Cells were harvested and the KGDHC activity was measured in cell lysates. Detailed methods are described in supplemental Method S1. After treatment, cell pellets were processed differently for different measurements (see detailed procedures in each section below).
KGDHC Activity Measurement
KGDHC catalyzes the following reaction: α-ketoglutarate + NAD+ + CoA → succinyl CoA + CO2 + NADH. Two independent, well established assays were used to measure KGDHC activities. Both methods use the specific substrate α-ketoglutarate (α-KG) to distinguish NADH production from KGDHC to that from other enzymes (i.e. substrate specificity). One method assays activity in purified enzyme or cell lysates whereas the other assesses the activity of the enzyme in a clear native gel.
KGDHC Activity Measured in Purified Enzyme and Cell Lysates
Purified enzyme samples were used directly for KGDHC activity measurements. Cell pellets were immediately resuspended with 500 μl of lysis buffer (50 mm Tris-HCl, pH 7.2, 0.4% Triton X-100, 0.2 mm EGTA, 50 μm leupeptin, and 1 mm DTT) for 30 min with shaking at 4 °C. After the 30-min incubation, cell lysates were disrupted by repeated aspiration through a 21-gauge needle for 12 times on ice. KGDHC activity was assayed as described previously (11).
KGDHC Activity by In-gel Activity Staining
The basis of the in-gel activity staining is very similar to the lysis method except that the reducing equivalents are coupled to nitroblue tetrazolium (NBT) (33). The specificity again depends upon the substrate. Equal amount of samples (10 μg) were loaded to clear native gels. After electrophoresis, the clear native gel was washed once with the reaction mixture (50 mm Tris, pH 7.6; 1 mm MgCl2, 0.1 mm CaCl2, 0.05 mm EDTA, 0.2% Triton X-100, 0.3 mm TPP, 5 μg/ml rotenone in ethanol, and 3.5% polyvinylalcohol). Then 10 ml of assay mix (3 mm β-NAD, 1 mm CoA, 3 mm α-KG, 0.75 mm NBT, 0.05 mm phenazine methosulfate (PMS) in reaction mixture) was added to the gel to stain the KGDHC activity. An assay mix without CoA and α-KG was added to a duplicate gel and used as the blank. After a 5-min incubation at room temperature, the reaction was stopped by washing the gel with distilled water.
SDS-gel, Blue, or Clear Native Gel Electrophoresis and Western Blotting
For SDS-PAGE, a 5× SDS-sample loading dye (50% glycerol, 250 mm Tris (pH 6.8), 10 mm EDTA, 10% SDS, and 0.04% bromphenol blue) was added to either purified enzyme or cell lysates to achieve 1× final concentration. Electrophoresis was done with a 4–20% Tris-glycine gel (Invitrogen, Carlsbad, CA) using 120 volts for 2.5 h.
For Blue-native gel electrophoresis, NativePAGETM Sample buffer (4×) was added to samples to make 1× final concentration. About 6 μg of protein was loaded into each well, and the electrophoresis was done with a NativePAGETM Novex® 3–12% Bis-Tris gel (Invitrogen) using 50 volts for overnight according to the manufacturer's instruction. Unlike Blue-native gel electrophoresis, the cathode buffer for clear native gel electrophoresis contains no NativePAGETM 20X Cathode Buffer Additive (0.4% Coomassie® G-250). Western blotting was performed as described in supplemental Method S2.
Mass Spectrometry
The protein gel bands were excised from the SDS or Blue-Native gels. The gel bands were reduced with 10 mm of DTT and alkylated with 55 mm iodoacetamide, and then digested with Sequence Grade Modified Trypsin (Promega, Madison, WI) in ammonium bicarbonate buffer at 37 °C overnight. The digestion products were extracted twice with 0.1% trifluoroacetic acid and 50% acetonitrile and 1.0% trifluoroacetic acid, respectively. The extracted mixture was dried by SpeedVac and dissolved in 10 μl of 0.1% tri-fluoroacetic acid. Half of the extracts were injected for LC-MS/MS analysis. For LC-MS/MS analysis, each digestion product was separated by a 60-min gradient elution with the Dionex U3000 capillary/nano-HPLC system (Dionex; Sunnyvale, CA) at a flow rate of 0.250 μl/min that is directly interfaced with the Thermo-Fisher LTQ-Obritrap mass spectrometer (Thermo Fisher, San Jose, California) operated in data-dependent scan mode. The analytical column was a home-made fused silica capillary column (75 μm ID, 100 mm length; Upchurch, Oak Harbor, Washington) packed with C-18 resin (300 A, 5 μm, Varian, Palo Alto, California). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. The 60 min gradients with 250 nl/min flow rate for B solvent went from 0 to 55% in 34 min and then in 4 min to 80%. The B solvent stayed at 80% for another 8 min and then decreased to 5% in 8 min. Another 6 min was used for equilibration, loading and washing. The mass acquisition method was one FT-MS scan followed by 6 subsequent MS/MS scan in the ion trap. The FT-MS scan was acquired at resolution 30,000 in the Orbi-trap. The six most intense peaks from the FT full scan were selected in the ion trap for MS/MS. Data were analyzed using the MASCOT software search algorithm and manual inspection. Nitrated peptides and un-nitrated peptides are chemically similar to one another, and it is reasonable to expect that they exhibited similar ionization efficiencies. The relative quantification of nitrated tyrosines and un-nitrated tyrosines was accomplished by comparing the peak intensity of the corresponding nitrated peptides and unnitrated peptides.
Purification of KGDHC from N2a
All procedures were carried out at 4 °C unless otherwise indicated. Mitochondria were isolated from N2a cells by modification of a previous method (34). Detailed methods are described in supplemental Method S3. Mitoplast preparation (35) and KGHDC purifications are described in supplemental Method S4.
