Abstract
HIV-1 integrase (IN) is a key viral enzymatic protein acting in several viral replication steps, including integration. IN has been shown to be an unstable protein degraded by the N-end rule pathway through the host ubiquitin-proteasome machinery. However, it is still not fully understood how this viral protein is protected from the host ubiquitin-proteasome system within cells during HIV replication. In the present study, we provide evidence that the host protein Ku70 interacts with HIV-1 IN and protects it from the Lys48-linked polyubiquitination proteasomal pathway. Moreover, Ku70 is able to down-regulate the overall protein polyubiquitination level within the host cells and to specifically deubiquitinate IN through their interaction. Mutagenic studies revealed that the C terminus of IN (residues 230–288) is required for IN binding to the N-terminal part of Ku70 (Ku70(1–430)), and their interaction is independent of Ku70/80 heterodimerization. Finally, knockdown of Ku70 expression in both virus-producing and target CD4+ T cells significantly disrupted HIV-1 replication and rendered two-long terminal repeat circles and integration undetectable, indicating that Ku70 is required for both the early and the late stages of the HIV-1 life cycle. Interestingly, Ku70 was incorporated into the progeny virus in an IN-dependent way. We proposed that Ku70 may interact with IN during viral assembly and accompany HIV-1 IN upon entry into the new target cells, acting to 1) protect IN from the host defense system and 2) assist IN integration activity. Overall, this report provides another example of how HIV-1 hijacks host cellular machinery to protect the virus itself and to facilitate its replication.
Keywords: HIV, Integrase, Protein Degradation, Protein-Protein Interactions, Ubiquitination, DNA Repair Protein Ku70, Ubiquitin-Proteasome Pathway
Introduction
Integration is an obligatory step in the life cycle of all of the retroviruses and is performed by the viral enzyme integrase (IN).5 During HIV-1 integration, IN catalyzes the insertion of newly reverse-transcribed ∼10-kb viral DNA into the host genome. In addition, IN plays important roles in other viral replication steps, such as reverse transcription, the nuclear import of preintegration complexes (PICs), and chromatin targeting. By interaction with the host chromatin-tethering factor LEDGF/p75, IN preferentially targets viral DNA into transcriptionally active sites in the host genome to optimize the transcription and translation of its gene products (1–3). Cellular proteins are recruited to assist IN to accomplish integration from different pathway, including nuclear import, shielding IN from proteasomal degradation, integration site selection, and gap repair (4). Recently, considerable interest has been focused on the functional interaction between IN and host cellular proteins in the hope of disrupting their interactions, thereby blocking HIV-1 replication. In an attempt to identify host cellular partners for IN, several research groups have identified a number of IN cofactors using the yeast two-hybrid system, coimmunoprecipitation (co-IP) assays, or in vitro reconstitution of the enzymatic activity of salt-stripped PICs (5–11). A recent study by Studamire et al. (5) found that 12 cellular proteins, including Ku70, could bind to the INs of both the Moloney murine leukemia virus (MMLV) and HIV-1 through screening with a yeast two-hybrid system. However, whether these cellular cofactors are associated with HIV-1 IN during HIV replication and their functional relevance remain unknown.
Ku70 is an evolutionarily conserved protein; it is found ubiquitously in eukaryotes and some prokaryotes, such as Archaea and Bacteria (12–14). It is well known as a DNA repair protein and is part of the nonhomologous end-joining (NHEJ) pathway. Ku70 has also been implicated in many cellular processes, including antigen-receptor gene rearrangement, mobile genetic element biology, V(D)J recombination of immunoglobulins, telomere maintenance, DNA replication, transcription, cell cycle control, and apoptosis (13, 15). As a DNA repair protein, Ku70 can bind to any double-stranded DNA irrespective of sequence specificity or end configuration, including 5′ overhangs, 3′ overhangs, or blunt ends (for a review, see Ref. 15). Ku70 can also bind specific DNA sequences to affect gene transcription (16). For most biological functions in which Ku70 participates, Ku functions as a heterodimer consisting of Ku70 and Ku80, named according to their respective molecular masses of 70 and 80 kDa. Two regions of Ku70 amino acids 1–115 and 430–482 are responsible for its heterodimerization with Ku80 (17). Successful HIV-1 integration requires gap repair between viral DNA and host genome, which is believed to be performed by host DNA repair enzymes (18). Two different host DNA repair pathways have been suggested to fill in the gap during HIV-1 infection: the NHEJ and DNA damage-sensing pathways (19–21). The NHEJ pathway begins with the recruitment of the Ku70/80 heterodimer, followed by the catalytic subunit of DNA-dependent protein kinase or DNA-PKcs, Xrcc4, and DNA ligase IV. Studies have shown that the NHEJ pathway is important for retroviral transduction or infection and for the cell survival of infected or transduced cells (20, 22–25). For example, HIV-1-based vector transduction or infection was markedly reduced in cells deficient in Ku80, DNA-PKcs, Xrcc4, or ligase IV (22, 24). Moreover, NHEJ activity is required for two-long terminal repeat (2-LTR) circle formation, and Ku70 has been detected in MMLV PICs (24, 26–28). Ku80 was also shown to suppress HIV transcription by specifically binding to a negative regulatory element within the LTR (29). All of these observations suggest that Ku70 or the K70/80 heterodimer may be involved in HIV-1 infection by affecting multiple steps of the viral replication cycle, such as integration. In addition, a novel deubiquitinating enzymatic activity of Ku70 was recently described in which Ku70 has a regulatory effect on Bax-mediated apoptosis by decreasing the ubiquitination of Bax and blocking Bax from proteasomal degradation (30). However, whether Ku70 also exerts a deubiquitinating effect on other identified binding partners of Ku70 and how Ku70 interacts with the ubiquitin-proteasome pathway to deubiquitinate protein substrates are still unclear.
In this study, we investigated the interaction between Ku70 and HIV-1 IN and the potential roles of Ku70 during HIV-1 replication using cell-based coimmunoprecipitation and short hairpin RNA (shRNA)-mediated knockdown approaches. Interestingly, our results provide evidence that Ku70 is able to protect HIV-1 IN from Lys48-linked polyubiquitination and degradation by down-regulation of the overall protein polyubiquitination level within the host cells and by specific IN deubiquitination through its binding to IN. Moreover, our study showed that Ku70 depletion in both virus-producing and target cells drastically inhibited HIV-1 replication and blocked 2-LTR formation and integration in the real-time PCR analysis. Our data also showed that, mediated by HIV-1 IN, Ku70 was incorporated into the progeny virus. All of these results suggest that Ku70 may interact with IN during viral assembly and accompany HIV-1 IN into newly infected cells to assist IN integration activity and protect IN from host-mediated degradation.
