Abstract
Protein phosphatase 2A (PP2A) is regulated through a variety of mechanisms, including post-translational modifications and association with regulatory proteins. Alpha4 is one such regulatory protein that binds the PP2A catalytic subunit (PP2Ac) and protects it from polyubiquitination and degradation. Alpha4 is a multidomain protein with a C-terminal domain that binds Mid1, a putative E3 ubiquitin ligase, and an N-terminal domain containing the PP2Ac-binding site. In this work, we present the structure of the N-terminal domain of mammalian Alpha4 determined by x-ray crystallography and use double electron-electron resonance spectroscopy to show that it is a flexible tetratricopeptide repeat-like protein. Structurally, Alpha4 differs from its yeast homolog, Tap42, in two important ways: 1) the position of the helix containing the PP2Ac-binding residues is in a more open conformation, showing flexibility in this region; and 2) Alpha4 contains a ubiquitin-interacting motif. The effects of wild-type and mutant Alpha4 on PP2Ac ubiquitination and stability were examined in mammalian cells by performing tandem ubiquitin-binding entity precipitations and cycloheximide chase experiments. Our results reveal that both the C-terminal Mid1-binding domain and the PP2Ac-binding determinants are required for Alpha4-mediated protection of PP2Ac from polyubiquitination and degradation.
Keywords: Adaptor Proteins, Phosphatase, Protein Degradation, Protein Phosphatase, Protein Structure, Protein Turnover, Ubiquitin, Ubiquitin Ligase
Introduction
PP2A3 is a ubiquitous serine/threonine phosphatase involved in the regulation of numerous cell signaling pathways and cellular functions, including proliferation, cytoskeletal rearrangement, apoptosis, and cell migration (1–3). Several pathologies have been linked to dysregulation of PP2Ac, including Alzheimer disease, cancer, and diabetes (4–8). The activity of PP2Ac is tightly controlled in vivo via association with regulatory subunits, interactions with other cellular proteins, and various post-translational modifications (9–12). PP2A regulatory subunits play a critical role in determining phosphatase activity and substrate selectivity, as well as directing the subcellular localization of the PP2A holoenzyme (3). PP2A exists primarily as a heterotrimeric holoenzyme consisting of a structural A-subunit, a variable regulatory B-subunit, and PP2Ac. However, an atypical pool of PP2Ac exists in complex with the regulatory subunit Alpha4 that binds directly to PP2Ac in the absence of the A- and B-subunits (13–16). Recent studies have shown that Alpha4 plays a crucial role in the control of PP2A ubiquitination and stability (12, 17, 18).
Alpha4, a multidomain protein with similarity to Tap42 from yeast, was initially discovered as a 52-kDa phosphoprotein in B-cell receptor complexes (16, 19). Both Alpha4 and Tap42 consist of an N-terminal domain that contains the residues important for PP2Ac binding (20) and a C-terminal domain that is protease-sensitive and intrinsically disordered (21). The C-terminal domain of Alpha4 binds Mid1, a putative E3 ligase (12, 22). Alpha4 regulates all three type 2A protein phosphatases (PP2Ac, PP4, and PP6), modulating both catalytic activity and expression levels (13, 14, 17, 23). In addition to its association with PP2A family members, Alpha4 associates and co-localizes with Mid1, a putative E3 ubiquitin ligase thought to facilitate PP2Ac polyubiquitination (12, 22). The C terminus of Alpha4 and the B-box1 domain of the Mid1 protein mediate the association between Mid1 and Alpha4 (12, 22). Mutations in Mid1 have been linked to Opitz syndrome, a developmental disorder (24, 25). At the cellular level, mutations in Mid1 lead to decreases in ubiquitination and degradation of PP2Ac, especially microtubule-associated PP2Ac (12, 26).
Alpha4 serves as a scaffold for PP2Ac and Mid1 and protects PP2Ac from polyubiquitination (12, 18). The protective effect of Alpha4 is abolished in UIM-deficient mutants, leading to the hypothesis that the consensus UIM, located at residues 46–60, has a role in Alpha4-mediated regulation of PP2Ac ubiquitination (18). This led to a model in which Alpha4 protected PP2Ac from polyubiquitination via a capping mechanism in which the consensus UIM interacted with ubiquitin (18). Here, to gain insight into its possible mechanisms of action, we have determined the structure of the N-terminal portion of murine Alpha4 and evaluated the role of the Mid1- and PP2Ac-binding domains in the regulation of PP2Ac ubiquitination and degradation.
EXPERIMENTAL PROCEDURES
Plasmids
The HA-ubiquitin plasmid was a gift from H. Moses (Vanderbilt University), the Myc-Mid1/pCMV-tag3A construct was a gift from S. Schweiger (University of Dundee, Dundee, Scotland, United Kingdom), and the HA3-PP2Ac construct was a gift from D. Brautigan (University of Virginia, Charlottesville, VA). Construction of the FLAG-Alpha4/pcDNA5TO, FLAG-Alpha4ΔC/pcDNA5TO, and FLAG-Alpha4_ED/pcDNA5TO constructs was described previously (18, 27). Murine Alpha4ΔC was amplified by PCR from the Alpha4/pGEX4T2 vector and then inserted into the pET28a vector using the BamHI and NdeI restriction sites to create a N-terminal His6-Alpha4ΔC construct. Murine Alpha4ΔC mutants were created using the QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: Alpha4ΔC_AA (R156A/K159A), 5′-GCTATGGCATCTCAAGCACAGGCTGCAATGAGAGATACAAGC (forward) and 5′-GCTTGTATCTCTCTATTGCAGCCTGTGCTTGAGATGCCATAGC (reverse); Alpha4ΔC_CF (C117S/C119S), 5′-CGTACATTTCTTAACTCAGAGTCATAGCTATCATGTGGCAGAG (forward) and 5′-CTCTGCCACATGATGCTATGACTCTGAGTTAAGAAATGTACG (reverse); K98C, 5′-CAAGTCAACCCCAGCTGTCGTCTAGATCATTTGC (forward) and 5′-GCAAATGATCTAGACGACAGCTGGGGTTGACTTG (reverse); and Y146C, 5′-GCTCCTCCATGGCCTGTCCAAATCTCGTTGC (forward) and 5′-GCAACGAGATTTGACAGGCCATGGAGGAGC (reverse). All constructs were verified by DNA sequencing analysis.