Immunoprecipitation of Nitrated KGDHC from N2a Cells
Immunoprecipitation (IP) was utilized to pull down nitrated KGDHC from N2a cells treated with peroxynitrite. Cell pellets (see “Cell Culture and Treatment”) were resuspended in 2 ml of ice-cold RIPA buffer (50 mm Tris HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) and incubated at 4 °C for 30 min. Then cells were disrupted by repeated aspiration through a 21-gauge needle. Cellular debris were pelleted by centrifugation at 10,000 × g for 10 min at 4 °C. Supernatants were saved, and the pellets were resuspended with 0.5 ml of RIPA buffer and subjected to above steps to re-extract cellular proteins. Supernatants combined from two centrifugations were precleared by adding 1.0 μg of normal mouse IgG and 20 μl of resuspended protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) followed by incubation at 4 °C for 30 min. The agarose beads were pelleted by centrifugation at 1,000 × g for 5 min at 4 °C. Supernatants (1.2 ml) were transferred to a new 1.5 ml tube and 12.5 μl of nitrotyrosine antibody (Abcam; Cambridge, MA, 0.25 μg/μl) was added. After incubation at 4 °C for 1 h, 20 μl of resuspended protein A/G PLUS-agarose was added, and the mixture was incubated at 4 °C on a rocking platform overnight. Immunoprecipitate was collected by centrifugation at 1,000 × g for 5 min at 4 °C. The pellets were washed three times with 1 ml of RIPA buffer by incubating the resuspension for 10 min with gently rocking in the cold room followed by centrifugation (1,000 × g/5 min). After the final wash, the pellets were resuspended in 40 μl of 1× electrophoresis sample buffer and subjected to SDS-PAGE and Western blotting analyses.
Real-time PCR
Total RNA was isolated from cell pellets by an RNesay Plus Micro kit (Qiagen, Valencia, CA) according to the manufacturer's instruction. First-strand cDNA was synthesized from the isolated total RNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Indianapolis, IN) with anchored oligo(dT)18 as primer. Real-time PCR of E1k, E2k, and E3 was performed using an Applied Biosystems 7500 Fast Real-Time PCR system with pre-designed Taqman® gene expression assays (Applied Biosystems, Foster City, CA) by modification of a published method (36) as described in supplemental Method S5.
RESULTS
Inhibition of Purified KGDHC by Peroxynitrite
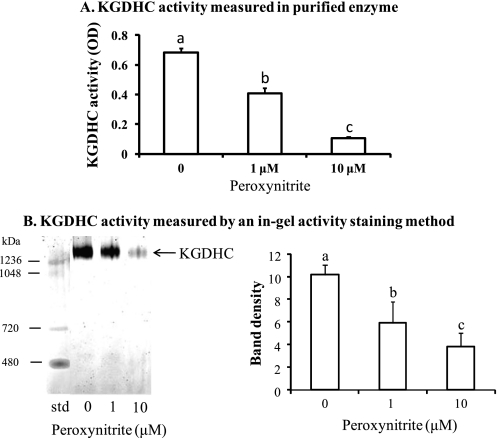
Treatment of purified KGDHC with peroxynitrite inhibited KGDHC activity in a dose-dependent manner (Fig. 1). Peroxynitrite, as low as 1 μm, diminished KGDHC activity by 40% (Fig. 1A). Similarly, 1 μm peroxynitrite reduced KGDHC activity by 42% as measured by in gel activity staining (Fig. 1B).
FIGURE 1.
Peroxynitrite inhibited KGDHC activity in a dose-dependent manner. The effects of peroxynitrite on the KGDHC activity were measured in a plate reader (A) and in a Clear Native gel (B). B includes a representative in-gel activity staining of KGDHC (left panel) and a bar graph (right panel). Values are means ± S.E. from three independent experiments done in triplicate. Values with different letters vary significantly (p < 0.05) as determined by a one-way ANOVA followed by a Student-Newman-Keul's test.
Nitration of Purified KGDHC by Peroxynitrite
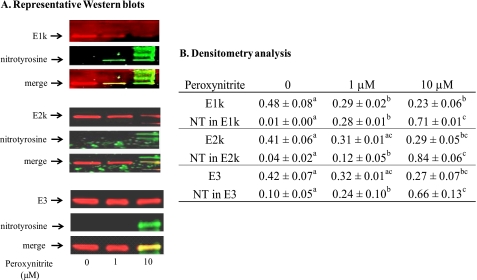
Whether peroxynitrite induces nitration of KGDHC was determined by SDS gel electrophoresis followed by Western blotting using an anti-nitrotyrosine antibody (Fig. 2A). The lower concentration of peroxynitrite (1 μm) significantly reduced the immunoreactivity of E1k only (Fig. 2B). While the immunoreactivities of E1k, E2k, and E3 were all decreased significantly in response to the higher concentration of peroxynitrite (10 μm) (Fig. 2B). In contrast, the immunoreactivity of nitrotyrosine increased with increasing concentration of peroxynitrite (Fig. 2, A and B).
FIGURE 2.
Western blots of KGDHC separated by SDS-gel and analyzed by densitometry. Purified KGDHC was treated with different concentrations of peroxynitrite, and subjected to SDS-PAGE followed by Western blotting. A, blots were probed with antibodies to E1k, E2k, or E3 (red) and to nitrotyrosine (green), respectively. The overlap is represented by yellow. B, densitometry analysis of E1k, E2k, E3 and nitrated subunits of KGDHC from at three independent Western blots by the Odyssey Infrared Imaging System. a–c, values with different letters in each row are significantly different from each other (p < 0.05) as determined with a one-way ANOVA followed by a Student-Newman-Keul's test.