EXPERIMENTAL PROCEDURES
Cell Lines and Transfection
Human embryonic kidney 293T and HeLa cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin. Human CD4+ C8166 T-lymphoid cells were maintained in RPMI 1640 medium supplemented with 10% FCS and 1% penicillin/streptomycin. For the transfection of 293T cells and HeLa cells, the standard calcium phosphate precipitation technique was used, as described previously (31).
Plasmids and Reagents
To achieve high level IN expression, a codon-optimized IN (INopt) cDNA was synthesized and cloned into the pUC57 vector (GenScript Co., Ltd.). To construct pAcGFP-INopt, the INopt fragment was excised from pUC57-INopt with BamHI and cloned in frame at the 3′ end of the pAcGFP1-C vector (Clontech) with the same restriction enzyme. To construct pAcGFP-INwt/mut, each of the INwt/mut coding sequences, including 1–230, 1–250, 1–270, 50–288, 112–288, and K186A/R187A, was amplified by PCR-based mutagenesis and subcloned into the pAcGFP1-C vector (Clontech) in frame with the GFP coding sequence at the BglII and BamHI restriction sites (32). Plasmids IN-YFP and MA-YFP were described previously (33, 34). Untagged human full-length Ku70 cDNA in the pCMV6-XL5 vector was purchased from OriGene Technologies Inc. To construct SVCMV-T7-Ku70, a Ku70 cDNA without a start codon was amplified and cloned into the SVCMVin-T7 vector at the BamHI and NotI restriction sites. The T7-Ku70 truncation mutants (1–263, 1–430, 226–609, and 430–609) were obtained using the same strategy. The nucleotide sequences of the mutagenic oligonucleotides are as follows: 5′Ku70-BamHI, 5-TAGCCGGATCCTCAGGGTGGGAGTCATATTA-3; 3′Ku70-NotI, 5-TATATGCGGCCGCTCAGTCCTGGAAGTGCTT-3; Ku70–263-NotI, 5-AGATGCGGCCGCCTAGAGCTTCAGCTTT-3; Ku70–430-NotI, 5-TATGCGGCCGCTCATGGAGGAGTCACCTGAAT-3; Ku70–226-BamHI, 5-TATGGATCCGATGAGGACCTCA-3; Ku70–430-BamHI, 5-ATATGGATCCCCAGGCTTCCAGCT-3. HA-tagged ubiquitin (HA-Ub) and mutants HA-UbK48R and HA-UbK63R were described previously (35). The HIV-1 proviruses pNL4.3-GFP, HxBru, and HxBru-IN-HA were described earlier (32, 36). For single cycle HIV virus, RT/IN/Env gene-deleted NL4.3lucΔBglΔRI provirus and CMV-Vpr-RT-IN were described previously (34, 37, 38). CMV-Vpr-RT has two stop codons TAGTGA in place of the first six nucleotides in IN sequences, and the sequence was confirmed by sequencing. To construct CMV-Vpr-RT-IN-ProLabel (Vpr-RT-IN-PL) plasmid, a two-step-based PCR method was used. ProLabel tag sequence was amplified from the pProLabel-C vector from ProLabelTM Detection Kit II (Clontech) and inserted after the IN sequence in the CMV-Vpr-RT-IN plasmid with the IN stop codon and ProLabel start codon removed. The following primers were used: RT NheI 5′, 5-GCAGCTAGCAGGGAGACTAA-3; R-RT-IN-ProLabel pstI 3′, 5-GTCGACTGCAGAATTCGAAGCTTATTC-3; IN-ProLabel 5′, 5-AGACAGGATGAGGATAGCTCCAATTCACTG-3; IN-ProLabel 3′, 5-CAGTGAATTGGAGCTATCCTCATCCTGTCT-3.
Antibodies and Reagents
The rabbit anti-GFP polyclonal antibody (Molecular Probes Inc.), the rabbit anti-HA antibody (Sigma), and mouse anti-T7 antibody (Novagen) were used for immunoprecipitation. The antibodies for Western blot (WB) were as follows: the mouse monoclonal anti-β-actin antibody (Abcam), mouse anti-α-tubulin (Sigma), mouse anti-Ku70 (Abcam), mouse anti-Ku80 (Abcam), horseradish peroxidase (HRP)-conjugated anti-GFP antibody (Molecular Probes), HRP-conjugated anti-HA antibody (Miltenyi Biotec), and HRP-conjugated anti-T7 antibody (Novagen). As the secondary antibodies, the ECLTM HRP-conjugated donkey anti-rabbit IgG and sheep anti-mouse IgG were purchased from Amersham Biosciences. The WB detection ECL kit was purchased from PerkinElmer Life Sciences (Boston, MA). Nonidet P-40 was from Roche Applied Science. Proteasome inhibitor MG-132 and puromycin were obtained from Calbiochem. Subtilisin was purchased from Sigma.
Transient and Stable Knockdown of Ku70 in 293T, HeLa Cells, and C8166 T Cells
To test the effect of Ku70 levels on the stability of IN, siRNA targeting human Ku70 (GenBankTM accession number NM_001469) was used to transiently knock down Ku70 expression in 293T cells and HeLa cells using the LipofectamineTM RNAiMAX transfection reagent (Invitrogen). The sense primer for this siRNA is 5′-GAUCCAGGUUUGAUGCUCAtt-3′, targeting Ku70 nucleotides 1094–1112. In parallel, a scrambled siRNA (Invitrogen) was used as a negative control (siNC). After 5 nm siKu70 or siNC oligonucleotide was transfected into cells for 12 h, cells were transfected again with 5 nm siKu70 to maximize knockdown efficiency.
To produce a stable Ku70-knockdown (KD) 293T and C8166 CD4+ T cell line, lentivirus-like particles harboring Ku70 shRNA were produced by cotransfecting the shRNA pLKO.1 vector containing shRNA targeting the Ku70 mRNA (5′-CCGGCGACATAAGTCGAGGGACTTTCTCGAGAAAGTCCCTCGACTTATGTCGTTTTTG-3′ (Oligo ID: TRCN0000039608; purchased from Open Biosystems)), packaging plasmid Δ8.2, and vesicular stomatitis virus G (VSV-G) expressor into the 293T cells. After 48 h, shRNA pLKO.1 vector particles were pelleted by ultracentrifugation (32,000 rpm at 4 °C for 1 h) and used to transduce cells for 48 h, followed by selection with 2 μg/ml puromycin for 1 week. Ku70-KD efficiency was determined by WB analysis with anti-Ku70 antibody. Endogenous β-actin was used to normalize sample loading. The pLKO.1 vector without the shRNA sequence (empty vector) was introduced into cells by the same method as a negative control.
