Abstract
Much progress has been made concerning histone function in the nucleus; however, following their synthesis, how their marking and subcellular trafficking are regulated remains to be explored. To gain an insight into these issues, we focused on soluble histones and analyzed endogenous and tagged H3 histones in parallel. We distinguished six complexes that we could place to account for maturation events occurring on histones H3 and H4 from their synthesis onward. In each complex, a different set of chaperones is involved, and we found specific post-translational modifications. Interestingly, we revealed that histones H3 and H4 are transiently poly(ADP-ribosylated). The impact of these marks in histone metabolism proved to be important as we found that acetylation of lysines 5 and 12 on histone H4 stimulated its nuclear translocation. Furthermore, we showed that, depending on particular histone H3 modifications, the balance in the presence of the different translocation complexes changes. Therefore, our results enabled us to propose a regulatory means of these marks for controlling cytoplasmic/nuclear shuttling and the establishment of early modification patterns.
Keywords: Histone Methylation, Histone Modification, Histones, Nuclear Translocation, Protein Purification
Introduction
Eukaryotic DNA in the form of chromatin utilizes two copies of each of the four histones, H2A, H2B, H3, and H4, to wrap 147 bp of DNA for each basic unit, the core nucleosome (1–3). Besides compaction of the genetic material, the nucleosome also provides information that can regulate many functions operating on the DNA. Part of this information is conveyed via the choice of histone composition, as histone variants, and through distinct combinations of post-translational modifications that can mark specific chromatin domains, enabling key regulatory regions to be identified (4, 5).
In mammals, histone H3 is present in five major forms. H3.1 and H3.2, the replicative histones, are expressed in the S phase and provide the main supply for chromatin assembly during DNA replication. Histone H3.1 is incorporated into chromatin in a DNA replication-dependent manner by the histone chaperone CAF-13 (chromatin assembly factor 1) (6). In contrast, H3.3, the replacement histone, is synthesized throughout the cell cycle and is incorporated into the chromatin independently of DNA replication by the histone chaperones HIRA (histone regulation A) (7, 8) and Daxx (death-associated protein) (8–10), and it is enriched in marks associated with transcriptional activation (6). Much has been learned about the nuclear complexes containing H3; however, a current challenge is to gain an understanding of how these histones are taken care of before becoming incorporated into these complexes to obtain a clearer picture concerning the cellular trafficking of histones.
After synthesis in the cytoplasm histone proteins are transported into the nucleus. Thus, a cytoplasmic/nuclear shuttling mechanism should be at work. Accordingly, in yeast, the nuclear localization signals (NLS) of histones H3 and H4, located at their N termini, are sufficient for both nuclear accumulation and interactions with the nuclear import proteins Kap123p (karyopherin 123) and Kap121p (karyopherin 121) (11). In humans, a recent report showed that after the tagged histone H3.1 is synthesized it sequentially passes through four different cytosolic complexes to assemble the H3-H4 dimers, to acquire the H4 acetylation pattern of newly synthesized H4, that is, the acetylation of lysines 5 and 12, and finally to interact with the nuclear translocation protein Importin4 (12). Interestingly, we previously reported that nucleosomal histone H3 modification patterns can be influenced by an earlier modification that occurs before histones are incorporated into the chromatin, i.e. predeposition histones (13). Although some of the marks present in predeposited histones have already been characterized, very little is known about their functional importance. As an example, we showed that histone H3 monomethylated at the residue K9 (H3K9me1), the only methylation mark detectable on soluble histone H3, is critical for the formation of the heterochromatic H3K9me3 marking (14). Therefore, the role of the other soluble post-translational modifications (PTMs) should be explored, and this is particularly crucial for our understanding of how these marks may impact on the regulation of histone traffic in the cell.
Here, we have been able to distinguish six different endogenous soluble histone H3 and H4 complexes. We assigned specific marks for each of them. We also examined how these marks influenced the importin/histone interaction that we uncovered. This enabled us to propose a regulatory means of these marks for controlling cytoplasmic/nuclear shuttling.
EXPERIMENTAL PROCEDURES
Antibodies
ASF1a/b (15), CAF-1/p150 (Abcam ab7655), Daxx (Santa Cruz Biotechnology sc-7152), HA (Roche Applied Science 1-867-423), histone acetyltransferase 1 (HAT1) (Abcam ab12164), His (BD Biosciences 8916-1), histone H3 (Abcam ab7834), H3K9K14ac (Upstate 06-599), H3K9me1 (Upstate 07-450), histone H4 (Abcam ab10158), H4K12ac (Upstate 06-761), heat shock cognate protein 70 (Hsc70) (Abcam ab19136), heat shock protein 70 (Hsp70) (Cell Signaling 4876), Hsp90 (Santa Cruz Biotechnology sc-7947), Importin4 (Abcam ab283887), Importin5 (Santa Cruz Biotechnology sc-11369), and poly(ADP-ribose) (Upstate mAB3192) were used. For Western blot analysis the primary antibodies were detected with a horseradish peroxidase-conjugated secondary antibody, developed with enhanced chemiluminescence (ECL, Pierce), and exposed onto an x-ray film. For Figs. 3A and 4A, the Western blotting was carried out with the histone H3 antibody followed by the histone H4 antibody and developed together.
