Abstract
The dysregulation of EGF family ligand cleavage has severe consequences for the developing as well as the adult organism. Therefore, their production is highly regulated. The limiting step is the ectodomain cleavage of membrane-bound precursors by one of several a disintegrin and metalloprotease (ADAM) metalloproteases, and understanding the regulation of cleavage is an important goal of current research. We have previously reported that in mouse lung epithelial cells, the pro-EGF ligands TGFα, neuregulin 1β (NRG), and heparin-binding EGF are differentially cleaved depending on the cleavage stimulus (Herrlich, A., Klinman, E., Fu, J., Sadegh, C., and Lodish, H. (2008) FASEB J.). In this study in mouse embryonic fibroblasts that lack different ADAMs, we show that induced cleavage of EGF ligands can involve the same substrate-specific metalloprotease but does require different stimulus-dependent signaling pathways. Cleavage was stimulated by phorbol ester (12-O-tetradecanoylphorbol-13-acetate (TPA), a mimic of diacylglycerol and PKC activator), hypertonic stress, lysophosphatidic acid (LPA)-induced G protein-coupled receptor activation, or by ionomycin-induced intracellular calcium release. Although ADAMs showed substrate preference (ADAM17, TGFα and heparin-binding EGF; and ADAM9, NRG), substrate cleavage differed substantially with the stimulus, and cleavage of the same substrate depended on the presence of different, sometimes multiple, PKC isoforms. For instance, classical PKC was required for TPA-induced but not hypertonic stress-induced cleavage of all EGF family ligands. Inhibition of PKCζ enhanced NRG release upon TPA stimulation, but it blocked NRG release in response to hypertonic stress. Our results suggest a model in which substantial regulation of ectodomain cleavage occurs not only on the metalloprotease level but also on the level of the substrate or of a third protein.
Keywords: G protein-coupled Receptors (GPCR), Metalloprotease, Protein Kinase C (PKC), Signal Transduction, Transforming Growth Factor-α (TGFα)
Introduction
Metalloprotease-mediated ectodomain cleavage of transmembrane proteins such as EGF ligand precursors is involved in the regulation of many physiological signaling pathways, and its dysregulation can cause kidney disease, heart disease, and cancer (1, 2). For example, in the kidney, many of the deleterious effects of chronic exposure to the G-protein-coupled receptor (GPCR)2 agonist angiotensin II are mediated by metalloproteinase (ADAM17)-dependent release of the EGF ligand TGFα and subsequent epidermal growth factor receptor activation. Angiotensin II is the main effector of the renin-angiotensin system and has important roles in the regulation of blood pressure and aldosterone secretion (3).
Although several ADAM gene disruptions are lethal in the mouse, metalloprotease inhibitors have shown therapeutic potential; however, not unexpectedly, the essential role of ADAMs in normal physiology as well as the broad spectrum nature of currently available inhibitors are probably the cause for a number of their side effects. In addition, concerns have been raised that some metalloproteases act as tumor suppressors (4, 5). A more precise inhibition of specific metalloproteases and/or of the cleavage of specific disease-involved substrates could circumvent these problems.
Unfortunately, how metalloprotease cleavage is regulated is mostly unknown. Much of the previously published data suggested that the regulation occurs predominantly on the metalloprotease level. As such, phorbol ester (TPA)-induced protein kinase C (PKC) activation has been linked to the activation of ADAM17, whereas ionomycin-induced intracellular calcium increase has been proposed to activate ADAM10 and -17 (6, 7). However, after stimulation of GPCRs, several ADAMs, including ADAM10, -12, and -17, have been found activated, depending on the cell type and substrate studied (2, 8, 9). Hypertonic stress, another cleavage-inducing stimulus, has been associated with activation of ADAM17 in cancer cells (8). The question thus is, how is substrate specificity ascertained and regulated?
In previous studies in mouse lung epithelial (MLE) cells using a newly developed high throughput FACS-based cleavage assay, we reported that cleavage induction by phorbol esters, GPCR activation, and hypertonic stress engages distinct signaling pathways that in part involve the action of distinct protein kinase C isoforms. Moreover, within the same cells, EGF ligand precursors showed differential cleavage responses depending on the cleavage stimulus used, suggesting that regulation of cleavage may occur at least in part at the substrate level (11).
In this study, we were interested in how specificity of EGF ligand release is regulated. In a simplified model, if specificity of cleavage were regulated on the metalloprotease level, we would expect particular metalloproteases to be activated by particular stimuli and possibly cleave a large number of different substrates. If specificity of cleavage were regulated on the substrate level, we would expect that different stimuli acted on the substrate (or a third protein affecting the substrate) and potentially caused different modifications that would regulate accessibility by or strength of interaction with a constitutively active metalloprotease. To examine these possibilities, we studied TPA-, hypertonic stress-, LPA-, and IM-induced signaling pathways that regulate cleavage-dependent release of three different EGF ligands in MEF cells as follows: transforming growth factor-α (TGFα), NRG, and heparin-binding epidermal growth factor (HB-EGF). We then determined which ADAM metalloproteases were responsible for cleavage of the different substrates in response to the same four stimuli. Together with our previously published data in MLE cells (11), these results suggest that induced release of EGF ligands is rendered specific by PKC-dependent and -independent signaling pathways that regulate substrate cleavage by acting not only on the metalloprotease but also on the substrate or a third linker protein.
MATERIALS AND METHODS
Antibodies
The following antibodies and dilutions were used: monoclonal anti-FLAG antibody M2 for FACS (1:100), immunoprecipitation (1:100), and Western blot (1:1000); monoclonal anti-MYC 9E10 for FACS (1:100 dilution; Covance); monoclonal anti-HA11.1 antibody for FACS (1:100 dilution; Covance); allophycocyanin-coupled goat anti-mouse antibody for FACS (1:150 dilution; BD Biosciences); and polyclonal anti-phospho-specific PKC substrate antibody (1:1000 dilution; Cell Signaling).
Reagents
The following reagents were used: TPA, LPA, and sorbitol (Sigma); ionomycin (Cell Signaling); Polybrene (Sigma); FuGENE 6 (Roche Applied Science); RPMI 1640 medium, DMEM, and FCS (Invitrogen); propidium iodine (Sigma); BB94 (batimastat; Tocris Biosciences); bisindolylmaleimide (Calbiochem 203293) and myristoylated PKZζ/ι pseudosubstrate inhibitor (Calbiochem 539624); and protein G-agarose (Sigma).