Direct Immunofluorescence Assay
To test the effect of Ku70-KD level on the expression of IN, HeLa cells were first transfected with Ku70-specific siRNA oligonucleotides or nontargeting random siRNA (siNC) for 48 h and further transfected with GFP-INopt for another 48 h with or without MG-132 (10 μm) treatment. GFP fluorescence-positive cells were imaged by microscopy under a ×20 objective lens (Carl Zeiss).
Coimmunoprecipitation Assay in 293T Cells and in HIV-1-infected C8166T Cells
To detect the interaction between GFP-INwt/mut and T7-Ku70wt/mut and to identify their mutual binding regions, the cell-based co-IP assay was performed as described previously (37). Briefly, GFP or GFP-INwt/mut plasmid was cotransfected with pCMV-Ku70 or T7-Ku70, respectively, into 293T cells for 48 h. To increase GFP-IN stability, 10 μm MG-132 was added 12 h prior to cell lysis for co-IP. Then 90% of the transfected cells were lysed in 0.25% Nonidet P-40 prepared in 199 medium containing a mixture of protease inhibitors (Roche Applied Science) and clarified by centrifugation at 14,000 rpm for 30 min at 4 °C. Supernatant was precleared with Protein G-agarose on a rotator for 2 h at 4 °C and subsequently subjected to IP with a rabbit anti-GFP antibody and Protein A-Sepharose overnight. The IN-bound proteins were detected by WB using anti-Ku70 or anti-T7 antibodies. The same nitrocellulose membrane was then stripped and probed with anti-GFP antibody to detect GFP-INwt/mut or GFP expression. Meanwhile, 5% of the transfected cells were lysed in 0.5% Nonidet P-40, and the lysates were used to detect the expression of GFP-INwt/mut and Ku70 by WB using their corresponding antibodies.
To examine the IN/Ku70 interaction in HIV-1-infected cells, HIV-1 (HxBru or HxBru-IN-HA)-infected C8166 T cells were lysed with 0.25% Nonidet P-40 and immunoprecipitated with anti-HA antibody followed by WB with anti-Ku70 antibody to detect IN-bound Ku70.
Detection of Ubiquitination of IN in the Absence or Presence of Ku70
To determine the ubiquitination level of HIV-1 IN in the absence and presence of Ku70, 293T cells were cotransfected with GFP-IN and HA-Ub wild type or mutants K48R and K63R with and without T7-Ku70wt or T7-Ku70(1–430). After 48 h, cells were lysed in 199 medium containing 0.25% Nonidet P-40 and a protease/inhibitor mixture and immunoprecipitated with anti-GFP antibody. Then the precipitated complexes were run on a 10% SDS-polyacrylamide gel and analyzed for the presence of HA-Ub by WB with HRP-conjugated anti-HA antibody. Simultaneously, GFP-IN was detected by immunoblotting the same membrane with HRP-conjugated anti-GFP antibody. And protein band intensity was quantified using Quantity One 1-D analysis software (Bio-Rad).
Virus Production and Infection
To study the effect of Ku70-KD on HIV-1 replication, equal amounts (quantified by HIV-1 p24 antigen) of pNL4.3-GFP virus were used to infect Ku70-KD or empty vector-transduced C8166 T cells for 2 h; cells were then washed and cultured in a 37 °C incubator. At different time points, viral replication levels were monitored by the measurement of p24 levels using an HIV-1 Gag-p24 ELISA. To test the infectivity of progeny virus produced from the Ku70-KD cells, empty-vector and Ku70-KD C8166 T cells were infected with the same amounts of pNL4.3-GFP. Progeny viruses were collected by ultracentrifugation after 4 days of infection, and equal amounts of viruses (quantified by HIV-1 p24 antigen) were used to infect empty vector or Ku70-KD C8166 T cells. Viral infection was examined at 3 days postinfection by monitoring HIV p24 levels in the supernatant.
Quantitative Real-time PCR
1.5 × 106 stable C8166 T cell lines with Ku70-KD or empty vector-transduced were infected with the pNL4.3-GFP virus as described above. Heat-inactivated virus (70 °C for 30 min) was used as a negative control for infection. After 4 h of infection, cells were washed and cultured in fresh RPMI medium. At 24 h postinfection, cells were harvested and washed with PBS twice. DNA was isolated using a QIAamp blood DNA minikit (Qiagen). The total levels of HIV-1 DNA, 2-LTR circles, and integrated DNA were quantified following the same procedure in an Mx3000P real-time PCR system (Stratagene) as described (32).
Virus Composition and Incorporation of Cellular Protein into HIV-1 Virion
To examine the viral protein compositions, the pNL4.3-GFP viruses from empty vector-transduced and Ku70-KD C8166 T cells were pelleted through a 20% sucrose cushion at 35,000 rpm for 1.5 h at 4 °C. Then equal amounts of viruses (normalized by p24 values) were lysed with 4× Laemmli buffer and directly loaded onto an SDS-PAGE gel and analyzed for IN and p24 expressions using their corresponding antibodies. The reverse transcription activity from the purified viruses was analyzed by a reverse transcription assay using a commercial RT assay kit (Roche Applied Science) according to the manufacturer's instructions.
To detect the presence of Ku70 in the HIV-1 particles, 15 × 106 CD4+ C8166 T cells were mock-infected or infected with pNL4.3-GFP for 3 days. Then supernatants from both cell cultures were ultracentrifuged at 35,000 rpm for 1.5 h through a 20% sucrose cushion. The pellets were dissolved in the same volume of radioimmune precipitation assay buffer and mixed with 20% (v/v) TCA, followed by precipitation on ice for 30 min and acetone washing. Protein precipitates were dissolved in 4× Laemmli buffer and directly loaded onto a 10% SDS-polyacrylamide gel. Virus-associated Ku70 and p24 were then examined by WB using the corresponding antibodies.
Subtilisin treatment of purified HIV-1 virions. The subtilisin assay was performed according to the protocol as described (39). The Vpr-RT-IN or Vpr-RT expressor was cotransfected with VSV-G and NL4.3lucΔBglΔRI to produce single cycle IN+ and IN− virus. The viruses were first ultracentrifuged through 20% sucrose at 35,000 rpm for 2 h and then mock-treated or treated with 0.1 mg/ml of subtilisin (Sigma) for 20 h at a 37 °C incubation. Subtilisin was inactivated by phenylmethylsulfonyl fluoride. Virus was then repelleted as described above, lysed in radioimmune precipitation assay buffer, and loaded onto SDS-polyacrylamide gel followed by WB. Blots were sequentially probed with anti-Ku70, anti-IN, and p24 antibodies.