Antibodies
The mouse anti-PP2Ac monoclonal antibody was from BD Transduction Laboratories. The rabbit anti-Myc tag monoclonal antibody was from Cell Signaling Technology, Inc. (Danvers, MA). The rabbit anti-FLAG polyclonal antibody was from Sigma. The rabbit anti-Alpha4 polyclonal antibody was from Bethyl Laboratories (Montgomery, TX). The rabbit anti-ubiquitin polyclonal antibody was from Dharmacon (Lafayette, CO). The rabbit anti-His6 polyclonal and mouse anti-HSP90 monoclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
Expression and Purification of Alpha4ΔC
Protein expression was performed at 20 °C overnight in BL21(DE3) cells. Alpha4ΔC was purified using metal affinity chromatography, cleaved overnight with thrombin to remove the N-terminal hexahistidine tag, and dialyzed into gel filtration buffer (10 mm Tris-HCl, 150 mm NaCl, and 1 mm sodium azide, pH 7.5). The protein was further purified by size exclusion gel filtration on an S200 column (Amersham Biosciences) in gel filtration buffer. Selenomethionine-labeled protein was grown in minimal medium (28) in BL834(DE3) auxotrophic cells.
Crystallization and Structure Determination of Alpha4ΔC
Purified Alpha4ΔC was concentrated to ∼17 mg/ml in gel filtration buffer, and crystallization trials were conducted by hanging drop vapor diffusion, performed by mixing 2 μl of protein and 2 μl of mother liquor. Crystals were obtained in two conditions, both at 18 °C: in 1.6 m ammonium sulfate, 2% PEG 400, and 0.1 m BisTris, pH 6.0, and in 25% PEG 1500. Crystals were cooled in liquid nitrogen, and diffraction data were collected to 2.35 Å at the Northeastern Collaborative Access Team ID-C beamline and the Southeastern Collaborative Access Team BM-22 beamline at the Argonne National Laboratory. Selenomethionine-labeled crystals were produced in the ammonium sulfate conditions, and an initial model was produced with data from these crystals. The data from these crystals were severely anisotropic (with average diffraction intensities three times greater in one dimension than the other two), but selenium positions were found using SHELX (29) and refined using Sharp (30), and density modification was done using Solomon (31) as implemented in autoSHARP (30). The structure was built using COOT (32) and refined using PHENIX (32) and CNS (33). Data from crystals grown in 25% PEG 1500 were used in the final structure refinement. Phasing and refinement statistics are given in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
Values in parentheses are for the highest resolution bin. SeMet, selenomethionine; APS, Advanced Photon Source; FOM, figure of merit; r.m.s.d., root mean square deviation.
| Parameter | Native crystal | SeMet |
|---|---|---|
| Space group | P3221 | P3221 |
| a = b (Å) | 76.6 | 80.6 |
| c (Å) | 72.7 | 73.4 |
| X-ray source | APS 24 ID-C | APS 22-BM |
| Wavelength (Å) | 0.97949 | λ1, 0.97625; λ2, 0.97949; λ3, 0.9826 |
| Resolution range (Å) | 50–2.35 | 50–2.5 |
| No. observed reflections | 258,546 | λ1, 96,531; λ2, 108,359; λ3, 95,973 |
| No. unique reflections | 12,891 | λ1, 9876; λ2, 9843; λ3, 9972 |
| Completeness (%) | 95.61 (91) | λ1, 99.9 (100); λ2, 99.9 (100); λ3, 99.9 (100) |
| Redundancy | 6.9 (7.3) | λ1, 5.2 (3.9); λ2, 5.9 (5.9); λ3, 5.9 (3.8) |
| Rmerge | 5.6 (28.0) | λ1, 5.5 (38.9); λ2, 5.3 (77.6); λ3, 4.3 (51.9) |
| FOM (50-2.8 Å) | 0.44 | |
| FOM after density modification (50-2.5 Å)a | 0.86 | |
| I/σ | 22 (3.8) | λ1, 23.5 (1.8); λ2, 23.6 (1.9); λ3, 28.9 (1.4) |
| No. reflections used in refinement (N) | 10,142 | |
| No. reflections used in Rfree | 1011 | |
| No. water molecules | 38 | |
| Protein atoms | 1537 | |
| Rcrystal (%) | 20.7 (29.6) | |
| Rfree (%) | 26.5 (36.9) | |
| Wilson B-factor | 54 | |
| Average B-factor | 67 | |
| r.m.s.d. bond lengths (Å) | 0.008 | |
| r.m.s.d. bond angles | 1.044° | |
| Ramachandran (%) | ||
| Favored | 92.0 | |
| Allowed | 6.8 | |
| Outliers | 1.1 |
a Density modification was performed using Solomon as described under “Experimental Procedures.”