Cell Lines
ADAM9 (12) and ADAM17 (6) knock-out mouse embryonic fibroblasts (MEF cells) were a kind gift from Dr. Carl Blobel (Hospital for Special Surgery, New York). ADAM10 knock-out cells (13) were a kind gift from Dr. Paul Saftig (University of Kiel, Germany). Mouse lung epithelial cells (originally generated by Jeffrey Whitsett) and HEK 293T cells were from ATCC.
Cloning
The cloning of the epitope-tagged cDNAs for NRG1β (FLAG tag), HB-EGF (Myc tag), and TGFα (HA tag) is described in detail in Herrlich et al. (11).
Generation or Reporter Cell Lines
Retroviral EGF ligand reporter constructs were co-transfected with pCLEco (14) into 293T cells, and the resultant retrovirus was used to infect MEF cells or MLE cells at 50% confluence with 4 μg/ml Polybrene.
FACS Assay
For the inhibitor experiments, cells were preincubated with either 10 μm BB94, 1 μm bisindolylmaleimide, and 20 μm myristoylated PKZζ/ι pseudosubstrate inhibitor for 30 min prior to cleavage stimulation. Reporter cells were either control-treated or stimulated with 400 mosm sorbitol (400 mosm final gradient between extracellular and intracellular space), 1 μm TPA, 20 μm LPA, or with 2.5 μm IM for times indicated in the individual figures. Stimulation was stopped by adding 1 ml of a cold PBS-based enzyme-free proprietary cell dissociation solution (Millipore S-014-B) and by placing cells on ice. Cells were dissociated, resuspended, washed with cold PBS, 3% FCS, and subsequently incubated at 4 °C for 1 h with the respective anti-epitope primary antibody. After washing three times with cold PBS, 3% FCS, cells were incubated at 4 °C for 1 h with anti-mouse allophycocyanin-coupled secondary antibody. Finally, cells were again washed three times with cold PBS, 3% FCS and then incubated with a PBS-based enzyme-free proprietary cell dissociation solution (Millipore S-014-B) containing 2 μg/ml propidium iodine. FACS analysis was performed with a BD Biosciences FACSCalibur machine as detailed previously (11).
Immunoprecipitation and Western Blot
MEF cells expressing the FLAG-NRG-EGFP reporter were pretreated for 30 min with batimastat (BB94, 20 μm) and either control (DMSO) or the classical PKC inhibitor BIM1 (1 μm). Cells were then either control-treated (DMSO) or stimulated with TPA (1 μm). After stimulation, cells were washed with cold PBS and incubated on ice with anti-FLAG M2 antibody (1:100) in PBS, FCS 3% to capture only cell-surface located reporter. Cells were then lysed on ice with lysis buffer containing 1% Triton, protease, and phosphatase inhibitors. Lysates were harvested; debris was cleared by centrifugation, and anti-FLAG immunocomplexes were precipitated with protein G-agarose and washed several times. Immunocomplexes were then subjected to SDS-PAGE and Western blotting. After transfer to a nitrocellulose membrane and blocking for 1 h at room temperature with 5% milk/TBST (TBST: Tris-buffered saline 50 mm, Tris-HCl, pH 7.4, 150 mm NaCl, with 0.1% Triton), antibodies were incubated overnight at 4 °C in 5% milk/TBST (anti-FLAG M2 antibody, 1:1000) or 5% BSA/TBST (anti-phospho-PKC substrate antibody, 1:1000). Membranes were then washed three times with TBST and incubated for 1 h at room temperature with secondary antibody in 5% milk/TBST.
RESULTS
Cell Type Specificity of EGF Ligand Release
Using our previously published FACS-based cleavage assay (11) that allows monitoring of substrate cleavage in single living cells, we first analyzed cleavage of three different EGF ligand precursors, TGFα, NRG, and HB-EGF in MEFs. To this end, we generated MEF cells stably overexpressing pro-EGF ligands tagged in the N-terminal ectodomain with an epitope tag and fused to green fluorescent protein (GFP) at the C terminus. When stained with an anti-epitope tag antibody coupled to red fluorescence, cells show a specific red:green fluorescence ratio in the uncleaved state (which in the ideal case of similar strength of fluorescence would be 1:1). Cleavage of the pro-EGF ligand substrates decreases this red:green fluorescence ratio. We confirmed cleavage as measured by FACS with Western blots for each of the reporter constructs (11). Cleavage was induced with phorbol ester (TPA 1 μm, 0.65 ng/ml; a diacylglycerol mimic and activator of classical PKC isoenzymes), hypertonic stress using sorbitol (400 mosm gradient between intracellular and extracellular space), activation of the G-protein-coupled LPA receptor (20 μm), or by triggering an increase of the intracellular calcium concentration with ionomycin (IM 2.5 μm). Cells were control-treated or stimulated with one of the four stimuli for 5, 15, and 30 min and subjected to FACS to determine red and green fluorescence of the cells. In Fig. 1, we compare our results obtained for EGF ligand cleavage in wild type MEF cells with our previously published dataset obtained in MLE cells (11), using the same cleavage stimuli and EGF ligands. Table 1 provides a summary of the data by focusing on the time points of maximal ligand cleavage.
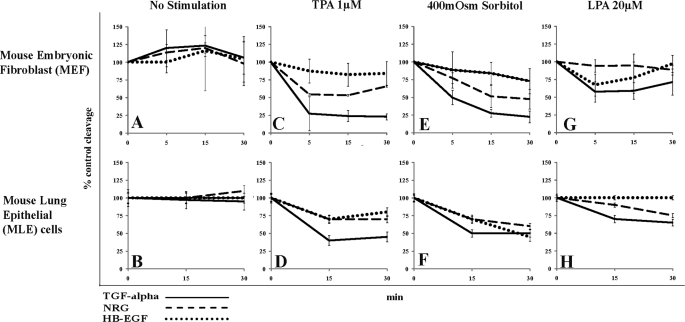
FIGURE 1.
Comparison of pro-EGF ligand cleavage in MEF and MLE cells. Cleavage of pro-EGF reporter ligand was detected by changes in the cellular red:green fluorescence ratio as measured by FACS (details see text). Red:green fluorescence ratio was plotted over time and compared with % control in MEF cells or MLE cells stably overexpressing precursors of either TGFα (solid lines), NRG (dashed lines), or HB-EGF (dotted lines). MEF or MLE cells were either left unstimulated (A and B) or were incubated with 1 μm TPA (C and D), 400 mosm sorbitol (E and F), or 20 μm LPA (G and H) and monitored by FACS at indicated time points. Data are from at least three independent experiments performed in triplicate. Data from MLE cells were published previously (11) and are included here to make comparison of the data sets easier.
TABLE 1.