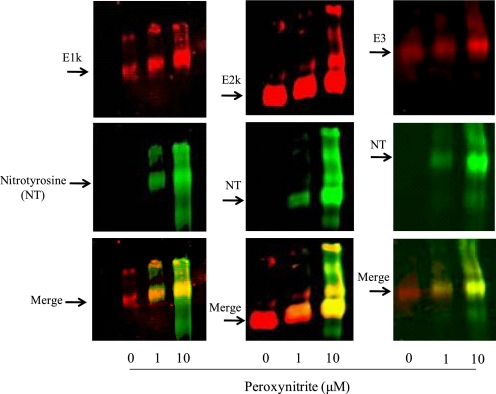
Western blotting following blue-native gel electrophoresis was also utilized to determine the nitration of KGDHC. Higher shifted bands occurred in all three subunits of KGDHC that were treated with peroxynitrite as compared with untreated ones (Fig. 3). Multiple up shifted bands (greater than the 1263-kDa band) were mainly observed in 10 μm peroxynitrite-treated KGDHC. They are likely the up-shifted subunits that are nitrated by peroxynitrite. Similarly, the immunoreactivity of nitrotyrosine increased with increasing concentration of peroxynitrite.
FIGURE 3.
Western blots of KGDHC separated by Blue-native gel. Purified KGDHC was treated with different concentrations of peroxynitrite, and subjected to a Blue-native gel followed by Western blotting. The blots were probed with antibodies to E1k, E2k or E3 (red) and to nitrotyrosine (green), respectively. The overlap is represented by yellow.
Tryrosine 175, 556 of E1k, 313 of E2k, and 352, 416 of E3 Are the Targets of Peroxynitrite
The modification sites in KGDHC were further investigated by nano-LC-MS/MS mass spectrometry. Purified KGDHC was incubated with peroxynitrite and then processed for SDS and/or Blue native gel electrophoresis. The bands were cut and analyzed by nano-LC-MS/MS mass spectrometry.
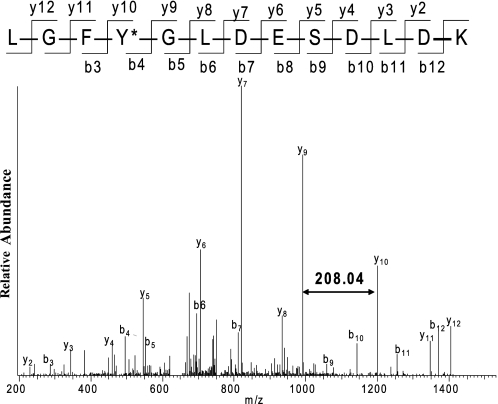
The tyrosine in positions Tyr-175 and Tyr-556 in E1k of KGDHC were nitrated by peroxynitrite as determined by both MS and MS/MS analysis. For example, the mass of a peptide ion at m/z 758.85 matches to the mass of the nitrated 172LGFY*GLDESDLDK184, in which the peptide mass has the 45 dalton mass increase corresponding to the incorporation of a -NO2 group in the peptide. The MS/MS spectrum of this peptide ion at 758.85 allowed us to determine that Tyr-175 was nitrated as shown in Fig. 4. Following a similar procedure, the residue Tyr-313 in E2k (supplemental Fig. S1A) and the residues Tyr-352 and Tyr-416 (supplemental Fig. S1B) in the E3 subunit of KGDHC were also found to be nitrated by peroxynitrite. Furthermore, the percent nitrated tyrosine in the three subunits of KGDHC was calculated by using a relative quantitative method (see “Experimental Procedures”). MS-identified nitration sites and the percentage of nitrated tyrosine in each subunit are summarized in Table 1. The percent of nitration in Tyr-175 and Tyr-556 of E1k is 19 and 23%, respectively. The percent of nitration in Tyr-313 of E2k is 39%. The percent nitration in Tyr-352 and Tyr-416 in E3 is 13 and 46%, respectively.
FIGURE 4.
The selected MS/MS spectrum of a nitrated peptide 172LGFY*GLDESDLDK184 of E1k at m/z 758.85. Purified KGDHC was treated with 10 μm peroxynitrite followed by SDS gel electrophoresis as described under “Experimental Procedures.” Protein bands corresponding to the E1k were cut and subjected to trypsin digestion followed by nano-LC-MS/MS analysis. The mass difference (208.04) between y9 and y10 corresponding to nitrated tyrosine residue (163.06 + 45) indicated the nitration of tyrosine 175 in the E1k subunit of KGDHC. The fragment ions match all of the y and b ions of the peptide, in which the tyrosine (Y) residue was nitrated with a mass difference of 45Da.
TABLE 1.
Summary of peroxynitrite-induced modification in purified KGDHC
Peroxynitrite (10 μm)-treated KGDHC was subjected to either SDS-gel or Blue-native gel electrophoresis. Protein bands that were cut from gels were digested with trypsin and subjected to nano-LC-MS/MS to identify modification sites. Tyrosine 175, 556 of E1k, 313 of E2k, and 352, 416 of E3 were nitrated by peroxynitrite. The relative quantification of nitrated tyrosines and un-nitrated tyrosines was accomplished by comparing the peak intensity of the corresponding nitrated peptides and un-nitrated peptides.