EPR Spectroscopy
The double-cysteine point mutant K98C/Y146C was created in the cysteine-free Alpha4ΔC_CF background and then expressed and purified as described above. The protein concentration after elution was measured by absorbance at 280 nm, and a 10-fold excess of (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate (Toronto Research Chemicals) was added to ∼10 mg of the protein. The protein was incubated in the dark at room temperature for 2 h before being placed overnight in the dark at 4 °C. After overnight incubation, the protein was further purified by size exclusion gel filtration on an S200 column in gel filtration buffer. This purified protein was concentrated to 200 μm with 30% (w/v) glycerol and frozen at −80 °C. Samples were analyzed using a previously described DEER protocol (34, 35).
Cell Culture and Transfection
HEK293FT cells were grown at 37 °C in a humidified atmosphere with 5% CO2 in DMEM supplemented with 10% fetal bovine serum and 2 mm l-glutamine. Cells were transfected using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's directions.
Binding Assays
Binding assays were conducted using 40 μg of either purified recombinant His6-Alpha4ΔC or His6-Alpha4ΔC_RK_AA protein and 100 μl of whole cell lysate from HEK293FT cells lysed with 500 μl of radioimmune precipitation assay buffer (20 mm sodium phosphate, pH 8.0, 150 mm NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, and 0.1% SDS) per 10-cm plate. The lysate was incubated with the purified recombinant protein for 30 min at 4 °C before adding 40 μl of a 50% slurry of cobalt-nitrilotriacetic acid resin (Talon) and incubating for an additional 30 min at 4 °C. The resin was washed three times with 1 ml of 20 mm sodium phosphate, pH 8.0, 150 mm NaCl, and 20 mm imidazole. Bound proteins were eluted with 200 mm imidazole and analyzed by SDS-PAGE and immunoblotting for PP2Ac and the hexahistidine tag.
Immunoprecipitations
Cells were lysed in immunoprecipitation buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% IGEPAL, 5 μg/ml aprotinin, 1 μg/ml pepstatin, 1 mm PMSF, and 1 μg/ml leupeptin) and centrifuged at 12,000 × g for 10 min. Clarified lysates were incubated overnight with 20 μl of a 50% slurry of anti-FLAG M2-agarose (Sigma) or 20 μl of a 50% slurry of anti-HA-agarose (Roche Applied Science). Immunoprecipitates were washed three times with 1 ml of immunoprecipitation buffer, and bound proteins were eluted in SDS sample buffer and subjected to Western analysis.
Cycloheximide Chase Experiments
HEK293FT cells (seeded in 6-well tissue culture plates at 300,000 cells/well) were transfected either with HA3-PP2Ac alone or with HA3-PP2Ac and FLAG-Alpha4, FLAG-Alpha4ΔC, or FLAG-Alpha4_ED. At 48 h post-transfection, cells were treated with 100 μg/ml cycloheximide (Sigma) for the indicated times. Cell lysates were prepared and subjected to Western analysis using antibodies recognizing PP2Ac, Alpha4, and HSP90 (used as a loading control).
TUBE Isolations
HEK293FT cells were lysed in immunoprecipitation buffer and centrifuged at 12,000 × g for 10 min. Clarified lysates were incubated with 20 μl of a 50% slurry of TUBE2-agarose (LifeSensors) overnight at 4 °C. TUBE2 complexes were washed three times with 1 ml of immunoprecipitation buffer, and bound proteins were eluted in SDS sample buffer and subjected to Western analysis.
Western Analysis
SDS-solubilized protein samples were separated by SDS-PAGE (4–12% BisTris gradient acrylamide gels or 10% Tris/glycine acrylamide gels) and transferred to 0.45-μm nylon-supported nitrocellulose membranes. Membranes were blocked in Odyssey buffer (LI-COR, Lincoln, NE). All primary antibodies were used at 1:1000 dilution in a 1:1 mixture of Odyssey buffer and Tween/Tris-buffered saline. For detection with the Odyssey infrared imaging system, appropriate secondary fluorophore-conjugated antibodies were used at 1:20,000 dilution in a 1:1 mixture of Odyssey blocking buffer and Tween/Tris-buffered saline. Bound antibodies were visualized using the Odyssey infrared imaging system and Odyssey software (LI-COR).
RESULTS
Structural Analysis
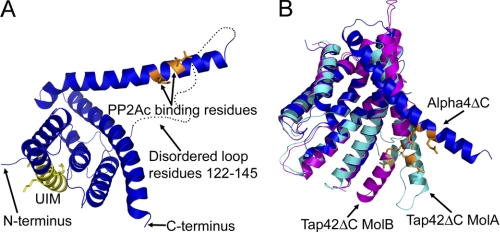
To gain insight into the molecular mechanism of Alpha4-mediated inhibition of PP2Ac polyubiquitination (18), we determined the structure of a mammalian version of Alpha4 that contains the UIM consensus sequence. Given the intrinsically disordered and proteolytically sensitive nature of the C-terminal 120 residues of Alpha4, also characterized as the Mid1-binding domain, we created a hexahistidine-tagged construct of N-terminal amino acids 1–223 of murine Alpha4 for crystallization purposes (hereafter referred to as Alpha4ΔC). We crystallized Alpha4ΔC and determined the structure to a resolution of 2.35 Å (Fig. 1A). Statistics for data collection and structure refinement are provided in Table 1. Alpha4ΔC is an all α-helical protein with dimensions of 71 × 42 × 29 Å, similar to the dimensions found for Tap42ΔC of 65 × 35 × 25 Å (20) and the 72 Å measured by scattering studies for the largest dimension (21). A large flexible loop composed of residues 122–144 joins helices 4 and 5 and it is not observed in the crystal structure of Alpha4ΔC. A search in DALI (36) for structures similar to Alpha4ΔC revealed both TPR and 14-3-3 proteins, with the closest match being the yeast homolog of Alpha4, Tap42, and the next closest proteins being the TPR domain of prolyl 4-hydroxylase and 14-3-3 protein (Table 2).