Comparison of pro-EGF ligand cleavage in wild type MEFs and wild type MLE cells
We previously published data examining the cleavage of pro-TGF-α, pro-NRG, and pro-HB-EGF in mouse lung epithelial cells using the same reporter assay and the same cleavage stimuli (11). Here, we compare these data to our current dataset in MEF cells at points of maximal cleavage, using % control of the red:green fluorescent ratio. Data are from at least three independent experiments performed in triplicate.
| Stimulus | % Control of red:green fluorescence ratio at point of maximal cleavage |
|||||
|---|---|---|---|---|---|---|
| TGF-α at 30 min |
NRG at 30 min |
HB-EGF at 15 min (at 30 min) |
||||
| MEF cells | MLE cells | MEF cells | MLE cells | MEF cells | MLE cells | |
| TPA 1 μm | 75 | 50 | 40 | 30 | 15 (15) | 25 (20) |
| Sorbitol 400 mosm | 75 | 50 | 50 | 30 | 15 (20) | 25 (50) |
| LPA 20 μm | 30 | 30 | 0 | 25 | 25 (0) | 0 (0) |
Both MEF and MLE cells endogenously express the metalloproteases ADAM9, -10, and -17 (quantitative PCR data not shown). Yet EGF ligand cleavage proceeds very differently in both cell types, depending on the cleavage stimulus and substrate concerned. We observed at least three discernible patterns of cleavage, as illustrated in Fig. 1 and summarized in Table 1.
Same Stimulus Causes Different Degrees of Ligand Release Depending on the Cell Type and the Substrate
In MEF cells, as compared with MLE cells, TPA and hypertonic stress induce much stronger cleavage of TGFα (Fig. 1, compare solid lines in C with D and in E with F; Table 1) and NRG (Fig. 1, compare dashed lines in C with D and in E with F; Table 1). LPA-induced TGFα cleavage was weaker but at a similar level in both cell types (Fig. 1, compare solid lines in G with H; Table 1). HB-EGF release was comparably low in both cell types, except for hypertonic stress-induced release, which was much stronger in MLE cells (Fig. 1, compare dotted lines in E and F; Table 1).
Different Stimuli Cause Different Degrees of Release of the Same Ligand
In both MEF and MLE cells, TPA- and hypertonic stress-induced release of TGFα and NRG is much stronger than LPA-induced release of the same ligands (Fig. 1, compare solid lines and dashed lines in C and D and E and F with G and H; Table 1).
Same Stimulus Is Able to Induce Cleavage of a Particular Substrate in Only One Cell Type
LPA induced strong cleavage of NRG in MLE cells but not in MEF cells (Fig. 1, compare dashed lines in G and H; Table 1). In contrast, LPA induced release of HB-EGF only in MEF cells but not in MLE cells (Fig. 1, compare dotted lines in G and H; Table 1). These results suggest significant cell type-specific differences in the regulation of EGF ligand cleavage.
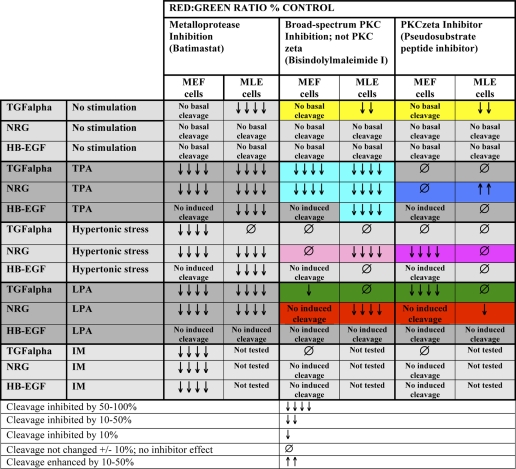
Substrate- and Cell Type-specific Regulation of Cleavage by PKC-dependent and -independent Pathways
FACS cleavage experiments using PKC and metalloprotease inhibitors in MLE cells had suggested that the activity of different PKC isoforms was involved in regulating pro-EGF ligand cleavage (11); these MLE cell data are summarized and compared with our new results in MEF cells in Table 2. By extending our PKC inhibitor studies from MLE to MEF cells (Fig. 2; Table 2) we attempted to answer the following two major questions. 1) Is PKC activity generally involved in the regulation of EGF ligand cleavage? 2) Are differences in EGF ligand cleavage observed in MLE versus MEF cells related to cell type-specific differences in signaling pathways initiated by the different stimuli? Because we detected only low level HB-EGF cleavage overall, we focused our inhibitor studies in MEF cells on TGFα and NRG.
TABLE 2.
Comparison of the effect of PKC isoform and metalloprotease inhibition on pro-EGF ligand release in MEF and MLE cells
MLE cell data are from Herrlich et al. (11). Data are from at least three independent experiments performed in triplicate. Highlighted panels are mentioned in the text.
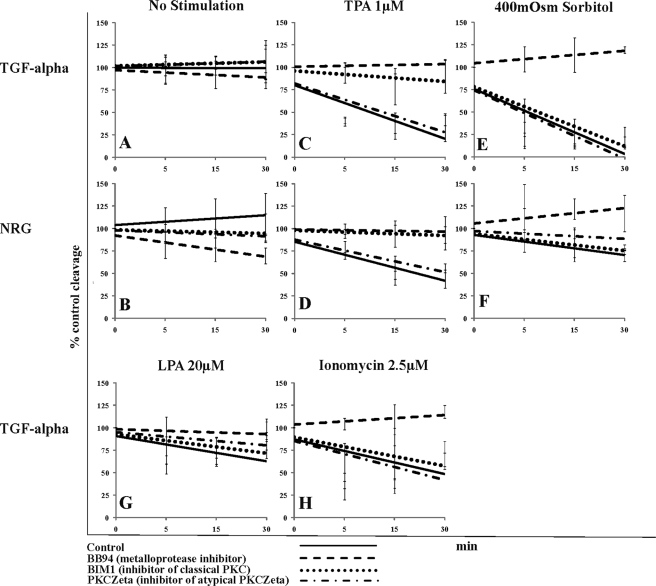
FIGURE 2.
Effect of metalloprotease and protein kinase C inhibitors on TGFα and NRG release in MEF cells. Shown are trend lines of % control red:green fluorescent ratio as determined by FACS. MEF cells were either control-treated or pretreated with the indicated inhibitor. Subsequently, cells were either left unstimulated (No stimulation) or stimulated with one of the indicated stimuli. LPA and IM do not induce NRG cleavage in MEF cells. Solid line, control-treated cells; dashed line, metalloprotease inhibitor BB94 (10 μm); dotted line, classical PKC inhibitor bisindolylmaleimide (1 μm); dotted-dashed line, PKCζ pseudosubstrate inhibitor (10 μm); TPA, phorbol ester; sorbitol 400 mosm, hypertonic stress, 400 mosm gradient; LPA, lysophosphatidic acid (20 μm); IM, ionomycin (2.5 μm). Data are from at least three independent experiments performed in triplicate.