| Subunit | Modification | aa | % Nitration | Type of Gel/digestion |
|---|---|---|---|---|
| E1k | Nitration | Y175 | 19 | SDS gel/Blue-native gel/trypsin |
| Y556 | 23 | SDS gel/Blue-native gel/trypsin | ||
| E2k | Nitration | Y313 | 39 | SDS-gel/Blue-native gel/trypsin |
| E3 | Nitration | Y352 | 13 | SDS gel/trypsin |
| Y416 | 46 | SDS gel/trypsin |
Denitration of the Nitrated Purified KGDHC
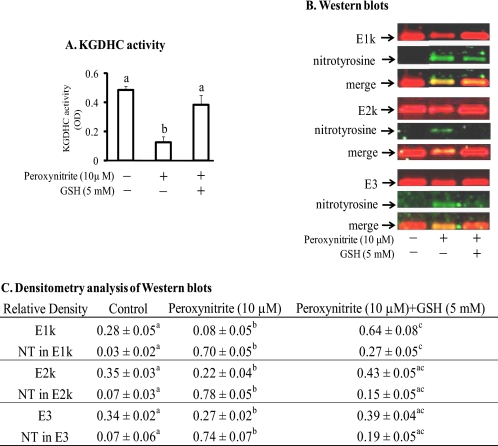
If the nitration serves as a signal, then it should be reversible. KGDHC that is inactivated by H2O2 or oxidant can be reactivated with glutathione (8). Thus, reduced glutathione (GSH) was added after treatment of KGDHC with peroxynitrite to determine whether glutathione would alter the nitration. The consequences on KGDHC were assessed by measuring the change in KGDHC activity and by Western blotting with the anti-nitrotyrosine antibody. Peroxynitrite (10 μm) reduced KGDHC activity by 74% (Fig. 5A). Addition of GSH after incubation of KGDHC with peroxynitrite restored the activity to levels comparable to those of the untreated KGDHC (Fig. 5A). Incubation of peroxynitrite-treated KGDHC with GSH reduced the immunoreactivity of nitrotyrosine (Fig. 5, B and C).
FIGURE 5.
Denitration of KGDHC by GSH following peroxynitrite. Purified KGDHC was treated with 10 μm peroxynitrite for 15 min followed by 5 mm GSH for another 15 min. KGDHC activity was measured. SDS-PAGE followed by Western blotting was performed. A, KGDHC activity. Values are means ± S.E. of three independent experiments done in duplicate. Values with different letters vary significantly (p < 0.05) as determined by a one-way ANOVA followed by a Student-Newman-Keul's test. B, representative blots of E1k, E2k, E3 (red) and nitrotyrosine (green). The yellow reflects overlap. C, densitometry analysis of E1k, E2k, E3 and nitrated subunits of KGDHC by the Odyssey Infrared Imaging System from at least three independent experiments. a–c, values with different letters in each row are significantly different from each other (p < 0.05) as determined with a one-way ANOVA followed by a Student-Newman-Keul's test.
Mass spectrometry analysis revealed very specific changes in the reversal of nitration following incubation with GSH. Peroxynitrite induced a 13 and 46% nitration in the Tyr-352 and Tyr-416, respectively, in E3 (Table 2). The percent nitration in these two tyrosine residues was reduced to zero following GSH incubation (Table 2). While the nitration of E2k (34%) was reduced by about half after subsequent GSH incubation (15%) (Table 2).
TABLE 2.
Denitration of KGDHC by GSH following peroxynitrite demonstrated by LC-MS/MS
KGDHC was treated with peroxynitrite or peroxynitrite + GSH. After treatment, samples were subjected to SDS- PAGE. Protein bands corresponding to the subunits of KGDHC were cut and further analyzed by mass spectrometry. About half of the nitration of Tyr-313 in E2k was removed after subsequent addition of GSH following peroxynitrite. Nitration of Tyr-352 and Tyr-416 in E3 induced by peroxynitrite were completely removed following GSH. Underlined Y indicates nitration.
| Subunit | Untreated | Peroxynitrite | GSH following peroxynitrite |
|---|---|---|---|
| E2k | EVVY(313)R | EVVY(313)R | EVVY(313)R |
| % nitration | 0% | 34% | 15% |
| E3 | IPNIY(352)AIGD | IPNIY(352)AIGD | IPNIY(352)AIGD |
| % nitration | 0% | 13% | 0% |
| SEEQLKEEGIEY(416)K | SEEQLKEEGIEY(416)K | SEEQLKEEGIEY(416)K | |
| % nitration | 0% | 46% | 0% |
Nitration of KGDHC in N2a Cells by Peroxynitrite
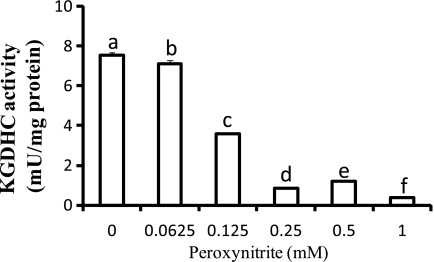
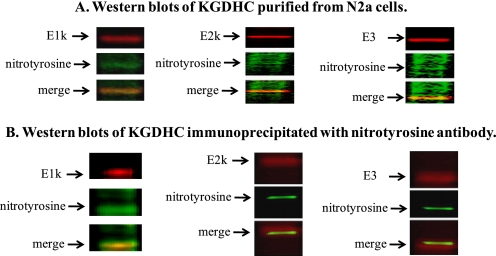
Peroxynitrite decreased KGDHC activity in a dose-dependent manner in N2a cells. About a 50% reduction in the activity was found in cells treated with 0.125 mm (Fig. 6). Moreover, Western blots of KGDHC purified from cell lysates suggested a colocalization of KGDHC with nitrotyrosine (Fig. 7A). The purity of KGDHC was demonstrated by its significantly higher specific activity (>59 fold) than the starting homogenate (cell lysate). Immunoprecipitation (IP) of cells after peroxynitrite treatment with a nitrotyrosine antibody further confirmed the colocalization of KGDHC with nitrotyrosine (Fig. 7B).
FIGURE 6.
Peroxynitrite inhibited KGDHC activity in N2a cells. N2a cells were treated with different concentrations of peroxynitrite (0, 0.0625, 0.125, 0.25, 0.5, and 1 mm) for 5 min and allowed to recover in normal medium for 24 h. Then cells were harvested, and the KGDHC activity was measured in cell lysates. Values are means ± S.E. of at least two independent experiments done in triplicate. Values with different letters are significantly different from each other (p < 0.05) as determined by a one-way ANOVA followed by a Student-Newman-Keul's test.
FIGURE 7.
Colocalization of KGDHC and nitrotyrosine following treatment of N2a cells with peroxynitrite. A, representative Western blots of KGDHC purified from N2a cells after peroxynitrite treatment. B, representative Western blots of N2a cells immunoprecipitated with nitrotyrosine antibody. E1k, E2k, or E3 (red), nitrotyrosine (green), and the yellow reflects overlap.