FIGURE 3.
Histones H3 and H4 interact with Importin4. A, top, scheme illustrating the Importin pulldown assay. Bottom, Western blot of the pulldown assay. B, top, scheme illustrating the GST-H4 pulldown assay in the presence of Importin4. Bottom, Western blot of the pulldown assay. Ponceau Red staining provided the loading control for the reaction. C, representative images of the transfection of HeLa cells with the constructs EYFP, H3-NLS-EYFP, and H4-NLS-EYFP, visualized by fluorescence microscopy. Left, images of the cells expressing EYFP and H3-NLS-EYFP or H4-NLS-EYFP quantified to obtain the number of cells that showed enrichment of fluorescence in the nucleus (N) or in the cytoplasm (C). Right, N/C ratio values of H3-NLS-EYFP and H4-NLS-EYFP normalized with the EYFP N/C ratio. The S.D. was derived from the quantification of at least 300 cells for each construct. D, top, scheme illustrating the Importin4 immunoprecipitation assay in the presence of the Ran mutant RanQ69L, which was unable to hydrolyze GTP. Bottom, Western blot of the sample from the immunoprecipitation assay.
FIGURE 4.
Function of the soluble histone H3 and H4 PTMs. A, top, scheme illustrating the Importin pulldown assay. Prebound Ni2+-agarose beads-Importin4 were incubated with increasing amounts of the unmodified or tetraacetylated (K5K8K12K16) histone H4 peptide (residues 2–20), followed by incubation with the H3-H4 histones. Bottom, Western blot of the pulldown assay. B, nuclear import assay performed in the presence of H4-NLS-EYFP (left) and the mutant H4K5K12Q-NLS-EYFP (right), in the absence (top) or presence (bottom) of the reticulocyte extract. Middle, images of the cells incubated with H4-NLS-EYFP (WT) and the mutant H4K5K12Q-NLS-EYFP (K5K12Q) quantified to obtain the mean fluorescence intensity of the nucleus. The enrichment fold was calculated taken the fluorescence intensity of H4-NLS-EYFP as 1. The S.D. was derived from the quantification of at least 50 cells. The graph is a representation of three independent experiments. Right, Coomassie Blue-stained gel of increasing amounts of H4-NLS-EYFP and the mutant H4K5K12Q-NLS-EYFP proteins. C, methylation assay performed using H3-NLS-EYFP and H3K14QK18Q-NLS-EYFP proteins and recombinant SetDB1 and detected by antibodies against H3K9me1. Ponceau Red is shown as the loading control. D, model for the establishment of soluble H3 and H4 histone modifications. PAR, poly(ADP-ribose).
Cell Lines
FLAG-HA-H3.1- and H3.3-expressing HeLa S3 cell lines were grown as described in Ref. 7. HeLa cells grown in DMEM containing 10% FBS and penicillin/streptomycin were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
FLAG-tagged Complex Purification
The H3.1 and H3.3 complexes were purified from a cytosolic extract (16) by immunoprecipitation using anti-FLAG antibody-conjugated agarose beads. Peptides derived from the trypsin digestion of the complexes were analyzed by mass spectrometry. For density gradient sedimentation, the FLAG peptide-eluted material was loaded onto 5 ml of a 10–40% glycerol gradient and spun at 45,000 rpm in a Beckman SW55Ti rotor for 16 h. Then, 250-μl fractions were collected.
Endogenous Complex Purification
Approximately 25 mg of cytosolic HeLa extracts (donated by D. Reinberg) were loaded onto a Tosoh Bioscience TSK-DEAE-5PW (7.5-mm × 7.5-cm) column, equilibrated with a buffer containing 50 mm KCl. The column was washed with the same buffer and eluted with a linear gradient of salt from 50 to 700 mm KCl. The peak of each complex was loaded onto a GE Healthcare Superdex 200 10/300 GL column equilibrated with a buffer containing 500 mm KCl plus 0.01% Nonidet P-40.
Plasmids
The H3- and H4-NLS-EYFP reporter constructs were made by cloning the histone H3 and H4 NLS residues 1–28 and 1–20, respectively, by PCR into the pEYFP-N1 construct. For bacterial expression, the H3- and H4-NLS-EYFP sequences were subcloned to the pET28a(+) vector. Mutations to histones H3- and H4-NLS were performed using the QuikChangeTM Site-directed Mutagenesis kit (Stratagene).
Protein Purification
Bacterially expressed His-tagged Importin4 (construct from D. Görlich) and His-tagged H3- and H4-NLS-EYFP were purified by standard nickel affinity purification. Recombinant FLAG-tagged SET domain bifurcated 1 (SetDB1) was expressed from baculovirus (donated by D. Schultz) in Sf9 cells and purified by standard M2 affinity purification.