Basal Cleavage Is Cell Type- and Substrate-specific and Involves PKC Activity
In unstimulated MLE cells, metalloprotease inhibition (BB94) caused an increase in the level of cell surface TGFα, thus revealing a significant degree of TGFα release under basal (unstimulated) conditions (yellow highlight; Table 2) that was not readily visible without the inhibitor (Fig. 1B). Specifically, basal TGFα cleavage was reduced by an inhibitor (bisindolylmaleimide 1 (BIM1)) of classical TPA- and calcium-inducible PKC isoenzymes (which includes PKCα, for example), as well as inhibition of atypical (not TPA- and calcium-inducible) PKCζ (Table 2). In contrast, MEF cells did not show a significant accumulation of TGFα or of NRG on the cell surface in the presence of BB94 (Fig. 2, A and B; Table 2), suggesting that there is very little cleavage of these substrates in the unstimulated state.
TPA-induced Cleavage Depends on Classical PKCs in Both MLE and MEF Cells
Similar to what we previously showed using MLE cells (11), TPA-induced cleavage of any EGF ligand studied in MEF cells was sensitive to inhibition of classical PKCs by BIM1 (dotted lines in Fig. 2, C and D; light blue highlight in Table 2). Only in MLE cells was TPA-induced release of NRG moderately enhanced by inhibition of PKCζ (dark blue highlight in Table 2).
Depending on the Substrate and Cell Type Hypertonic Stress-induced Cleavage Can Require PKC Activity
Hypertonic stress-induced cleavage of any studied EGF ligand in MLE and MEF cells was predominantly independent of PKC activity; only NRG cleavage in response to hypertonic stress required PKC activity. In MLE cells NRG release was strongly inhibited by BIM1, the generic PKC inhibitor, whereas this inhibitor had no effect on NRG cleavage in MEF cells (Fig. 2, dotted lines in E and F; light pink highlight in Table 2). In contrast, PKCζ inhibition partially blocked hypertonic stress-induced NRG cleavage only in MEF cells but not in MLE cells (dashed-dotted lines in Fig. 2, E and F; dark pink highlight in Table 2). Thus, different PKC isoforms regulate hypertonic stress cleavage of NRG in different cell types.
Depending on the Substrate and Cell Type LPA-induced Cleavage Can Require PKC Activity
LPA induced different PKC-dependent and -independent pathways in MLE and MEF cells that modulated cleavage in a substrate-specific manner. In MLE cells, LPA-induced release of NRG was strongly blocked by BIM1 and to a minor degree by the PKCζ-specific inhibitor (red highlight in Table 2). LPA-stimulated TGFα release did not require PKC activity in MLE cells. In contrast, in MEF cells, LPA-induced cleavage of TGFα was dependent on PKC activity; cleavage was strongly inhibited by the PKCζ-specific inhibitor and to a minor degree by the classical PKC inhibitor BIM1 (Fig. 2G, green highlight in Table 2).
IM-induced Cleavage Is Independent of PKC Activity
IM-induced TGFα cleavage (a stimulus not tested in MLE cells) was not dependent on PKC (Fig. 2H; Table 2).
TPA-induced Serine Phosphorylation on NRG Requires PKC Activity
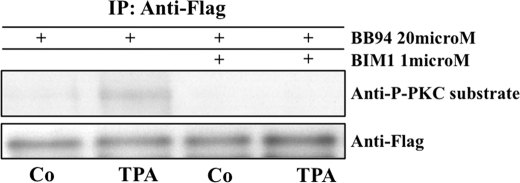
To determine whether PKC activity was required for regulation of cleavage on the substrate level, we exemplarily tested for the presence of induced serine or threonine phosphorylations on NRG, which contains several serines in its C-terminal tail. We blocked metalloprotease cleavage of NRG reporter-expressing MEF cells by preincubation with batimastat (BB94; 20 μm) and then control-treated or stimulated the cells with TPA (1 μm). Following this, we immunoprecipitated the cell surface fraction of the reporter with anti-FLAG ectodomain antibody. TPA stimulation indeed led to an accumulation of serine phosphorylation(s) on uncleaved, full-length cell-surface NRG, as measured by Western blot with an anti-phosphoserine antibody specific to PKC phosphorylation sites. This could be blocked by the classical PKC inhibitor BIM1 (Fig. 3).
FIGURE 3.
TPA induces serine phosphorylation on NRG that can be blocked by PKC inhibition. MEF cells were preincubated with the metalloproteinase inhibitor batimastat (BB94, 20 μm) and with either control (DMSO) or with an inhibitor of classical PKC isoforms bisindolylmaleimide I (BIM1, 1 μm). Subsequently, cells were stimulated for 30 min with either control medium (DMSO) or TPA (1 μm). After stimulation, cells were placed on ice and incubated with anti-FLAG antibody (on plate), prior to cell lysis, to capture only the cell surface fraction of the overexpressed reporter FLAG-NRG-GFP. Anti-FLAG-NRG complexes were immunoprecipitated (IP) with protein G-agarose and resolved by SDS-PAGE with the antibodies indicated. Anti-P-PKC-substrate antibody recognizes phosphorylated serine within a consensus PKC phosphorylation site (arginine or lysine in position −2 and +2 and a hydrophobic amino acid at position +1 relative to the serine). Shown is one of four identical experiments with the same outcome.
In summary, our inhibitor experiments suggest that PKC-dependent and -independent signaling pathways modulate EGF ligand cleavage in both MLE and MEF cells in a substrate- and cell type-specific way. Cleavage of some pro-EGF ligands requires the activity of several PKC isoforms depending on the cleavage stimulus. TPA-induced serine phosphorylation of the C-terminal tail of NRG is PKC-dependent and possibly participates in regulation of substrate cleavage.
Specificity of EGF Ligand Cleavage in ADAM Knock-out Cells
Substrate specificity of metalloproteases may represent one level of regulation of substrate cleavage that could explain our observed differences in EGF ligand cleavage. Substrate-specific signaling pathways may in principle act on the metalloprotease or the substrate. Therefore, as a next step, we determined the contribution of different ADAM metalloproteases to EGF ligand cleavage in MEF cells, using MEF wild type cells and MEF cells that were isolated from mice with a knock-out of either ADAM9, -10, or -17 (6, 12, 13). A summary of the data is provided in Table 3.
TABLE 3.