FIGURE 1.
Structure of Alpha4ΔC. A, ribbon diagram of Alpha4ΔC, with residues important for PP2Ac binding shown in orange and the consensus UIM shown in yellow. B, comparison of the Alpha4ΔC structure (blue) with the Tap42ΔC structures (cyan and magenta) showing the variable positions of the extended helix (residues 147–182). PyMOL was used to depict all molecular structures (48). MolA and MolB, molecules A and B, respectively.
TABLE 2.
Highest structural similarity matches to Alpha4ΔC defined by DALI
PDB, Protein Data Bank.
| Protein | PDB code | Z-score | r.m.s.d.a | No. aligned and matched residues | Sequence ID |
|---|---|---|---|---|---|
| Å | % | ||||
| Tap42 | 2V0P | 17.8 | 2.6 | 164 | 23 |
| P4HA1 | 2V5F | 10.9 | 2.0 | 93 | 14 |
| 14-3-3 | 3EFZ | 10.1 | 4.1 | 109 | 10 |
| APC7 | 3FF1 | 9.5 | 2.2 | 93 | 8 |
| SycD | 2VGY | 9.1 | 2.3 | 90 | 10 |
| PP5 | 1WAO | 9.0 | 3.1 | 88 | 6 |
| TOM20-3 | 1ZU2 | 9.0 | 4.3 | 109 | 14 |
a Root mean square deviation (r.m.s.d.) was calculated using the matched residue's C-α atoms.
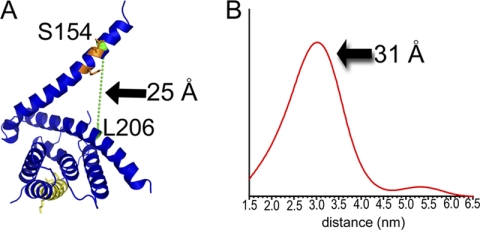
Comparisons between Alpha4ΔC and Tap42ΔC indicate that helix 5 (residues 145–182) of Alpha4ΔC adopts multiple conformations (Fig. 1B). In the structure presented here, helix 5 protrudes away from the rest of the molecule. However, in the crystal lattice, helix 5 interacts with helix 2 of a neighboring molecule, indicating that crystal lattice contacts might alter the position of helix 5. To determine the relative orientation of helix 5 in the absence of the crystal lattice, DEER spectroscopy was performed on Alpha4ΔC labeled with (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate at residues 98 and 146, 206 and 154, and 98 and 154. All three distance measurements support an open conformation (Fig. 2 and supplemental Fig. 1), similar to that seen in the Alpha4 crystal structure, as being the predominant conformation seen in solution, although the specific distance distribution is likely influenced by PP2Ac. The DEER data also support the idea that the protein exists in multiple conformational states, as the multiple peaks present within the distance distribution are indicative of flexibility of Alpha4 either in the position of the helix or in the conformation of the backbone (supplemental Fig. 1). Helix 5 contains residues shown to be important for binding PP2Ac: Arg-156 and Lys-159 (20). These residues face toward the main body of Alpha4 and are in an open conformation, allowing a high degree of accessibility to this interface for the globular PP2Ac subunit (supplemental Fig. 2).
FIGURE 2.
Distances between the spin label pair L206C/S154C computed via DEER/pulsed EPR studies. A, ribbon diagram showing the locations of spin labels L206C and S154C (green) and the distance between β-carbons. The UIM is shown in yellow, and PP2Ac-binding residues are shown in orange. B, distance distribution profiles corresponding to the best fit (shown in supplemental Fig. 1, A and B), showing a major distance distribution of ∼31 Å compared with ∼25 Å in the crystal structure.
A key difference between mammalian Alpha4 and non-mammalian homologs, such as Tap42, is the presence of an identifiable UIM consensus sequence, composed of residues 46–60, which has been shown to be functionally important in Alpha4 (supplemental Figs. 3, 4, and 6) (18). It remains to be seen if such a sequence is important in non-mammalian homologs. The UIM is within helix 2 of the structure and on the opposite face of Alpha4, relative to the PP2Ac-binding site (Fig. 1A). Overlaying the UIM consensus sequence with a known UIM-ubiquitin structure (Protein Data Bank code 2D3G) revealed that the UIM-containing helix within Alpha4 must rotate if it is to bind ubiquitin (supplemental Fig. 4) and that this rotation would likely perturb the structure in this region of Alpha4. The multiple conformations observed for helix 5 (and expected for helix 2) indicate that Alpha4 is a flexible molecule and that this flexibility may be functionally important.
Alpha4ΔC is a TPR-like protein with similarities to both TPR-containing and 14-3-3 proteins, but important topological differences create a possible binding site for PP2Ac. Both TPR and 14-3-3 proteins are scaffolding proteins, which mediate protein-protein interactions (37–39). TPR proteins are highly flexible molecules, with many TPR domains partially unstructured when not bound to their cognate ligands (40). Structural analysis using PISA (protein interactions, surfaces, and assemblies) (41) indicated that Alpha4ΔC has a high percentage of hydrophilic intramolecular interactions relative to α-helical proteins in general but similar to those found in TPR motifs from other proteins (supplemental Fig. 5), consistent with our findings that Alpha4 is a conformationally flexible molecule.