Importin/Histone Binding Assay
Immobilized recombinant His-tagged Importin4 on Ni2+-agarose beads was incubated with histones H3/H4 for 1 h, after which the beads were washed as indicated. Bound proteins were loaded directly onto a 5–18% gradient SDS-PAGE and analyzed by Western blotting.
Competition Assay
10 pmol of immobilized recombinant His-tagged Importin4 on Ni2+-agarose beads was incubated with 2000 or 3000 pmol of the unmodified or 1000, 2000, or 3000 pmol of the acetylated histone H4 peptides at lysine 5, 8, 12, and 16, corresponding to amino acids 2–20 (Upstate) for 30 min at 4 °C. Then beads were recovered and incubated with 200 pmol of (H3-H4)2 for 1 h at 4 °C. Beads were then washed twice with 500 mm KCl, 0.15% Nonidet P-40, and 25 mm imidazole, once with 50 mm KCl, and analyzed by Western blotting.
Histone Methyltransferase Assay
This assay was performed using recombinant SetDB1 and either 4 μg of wild-type H3-NLS-EYFP or 4 μg of the mutant H3K14K18Q-NLS-EYFP.
Nuclear Translocation Assay
The nuclear translocation assay was performed according to Ref. 17. In brief, HeLa cells grown on a Lab-Tek® Chamber SlideTM were washed in cold translocation buffer (20 mm Hepes, pH 7.3, 110 mm potassium acetate, 5 mm sodium acetate, 2 mm DTT, 1 mm EGTA, 1 mm ATP, 5 mm creatine phosphate, 20 units/ml creatine phosphokinase, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) and permeabilized with 40 μg/ml digitonin for 5 min. After washing, the cells were incubated with 50% rabbit reticulocyte extract (Promega), 1 μg of Importin4, and either 1 μg of wild-type H4-NLS-EYFP or 1 μg of the mutant H4K5K12Q-NLS-EYFP for 20 min at 28 °C. The cells were then washed and fixed with 3.7% formaldehyde for 15 min at room temperature and observed using an Olympus IX71 microscope.
RESULTS
Isolation of Cytosolic Tagged H3.1 and H3.3 Complexes
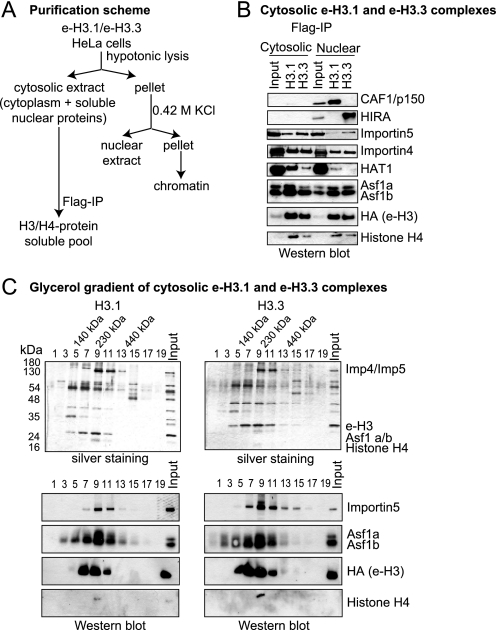
We used classical biochemical fractionation to generate cytosolic and nuclear extracts from epitope-tagged H3.1 and H3.3 histones expressing cell lines (e-H3) from a constitutive viral promoter expressing low levels of histones, as previously described (13). This way, we could focus on the cytosolic fraction as a means of identifying novel partners and important modifications prior to incorporation into chromatin in the early stages after histone biogenesis. We isolated e-H3.1 or e-H3.3 cytosolic complexes by affinity purification (Fig. 1, A and B). Combined analysis of Western blot and Coomassie Blue staining of the isolated complexes followed by mass spectrometry allowed the identification of different e-H3-associated polypeptides (Fig. 1B, supplemental Fig. 1A, and data not shown). In terms of the histones, they only contained histone H4 and the tagged version of the variant, indicating that dimeric forms of H3-H4 are established prior to the formation of the predeposition complex. We also identified the cytoplasmic/nuclear shuttling proteins Importin4 and Importin5, the heat shock chaperones Hsp90 and Hsc70, the histone acetyltransferase HAT1, and the histone chaperone Asf1a/b (anti-silencing function 1). In agreement with a recent report (8), we found the Daxx protein to be exclusively associated with histone H3.3 (supplemental Fig. 1A). Interestingly, the cytosolic and nuclear H3 complexes were different; whereas nuclear histone H3.1 associated with the chaperone CAF-1 and H3.3 with HIRA, the cytosolic counterparts did not (Fig. 1B), suggesting distinct functions. To confirm the stable association of the isolated H3 complexes, FLAG immunoprecipitates were run on a glycerol gradient. Silver staining and Western blots of samples derived from the gradient corroborated the co-migration of Importin, Asf1a/b, e-H3, and H4 at a size of ∼250 kDa (Fig. 1C, fraction 9). Interestingly, the elution of e-H3 was broader than the other proteins analyzed, suggesting the existence of more than one e-H3 complex. Thus, we concluded that cytosolic histone H3.1 and H3.3 complexes are different from the nuclear complexes and are likely to be involved in the processing of histones and translocation to the nucleus. For simplicity, from now on we will call this histone pool “soluble histones.”