Summary of pro-EGF ligand cleavage in MEFs
EGF reporter ligand cleavage was compared at points of maximal cleavage based on the results shown in Fig. 3. Maximal cleavage of TGF-α and NRG occurred at approximately 30 min, and maximal cleavage of the HB-EGF substrate was observed at approximately 15 min. Cleavage is reported as % control. Negative values indicate an accumulation rather than cleavage of the substrate. ∅ = lack of cleavage.
| Stimulus | ADAM | % control of red:green fluorescence ratio at maximal cleavage |
||
|---|---|---|---|---|
| TGF-α at 30 min | NRG at 30 min | HB-EGF at 15 min | ||
| No stimulation | WT | ∅ | ∅ | ∅ |
| 9−/− | −20 | −20 | −20 | |
| 10−/− | 20 | 10 | −10 | |
| 17−/− | 10 | 10 | −10 | |
| TPA 1 μm | WT | 80 | 40 | 20 |
| 9−/− | 80 | ∅ | 20 | |
| 10−/− | 90 | 50 | ∅ | |
| 17−/− | ∅ | 30 | ∅ | |
| Sorbitol 400 mo sm | WT | 80 | 50 | 20 |
| 9−/− | 80 | ∅ | 20 | |
| 10−/− | 80 | 40 | ∅ | |
| 17−/− | 75 | ∅ | 20 | |
| LPA 20 μm | WT | 30 | 10 | 20 |
| 9−/− | 40 | ∅ | 20 | |
| 10−/− | 60 | 40 | 20 | |
| 17−/− | ∅ | ∅ | ∅ | |
| Ionomycin 2.5 μm | WT | 60 | 10 | 10 |
| 9−/− | 50 | ∅ | 10 | |
| 10−/− | 60 | 40 | ∅ | |
| 17−/− | ∅ | 30 | ∅ | |
TGFα Cleavage
Basal TGFα Cleavage
Under basal conditions, we detected no TGFα cleavage in wild type and ADAM9 knock-out MEF cells (Fig. 4, A and B, solid lines; Table 3). ADAM10 and -17 knock-out cells however, showed an ∼10–20% increase in basal TGFα cleavage over control-treated cells that could be inhibited by a broad spectrum metalloprotease inhibitor (BB94, 10 μm; data not shown) (Fig. 4, C and D, solid lines; Table 3), suggesting that ADAM10 and -17 knock-out cells may have other adaptive changes that influence ligand cleavage beyond their ADAM knockdown.
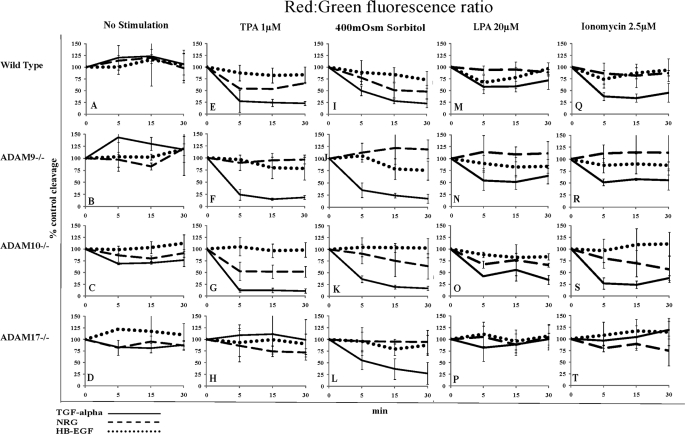
FIGURE 4.
Comparison of pro-EGF ligand cleavage in wild type MEFs and MEFs knock-out for either ADAM9, -10, or -17. Cleavage of pro-EGF reporter ligands was detected by changes in the cellular red:green fluorescence ratio as measured by FACS (details see text). Red:green fluorescence was plotted over time and compared with % control in wild type or ADAM knock-out mouse embryonic fibroblasts (lacking either ADAM9, -10, and -17) stably overexpressing precursors of either TGFα (solid lines), NRG (dashed lines), or HB-EGF (dotted lines). Plots for each different cell line are shown in horizontal rows, whereas plots for different cell lines subjected to the same cleavage stimulus are arranged in vertical columns. Cells were either control-treated (A–D) or incubated with TPA 1 μm (E–H), sorbitol 400 mosm (I–L), LPA 20 μm (M–P), or ionomycin 2.5 μm (Q–T) and monitored by FACS at 5, 15, and 30 min.
TPA-, LPA-, and IM-induced TGFα Cleavage Depends on ADAM17
Induced cleavage of TGFα was maximal within 5–15 min of stimulation and was maintained for over 30 min. TPA stimulated 80% cleavage of TGFα in wild type and MEF cells lacking ADAM9 or -10 (Fig. 4, E–G, solid lines; Table 3). However, cleavage was completely inhibited in ADAM17 knock-out cells (Fig. 4H, solid line; Table 3). LPA- and IM-induced cleavage of TGFα in wild type and ADAM9 and -10 knock-out cells was weaker (30–60% of control-treated cells) than TPA-induced cleavage. Importantly, cleavage in response to both of these stimuli was also strongly reduced in the absence of ADAM17 (Fig. 4, M–P and Q–T, solid lines; Table 3).
Hypertonic Stress-induced TGFα Cleavage Depends on a Metalloprotease but Not ADAM9, -10, or -17
In contrast to the results observed with TPA, LPA, and IM (Fig. 4, E–H, M–P, and Q–T, solid lines; Table 3), hypertonic stress-induced cleavage of TGFα (70–80% of control-treated cells) was independent of ADAM9, -10, and -17 (Fig. 4, I–L, solid lines; Table 3) but could be blocked by the broad spectrum metalloprotease inhibitor, BB94 (Fig. 2E).
NRG Cleavage
In MEF cells, the pro-EGF ligand NRG showed a notably different cleavage pattern than TGFα using the same cleavage stimuli. NRG did not show any significant basal release in MEF cells (Fig. 4, A–D, dashed lines; Table 3), and opposite to their effects on TGFα, LPA and IM were unable to induce NRG release (Fig. 4, M and Q, dashed lines; Table 3).
TPA-induced NRG Cleavage Depends on ADAM9
Although induced TGFα cleavage was maximal after 5 min, NRG required 30 min of stimulation to achieve maximal cleavage. TPA-induced NRG cleavage (Fig. 4, E–H, dashed lines; Table 3), unlike that of TGFα, was only mildly reduced in ADAM17 knock-out cells (Fig. 4H, compare dashed and solid line; Table 3) but completely blocked in the absence of ADAM9 (Fig. 4F, dashed line; Table 3).