ProLabel Detection Assay
To test the effect of Ku70 on IN during HIV infection, VSV-G pseudotyped HIV single cycle virus containing ProLabel tag fused to the C terminus of IN was generated to quantify IN expression under HIV infection. NL4.3lucΔBglΔRI was cotransfected with Vpr-RT-IN-PL and VSV-G expressor into 293T cells to generate VSV-G pseudotyped HIV-1 single cycle IN-PL virus. The viruses were used to infect shKu70-KD or empty vector-transduced C8166T cells for 3 h. The cells were washed three times and kept in fresh medium and then lysed with lysis/complementation buffer at 8 h p.i. IN-ProLabel activity in the cell lysate was measured according to the manufacturer's instructions from the assay kit (ProLabelTM detection kit II, Clontech).
Statistical Analysis
The statistical significance was calculated using Student's t test, and a p value of ≤0.05 was considered significant.
RESULTS
Cellular Protein Ku70 Protects HIV-1 IN from Proteasomal Degradation
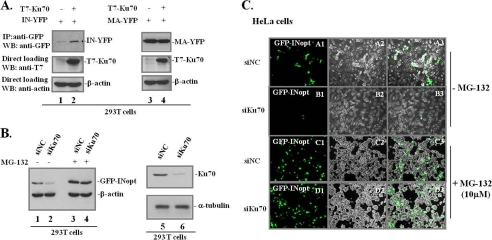
As a part of the NHEJ machinery, the host protein Ku70 has been shown to participate in HIV integration and in the circularization of unintegrated viral DNAs (25, 27). Surprisingly, based on the results of a yeast two-hybrid assay, a recent study indicated that HIV-1 IN may bind to Ku70 (5), suggesting a direct association between HIV-1 IN and Ku70. To further investigate this viral/host protein interaction, we coexpressed Ku70 (T7-tagged Ku70) and HIV-1 IN (IN-YFP) in 293T cells and analyzed their interaction after 48 h of transfection. Noticeably, our results revealed that T7-Ku70 overexpression significantly increased IN expression (Fig. 1A, lanes 1 and 2). However, the coexpression of Ku70 with another HIV-1 protein, MA (MA-YFP), did not change the MA expression level (Fig. 1A, lanes 3 and 4). This suggests that Ku70 is able to increase IN expression. Alternatively, Ku70 could protect the IN protein from degradation (40).
FIGURE 1.
Effects of Ku70 on the stability of HIV-1 IN. A, 293T cells were cotransfected with IN-YFP or MA-YFP and T7-Ku70wt for 48 h. Expression of IN-YFP (lanes 1 and 2) and MA-YFP (lanes 3 and 4) were detected by IP with rabbit anti-GFP antibody and anti-GFP-HRP antibody in a WB (top panel). An aliquot of cells (about 5%) was collected and checked for T7-Ku70wt and β-actin expression (middle and bottom panels). B, proteasome inhibitor MG-132 treatment restores IN expression in Ku70-KD 293 T cells. After transfection with 5 nm siKu70 or the siNC for 48 h, 293T cells were transfected with optimized IN (GFP-INopt) for another 48 h. The control and KD cells were treated with or without 10 μm MG-132 for 12 h prior to harvesting. The expression of GFP-INopt was detected by direct loading of protein samples onto a 10% SDS-polyacrylamide gel using HRP-conjugated anti-GFP antibody in a WB (lanes 1–4), and the same membrane was probed with anti-β-actin antibody to assess protein loading. Ku70 knockdown efficiency was detected by mouse anti-Ku70 antibody in WB (lanes 5 and 6), and α-tubulin was used as the protein-loading control in each sample. C, the effects of Ku70 on HIV-1 IN expression were confirmed in HeLa cells by direct fluorescence. HeLa cells were treated with 5 nm siKu70 or siNC for 48 h prior to transfection with GFP-INopt. After another 48 h, GFP-positive HeLa cells without MG-132 (A1–A4 and B1–B4) or with 12-h treatment of 10 μm MG-132 (C1–C4 and D1–D4) were examined by fluorescence microscopy.
To further test whether endogenous Ku70 could exert the same activity and whether it is due to a protective effect, we first knocked down the Ku70 expression using specific siRNA in 293T (Fig. 1B) or HeLa cells (Fig. 1C) and checked the level of GFP-IN expression by WB or fluorescence microscopy (Fig. 1, B and C). To increase IN expression under normal conditions, we used a pAcGFP-IN with a codon-optimized IN sequence (GFP-INopt). The results showed that IN expression in Ku70-KD cells was significantly decreased when compared with IN expression in siNC cells (Fig. 1, B (compare lanes 1 and 2) and C (compare A1–A3 and B1–B3)). Intriguingly, in the presence of the specific proteasome inhibitor MG-132 (10 μm), IN expression in Ku70-KD cells was remarkably increased, reaching levels similar to those in siNC-transfected 293T and HeLa cells (Fig. 1, B (compare lanes 4 and 3) and C (compare C1–C3 and D1–D3)). Thus, these results clearly indicate that Ku70 is able to protect IN from proteasomal degradation.
Ku70 Is Able to Interact with HIV IN in both 293T Cells and HIV-1-infected T Cells
Provided that Ku70 is able to protect HIV-1 IN from proteasomal degradation, we next tried to reveal the molecular mechanisms underlying this effect. Because Ku70 has been implicated as an HIV-1 IN cofactor (5), it is possible that IN could escape from the host proteasomal degradation machinery by directly interacting with Ku70. Therefore, we further investigated the interaction between HIV IN and host protein Ku70 under more physiological conditions by using a co-IP approach in 293T cells and HIV-1-infected CD4+ C8166 T cells.