FIGURE 1.
Isolation of the cytosolic e-H3.1 and e-H3.3 complexes. A, scheme illustrating the purification procedure for obtaining the cytosolic e-H3.1 and e-H3.3 complexes. B, Western blots of the cytosolic and nuclear e-H3.1 and e-H3.3 complex subunits, as indicated. Input corresponded to 10% of the extract utilized on immunoprecipitation (IP). C, silver staining (top) and Western blot (bottom) analysis of the fractions derived from the glycerol gradient of cytosolic e-H3.1 (left) and e-H3.3 (right) complexes. Fraction 1 corresponded to the top of the gradient and fraction 19 to the bottom. It should be pointed out that histone H4 does not stain well in silver staining, and the HA antibody used to detect e-H3 is more sensitive than the one used to detect histone H4, explaining why histone H4 is only detected in fraction 9 of the glycerol gradient.
Endogenous Soluble Histone H3 and H4 Complexes
With the idea of analyzing endogenous histone H3 complexes and corroborating our findings with the tagged version of histone H3, we fractionated cytosolic HeLa extracts over an anion-exchange resin, and the bound material was eluted with a linear gradient of salt (Fig. 2A). We then performed Western blot analyses of histones H3 and H4 and of the proteins identified in the tagged-histone H3 purification of samples derived from the fractionation (Fig. 2B). To our surprise, we found five peaks of histone H3 eluting at different salt concentrations, and each peak had a different set of associated proteins. We confirmed the existence of the complexes by running them through a gel-filtration column (Fig. 2C). Based on the associated proteins and the modifications observed in histones H3 and H4, we deduced the existence of a temporal order of processing of histones H3 and H4. The identified complexes would explain the proposed cascade of events that would occur to the histones while they are in the cytoplasm. The events start from their protein synthesis through to their translocation to the nucleus. Although our model is based on indirect evidence, it is consistent with the study of Campos et al. describing the existence of several cytosolic histone H3 complexes (12). For simplicity, we will thus use the same nomenclature system to refer to the complexes that we identified in the present study.
FIGURE 2.
Isolation of soluble endogenous histone H3 and H4 complexes. A, scheme illustrating the purification procedure for obtaining the endogenous soluble histone H3 and H4 complexes. B, Western blots of the fractions derived from the DEAE-5PW column, as indicated. The elution of the six different histone complexes is indicated, with their corresponding names. Below, all of the fractions between fraction 17 and fraction 23 were Western blotted to resolve Complex IVa and Complex IVb. C, Western blot analysis of the fractions derived from the gel-filtration column: Complexes Ia, Ib, II, and III, blotted as indicated. On top, elution of the molecular markers is indicated. D, Western blot analysis of fractions derived from the DEAE-5PW column, blotted as indicated. PAR, poly(ADP-ribose). Dotted lines indicate different autoradiograms.
We first identified Complex Ia (Fig. 2B, fraction 13, Fig. 2C, fraction 27, and supplemental Fig. 1B), composed of histone H3 and the chaperone Hsc70, with a size of about 250 kDa. Interestingly, Importin4 is not present in the complex. Then, we identified Complex II (Fig. 2B, fraction 37, and Fig. 2C, fraction 29), composed of histone H3, histone H4, and the chaperone Hsp90, with a size of about 250 kDa. Although Hsc70 was detected on the analyzed fractions, its elution profile did not correlate with the elution of the complex (Fig. 2C, fraction 29). Complex III, on the other hand (Fig. 2B, fraction 19, and Fig. 2C, fraction 33), was of a size of about 180 kDa and was composed of histone H3, histone H4, and HAT1. In contrast to the previous report (12), we also identified the chaperone Asf1a, but not Asf1b. This was missed most likely because Campos et al. used an antibody specific for Asf1b. Finally, we identified two types of Complex IV; Complex IVa eluted at a salt concentration of 200 mm KCl (Fig. 2B, bottom, fraction 18) and was composed of Importin4, Asf1a, and histones H3 and H4. On the other hand, Complex IVb eluted at a salt concentration of 180 mm KCl (Fig. 2B, bottom, fraction 17) and was formed by Importin4, Asf1b, and histones H3 and H4. Interestingly, the H3 histones on Complexes IVa and IVb had different post-translational modifications (see below).
Given that Complex Ia only had histone H3 and did not show a significant amount of histone H4, we hypothesized the existence of a similar complex for histone H4, in which histone H3 would not be present. Therefore, we carefully looked at the fractionation using the DEAE-5PW column and found a complex with these properties in fraction 29 (Fig. 2B). On a gel-filtration column, Complex Ib eluted at a size of about 250 kDa (Fig. 2C, fractions 25–29), apart from Importin4 (Fig. 2C, fraction 17). Mass spectrometry on fraction 29 identified the heat shock chaperones Hsp90 and Hsp70 as part of the complex (data not shown), which was confirmed by Western blot analysis on fractions derived from the gel-filtration column (Fig. 2C). The fact that the endogenous histone H3 complexes had similar sizes ranging between 180 and 250 kDa explains why we did not split the five histone H3 complexes when we purified the tagged histone H3 complex over the glycerol gradient.