Reversal of Nitrated KGDHC in N2a Cells
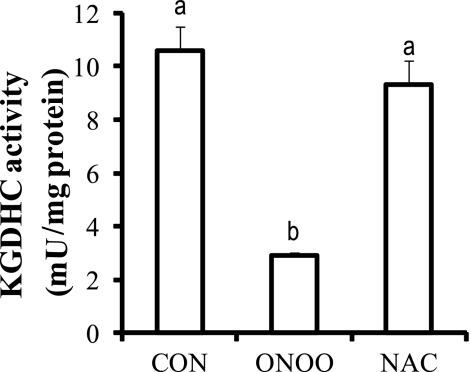
Subsequent experiments tested whether the reduction in KGDHC activity following peroxynitrite could be reversed in cultured N2a cells by N-acetylcysteine (NAC). NAC is the most bioavailable precursor of glutathione, and it has been used widely to increase GSH level within cells. NAC readily enters cells and is hydrolyzed to cysteine for synthesizing GSH in cells. The reduced form of glutathione (GSH) has short half-life of less than 1 h in culture medium (37). The reversal experiments require a 24-h incubation of cells with glutathione. Thus, the short half-life of less than 1 h excluded GSH as an optimal thiol compound in this experiment. Peroxynitrite diminished KGDHC activity by 74% (Fig. 8). Addition of NAC to peroxynitrite-treated cells rescued KGDHC activity back to control values (Fig. 8).
FIGURE 8.
Inactivation of KGDHC by peroxynitrite in N2a cells was reversed by NAC. N2a cells were first treated with 0.5 mm peroxynitrite for 5 min and the medium was replaced with fresh medium containing 1 mm NAC. The cells were maintained for another 24 h before harvesting for KGDHC activity measurement. Values (means ± S.E.) are from at least two independent experiments done in triplicate. Values with different letters are significantly different from each other (p < 0.05) as determined by a one-way ANOVA followed by a Student-Newman-Keul's test.
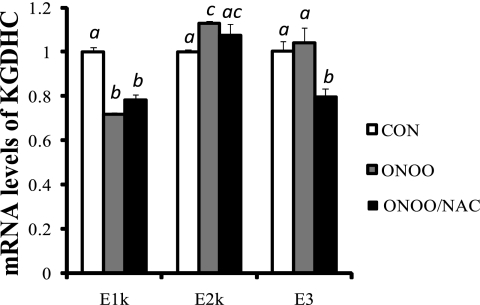
The reversal of diminished KGDHC by NAC was not mediated at the level of transcription. The levels of mRNA of the three subunits of KGDHC measured by real-time qPCR showed a 30% reduction in E1k, a 12% increase in E2k and no change in E3 in response to peroxynitrite. Addition of NAC to peroxynitrite-treated cells did not alter the mRNA levels of E1k and E2k as compared with peroxynitrite-treated cells, but NAC reduced mRNA level of E3 by 20% (Fig. 9).
FIGURE 9.
mRNA levels of the subunits of KGDHC in N2a cells treated with peroxynitrite and peroxynitrite/N-acetylcysteine. Cells were subjected to total RNA isolation, cDNA synthesis and quantitative real-time RT-PCR. β-Microglobulin (β2m) was used as an internal control. Values are the means ± S.E. of relative changes over controls from two independent experiments done in triplicate after normalization to β2m. Values with different letters vary statistically from each other (p < 0.05) as determined by a one-way ANOVA followed by a Student-Newman-Keul's test.
DISCUSSION
The present study is the first to identify distinct modification sites in purified KGDHC in response to peroxynitrite. The closest reported study on KGDHC modification in rat heart mitochondria only suggests that KGDHC is nitrosated by NO but without identification of modified residues (38). Glutathionylation of KGDHC has been demonstrated in cardiac mitochondria treated with H2O2. Lipoic acid, a required cofactor covalently attached to the E2k subunit, is the site of glutathionylation (39). KGDHC activity is diminished in a number of neurodegenerative diseases (11, 13, 40–42). The mechanism of inhibition is unknown in the diseases. Post-translational modification of the enzyme mediated by oxidative stress, which is associated with these age-related diseases, is likely to contribute to the inhibition under the disease conditions. The present study explored the underlying mechanism of peroxynitrite induced inhibition and reactivation of KGDHC following peroxynitrite treatment of either the purified enzyme or cells in culture.
The present study, for the first time, utilized a Blue-native gel electrophoresis system to study the complex and show that it shifts with stress. The molecular mass of the KGDHC is unknown in mammalian systems. In E. coli, KGDHC is approximately of 5–10 × 106 Da based on ratio of 24:24:12 (E1k:E2k:E3) (43, 44). The three subunits of KGDHC are bound together tightly through non-covalent bonds. The nature of the dynamic composition of the complex makes it difficult to determine its MW in different species precisely (45). In addition, the cellular physiological state may alter the ratio of the three subunits of the complex. The shift in bands of KGDHC (Fig. 3) in response to peroxynitrite suggests that the complex is very dynamic. Although the physiological consequences are still unknown, the changes seem to be associated with diminished activity.
Our previous studies showed that KGDHC is sensitive to peroxynitrite (24), but the precise modification sites in KGDHC have not been assessed. Peroxynitrite is increased in many neurodegenerative disorders including AD, Parkinson Disease, multiple sclerosis, and amyotrophic lateral sclerosis (46). The combination of Blue-native and SDS-gel electrophoresis with mass spectrometry analysis revealed that peroxynitrite-induced nitration occurred only in two tyrosine residues in E1k and E3 subunit, and one tyrosine in E2k even though there are multiple tyrosine residues in KGDHC (31 tyrosine in E1k, 5 in E2k, and 11 in E3). PTM of a specific protein does not occur randomly. The abundance of a protein or the number of tyrosine residues in a protein cannot predict whether it is a target for PTM (47–49). For example, only 2 out of 18 tyrosines are nitrated in human serum albumin following treatment with peroxynitrite (50). The sites of PTM are likely to be determined by the protein secondary structure and the local environment of the tyrosine residue (48, 49).