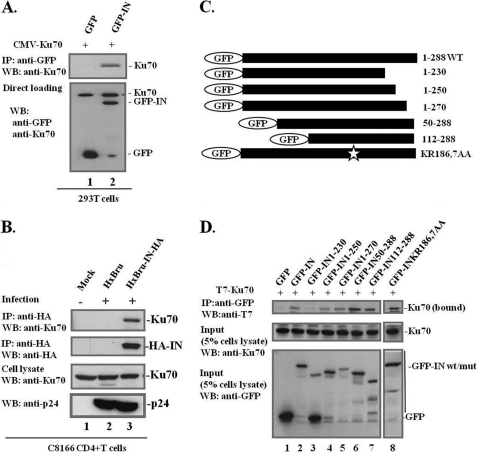
First, a CMV-Ku70 expressor and GFP or GFP-INwt plasmid were cotransfected into 293T cells. In order to prevent IN degradation, MG-132 (10 μm) was added to the cells at 12 h before cell lysis. The cells were then lysed and immunoprecipitated with anti-GFP followed by WB using anti-Ku70. We found that GFP-IN, but not GFP, was able to pull down Ku70 (Fig. 2A). Given that both Ku70 and IN are DNA-binding proteins, we added DNase I in the cell lysate during the co-IP assay and found that GFP-IN still bound to Ku70 with DNase I treatment, suggesting a direct protein-protein interaction (data not shown). Considering that the protein overexpression system might not necessarily reflect normal functional binding of IN/Ku70, we also tested the authentic interaction of IN/Ku70 in HIV-1-infected cells (Fig. 2B). To do this, C8166 CD4+ T cells were infected with HIV-1 HxBru or HxBru-IN-HA viruses. In the provirus HxBru-IN-HA, an HA tag was inserted at the C terminus of IN (33), allowing us to pull down HIV-1 IN-associated cellular proteins by using anti-HA antibody in the co-IP assay. At 72–96 h postinfection, the C8166T cells were lysed, and the cell lysates were subjected to a co-IP assay to detect IN-bound endogenous Ku70 in the infected cells. The results showed that Ku70 was coprecipitated with IN-HA from HxBru-IN-HA-infected cells but not from mock C8166T cells or HIV HxBru-infected C8166 T cells (Fig. 2B, top panel). The same immunoblot was reprobed with anti-HA to detect the immunoprecipitated level of IN-HA (Fig. 2B, second panel). Similar levels of endogenous Ku70 and viral Gag-p24 were detected in HxBru- and HxBru-IN-HA-infected cells (Fig. 2B, third and fourth panel). Together, these results clearly indicated that the DNA repair protein Ku70 is an authentic, newly described host cofactor for IN.
FIGURE 2.
IN interacts with Ku70 in mammalian cell lines and in HIV-1-infected CD4+ T-lymphocytes, and the interaction is through the C terminus of IN. A, interaction of Ku70 with GFP-IN in 293T cells. GFP or GFP-IN was coexpressed with CMV-Ku70 as indicated into 293T cells for 48 h, and cells were treated with the proteasome inhibitor MG-132 (10 μm) for 12 h prior to co-IP analysis. Co-IP was performed using rabbit anti-GFP antibody for immunoprecipitation and WB using anti-Ku70 to detect IN-bound Ku70 (top panel). GFP-IN and untagged pCMV-Ku70 were detected with the corresponding antibodies to assess the expression levels of IN and Ku70 (bottom panels of lanes 1 and 2). B, C8166 CD4+ T cells were mock-infected or infected with HIV-1 HxBru or HxBru-IN-HA, as indicated. Cells were collected at 72 h postinfection, and the cell lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Ku70 antibody to detect IN-associated Ku70 in HIV-1-infected T cells (top panel). The same membrane was then reprobed with anti-HA antibody to detect IN-HA expression (second panel). The same amount of cells was assessed for Ku70 expression prior to co-IP, serving as the immunoprecipitation input (third panel). The p24 levels in the infected cells were checked with anti-p24 antibody (bottom panel). C, schematic representation of the GFP-IN wild type, five IN deletion mutants, and point mutant K186A/R187A. D, the C terminus of IN is required for IN/Ku70 interaction. GFP or various GFP-IN wild type/mutants were cotransfected with T7-Ku70 (lanes 1–8) into 293T cells, and their interactions were studied by a co-IP assay. Cells were treated with MG-132 at a concentration of 10 μm for 12 h prior to co-IP analysis. IN-bound Ku70 was detected by IP with anti-GFP antibody and WB with anti-Ku70 antibody (top panel). Total cell lysates were analyzed for GFP-INwt/mutants and Ku70 expression (middle and bottom panels). Input, 5% of total cell extract).
To delineate the Ku70-binding domain of IN, five previously described IN N-terminal or C-terminal deletion mutants, including GFP-IN(1–230), GFP-IN(1–250), GFP-IN(1–270), GFP-IN(50–288), and GFP-IN(112–288), and one substitution mutant, GFP-IN K186A/R187A (32, 37, 38), were used to test Ku70 binding ability (Fig. 2C). The results revealed that GFP and GFP-IN(1–230) did not bind to Ku70 (Fig. 2D, lanes 1, 3, and 8), whereas other truncated GFP-IN mutants (residues 1–250, 1–270, 50–288, and 112–288) and the point mutant K186A/R187A still retained their Ku70-binding ability (Fig. 2D, lanes 2, 4–7, and 9). K186A/R187A, a well characterized oligomerization-defective mutant of IN (41, 42), still bound Ku70, suggesting that multimerization of IN is not required for IN/Ku70 interaction (Fig. 2D, lane 9). Overall, our analysis suggested that the C-terminal half of IN (IN(112–288)) is sufficient to bind to Ku70.
The Ku70 Truncated Mutant Ku70(1–430) Interacts with HIV IN but Cannot Form a Heterodimer with Ku80
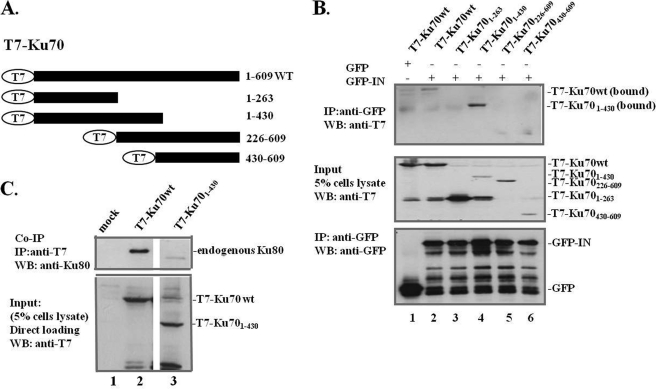
To define the IN-binding region within Ku70, expressors for four T7-tagged Ku70 deletion mutants (residues 1–263, 1–430, 226–609, and 430–609) were constructed (Fig. 3A) and cotransfected with GFP-IN or the expressor into 293T cells. Based on a previous mutational analysis indicating that the minimum region for DNA binding within the Ku70 core region was estimated to be around aa 263–430, 1–263 and 1–430 mutants were constructed; Ku70(226–609) was shown to be defective for DNA-PK activity and DNA binding in the same study (43). The Ku70(430–609) mutant was sufficient for the heterodimerization of Ku70/80 but lost DNA end binding ability in the two-hybrid analysis (44). In parallel, 293T cells cotransfected with T7-Ku70 and GFP plasmids were used as a negative control. The results showed that T7-Ku70wt and deletion mutant T7-Ku70(1–430) were coimmunoprecipitated with GFP-IN (Fig. 3B, top panel). Interestingly, T7-Ku70(1–430) displayed a higher binding affinity for IN than T7-Ku70wt (Fig. 3B, top panel, compare lane 4 with lane 2). Because the N-terminal truncation (aa 1–263) of Ku70 did not interact with IN (Fig. 3B, top panel, lane 3), whereas T7-Ku70(1–430) showed a strong binding affinity, the aa 263–430 region of Ku70 is probably necessary but not sufficient for IN interaction. However, another Ku70 mutant, residues 226–609, which also encodes the aa 263–430 region, failed to interact with GFP-IN (Fig. 3B, top panel, lane 5), suggesting that another important binding domain might exist within the N terminus (residues 1–226) of Ku70. Therefore, both the N-terminal domain and the core domain of Ku70 are suggestive of binding surface for IN.