Stepwise Establishment of Soluble Histone H3 and H4 PTMs
We previously characterized the PTMs of the predeposited H3 and H4 histones and showed that they are both acetylated and that histone H3 is methylated at the residue lysine 9 (13). Therefore, we investigated the histone marks in the different soluble complexes (Fig. 2, B and D). Interestingly, we found that each complex had a different set of modifications, of which H3K9me1 was the first mark that we detected, which had already been observed in Complex Ia (Fig. 2B). A closer look at this band using Western blotting for histone H3 and its comparison with the H3 bands derived from the other complexes showed an intriguing fact: it was a lower migrating form, indicating that histone H3 from Complex Ia was slightly heavier than in the rest of the complexes (Fig. 2D). We performed Western blot analysis of several marks that could possibly explain the difference in the migration on SDS-polyacrylamide gels and found that histone H3 in Complex Ia is poly(ADP-ribosylated), which is a novel predeposition mark (Fig. 2D, fraction 13). Even more interestingly, histone H3 derived from Complex II was still poly(ADP-ribosylated), but the band had migrated as usual, indicating that histone H3 had lost some of its ADP-riboses (Fig. 2D, fraction 37). This modification was not observed in the following complexes (III and IV). Histone H4, on the other hand, was also found to be poly(ADP-ribosylated) in Complex Ib (Fig. 2C), and this modification was undetectable in Complex II (data not shown). Therefore, the data indicated that poly(ADP-ribosylation) is present in the H3 and H4 histones derived from Complex I and that it is removed when histones H3 and H4 get together. We then continued the analysis and found that histone H3 was acetylated in Complex II (Fig. 2D). Given that the antibody used was raised against acetylated histone H3K9 and lysine 14 and the non-nucleosomal histone H3 is acetylated at residues lysines 14 and 18 (13, 18), the H3 acetylation observed corresponded to H3K14ac. Although we performed Western blot analysis on the sizing column of Complex II, we could not detect H3K14ac because it was below the detection limit. However, Fig. 2D showed that H3 is acetylated in fraction 37, which corresponded to the input of the sizing column, indicating that histone H3 in Complex II is acetylated at lysine 14. Then, we found that histone H4 established its typical H4K5K12ac predeposition pattern in Complex III, where H4 interacts with the enzyme HAT1 (Fig. 2B). From Complex III, the histones passed to Complex IV, where they finally interacted with Importin4 for their nuclear translocation. However, we found two types of Complex IV containing different modification patterns: Complex IVa contained H3K14ac, H3K9me1, and H4K5K12ac, whereas Complex IVb contained H3K9me1 and H4K5K12ac (Fig. 2B and supplemental Fig. 1), suggesting the existence of different nuclear translocation complexes for H3K14ac and H3K9me1. Taken altogether, we concluded that the establishment of the histone H3 and H4 marks occurs in a sequential manner from one complex to the next, in a cascade of maturation events that start at the time of histone synthesis.
Histones H3 and H4 Interact Directly with Importin4
The presence of Importin4 in the soluble H3 Complex IV suggested that the function of this complex is to translocate histones H3 and H4 to the nucleus. To investigate this, we first analyzed how histones and Importin interact. Pulldown experiments using His-Importin as bait and either the cytosolic extract or the tagged complexes showed that histone H3 interacts with Importin4 and Importin5 (supplemental Fig. 2A). Pulldown analysis using His-Importin and recombinant histones H3 and H4 demonstrated that there is a direct interaction between Importin4 and Importin5 and histones H3 and H4 (Fig. 3A and supplemental Fig. 2B). In the case of histone H4, the interaction occurred through the N-terminal region (Fig. 3B and supplemental Fig. 2C). Accordingly, in yeast, the NLS corresponds to amino acids 1–28 and 1–21 of histones H3 and H4, respectively (11). Given that these regions are identical in yeast and mammals, we tested whether or not they would correspond to the mammal NLS. With this aim, we expressed amino acids 1–28 and 1–21 of histones H3 and H4, respectively, as fusion proteins with the EYFP in HeLa cells. In contrast to the cellular distribution of EYFP, both H3- and H4-NLS efficiently accumulated the EYFP protein in the nucleus (Fig. 3C). Quantification of the data showed that the nuclear/cytoplasmic ratio of H3- and H4-NLS was 6 and 2, respectively. Taken altogether, our results indicated that, similar to yeast, amino acids 1–28 and 1–21 of histones H3 and H4, respectively, are sufficient for nuclear translocation in human cells and correspond to the NLS.