Whether these nitration sites in KGDHC can be directly coordinated with the loss of KGDHC activity is unknown. The present studies did not reveal any modification of the active sites of the three proteins of the complex. Active site studies of all three of these large proteins with multiple copies in the complex are difficult. Most of the active sites studies are performed with KGDHC purified from E. coli. For example, histidine 260, 298, and 729 as well as serine 321 in E1k appear to confer substrate specificity and the capacity to accommodate the TCA metabolite oxaloacetate (51). Histidine 375, threonine 323, and aspartate 379 are involved in E2k active site (52). The denitration of the nitrated tyrosine residues in KGDHC correlates well with the reversal of diminished KGDHC activity. This suggests the importance of these tyrosine residues. Thus, site-directed mutagenesis studies on KGDHC are essential to pinpoint whether nitration of the specific residue of KGDHC we identified are responsible for the loss of activity.
The present studies provide the first evidence that KGDHC can be nitrated in cells. The nitration correlates with the changes in activity. Whether peroxynitrite also induces nitration of KGDHC in N2a cells was first tested by immunodetection of nitrated KGDHC from highly purified KGDHC from treated N2a cells and then further confirmed by immunoprecipitation of nitrated KGDHC with the nitrotyrosine antibody. Colocalization of KGDHC and nitrotyrosine demonstrated by these two means provides strong evidence that nitration of KGDHC occurs in cellular system upon peroxynitrite exposure and correlates with the changes in activity. The 30% reduction in E1k mRNA level in response to peroxynitrite may also contribute to the reduction in E1k protein and the overall KGDHC activity in addition to the nitration of KGDHC. Moreover, other TCA cycle enzymes especially the pyruvate dehydrogenase complex (PDHC), which shares the E3 subunit with KGDHC, are also likely to be nitrated in response to peroxynitrite and their activities may be affected. Thus, nitration and/or other modifications of these enzymes may contribute to the decline in the oxidative metabolism in brains from patients with neurodegenerative diseases.
Reversal of the nitration of KGDHC would suggest that nitration may be a normal part of cell signaling. Whether the inhibition of KGDHC activity by peroxynitrite can be reversed was tested in the semi-purified enzyme as well as in cells. With purified enzyme, the loss of KGDHC activity is directly correlated to the increased immunoreactivity of nitrotyrosines in response to peroxynitrite. Diminished immunoreactivity of nitration by addition of GSH following peroxynitrite parallels the recovery of KGDHC activity. This supports the suggestion that nitration of KGDHC likely contributes to its inhibition. Moreover, mass spectrometry analysis showed nitration of tyrosine residues (Tyr-352 and Tyr-416) in the E3 subunit of KGDHC induced by peroxynitrite was completely removed by subsequent addition of GSH. About half of the nitration of Tyr-313 in E2k was removed. The nitration of E1k was not observed by MS in the reversal experiments with peroxynitrite and peroxynitrite/GSH. This was surprising because the nitration of the two tyrosine residues under the conditions used in Table 1 was highly reproducible by Western and MS. The treatment conditions were different in the two experiments. In the GSH reversal experiment, the samples were incubated for another extra 15 min after addition of peroxynitrite as compared with the peroxynitrite inhibition experiment in which samples were incubated with peroxynitrite for only 15 min. The extra 15 min of incubation of KGDHC would increase the time for denitration (see the following paragraph). A technical factor may also be involved. The percent nitration of Tyr-175 and Tyr-556 (19 and 23%, respectively) in E1k is low. Thus, the peptides with nitrated tyrosine have far less chance to be recovered from pool of the same peptides without nitration.
Similarly, peroxynitrite inhibited KGDHC activity in N2a cells. Further treatment of cells with NAC, a precursor of GSH, rescued KGDHC activity to near control level. Reversal of KGDHC activity in both isolated enzyme and in cells by glutathione following peroxynitrite indicates a direct interaction of the enzyme complex with GSH. No changes in E1k, E2k mRNA levels and a slight reduction in the E3 mRNA level after NAC treatment further support the direct interaction of KGDHC with GSH.
The partial removal of nitration of KGDHC is consistent with the presence of a denitrase activity since it is difficult to chemically remove a nitro group. This is supported by our results that GSH is not able to diminish the nitration of actin induced by peroxynitrite (data not shown). Hence, the de-nitration of KGDHC is not a simple chemical removal of the nitro group by GSH. Interestingly, E3 of KGDHC in the presence of biological reducing agents represents a possible reducing system capable of decreasing the nitrative damage of DNA and proteins. For example, the E3 component of KGDHC can act as a denitrase in the presence of cofactors of NAD(P)H or dihydrolipoic acid (53). In the absence of E3, these substrates could not act as denitrases. The efficiency of the E3 denitrase activity could be increased by other compounds such as lipoic acid, or by including an NAD(P)H regenerating system. Whether glutathione can directly or indirectly act as a cofactor or activator has not been tested. Glutathione, a tripeptide composed of l-glutamate, l-cysteine, and glycine, is considered to be the most important endogenous intracellular antioxidant or reducing agent in mammalian cells (54, 55). Increasing GSH by NAC, a precursor of glutathione, in mammalian cells and animals has been shown to protect cells and animals from oxidative stress and toxicity (56, 57). Treatment of cultured HepG2 cells with 4-hydroxynonenal (HNE) resulted in a significant reduction of PDHC and KGDHC activities. Lipoyl compounds afforded protection from HNE-induced inhibition of PDHC. This protection was higher in the presence of cysteine and GSH (58). Thus, reversal of KGDHC activity in the presence of GSH in either purified enzyme or cells is consistent with denitration catalyzed by E3. In addition to its possible contribution to decreased metabolism, the diminished KGDHC activity in AD would also have lower denitrase activity, which is consistent with increased nitrated proteins in AD.