FIGURE 3.
The N terminus (aa 1–430) of Ku70 binds IN, and IN/Ku70 interaction is independent of the heterodimerization of Ku70/80. A, schematic diagrams depict the different T7-Ku70wt/mut constructs used in the domain-mapping experiments. The full length of T7-Ku70 is shown at the top, as indicated. B, interaction of GFP-IN with T7-Ku70wt/mut. GFP or GFP-INwt was cotransfected with T7-Ku70wt/mut in 293T cells for 48 h. MG-132 (10 μm) was added to the cells 12 h prior to cell lysis to enhance protein expression. The co-IP assay was performed to map the IN binding region in Ku70 using anti-GFP antibody IP, and a WB using anti-T7 antibody was performed to detect IN-bound T7-Ku70wt/mut (top). GFP, GFP-IN, and T7-Ku70wt/mut expression was checked by immunoblotting with anti-GFP or anti-T7 antibodies, respectively (middle and bottom panels). C, T7-Ku70(1–430) cannot form a heterodimer with Ku80. 293T cells were mock-transfected or transfected with T7-Ku70wt and the aa 1–430 truncation mutant for 48 h. Heterodimerization of Ku70/80 was determined by co-IP with anti-T7 antibody and WB using mouse anti-Ku80 antibody to detect T7-Ku70-bound endogenous Ku80 (top panel). About 5% of the cell lysates (input) were checked for the expression of T7-Ku70wt/mut by WB using anti-T7 antibody (bottom panel).
Ku70 forms a heterodimer with Ku80, a heterodimerization that has been shown to contribute to many cellular processes. For example, the heterodimerization of Ku70/80 is essential for activating DNA-PK and DNA repair and important for their nuclear translocation (43, 45). One might ask whether Ku80 is also involved in the IN/Ku70 interaction or if IN interacts with Ku70 indirectly through Ku70/80 heterodimerization. To address this question, we performed a co-IP assay in which T7-Ku70wt or T7-Ku70(1–430) was transfected into 293T cells. At 48 h post-transfection, cell lysates were immunoprecipitated with anti-T7 antibody followed by WB with an anti-Ku80 antibody. According to our previous findings, T7-Ku70wt was able to pull down endogenous Ku80 (Fig. 3C, lane 2, top panel). Interestingly, the deletion mutant T7-Ku70(1–430), which efficiently binds IN, could not form a heterodimer with Ku80 (Fig. 3C, lane 3, top panel). This result indicates that T7-Ku70(1–430) binding to IN was independent of Ku80. Thus, the heterodimerization of Ku70/80 may not be required for IN/Ku70 interaction.
Ku70 Protects IN from Degradation by Reducing the Total Ubiquitination Level in the Host Cells and by IN/Ku70(1–430) Binding
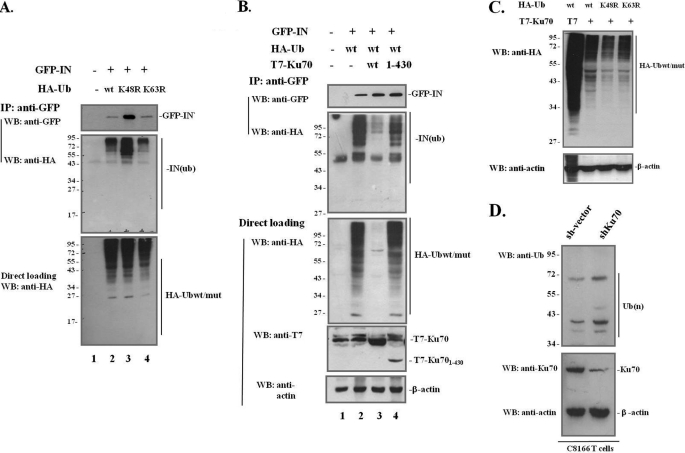
The results above showed that Ku70 binds and protects IN from proteasomal degradation; however, the detailed mechanisms underlying the degradation of IN through the ubiquitination-proteasome pathway remain unclear. Proteins tagged with ubiquitin can be monoubiquitinated or polyubiquitinated. The polyubiquitination chains are formed between the C-terminal residue glycine 76 of ubiquitin and any other internal lysine within the ubiquitin molecule (lysine 6, 11, 27, 29, 33, 48, or 63) through an isopeptide bond (46, 47). The two most important polyubiquitination chains are the Lys48- and Lys63-linked chains, with the Lys48-linked polyubiquitination chain recognized by the 26 S proteasomal pathway for degradation and the Lys63-linked polyubiquitination chain implicated in postreplicative DNA repair (48). In analysis of the ubiquitin-proteasome pathway involved in IN degradation, HA-Ub mutants K48R and K63R were included in our study, having mutations of Lys48 and Lys63 to arginine that were expected to disrupt Gly76-Lys48 and Gly76-Lys63 polyubiquitination chain formation, respectively. First, HA-Ubwt or mutant HA-UbK48R and HA-UbK63R were cotransfected with the IN expressor (GFP-IN) into 293T cells. At 48 h post-transfection, cells were lysed in 0.25% Nonidet P-40 and subjected to a co-IP assay using anti-GFP antibody to pull down GFP-IN, followed by WB with anti-GFP and anti-HA antibodies to detect HA-Ub-tagged IN-associated proteins (Fig. 4A, top and middle panels). Simultaneously, HA-Ub expression levels in the cells were also checked (Fig. 4A, bottom panel). The data showed that GFP-IN protein levels in the HA-UbK48R overexpression cells were much higher than in HA-Ubwt- and HA-UbK63R-transfected 293T cells (Fig. 4A, top panel; compare lane 3 with lanes 2 and 4). Similarly, the ubiquitination level of IN-associated protein expression was the highest in the HA-UbK48R-transfected sample (Fig. 4A, middle panel). The HA-tagged ubiquitination signal (Fig. 4A, middle panel) was from a pool of ubiquitinated IN and unknown IN-bound cellular proteins. The observation of increased levels of ubiquitinated IN-associated proteins in HA-UbK48R-cotransfected cells was expected given the facts that the immunoprecipitate input or GFP-IN (Fig. 4A, top panel) was the highest and that all of the IN-associated proteins subjected to the ubiquitin-proteasome system for degradation were accumulated due to the defective Lys48-linked polyubiquitination proteasome degradation pathway. However, similar levels of HA-Ubwt, HA-UbK48R, and HA-UbK63R were detected in the cell lysates (Fig. 4A, bottom panel). The highest IN expression was detected in the HA-UbK48R expression cells, clearly suggesting that GFP-IN is degraded though the Lys48-linked polyubiquitination proteasomal degradation pathway.