The Interaction between Importin and Histone Is Regulated by the GDP/GTP Ratio
In the cell, the interaction between Importin and its cargo protein is regulated by the small GTPase Ran, giving the directionality of the transport. Ran is predominantly bound to GTP in the nucleus and to GDP in the cytoplasm. In the nucleus, the association between Importins and RanGTP results in the dissociation of its cargo (19). To check whether or not the interaction between Importin and histones is regulated in this way, we performed an immunoprecipitation of the tagged H3 complex with antibodies against Importin5 in the presence and absence of the mutant RanQ69L/GTP, a constitutively GTP-bound form of Ran. We found that histone H3 co-immunoprecipitated with Importin antibodies (Fig. 3D). In contrast, only Importin was immunoprecipitated in the presence of the RanQ69L/GTP mutant, indicating that the RanGTP form competed with histone H3 for the binding of Importins (Fig. 3D). This finding provides support for the idea that the soluble H3 Complex IV is involved in nucleo-cytoplasmic transport and that histones in the nucleus are released from Importins by RanGTP.
Regulation of Nuclear Translocation by H4 Acetylation
The newly synthesized H4 histone is acetylated at residues 5 and 12 located within its NLS (13, 20); therefore, we asked whether or not acetylation regulated the interaction between histones and Importin and the subsequent nuclear translocation process. For this, we performed pulldown assays using His-Importin4 and recombinant H3 and H4 histones in the presence of histone H4 peptides corresponding to the amino acids 2–20 that were either unmodified or acetylated at lysines 5, 8, 12, and 16. We found that unmodified H4 peptides competed at some level with the full-length H3 and H4 histones for the binding to Importin4 (Fig. 4A). Interestingly, acetylated peptides showed a strong competition with the full-length H3 and H4 histones for the binding to Importin4 (Fig. 4A). This result indicated that the affinity of Importin4 for the acetylated H4 histone is stronger than for the unmodified H4 histone. We then set out to test whether or not acetylation of K5 and K12 on the H4 histone resulted in an improvement of its nuclear translocation process. For this, we performed an in vitro nuclear translocation assay in which HeLa cells were permeabilized with digitonin, which, at low concentrations, selectively perforates the plasma membrane releasing cytosolic components from the cell while the nuclear envelope remains intact (17). The NLS-dependent nuclear protein import could then be observed by complementation with the reticulocyte extract. We first evaluated the integrity of the nuclear envelope by incubating the permeabilized cells with the recombinant fusion protein H4-NLS-EYFP. The plasma membrane of digitonin-treated cells was freely permeable to the H4-NLS-EYFP protein, given that H4-NLS-EYFP surrounded the nuclear envelope (Fig. 4B). When the cells were incubated with the fusion protein in the presence of the reticulocyte extract and recombinant Importin4, H4-NLS-EYFP was enriched in the nucleus (Fig. 4B). This accumulation was ATP-dependent, because nuclear accumulation was reduced when the extract was depleted of ATP by apyrase (supplemental Fig. 3A). In addition, the removal of Importin4, the presence of RanQ69L, and treatment of the reticulocyte extract at 65 °C or incubation of the reaction at 4 °C inhibited nuclear translocation of the fusion protein, demonstrating the specificity of the assay (supplemental Fig. 3A). Finally, lectin wheat germ agglutinin, a specific inhibitor of nuclear transport (17), blocked the nuclear accumulation of the fusion protein (supplemental Fig. 3B).
Therefore, we then checked the effect of histone H4 lysine 5 and 12 acetylation on nuclear translocation. We analyzed a mutant H4-NLS-EYFP fusion protein in which residues 5 and 12 of the H4 histone were mutated to glutamine, which mimics acetylation (H4K5K12Q-NLS-EYFP). As with the wild-type H4-NLS, we found that the H4K5K12Q-NLS-EYFP protein added alone to the permeabilized cells surrounded the nuclear envelope (Fig. 4B), whereas it accumulated in the nucleus in the presence of the reticulocyte extract and Importin4 (Fig. 4B). The quantification of the nuclear fluorescence of several images from different experiments showed that the H4K5K12Q mutant was enriched by about 2-fold compared with the wild-type protein (Fig. 4B, graph) when the same amount of protein was added in the nuclear transport assay (Fig. 4B, right). Taken altogether, these results indicated that acetylation of the lysine 5 and 12 residues of the H4 histone stimulates nuclear import via increasing the affinity of histone H4 to Importin4.
Negative Cross-talk between H3K9me1 and H3K14K18ac
Soluble histone H3 is acetylated at lysine 14 and 18, and this acetylation is enriched in the histone variant H3.3, consistent with a role in transcriptional activation (13). In contrast, H3K9me1, enriched in the variant H3.1, is involved in the formation of the heterochromatic H3K9 methylation pattern (14). We showed here that there are two different H3 nuclear translocation complexes, in which the histone H3 in Complex IVa is both acetylated and K9me1, whereas in Complex IVb it is only K9me1. Based on all of these observations, we hypothesized the existence of a negative cross-talk between H3K9me1 and H3K14K18ac that regulates the establishment of these two modifications. To investigate this, we expressed and purified the wild-type H3-NLS-EYFP fusion protein and a mutant protein containing the lysine 14 and 18 mutated to glutamine, which mimics acetylation (H3K14K18Q-NLS-EYFP). With these two proteins, we performed an in vitro methylation assay with recombinant SetDB1, the enzyme responsible for the monomethylation of H3K9 (13). We observed that the wild-type H3-NLS-EYFP was strongly methylated at lysine 9, whereas methylation was inhibited on the mutant mimicking acetylation at lysine 14 and 18 (Fig. 4C). Therefore, we concluded the existence of a negative cross-talk between H3K9me1 and H3K14K18ac.