Hypertonic Stress-induced NRG Cleavage Depends on ADAM9 and -17
When stimulated with hypertonic stress, 50% of the NRG substrate was cleaved in wild type MEF cells and ADAM10 knockouts (Fig. 4, I and K, dashed line; Table 3). Both ADAM9 and ADAM17 knock-out cells (Fig. 4, J and L, dashed lines; Table 3) displayed complete absence of hypertonic stress-induced NRG cleavage.
HB-EGF Cleavage
HB-EGF Cleavage in MEF Cells Was Low but Depended on ADAM10 and -17
Interestingly, HB-EGF (Fig. 4, dotted lines) showed the weakest cleavage response in MEF cells as compared with the other EGF ligands (Fig. 1; Table 1) HB-EGF release in response to the same stimuli was much lower in comparison with TGFα, although both were a substrate of ADAM17. Basal cleavage of HB-EGF was negligible (Fig. 4, A–D, dotted lines; Table 3). Stimulation with any stimulus resulted in only about 20% of cleavage as compared with control cells (Fig. 4, E, I, M, and Q, dotted lines; Table 3). ADAM10 and/or -17 were required in the case of TPA- and IM-induced HB-EGF cleavage (Fig. 4, G and H and S and T; Table 3), although only ADAM10 was required for hypertonic stress-induced HB-EGF cleavage (Fig. 4K, dotted line; Table 3).
Taken together our results in MEF cells suggest that ADAM17 is the major sheddase for TGFα and HB-EGF, and ADAM9 is the major sheddase for NRG.
DISCUSSION
Here, we report that cleavage-dependent EGF ligand release is differentially regulated in different cell types. We found that in MEFs and in MLE cells, induced cleavage of the same pro-EGF ligands in response to the same cleavage stimuli can differ significantly. Experiments using inhibition of classical PKC isoforms or atypical PKCζ showed that substrate cleavage is regulated by PKC-dependent and -independent signaling pathways that share many common features but show some significant differences depending on the stimulus, substrate, or cell type. Finally, experiments in ADAM knock-out MEF cells revealed that, with some exceptions, only one substrate-specific metalloprotease was responsible for most cleavage events of a particular substrate in response to any stimulus. Taken together, we interpret these results to suggest that ectodomain cleavage is regulated not only on the metalloprotease level, as previously reported by others, but also on the substrate level.
In principal, regulation of substrate cleavage by ADAMs could be regulated in one of three ways. 1) For regulation of cleavage on the metalloprotease level, a particular stimulus activates one particular metalloprotease that recruits different substrates for cleavage. Differences in substrate affinity to this metalloprotease could explain differences in substrate cleavage. Such regulation could occur, for example by introducing modifications on the ectodomain or the C terminus of the metalloprotease. 2) For regulation on the substrate level, stimuli lead to modifications on the ectodomain or the C terminus of substrates that make it accessible to cleavage by one or several constitutively active metalloproteases, depending on substrate/metalloprotease specificity. Different stimuli may induce different modifications on the substrate, and this could explain differences in cleavage. 3) For regulation involving a third protein, cleavage stimuli could regulate, for example, linker proteins that promote access of metalloprotease and substrate to each other. These linker proteins could be differentially regulated by different signaling pathways and be metalloprotease- and/or substrate-specific.
Previous studies on shedding have been interpreted to suggest that regulation of ectodomain cleavage may occur predominantly on the metalloprotease level and that particular stimuli are connected to particular metalloproteases (1, 2, 9). ADAM10 and -17 are thought to be the TPA- and calcium-responsive sheddases, particularly in the context of EGF ligand cleavage (2, 6, 15) but also for most other shedding events studied to date (for a review see Ref. 2). In contrast, our results support the notion that ectodomain cleavage is not only regulated on the metalloprotease level but could also be regulated at the substrate level in at least three different ways.
Induced TGFα and HB-EGF Release Differ although Both are ADAM17 Substrates
TGFα cleavage in MEF cells is significantly stronger in response to TPA and hypertonic stress as compared with MLE cells. The opposite is true for HB-EGF, which is cleaved much stronger in MLE cells than in MEF cells in response to the same stimuli. LPA finally induced stronger cleavage of TGFα in MLE cells than in MEF cells and no cleavage of HB-EGF in MLE cells at all (Fig. 1 A and B, and Table 1). Both TGFα and HB-EGF are ADAM17 substrates (Table 3). For such differential cleavage to be regulated on the metalloprotease level, ADAM17 would need to undergo distinct modifications that modulate specificity for one or the other substrate. Because it has been reported that the C terminus of ADAM17 is not required for induced cleavage (6, 16, 17), it is difficult to imagine how intracellular signaling pathways could cause such modifications on the ADAM. Alternatively, regulation could, at least in part, lie on the substrate or the level of a third protein, which interacts either with the ADAM ectodomain or transmembrane domain and/or with the substrate. This would explain how very few metalloproteases very specifically cleave many different substrates expressed at the same time in the same cell.
The available evidence supports this conclusion. Because induced cleavage of surface proteins by TPA occurs within seconds to minutes, it seems unlikely that trafficking or processing regulates induced cleavage by ADAMs. Consistent with this, many studies have shown that cell-surface ADAMs are proteolytically processed and catalytically active (18–22). In addition, overexpression of ADAMs (whether endogenously or transiently by transfection) and substrates often results in significant basal cleavage in unstimulated samples (for example, in the study by Horiuchi et al. (6)). Importantly, overexpression of an ADAM9 mutant lacking the PKC binding domain resulted in basal cleavage and induced shedding of HB-EGF (24), suggesting that ADAM9 is constitutively active and does not require PKC interaction or modification to carry out cleavage. This in turn raises the possibility that PKC activity is required on the level of the substrate or of a third protein. Although the C terminus of ADAM17 may be dispensable for cleavage, the intact ectodomain of ADAM17 is required for TGFα cleavage (16, 17). This suggests that interactions on the level of the ectodomain could be important in the regulation of cleavage, whether mediated by the substrate or a third protein. Tissue inhibitors of metalloproteases require an interaction with the ADAM extracellular catalytic site to carry out their inhibitory function. Whether tissue inhibitors of metalloproteases interaction with ADAMs also affects substrate selectivity is unknown (9, 25). A direct association between the tetraspanin CD9, HB-EGF, and ADAM10 has been shown in bombesin-stimulated COS7 cells (26), but it is not known whether this interaction directs substrate-specific cleavage or modulates protease activity. A mechanism regulating substrate cleavage without affecting protease activity has just been described for ADAM10. ADAM10 is constitutively associated with the ephrin-A3 receptor. Interaction with its membrane-tethered ligand ephrin-A5 on a neighboring cell activates the receptor and induces a conformational change that permits ADAM10 to cleave Eph-A5 in trans (27). These results suggest that ADAM10 is constitutively active and that cleavage can be regulated by phosphorylation events on a third protein (here the receptor) that regulate substrate availability. Whether similar protein-protein interactions could regulate EGF ligand release is unknown.