FIGURE 4.
IN is degraded through the Lys48-linked polyubiquitination proteasomal pathway, and Ku70 protects IN by reducing the overall ubiquitination level in the cells and partially blocking ubiquitination of IN and its bound cellular proteins. A, IN is degraded through Lys48-linked polyubiquitination. 293T cells were mock-transfected or cotransfected with GFP-IN and HA-Ubwt, HA-UbK48R, or HA-UbK63R for 48 h, as indicated. The ubiquitination levels of IN and IN-associated proteins were checked by IP with anti-GFP antibody and anti-HA antibody in a WB (middle panel). The same membrane was reprobed with anti-GFP antibody to detect GFP-IN expression in each sample (top panel). About 5% of the cells prior to co-IP were lysed in 0.5% Nonidet P-40, and the protein contents were subjected to WB to detect total HA-Ub levels in the cells using anti-HA antibody (bottom panel). B, Ku70 protects IN from degradation by targeting the cellular ubiquitin-proteasome pathway and reducing ubiquitin binding to IN. 293T cells were mock-transfected (lane 1) or cotransfected with GFP-IN, HA-Ubwt and T7 vector, or T7-Ku70wt and T7-Ku70(1–430) (lanes 2–4). The co-IP assay was done at 48 h post-transfection using anti-GFP antibody to pull down IN and its associated proteins and using immunoblotting with anti-HA antibody to determine the ubiquitination level of IN and its associated proteins (second panel). The same membrane was reprobed with anti-GFP antibody to examine GFP-IN expression (first panel). Simultaneously, equal amounts of total cellular proteins (about 5% of the total cell lysates) were resolved on a SDS-PAGE gel and immunoblotted with anti-HA and anti-T7 antibodies to determine the expression levels of the transfected protein expressors (third and fourth panels). β-Actin was used as a loading control (bottom panel). C, reduction of ubiquitin level by Ku70 is independent of the Lys48- and Lys63-linked polyubiquitination proteasomal pathway. 293T cells were transfected with HA-Ubwt/mut with T7-Ku70 or T7 vector for 48 h. Cells were lysed and analyzed for HA-Ub expression using anti-HA antibody in a WB. On the same membrane, β-actin was used as a protein-loading control. D, endogenous ubiquitin levels were increased in Ku70-down-regulated cells. C8166 T cells were transduced with empty vector or lentiviral vector expressing an shRNA against human Ku70 and selected with 1 μg/ml puromycin. After 1 week of selection, equal amounts of control or stable Ku70-KD cell lines were collected and lysed. The expression levels of ubiquitin, Ku70, and β-actin were assessed by WB using specific antibodies.
To further investigate how Ku70 affects IN stability, we studied the ubiquitination level of IN in the presence of both T7-Ku70 and HA-Ub. Because T7-Ku70(1–430) showed a strong binding affinity with IN (Fig. 3), we also included this T7-Ku70 deletion mutant to determine if the interaction of Ku70/IN plays a role in the stability and ubiquitination of IN. As expected, GFP-IN expression in the cells transfected with T7-Ku70wt and T7-Ku70(1–430) deletion mutants was 1.47 ± 0.11- and 1.78 ± 0.24-fold increased compared with cells transfected with the empty vector (Fig. 4B, top panel; compare lanes 3 and 4 with lane 2). The total ubiquitin expression level detected in the whole-cell extract was dramatically reduced in the presence of wild-type Ku70 (Fig. 4B, lane 3, third panel). Consistently, the co-IP data showed that ubiquitinated IN-bound proteins also remarkably reduced by 4.58 ± 0.51-fold in the presence of wild-type Ku70 (Fig. 4B, lane 3, second panel). Interestingly, T7-Ku70(1–430) was still able to protect IN (Fig. 4B, first and third panels, lane 4) and significantly reduced the level of HA-tagged ubiquitination signal in IN-bound proteins in the presence of T7-Ku70(1–430) by 1.18 ± 0.19-fold (Fig. 4B, second panel; compare lane 4 with lane 2; the percentage is normalized by total HA-ubiquitin level in the third panel), although it did not affect the overall ubiquitin level in the cells (Fig. 4B, third panel; compare lane 4 with lane 2). These results thus indicate that although T7-Ku70(1–430) lacks the activity to reduce total ubiquitin level, as wild-type Ku70 does, it can protect IN by specifically reducing the ubiquitination of IN and its associated proteins. Based on these results, we conclude that Ku70 protects IN through two mechanisms; Ku70 reduces total ubiquitination level in the cells and reduces the ubiquitination of IN and IN-bound cellular proteins through their interaction.
We then tested whether Ku70 affects specific lysine-linked polyubiquitination proteasomal degradation pathways. The T7-Ku70 expressor was cotransfected with HA-Ubwt, HA-UbK48R, or HA-UbK63R into 293T cells. The results showed that, in the presence of Ku70, overall HA-Ub expression was still greatly reduced although Ub cannot form Lys48- or Lys63-linked polyubiquitin chains (Fig. 4C). This observation was confirmed by the fact that the down-regulation of Ku70 increased ubiquitin levels in the cells. We established a stable Ku70-KD CD4+ C8166 T cell line by transducing C8166 CD4+ T cells with a lentiviral vector carrying Ku70 shRNA. In parallel, the control cell line was transduced with an empty lentiviral vector. After puromycin selection, control cells and Ku70-KD cells were checked for Ku70 knockdown efficiency by WB (Fig. 4D, bottom panel). These cells were then used to detect endogenous ubiquitin levels. The results showed that the ubiquitin levels were higher in the Ku70-KD cells than in the empty vector-transduced cells (Fig. 4D, top panel). Taken together, we demonstrated here that Ku70 is able to protect HIV IN from degradation by down-regulating cellular ubiquitin levels and simultaneously preventing the ubiquitination of IN and its associated cellular proteins.