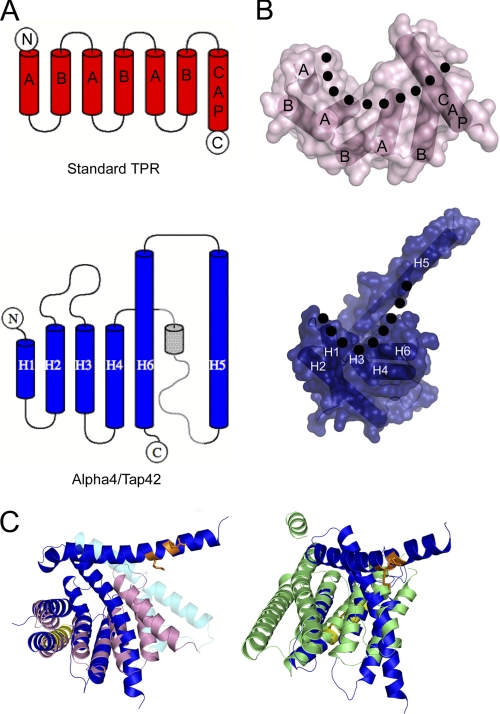
Although Alpha4ΔC adopts a TPR-like structure, it does not contain the TPR consensus residues (42), and the helices are longer and more irregular than a canonical TPR (Fig. 3). In addition, the overall topology of Alpha4ΔC differs from a canonical three-repeat TPR in the arrangement of the final three helices, and these differences allow greater flexibility (Fig. 3A). Both TPR and 14-3-3 proteins are composed of pairs of antiparallel helices stacked in parallel, with a twist, to create concave and convex faces (Fig. 3B). In TPR proteins, these pairs of helices are labeled A and B, with the concave face of the motif lined by the A helices (Fig. 3B). Many TPR proteins also contain a final capping helix that acts to extend the concave surface of the molecule (42). In Alpha4ΔC, the first four helices are arranged as pairs of antiparallel helices joined by a loop with the pairs stacking in parallel, as in a typical TPR or 14-3-3 protein (Fig. 3, A and B). Helix 6 occupies the position of the A-helix of a normal TPR motif, and helix 5 extends away from the body of the protein, analogous to the capping helix found in many TPR-containing proteins, but in an opposite orientation (Fig. 3, A–C). This extended helix (helix 5) extends away from the rest of the protein, unlike analogous capping helices, which pack against the concave surface. The distal portion of helix 5 is positioned above the concave binding surface such that known binding residues (Arg-156 and Lys-159) point toward the concave surface. This extension of helix 5 and its positioning disrupt the typical TPR fold and create a more closed concave face on the protein compared with standard TPR or 14-3-3 proteins (Fig. 3C). In the crystal structure of Alpha4ΔC, the residues connecting helices 4 and 5 are not observed, and the third TPR-like motif lacks a B-helix. In the crystal structure of Tap42ΔC, the loop between helices 4 and 5 is observed and includes a small helix in a similar position to the B-helix of a third TPR motif (Fig. 3C) (20). Alpha4ΔC differs from a canonical TPR repeat in the topology of the final TPR motif and the capping helix with the inclusion of a large loop and an inversion in orientation. This altered topology in Alpha4ΔC allows for opening and closing of the helix containing the PP2Ac-binding residues, creating a PP2Ac-binding site (supplemental Fig. 2).
FIGURE 3.
Comparison of Alpha4ΔC and TPR proteins. A, topology diagrams of TPR (upper) and Alpha4ΔC (lower) showing the altered topology of the final helices. The part represented in gray is based on the crystal structure of Tap42ΔC, as these residues are not observed in the crystal structure of Alpha4ΔC. The diagrams were created in TOPDRAW (49). B, structures and surface representations of TPR (upper) and Alpha4ΔC (lower) showing the configuration of helices and the formation of the concave and convex surfaces (with the outline of concavity denoted by the dotted line). C, superposition of Alpha4ΔC (colored blue, yellow, and orange as in Fig. 1) with the SycD TPR domain (pink; Protein Data Bank code 2VGY) and 14-3-3 (green; code 3EFZ) reveals similar tertiary structures but indicates that the concave face of Alpha4 is more closed than the canonical TPR and 14-3-3 proteins. The helices shown in cyan represent helices from Tap42ΔC that differ significantly in position from those in Alpha4ΔC.
Mutant Alpha4ΔC Is Capable of Binding to PP2Ac but Not to Mid1
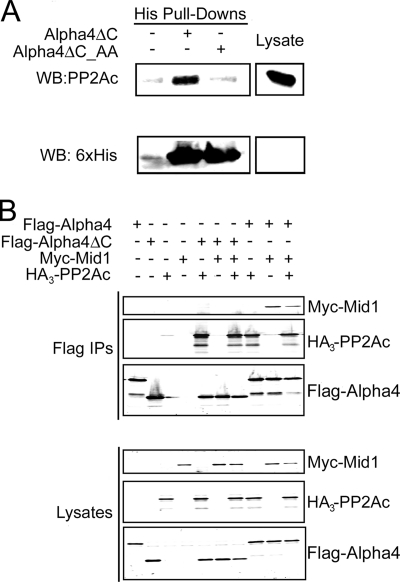
To determine whether the recombinant murine Alpha4ΔC used in our structural studies is capable of binding to PP2Ac and whether this interaction can be disrupted by mutations R156A and K159A (Alpha4ΔC_AA), we conducted in vitro binding assays using purified His6-Alpha4ΔC, His6-Alpha4ΔC_AA, and whole cell HEK293FT cell lysate. Charge reversal mutations of Arg-156 and Lys-159 were shown to abolish binding to PP2Ac in studies with full-length Alpha4 (20). Alpha4ΔC, but not Alpha4ΔC_AA, bound to endogenous PP2Ac (Fig. 4A), indicating that murine Alpha4ΔC is capable of binding to human PP2Ac and that Arg-156 and Lys-159 of Alpha4 mediate PP2Ac binding. Because Alpha4ΔC lacks the reported Mid1-binding region (12, 22) but retains the PP2Ac-binding determinants, we tested the ability of Alpha4ΔC to bind to both PP2Ac and Mid1. HEK293FT cells were transfected with full-length FLAG-Alpha4 or FLAG-Alpha4ΔC and HA3-PP2Ac, Myc-Mid1, or HA3-PP2Ac and Myc-Mid1. Western analysis of FLAG immunoprecipitations revealed that whereas full-length Alpha4 bound both PP2Ac and Mid1, Alpha4ΔC bound only HA3-PP2Ac (Fig. 4B).