Cleavage by the Same Stimuli Can Involve Different Metalloproteases Depending on the Substrate
In MEF cells, TGFα shedding in response to TPA and IM depends on ADAM17, although the same stimuli are connected to NRG cleavage by ADAM9. This is a novel observation, because so far only ADAM17 and ADAM10 had been connected to TPA- and calcium-induced shedding (6, 7). ADAM9 and ADAM17 were required for hypertonic stress-induced NRG cleavage and ADAM17 and ADAM10 for TPA- and IM-induced HB-EGF shedding (Table 3). These results speak against the clear assignment of particular metalloproteases to particular stimuli, but rather suggest that they respond to a variety of stimuli. Because cleavage in response to these stimuli is still differentially regulated in a substrate-specific way (yet involving the same metalloprotease), it is unlikely that this regulation occurs only on the metalloprotease level.
ADAM17 is the major sheddase for TGFα and HB-EGF in vivo (28), and ADAM10 is the GPCR-stimulated sheddase for HB-EGF release in several cell types (26, 29). In contrast, in MEF cells, ADAM17 was responsible for GPCR-induced HB-EGF cleavage stimulated by LPA (Table 3), suggesting a certain promiscuity of ADAMs for substrates. ADAM17 and -19 are known to release certain NRG isoforms, and TPA-induced cleavage of NRGβ1 and -2 is linked to ADAM17 (30). Our results in MEF cells identify ADAM9 as a major sheddase of NRG1β in response to TPA, hypertonic stress, LPA, and IM stimulation, with contributions of ADAM17 in the case of hypertonic stress and LPA stimulation. These findings together with ours support the following. First is the conclusion that there are major physiological sheddases specific to particular substrates independent of the stimulus, inferring specific regulation of cleavage beyond the metalloprotease level. Second, depending on the physiological context, some metalloproteases may be able to substitute for another. For example, ADAM10 can substitute in IM-induced cleavage of ADAM17 substrates when ADAM17 function is missing, but TPA-stimulated shedding of ADAM17 substrates could not be rescued with ADAM10 (7). We also found that ADAM17 was required for TPA-induced shedding of TGFα in MEF cells. In contrast, in our hands, IM-induced shedding of TGFα was completely blocked in ADAM17 knock-out cells and could not be rescued by the presence of endogenous ADAM10 in the same cells (quantitative PCR data not shown and Table 3). Third, our findings may imply that ADAMs potentially act in concert as a heterocomplex and that concerted action may be required for cleavage under certain circumstances. We observed a role for ADAM10 and -17 in both TPA- and IM-induced shedding of HB-EGF (albeit at a low rate) and a role for ADAM9 and -17 in hypertonic stress-induced NRG cleavage. This at least raises the possibility of sequential substrate cleavage events by two metalloproteases acting in a complex on the same substrate.
Cleavage of the Same Substrate Depends on the Activity of Different PKC Isoforms
Our inhibitor experiments show that PKC isoforms regulate cleavage in a substrate-, stimulus-, and cell type-specific way. Only in MLE cells, inhibition of atypical PKCζ-enhanced TPA induced release of NRG and inhibition of classical PKC blocked hypertonic stress-induced release of NRG. Only in MEF cells, LPA-induced cleavage of TGFα was partially sensitive to classical PKC inhibition and was completely blocked by the PKCζ inhibitor (Fig. 1, and Tables 1 and 2). Because we have to assume that the C-terminal tail of the ADAM (at least of ADAM17 (16, 17)) is not the target of these modifications, and because PKC does not act within the lipid bilayer or the extracellular space, these findings support the conclusion that signaling pathways introduce one or several modifications on the C termini of the substrate or of a third protein that influence specificity of substrate cleavage. The cytoplasmic domains of NRG, HB-EGF, and TGFα contain potential phosphorylation sites for tyrosine and serine/threonine kinases that could serve as sites for regulatory input. In other reports, PKC, MAPK, and Src kinase inhibitors indeed block induced cleavage of certain substrates in some cells (9, 31, 32). This is in line with our findings of a TPA-induced and PKC-dependent serine phosphorylation on NRG that accumulates before cleavage occurs (Fig. 3). Similarly, it was recently reported that PKC-dependent and ADAM17-mediated cleavage of L-selectin depends on a serine phosphorylation event within the C terminus of L-selectin (33). The cytoplasmic tail of HB-EGF is also phosphorylated on Ser-207 upon TPA, LPA, and IM stimulation, but in contrast to L-selectin, Ala substitution of Ser-207 had no effect on TPA-induced or constitutive ectodomain shedding (31).
The extracellular juxtamembrane domain carrying the substrate cleavage site of TGFα is necessary and sufficient for TPA-stimulated shedding (6), supporting the possibility that cleavage regulation and specificity are not predominantly conferred by substrate-ADAM ectodomain interactions beyond the cleavage site. But because no apparent consensus motif for cleavage of ADAM substrates has been identified (9, 34, 35), specificity of substrate cleavage must still require other determinants. The C terminus of TGFα can be exchanged for the C terminus of betacellulin (an EGF ligand not cleaved by ADAM17) without affecting TPA-induced ADAM17-catalyzed shedding of the TGFα chimera (6). This suggests at least that ADAM17 specificity for TGFα is not dependent on the C terminus of TGFα, but it does not exclude that interactions or modifications on the substrate C terminus regulate cleavage. This has, for example, been described for the cytoplasmic tail of pro-HB-EGF, where interaction with BAG-1 increases soluble HB-EGF release (10, 23).
In summary, we conclude that the major regulatory events occur not only on the level of the metalloprotease but also on the level of the substrate. Induced substrate cleavage probably does not involve the C terminus of the metalloprotease to any significant degree. It is likely that modifications on the metalloprotease, substrate, and/or a third protein act in concert to direct substrate and a constitutively active ADAM to each other. Third proteins could act extracellularly or intracellularly or themselves be class one transmembrane proteins. They may therefore address the ectodomains or the C termini of either substrate or metalloprotease and less likely the transmembrane sections. All mechanisms envisaged here are directed by intracellular signaling pathways that in part involve PKC isoforms. Further studies will need to be carried out to determine which specific serine residues within the C terminus of NRG are phosphorylated, whether there is similar regulatory events on other EGF ligands or other substrates, and how this affects the regulation of substrate cleavage.
Acknowledgments
We thank Dr. Paul Saftig and Dr. Carl Blobel for the kind provision of materials and Dr. Peter Herrlich for critical discussions and for input on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants K99-DK077731-01 and 4 R00 DK077731-03.