DISCUSSION
In the present work, we characterized the histone H3 and H4 line of production from histone synthesis through to their nuclear translocation and defined the steps where the establishment of the soluble modifications occurs. In addition, we investigated the impact of soluble marks on the metabolism of histones.
Soluble Histones H3 and H4 Exist in Different Complexes
Our data showed the existence of several soluble human histone H3 and H4 complexes, in agreement with a recent report (12). Furthermore, our analysis proposes the stepwise maturation events that occur on soluble histones. These maturation events involve an association with different chaperones that assist in histone folding. Thus, while histones H3 and H4 are being synthesized, polypeptides interact with the chaperones Hsc70 and Hsp90/70, respectively, to keep the histone polypeptides folded, avoiding aggregation (Complex I). We showed here for the first time that newly synthesized histone H4 associates first with the chaperone Hsp90/70 to form Complex Ib, in the absence of histone H3. Then, histones H3 and H4 come together to form dimers, assisted by the chaperones Hsp90 and, as shown in a recent report (12), by tNASP (nuclear autoantigenic sperm protein) (Complex II). Once the dimer is formed, the histones interact with the histone chaperone Asf1a, HAT1, and, as shown in Ref. 12, by sNASP (Complex III), from which the histones are delivered to Importin4 finally to translocate to the nucleus (Complex IV). It is worth noting that histones appear to be constantly under the surveillance of the specific chaperones involved in each step of histone metabolism, avoiding aggregation of individual polypeptides and supporting the formation of H3-H4 dimers, the establishment of PTMs, and nuclear translocation.
Sequential Establishment of Soluble H3 and H4 Marks
We showed that post-translational modifications of the soluble histones H3 and H4 occur in a sequential manner while passing through each one of the chaperone complexes, as illustrated in Fig. 4D. We observed the following marks.
Histones H3 and H4 Poly(ADP-ribosylation)
We showed here for the first time that soluble histones H3 and H4 are transiently poly(ADP-ribosylated). Intriguingly, we did not observe poly(ADP-ribosylation) on the tagged histone H3, suggesting that exogenous H3 is not poly(ADP-ribosylated) and might be missing a regulatory point. Interestingly, this mark was already present on Complex I, suggesting that it occurs while the histones are being synthesized. Although, in general, poly(ADP-ribosylation) is related with cell survival and cell death programs (21), we do not know the role of this mark. Given that poly(ADP-ribosylation) is imposed when histones are without their partner histone and that it is then removed when histones H3 and H4 get together and assemble the dimers, we speculate that poly(ADP-ribosylation) might help to keep histones H3 and H4 folded in the absence of the other histone, a hypothesis that should be explored in the future.
Histone H3K9me1
The H3K9me1 mark was also observed in Complex Ia, indicating that, like with poly(ADP-ribosylation), this modification occurs while histone H3 is being synthesized. We previously showed that this mark was enriched in the histone variant H3.1 (13) and that it is involved in the formation of the heterochromatic H3K9me3 pattern (14). Therefore, the reason why this modification occurs so early in the line of histone production is very intriguing. However, we also showed here the existence of negative cross-talk between H3K9me1 and H3K14K18ac; where H3K14K18ac is a mark associated with transcriptional activation (22). It is tempting to speculate that the competition between these two marks defines an early establishment of histone H3 PTM patterns. Indeed, the finding of nuclear translocation complexes (Complex IV) that differ in histone PTMs is in agreement with this idea.
Histone H3ac (Lysine 14 and 18)
Histone H3 was acetylated at the lysine 14 and 18 residues in Complex II. We do not know whether there is a role for this modification in the cytoplasm; at least, it does not affect nuclear translocation (data not shown). We speculate that as H3K9me1, H3K14K18ac helps to establish nucleosomal histone H3 PTM patterns. In addition, the findings that H3K9me1 is put onto histone H3 as the first modification and acetylation comes later fit with a recent report showing that the histone H3 associated with cytosolic Asf1b was found monomethylated at the lysine 9, whereas H3 acetylation (lysine 14 and 18) was stronger in the nuclear Asf1b complex (18).
Histone H4K5 and H4K12 Acetylation
The last modification to be imposed before histones translocate to the nucleus is the acetylation of lysines 5 and 12 of histone H4, a pattern that is conserved in several species and is characteristic of newly synthesized histone H4 (20). We showed here for the first time that this acetylation stimulates the interaction between histones and Importin4, which results in enhancement of the nuclear translocation.