FIGURE 4.
Alpha4ΔC binds PP2A but fails to bind Mid1. A, HEK293FT whole cell lysate was incubated with cobalt-nitrilotriacetic acid resin in the absence (−) or presence (+) of either Alpha4ΔC or Alpha4ΔC_AA (mutation of the PP2Ac-binding residues). Bound proteins were eluted with 200 mm imidazole, and the eluate was analyzed by SDS-PAGE and immunoblotting with antibodies recognizing PP2Ac (upper panel) and the His6 tag (lower panel). Data are representative of three independent experiments. WB, Western blot. B, HEK293FT cells were transfected with HA3-PP2Ac, FLAG-Alpha4, FLAG-Alpha4ΔC, Myc-Mid1, or a combination of the constructs. FLAG immune complexes (Flag IPs) were isolated from the cells and subjected to SDS-PAGE and Western analysis using anti-Myc, anti-PP2Ac, and anti-Alpha4 antibodies.
Both the Mid1-binding Domain and the PP2Ac-binding Residues of Alpha4 Are Essential for Regulation of PP2Ac Polyubiquitination
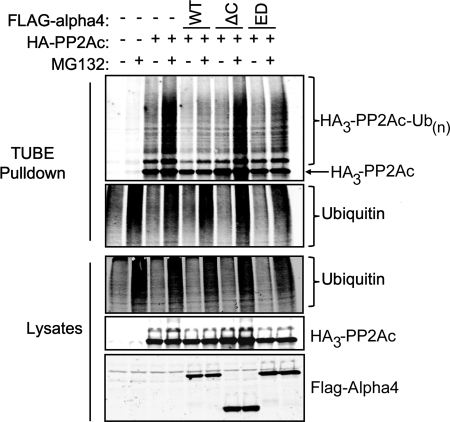
To investigate the role of the Mid1-binding domain of Alpha4 and its PP2Ac-binding residues in the control of PP2Ac polyubiquitination, we performed TUBE2 pulldown assays with lysates of cells transfected with HA3-PP2Ac and empty vector, full-length FLAG-Alpha4, FLAG-Alpha4ΔC, or FLAG-Alpha4_ED. These experiments involve using TUBEs linked to beads as a matrix to isolate ubiquitinated proteins from cells. In our experience, these matrices are superior to immunopurifications. As shown in Fig. 4, full-length FLAG-Alpha4, but neither FLAG-Alpha4ΔC nor FLAG-Alpha4_ED, prevented PP2Ac polyubiquitination. All cells that had been pretreated with a proteasome inhibitor (MG132) showed increased levels of polyubiquitinated proteins, and all showed similar levels of total polyubiquitinated proteins. These findings demonstrate that both the Mid1-binding domain and the PP2Ac-binding residues of Alpha4 are essential for the Alpha4-mediated protection of PP2Ac from polyubiquitination (Fig. 5).
FIGURE 5.
Both the Mid1-binding domain and PP2Ac binding are essential for Alpha4 inhibition of PP2Ac polyubiquitination. HEK293FT cells were transfected with HA3-PP2Ac and empty vector, full-length WT FLAG-Alpha4, FLAG-Alpha4ΔC, or FLAG-Alpha4_ED. At 48 h post-transfection, cells were treated with 25 μm proteasome inhibitor (MG132) for 4 h at 37 °C prior to lysis. Total polyubiquitinated proteins were isolated from the cell lysates using TUBE2-agarose beads. Protein expression and the polyubiquitination state of ectopic PP2Ac were analyzed via immunoblotting using anti-PP2Ac, anti-Alpha4, and anti-ubiquitin antibodies.
Both the Mid1-binding Domain and the PP2Ac-binding Residues of Alpha4 Are Required for Alpha4 to Protect PP2Ac from Degradation
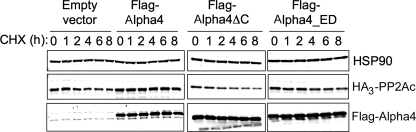
The Mid1-binding domain plays an essential role in protecting PP2Ac from polyubiquitination (Fig. 5). To examine the role of Mid1 and PP2Ac binding in the ability of Alpha4 to protect PP2Ac from degradation, we performed cycloheximide chase experiments to evaluate the half-life of HA3-PP2Ac when coexpressed with or without various Alpha4 constructs. Cells were treated with cycloheximide to inhibit protein production, and the levels of HA3-PP2Ac were monitored at various time points after cycloheximide treatment. As shown in Fig. 5, the cells that expressed HA3-PP2Ac alone showed a progressive decline in the level of ectopic PP2Ac over the 8-h time course, whereas the samples coexpressing wild-type FLAG-Alpha4 stabilized PP2Ac levels over this period. Cells coexpressing FLAG-Alpha4ΔC or FLAG-Alpha4_RK_ED (Fig. 6) did not promote this stabilization but rather showed a progressive decline in HA3-PP2Ac, similar to cells expressing HA3-PP2Ac alone. These results indicate that both the Mid1-binding domain and the PP2Ac-binding residues are essential for the PP2Ac-stabilizing effect of Alpha4. Our data (Figs. 5 and 6) indicate that the C-terminal domain has a more pronounced stabilizing effect, perhaps caused by incomplete disruption of PP2Ac binding by the Alpha4_RK_ED mutant.