- GPCR
- G protein-coupled receptor
- MLE
- mouse lung epithelial
- NRG
- neuregulin 1β
- MEF
- mouse embryonic fibroblast
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- HB-EGF
- heparin-binding EGF
- IM
- ionomycin
- LPA
- lysophosphatidic acid
- ADAM
- a disintegrin and metalloprotease.
REFERENCES
- 1. Murphy G. (2008) Nat. Rev. Cancer 8, 932–941 [DOI] [PubMed] [Google Scholar]
- 2. Blobel C. P., Carpenter G., Freeman M. (2009) Exp. Cell Res. 315, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lautrette A., Li S., Alili R., Sunnarborg S. W., Burtin M., Lee D. C., Friedlander G., Terzi F. (2005) Nat. Med. 11, 867–874 [DOI] [PubMed] [Google Scholar]
- 4. López-Otín C., Matrisian L. M. (2007) Nat. Rev. Cancer 7, 800–808 [DOI] [PubMed] [Google Scholar]
- 5. Arribas J., Bech-Serra J. J., Santiago-Josefat B. (2006) Cancer Metastasis Rev. 25, 57–68 [DOI] [PubMed] [Google Scholar]
- 6. Horiuchi K., Le Gall S., Schulte M., Yamaguchi T., Reiss K., Murphy G., Toyama Y., Hartmann D., Saftig P., Blobel C. P. (2007) Mol. Biol. Cell 18, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fischer O. M., Hart S., Gschwind A., Ullrich A. (2003) Biochem. Soc. Trans. 31, 1203–1208 [DOI] [PubMed] [Google Scholar]
- 9. Seals D. F., Courtneidge S. A. (2003) Genes Dev. 17, 7–30 [DOI] [PubMed] [Google Scholar]
- 10. Lin J., Hutchinson L., Gaston S. M., Raab G., Freeman M. R. (2001) J. Biol. Chem. 276, 30127–30132 [DOI] [PubMed] [Google Scholar]
- 11. Herrlich A., Klinman E., Fu J., Sadegh C., Lodish H. (2008) FASEB J., 22, 4281–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weskamp G., Cai H., Brodie T. A., Higashyama S., Manova K., Ludwig T., Blobel C. P. (2002) Mol. Cell. Biol. 22, 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 14. Bogan J. S., Hendon N., McKee A. E., Tsao T. S., Lodish H. F. (2003) Nature 425, 727–733 [DOI] [PubMed] [Google Scholar]
- 15. Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 16. Reddy P., Slack J. L., Davis R., Cerretti D. P., Kozlosky C. J., Blanton R. A., Shows D., Peschon J. J., Black R. A. (2000) J. Biol. Chem. 275, 14608–14614 [DOI] [PubMed] [Google Scholar]
- 17. Doedens J. R., Mahimkar R. M., Black R. A. (2003) Biochem. Biophys. Res. Commun. 308, 331–338 [DOI] [PubMed] [Google Scholar]
- 18. Black R. A., Rauch C. T., Kozlosky C. J., Peschon J. J., Slack J. L., Wolfson M. F., Castner B. J., Stocking K. L., Reddy P., Srinivasan S., Nelson N., Boiani N., Schooley K. A., Gerhart M., Davis R., Fitzner J. N., Johnson R. S., Paxton R. J., March C. J., Cerretti D. P. (1997) Nature 385, 729–733 [DOI] [PubMed] [Google Scholar]
- 19. Howard L., Maciewicz R. A., Blobel C. P. (2000) Biochem. J. 348, 21–27 [PMC free article] [PubMed] [Google Scholar]
- 20. Lum L., Reid M. S., Blobel C. P. (1998) J. Biol. Chem. 273, 26236–26247 [DOI] [PubMed] [Google Scholar]
- 21. Roghani M., Becherer J. D., Moss M. L., Atherton R. E., Erdjument-Bromage H., Arribas J., Blackburn R. K., Weskamp G., Tempst P., Blobel C. P. (1999) J. Biol. Chem. 274, 3531–3540 [DOI] [PubMed] [Google Scholar]
- 22. Schlöndorff J., Becherer J. D., Blobel C. P. (2000) Biochem. J. 347, 131–138 [PMC free article] [PubMed] [Google Scholar]
- 23. Higashiyama S., Nanba D. (2005) Biochim. Biophys. Acta 1751, 110–117 [DOI] [PubMed] [Google Scholar]
- 24. Izumi Y., Hirata M., Hasuwa H., Iwamoto R., Umata T., Miyado K., Tamai Y., Kurisaki T., Sehara-Fujisawa A., Ohno S., Mekada E. (1998) EMBO. J. 17, 7260–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brew K., Dinakarpandian D., Nagase H. (2000) Biochim. Biophys. Acta 1477, 267–283 [DOI] [PubMed] [Google Scholar]
- 26. Yan Y., Shirakabe K., Werb Z. (2002) J. Cell Biol. 158, 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janes P. W., Wimmer-Kleikamp S. H., Frangakis A. S., Treble K., Griesshaber B., Sabet O., Grabenbauer M., Ting A. Y., Saftig P., Bastiaens P. I., Lackmann M. (2009) PLoS Biol. 7, e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peschon J. J., Slack J. L., Reddy P., Stocking K. L., Sunnarborg S. W., Lee D. C., Russell W. E., Castner B. J., Johnson R. S., Fitzner J. N., Boyce R. W., Nelson N., Kozlosky C. J., Wolfson M. F., Rauch C. T., Cerretti D. P., Paxton R. J., March C. J., Black R. A. (1998) Science 282, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 29. Prenzel N., Zwick E., Daub H., Leserer M., Abraham R., Wallasch C., Ullrich A. (1999) Nature 402, 884–888 [DOI] [PubMed] [Google Scholar]
- 30. Horiuchi K., Zhou H. M., Kelly K., Manova K., Blobel C. P. (2005) Dev. Biol. 283, 459–471 [DOI] [PubMed] [Google Scholar]
- 31. Wang X., Mizushima H., Adachi S., Ohishi M., Iwamoto R., Mekada E. (2006) Cell Struct. Funct. 31, 15–27 [DOI] [PubMed] [Google Scholar]
- 32. Maretzky T., Zhou W., Huang X. Y., Blobel C. P. (2011) Oncogene 30, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Killock D. J., Ivetiæ A. (2010) Biochem. J. 428, 293–304 [DOI] [PubMed] [Google Scholar]
- 34. Caescu C. I., Jeschke G. R., Turk B. E. (2009) Biochem. J. 424, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turk B. E. (2009) Methods Mol. Biol. 539, 79–91 [DOI] [PubMed] [Google Scholar]