In summary, the present study revealed the precise nitration on the tyrosine residues in KGDHC treated with peroxynitrite. The nitration can be partially reversed by subsequent addition of GSH. KGDHC is likely to contribute to its reversal by acting as a denitrase in the presence of reduced glutathione. The data suggest similar changes occur in a neuronal cell line.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant PPG AG14930.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Methods S1–S5.
- TCA
- tricarboxylic acid cycle
- AD
- Alzheimer Disease
- KGDHC
- α-ketoglutarate dehydrogenase complex
- NAC
- N-acetylcysteine
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- E1k
- α-ketoglutarate dehydrogenase
- E2k
- dihydrolipoyl succinyltransferase
- E3
- dihydrolipoyl dehydrogenase
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- PDHC
- pyruvate dehydrogenase complex
- PTM
- post-translational modification.
REFERENCES
- 1. Au H. C., Scheffler I. E. (1998) Eur. J. Biochem. 251, 164–174 [DOI] [PubMed] [Google Scholar]
- 2. Miranda S., Foncea R., Guerrero J., Leighton F. (1999) Biochem. Biophys. Res. Commun. 258, 44–49 [DOI] [PubMed] [Google Scholar]
- 3. Piantadosi C. A., Carraway M. S., Babiker A., Suliman H. B. (2008) Circ. Res. 103, 1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chinopoulos C., Tretter L., Adam-Vizi V. (1999) J. Neurochem. 73, 220–228 [DOI] [PubMed] [Google Scholar]
- 5. Gibson G. E. (2002) Free Radic. Biol. Med. 32, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 6. Gibson G. E., Huang H. M. (2004) J. Bioenerg. Biomembr. 36, 335–340 [DOI] [PubMed] [Google Scholar]
- 7. Jeitner T. M., Xu H., Gibson G. E. (2005) J. Neurochem. 92, 302–310 [DOI] [PubMed] [Google Scholar]
- 8. Nulton-Persson A. C., Szweda L. I. (2001) J. Biol. Chem. 276, 23357–23361 [DOI] [PubMed] [Google Scholar]
- 9. Pettit F. H., Hamilton L., Munk P., Namihira G., Eley M. H., Willms C. R., Reed L. J. (1973) J. Biol. Chem. 248, 5282–5290 [PubMed] [Google Scholar]
- 10. Butterworth R. F., Besnard A. M. (1990) Metab. Brain Dis. 5, 179–184 [DOI] [PubMed] [Google Scholar]
- 11. Gibson G. E., Sheu K. F., Blass J. P., Baker A., Carlson K. C., Harding B., Perrino P. (1988) Arch. Neurol. 45, 836–840 [DOI] [PubMed] [Google Scholar]
- 12. Gibson G. E., Kingsbury A. E., Xu H., Lindsay J. G., Daniel S., Foster O. J., Lees A. J., Blass J. P. (2003) Neurochem. Int. 43, 129–135 [DOI] [PubMed] [Google Scholar]
- 13. Albers D. S., Augood S. J., Park L. C., Browne S. E., Martin D. M., Adamson J., Hutton M., Standaert D. G., Vonsattel J. P., Gibson G. E., Beal M. F. (2000) J. Neurochem. 74, 878–881 [DOI] [PubMed] [Google Scholar]
- 14. Park L. C., Albers D. S., Xu H., Lindsay J. G., Beal M. F., Gibson G. E. (2001) J. Neurosci. Res. 66, 1028–1034 [DOI] [PubMed] [Google Scholar]
- 15. Benson D. F., Kuhl D. E., Hawkins R. A., Phelps M. E., Cummings J. L., Tsai S. Y. (1983) Arch. Neurol. 40, 711–714 [DOI] [PubMed] [Google Scholar]
- 16. Markesbery W. R., Carney J. M. (1999) Brain Pathol. 9, 133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rapoport S. I., Horwitz B., Grady C. L., Haxby J. V., DeCarli C., Schapiro M. B. (1991) Adv. Exp. Med. Biol. 291, 231–248 [DOI] [PubMed] [Google Scholar]
- 18. Smith G. S., de Leon M. J., George A. E., Kluger A., Volkow N. D., McRae T., Golomb J., Ferris S. H., Reisberg B., Ciaravino J., et al. (1992) Arch. Neurol. 49, 1142–1150 [DOI] [PubMed] [Google Scholar]
- 19. Smith M. A., Rottkamp C. A., Nunomura A., Raina A. K., Perry G. (2000) Biochim. Biophys. Acta. 1502, 139–144 [DOI] [PubMed] [Google Scholar]
- 20. Bubber P., Haroutunian V., Fisch G., Blass J. P., Gibson G. E. (2005) Ann. Neurol. 57, 695–703 [DOI] [PubMed] [Google Scholar]
- 21. Abello N., Kerstjens H. A., Postma D. S., Bischoff R. (2009) J. Proteome. Res. 8, 3222–3238 [DOI] [PubMed] [Google Scholar]
- 22. Nauser T., Merkofer M., Kissner R., Koppenol W. H. (2001) Chem. Res. Toxicol. 14, 348–350 [DOI] [PubMed] [Google Scholar]
- 23. Padmaja S., Huie R. E. (1993) Biochem. Biophys. Res. Commun. 195, 539–544 [DOI] [PubMed] [Google Scholar]
- 24. Park L. C., Zhang H., Sheu K. F., Calingasan N. Y., Kristal B. S., Lindsay J. G., Gibson G. E. (1999) J. Neurochem. 72, 1948–1958 [DOI] [PubMed] [Google Scholar]
- 25. Bian K., Murad F. (2001) Free Radic. Biol. Med. 31, 421–429 [DOI] [PubMed] [Google Scholar]
- 26. Gow A. J., Duran D., Malcolm S., Ischiropoulos H. (1996) FEBS Lett. 385, 63–66 [DOI] [PubMed] [Google Scholar]