Different Translocation Complexes
The last step in the maturation assembly line of the soluble H3 and H4 histones is the association of histones with Importin4 for their translocation to the nucleus (Complex IV). Interestingly, we identified two types of Complex IV. We believed that they are different complexes because: (i) they showed a different behavior on the anion-exchange resin DEAE-5PW, as they eluted at different salt concentrations; (ii) they associated with different Asf1 isoforms; and (iii) the modifications of histone H3 were different. Complex IVa contained Asf1a, H3K9me1, and H3K14ac, whereas Complex IVb contained Asf1b and H3K9me1. This is interesting because a recent report showed that Asf1a and Asf1b have different behavior, so that Asf1b has a specific function in the proliferating capacity of the cell (23). Studies will need to characterize Asf1a and Asf1b complexes.
On the other hand, it has been reported that HIRA, the H3.3 chaperone, interacts with Asf1a, but not Asf1b (24). Therefore, there might be a link between Complex IVa (H3K14K18ac and Asf1a) and HIRA, and it is tempting to speculate that once Complex IVa is translocated to the nucleus, H3K14K18ac interacting with Asf1a will then interact with HIRA. This is consistent with the observation that nucleosomal H3.3 is enriched in acetylated forms (13, 25). Although we have not characterized the histone H3 variants present in the complexes, it is tempting to speculate that Complex IVa translocates the histone variant H3.3, and experiments will be designed to investigate this. Taken altogether, our results illustrate the possibility that histone chaperones might have more than a supporting role, by establishing, integrating and possibly transferring information.
Supplementary Material
Acknowledgment
We thank Adam Cook for helpful comments on the manuscript.
This work was supported by Fondo Nacional de Desarrollo Científico y Tecnológico Grant 1090270, Basal Project PFB16, and the Iniciativa Científica Milenio-Millenium Institute of Fundamental Applied Biology (to A. L.) and by grants from the Institut Curie/CNRS (to G. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- CAF-1
- chromatin assembly factor 1
- Asf
- anti-silencing function
- Daxx
- death-associated protein
- EYFP
- enhanced yellow fluorescent protein
- HAT
- histone acetyltransferase
- HIRA
- histone regulation A
- Hsc
- heat shock cognate
- Hsp
- heat shock protein
- NLS
- nuclear localization signal
- PTM
- post-translational modification
- SetDB1
- SET domain bifurcated 1.
REFERENCES
- 1. Kornberg R. D. (1974) Science 184, 868–871 [DOI] [PubMed] [Google Scholar]
- 2. Campos E. I., Reinberg D. (2009) Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 3. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 4. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J. (2011) Nature 470, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He H. H., Meyer C. A., Shin H., Bailey S. T., Wei G., Wang Q., Zhang Y., Xu K., Ni M., Lupien M., Mieczkowski P., Lieb J. D., Zhao K., Brown M., Liu X. S. (2010) Nat. Genet. 42, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loyola A., Almouzni G. (2007) Trends Biochem. Sci. 32, 425–433 [DOI] [PubMed] [Google Scholar]
- 7. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. (2004) Cell 116, 51–61 [DOI] [PubMed] [Google Scholar]
- 8. Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010) Genes Dev. 24, 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campos E. I., Reinberg D. (2010) Genes Dev. 24, 1334–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., Allis C. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., Pemberton L. F. (2002) J. Biol. Chem. 277, 862–868 [DOI] [PubMed] [Google Scholar]
- 12. Campos E. I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W. H., Seepany H., Gao Z., Day L. A., Greenblatt J. F., Reinberg D. (2010) Nat. Struct. Mol. Biol. 17, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G. (2006) Mol. Cell 24, 309–316 [DOI] [PubMed] [Google Scholar]
- 14. Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J. P., Imhof A., Nakatani Y., Dent S. Y., Almouzni G. (2009) EMBO Rep. 10, 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Groth A., Ray-Gallet D., Quivy J. P., Lukas J., Bartek J., Almouzni G. (2005) Mol. Cell 17, 301–311 [DOI] [PubMed] [Google Scholar]
- 16. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adam S. A., Marr R. S., Gerace L. (1990) J. Cell Biol. 111, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jasencakova Z., Scharf A. N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. (2010) Mol. Cell 37, 736–743 [DOI] [PubMed] [Google Scholar]
- 19. Wälde S., Kehlenbach R. H. (2010) Trends Cell Biol. 20, 461–469 [DOI] [PubMed] [Google Scholar]
- 20. Sobel R. E., Cook R. G., Perry C. A., Annunziato A. T., Allis C. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 22. Schiltz R. L., Mizzen C. A., Vassilev A., Cook R. G., Allis C. D., Nakatani Y. (1999) J. Biol. Chem. 274, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 23. Corpet A., De Koning L., Toedling J., Savignoni A., Berger F., Lemaître C., O'Sullivan R. J., Karlseder J., Barillot E., Asselain B., Sastre-Garau X., Almouzni G. (2011) EMBO J. 30, 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang Y., Poustovoitov M. V., Zhao K., Garfinkel M., Canutescu A., Dunbrack R., Adams P. D., Marmorstein R. (2006) Nat. Struct. Mol. Biol. 13, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKittrick E., Gafken P. R., Ahmad K., Henikoff S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1525–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.