FIGURE 6.
The Mid1-binding domain of Alpha4 is essential to protect PP2Ac from degradation. HEK293FT cells were transfected with HA3-PP2Ac alone or with HA3-PP2Ac and FLAG-Alpha4, FLAG-Alpha4ΔC, or FLAG-Alpha4_ED. Cells were treated with 50 μm cycloheximide (CHX) 48 h post-transfection and then lysed at the indicated time points post-treatment. The lysates were subjected to Western analysis using antibodies recognizing Alpha4, PP2Ac, and HSP90.
DISCUSSION
Alpha4ΔC adopts an α-helical structure that differs from canonical TPR proteins in length, irregularity, and topology of their helices. Comparison of the structure of Alpha4ΔC with those of its yeast homolog, Tap42, shows the extended helix containing the PP2Ac-binding determinants existing in multiple conformations, indicating that the PP2Ac-binding region is flexible. The structure of Tap42 was determined in two conformations, and Alpha4 adopts yet a third conformation, with most of the variation between the three structures occurring in the relative position of the extended helix containing the PP2Ac-binding residues (Fig. 1B). The Alpha4 structure contains the most open conformation of this helix, with the residues important for binding to PP2Ac in an open and accessible conformation. In the two structures of Tap42, these resides are less accessible, indicating that binding of a globular protein, such as PP2Ac, would require an opening of this helix in Tap42 to allow binding.
DEER studies interrogating the extended helix of Alpha4 indicate that an open conformation is the predominant conformation found in solution and that multiple conformations of this helix may exist based on the presence of multiple peaks within the DEER distance measurements (Fig. 2B and supplemental Fig. 1). The Alpha4 structure also has the critical PP2Ac-binding residues oriented such that they point toward the concave face of the molecule (supplemental Fig. 2). This is of note because structures of other TPR and 14-3-3 proteins with their cognate interacting proteins show that the interactions are mediated either by the concave face or via the interhelix loops (37, 43–47). Thus, it is likely that Alpha4 interacts with PP2Ac in a similar fashion.
Previous studies have revealed that Alpha4 acts to both inhibit and promote PP2Ac degradation (12, 17, 18). The initial model posited that Alpha4 was a scaffolding molecule that promoted polyubiquitination of PP2Ac by scaffolding PP2Ac to Mid1 (12). Subsequent studies showed that Alpha4 has a protective effect on PP2Ac degradation and polyubiquitination (17) and that Alpha4 contains a UIM, which plays a crucial role in protection of PP2Ac from polyubiquitination (18). In this study, we investigated the role of both the PP2Ac- and Mid1-binding domains in Alpha4 regulation of PP2Ac ubiquitination and degradation. Our data demonstrate that both of these domains are required for Alpha4 to protect PP2Ac from degradation. These findings indicate that the protective effects of Alpha4 cannot be entirely accounted for by a hypothesis that Alpha4 inhibits Mid1 function or sequesters Mid1 from PP2Ac and also raise questions about the role of Mid1 in PP2Ac polyubiquitination and degradation. The findings that both the PP2Ac- and Mid1-binding domains are required for Alpha4 to exert its protective effects on PP2Ac, along with the previous results indicating the importance of the consensus UIM, imply a more complex mechanism of Alpha4 inhibition of PP2Ac degradation that involves contributions from all of these domains.
Supplementary Material
Acknowledgments
We thank Hassane Mchaourab (Department of Molecular Physiology and Biophysics, Vanderbilt University School of Medicine) for access to EPR equipment and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM051366 and DK070787 (to B. E. W.) and T32 GM07628 (to M. L.-N. and G. R. W.). This work was also supported by American Cancer Society Grant IRG-58-009-49 (to B. W. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6 and additional references.
The atomic coordinates and structure factors (code 3QC1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PP2A
- protein phosphatase 2A
- PP2Ac
- PP2A catalytic subunit
- UIM
- ubiquitin-interacting motif
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DEER
- double electron-electron resonance
- TUBE
- tandem ubiquitin-binding entity
- TPR
- tetratricopeptide repeat.