- 27. Kamisaki Y., Wada K., Bian K., Balabanli B., Davis K., Martin E., Behbod F., Lee Y. C., Murad F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11584–11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuo W. N., Kanadia R. N., Shanbhag V. P. (1999) Biochem. Mol. Biol. Int. 47, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 29. Kuo W. N., Kanadia R. N., Shanbhag V. P., Toro R. (1999) Mol. Cell. Biochem. 201, 11–16 [DOI] [PubMed] [Google Scholar]
- 30. Smallwood H. S., Lourette N. M., Boschek C. B., Bigelow D. J., Smith R. D., Pasa-Tolić L., Squier T. C. (2007) Biochemistry 46, 10498–10505 [DOI] [PubMed] [Google Scholar]
- 31. Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 32. Ye Y., Quijano C., Robinson K. M., Ricart K. C., Strayer A. L., Sahawneh M. A., Shacka J. J., Kirk M., Barnes S., Accavitti-Loper M. A., Radi R., Beckman J. S., Estévez A. G. (2007) J. Biol. Chem. 282, 6324–6337 [DOI] [PubMed] [Google Scholar]
- 33. Park L. C., Calingasan N. Y., Sheu K. F., Gibson G. E. (2000) Anal. Biochem. 277, 86–93 [DOI] [PubMed] [Google Scholar]
- 34. Starkov A. A., Fiskum G., Chinopoulos C., Lorenzo B. J., Browne S. E., Patel M. S., Beal M. F. (2004) J. Neurosci. 24, 7779–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansson Petersen C. A., Alikhani N., Behbahani H., Wiehager B., Pavlov P. F., Alafuzoff I., Leinonen V., Ito A., Winblad B., Glaser E., Ankarcrona M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13145–13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Q., Karuppagounder S. S., Xu H., Pechman D., Chen H., Gibson G. E. (2007) Neurochem. Int. 50, 921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Golladay S. A., Park S. H., Aust A. E. (1997) Environ. Health. Perspect. 105, Suppl. 5, 1273–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chouchani E. T., Hurd T. R., Nadtochiy S. M., Brookes P. S., Fearnley I. M., Lilley K. S., Smith R. A., Murphy M. P. (2010) Biochem. J. 430, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Applegate M. A., Humphries K. M., Szweda L. I. (2008) Biochemistry 47, 473–478 [DOI] [PubMed] [Google Scholar]
- 40. Butterworth R. F., Kril J. J., Harper C. G. (1993) Alcoholism, Clinical Exp. Res. 17, 1084–1088 [DOI] [PubMed] [Google Scholar]
- 41. Mastrogiacomo F., Bergeron C., Kish S. J. (1993) J. Neurochem. 61, 2007–2014 [DOI] [PubMed] [Google Scholar]
- 42. Mizuno Y., Matuda S., Yoshino H., Mori H., Hattori N., Ikebe S. (1994) Ann. Neurol. 35, 204–210 [DOI] [PubMed] [Google Scholar]
- 43. Berg A., de Kok A. (1997) Biol. Chem. 378, 617–634 [PubMed] [Google Scholar]
- 44. Perham R. N. (1991) Biochemistry 30, 8501–8512 [DOI] [PubMed] [Google Scholar]
- 45. Perham R. N., Reche P. A. (1998) Biochem. Soc. Trans. 26, 299–303 [DOI] [PubMed] [Google Scholar]
- 46. Torreilles F., Salman-Tabcheh S., Guérin M., Torreilles J. (1999) Brain Res. Brain Res. Rev. 30, 153–163 [DOI] [PubMed] [Google Scholar]
- 47. Ischiropoulos H. (1998) Arch. Biochem. Biophys. 356, 1–11 [DOI] [PubMed] [Google Scholar]
- 48. Ischiropoulos H. (2003) Biochem. Biophys. Res. Commun. 305, 776–783 [DOI] [PubMed] [Google Scholar]
- 49. Souza J. M., Daikhin E., Yudkoff M., Raman C. S., Ischiropoulos H. (1999) Arch. Biochem. Biophys. 371, 169–178 [DOI] [PubMed] [Google Scholar]
- 50. Jiao K., Mandapati S., Skipper P. L., Tannenbaum S. R., Wishnok J. S. (2001) Anal. Biochem. 293, 43–52 [DOI] [PubMed] [Google Scholar]
- 51. Frank R. A., Price A. J., Northrop F. D., Perham R. N., Luisi B. F. (2007) J. Mol. Biol. 368, 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Knapp J. E., Carroll D., Lawson J. E., Ernst S. R., Reed L. J., Hackert M. L. (2000) Protein Sci. 9, 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen H. J., Chen Y. M., Chang C. M. (2002) Chem. Biol. Interact. 140, 199–213 [DOI] [PubMed] [Google Scholar]
- 54. Dringen R. (2000) Eur. J. Biochem. 267, 4903. [DOI] [PubMed] [Google Scholar]
- 55. Meister A. (1988) J. Biol. Chem. 263, 17205–17208 [PubMed] [Google Scholar]
- 56. Bajt M. L., Knight T. R., Lemasters J. J., Jaeschke H. (2004) Toxicol. Sci. 80, 343–349 [DOI] [PubMed] [Google Scholar]
- 57. Yeh S. T., Guo H. R., Su Y. S., Lin H. J., Hou C. C., Chen H. M., Chang M. C., Wang Y. J. (2006) Toxicology 223, 181–190 [DOI] [PubMed] [Google Scholar]
- 58. Korotchkina L. G., Yang H., Tirosh O., Packer L., Patel M. S. (2001) Free Radic. Biol. Med. 30, 992–999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.