REFERENCES
- 1. Kong M., Bui T. V., Ditsworth D., Gruber J. J., Goncharov D., Krymskaya V. P., Lindsten T., Thompson C. B. (2007) J. Biol. Chem. 282, 29712–29720 [DOI] [PubMed] [Google Scholar]
- 2. Kong M., Fox C. J., Mu J., Solt L., Xu A., Cinalli R. M., Birnbaum M. J., Lindsten T., Thompson C. B. (2004) Science 306, 695–698 [DOI] [PubMed] [Google Scholar]
- 3. Janssens V., Goris J. (2001) Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goedert M., Jakes R., Qi Z., Wang J. H., Cohen P. (1995) J. Neurochem. 65, 2804–2807 [DOI] [PubMed] [Google Scholar]
- 5. Eichhorn P. J., Creyghton M. P., Bernards R. (2009) Biochim. Biophys. Acta 1795, 1–15 [DOI] [PubMed] [Google Scholar]
- 6. Zolnierowicz S. (2000) Biochem. Pharmacol. 60, 1225–1235 [DOI] [PubMed] [Google Scholar]
- 7. Liu R., Wang J. Z. (2009) Pathophysiology 16, 273–277 [DOI] [PubMed] [Google Scholar]
- 8. Kowluru A. (2005) Biochem. Pharmacol. 69, 1681–1691 [DOI] [PubMed] [Google Scholar]
- 9. Bryant J. C., Westphal R. S., Wadzinski B. E. (1999) Biochem. J. 339, 241–246 [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., Martin B. L., Brautigan D. L. (1992) Science 257, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 11. Guo H., Damuni Z. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2500–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trockenbacher A., Suckow V., Foerster J., Winter J., Krauss S., Ropers H. H., Schneider R., Schweiger S. (2001) Nat. Genet. 29, 287–294 [DOI] [PubMed] [Google Scholar]
- 13. Chen J., Peterson R. T., Schreiber S. L. (1998) Biochem. Biophys. Res. Commun. 247, 827–832 [DOI] [PubMed] [Google Scholar]
- 14. Nanahoshi M., Tsujishita Y., Tokunaga C., Inui S., Sakaguchi N., Hara K., Yonezawa K. (1999) FEBS Lett. 446, 108–112 [DOI] [PubMed] [Google Scholar]
- 15. Prickett T. D., Brautigan D. L. (2004) J. Biol. Chem. 279, 38912–38920 [DOI] [PubMed] [Google Scholar]
- 16. Murata K., Wu J., Brautigan D. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong M., Ditsworth D., Lindsten T., Thompson C. B. (2009) Mol. Cell 36, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McConnell J. L., Watkins G. R., Soss S. E., Franz H. S., McCorvey L. R., Spiller B. W., Chazin W. J., Wadzinski B. E. (2010) Biochemistry 49, 1713–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inui S., Sanjo H., Maeda K., Yamamoto H., Miyamoto E., Sakaguchi N. (1998) Blood 92, 539–546 [PubMed] [Google Scholar]
- 20. Yang J., Roe S. M., Prickett T. D., Brautigan D. L., Barford D. (2007) Biochemistry 46, 8807–8815 [DOI] [PubMed] [Google Scholar]
- 21. Smetana J. H., Oliveira C. L., Jablonka W., Aguiar Pertinhez T., Carneiro F. R., Montero-Lomeli M., Torriani I., Zanchin N. I. (2006) Biochim. Biophys. Acta 1764, 724–734 [DOI] [PubMed] [Google Scholar]
- 22. Liu J., Prickett T. D., Elliott E., Meroni G., Brautigan D. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6650–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prickett T. D., Brautigan D. L. (2006) J. Biol. Chem. 281, 30503–30511 [DOI] [PubMed] [Google Scholar]
- 24. Quaderi N. A., Schweiger S., Gaudenz K., Franco B., Rugarli E. I., Berger W., Feldman G. J., Volta M., Andolfi G., Gilgenkrantz S., Marion R. W., Hennekam R. C., Opitz J. M., Muenke M., Ropers H. H., Ballabio A. (1997) Nat. Genet. 17, 285–291 [DOI] [PubMed] [Google Scholar]
- 25. Fontanella B., Russolillo G., Meroni G. (2008) Hum. Mutat. 29, 584–594 [DOI] [PubMed] [Google Scholar]
- 26. Schweiger S., Schneider R. (2003) BioEssays 25, 356–366 [DOI] [PubMed] [Google Scholar]
- 27. McConnell J. L., Gomez R. J., McCorvey L. R., Law B. K., Wadzinski B. E. (2007) Oncogene 26, 6021–6030 [DOI] [PubMed] [Google Scholar]
- 28. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 29. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 30. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 31. Abrahams J. P., Leslie A. G. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 34. Zou P., Bortolus M., McHaourab H. S. (2009) J. Mol. Biol. 393, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W. (2000) J. Magn. Reson. 142, 331–340 [DOI] [PubMed] [Google Scholar]
- 36. Holm L., Rosenström P. (2010) Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Das A. K., Cohen P. W., Barford D. (1998) EMBO J. 17, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Agarwal-Mawal A., Qureshi H. Y., Cafferty P. W., Yuan Z., Han D., Lin R., Paudel H. K. (2003) J. Biol. Chem. 278, 12722–12728 [DOI] [PubMed] [Google Scholar]
- 39. Obsilová V., Silhan J., Boura E., Teisinger J., Obsil T. (2008) Physiol. Res. 57, S11–S21 [DOI] [PubMed] [Google Scholar]
- 40. Cliff M. J., Williams M. A., Brooke-Smith J., Barford D., Ladbury J. E. (2005) J. Mol. Biol. 346, 717–732 [DOI] [PubMed] [Google Scholar]
- 41. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 42. D'Andrea L. D., Regan L. (2003) Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 43. Yang J., Roe S. M., Cliff M. J., Williams M. A., Ladbury J. E., Cohen P. T., Barford D. (2005) EMBO J. 24, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 45. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 46. Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995) Nature 376, 191–194 [DOI] [PubMed] [Google Scholar]
- 47. Lapouge K., Smith S. J., Walker P. A., Gamblin S. J., Smerdon S. J., Rittinger K. (2000) Mol. Cell 6, 899–907 [DOI] [PubMed] [Google Scholar]
- 48. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrodinger, LLC, New York [Google Scholar]
- 49. Bond C. S. (2003) Bioinformatics 19, 311–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.