Ku70 Knockdown Impairs HIV-1 Replication
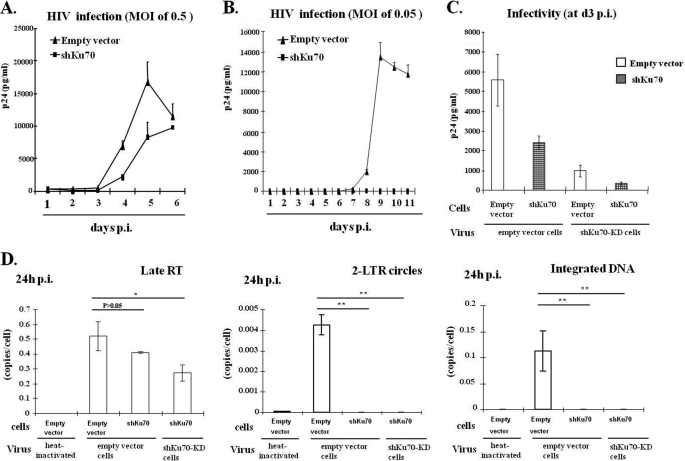
Because Ku70 is able to bind HIV IN and protects IN from degradation, we next tested whether and how Ku70 contributes to HIV-1 replication. To do so, the empty vector-transduced and Ku70-KD C8166 T cells were infected with pNL4.3-GFP+ viruses at an MOI of 0.5 (Fig. 5A) or 0.05 (Fig. 5B) for 2 h, and the viral replication kinetics were monitored. The infection was examined at different time intervals by harvesting virus-laden supernatant and checking for HIV-1 p24 antigen release. Fig. 5A shows that at the MOI of 0.5, HIV infection in Ku70-KD C8166 T cells was reduced by ∼50% at 4 and 5 days postinfection compared with the control cells (Fig. 5A). However, when Ku70-KD cells were infected with a lower MOI of 0.05, viral infection was undetectable by measuring HIV p24 levels up to 11 days. Nonetheless, in the control cells, viral replication peaked at day 9 (Fig. 5B). Interestingly, Ku70 knockdown completely inhibited low MOI HIV-1 infection but only reduced high MOI viral infection by 50%. This could be due to the fact that the infection of a large amount of viruses produced from normal T cells may at least partially overcome the shortage of Ku70 inside the target cells and establish an efficient first cycle of replication in Ku70-KD cells. If this is the case, we reasoned that viruses produced from Ku70-KD cells should have a significantly lowered infectivity compared with the virus produced from the normal cells. To test this possibility, we infected empty vector-transduced and Ku70-KD C8166 T cells with the same amounts of pNL4.3-GFP+ virus (normalized by p24 ELISA) produced from either empty vector or Ku70-KD C8166 T cells. At 3 days postinfection, virus-laden supernatants were harvested and checked for p24 antigen production by p24 ELISA. Consistent with the above results, when cells were infected with virus produced from empty vector-transduced cells (empty vector virus or E-virus), there was only a 2-fold difference in Ku70-KD cells compared with its infection in empty vector cells (Fig. 5C, compare bars 1 and 2). However, when empty vector-transduced cells were infected with either E-virus or Sh-virus produced from Ku70-KD cells, there was an ∼5-fold reduction of Sh-virus infection (Fig. 5C, compare bars 1 and 3). Strikingly, Sh-virus infection in Ku70-KD cells exhibited more severe impairment, with a 16-fold reduction in viral infectivity compared with the E-virus infection in empty vector-transduced cells (Fig. 5C, compare bars 1 and 4). Overall, this group of results indicates that down-regulation of Ku70 in both HIV-producing and target cells significantly impairs viral replication.
FIGURE 5.
Differential replication kinetics in Ku70-KD C8166 T cells with different titers of viral infection. A and B, HIV-1 replication kinetics in Ku70-KD or empty vector-transduced C8166 T stable cell lines at a high MOI of 0.5 (A) or a low MOI of 0.05 (B). Lentiviral shRNA targeting Ku70 or empty vector-transduced C8166 T stable cell lines were infected with different doses of pNL4.3-GFP virus for 2 h. At subsequent time intervals, the supernatants were collected, and viral replication was monitored by measuring HIV-1 p24gag levels. The data shown are the means and S.D. values (error bars) of p24 values from duplicate wells in one infection assay and are representative of three independent experiments. C, Ku70-KD inhibited the infectivity of progeny virus. pNL4.3-GFP viruses produced from empty vector-transduced or shKu70-KD C8166 T cells were normalized by p24 and used to infect both empty vector-transduced and shKu70-KD C8166 T cells. Viral replication was monitored by p24 ELISA at 3 days postinfection. D, Ku70-KD impaired 2-LTR formation and integration of proviral DNA. Stable Ku70-KD or empty vector C8166T cells were infected with the same amount of pNL4.3-GFP viruses produced from either empty vector-transduced C8166T cells or shKu70-KD cells for 24 h. The cellular genomic DNA was extracted and quantified for HIV-1 late RT, 2-LTR, and integrated DNA by real-time PCR. Infection with 70 °C heat-inactivated virus served as a negative control (bar 1). Data are representative of two independent experiments performed in duplicate, shown as mean ± S.D. The statistical significance is denoted as follows: *, p ≤ 0.05; **, p ≤ 0.01.
In order to investigate which early step(s) of viral replication is blocked by Ku70-KD, infected C8166 T cells as described above (Fig. 5C) were harvested at 24 h p.i., and DNA was isolated and assessed for late reverse transcription products (late RT), 2-LTR circles, and integrated DNA by quantitative PCR (32, 38). The results revealed that late RT products in the Ku70-KD C8166T cells did not show significant reduction when compared with normal empty vector-transduced cells (p > 0.05; Fig. 5D, left, compare bar 3 with bar 2), whereas late RT product was about 50% reduced compared with Ku70-KD cells infected with Sh-virus (p < 0.05; Fig. 5D, left, compare bar 4 with bar 2). Strikingly, 2-LTR and integrated DNA in shKu70-KD cells infected with either normal E-virus or Sh-virus were undetectable under current assay conditions (p < 0.01; Fig. 5D, middle and right panels, bars 3 and 4). 2-LTR circle formation is indicative of nuclear import (49), but it also requires proper circularization by host DNA repair enzymes (27, 50). For example, depletion of NHEJ pathway components (Ku80, XRCC4, and LigaseIV) resulted in undetectable or reduced 2-LTR formation (27, 50). The undetectable 2-LTR circle formation in Ku70-KD cell infection with both normal E-virus and Sh-virus could be due to insufficient DNA circularization by Ku70-KD in the target cells (Fig. 5D, middle panel, bars 3 and 4). All of these results indicate that the presence of Ku70 is required for an efficient viral reverse transcription and necessary for viral integration.
Host Protein Ku70 Is Incorporated into Viral Particles and Stabilizes IN Expression
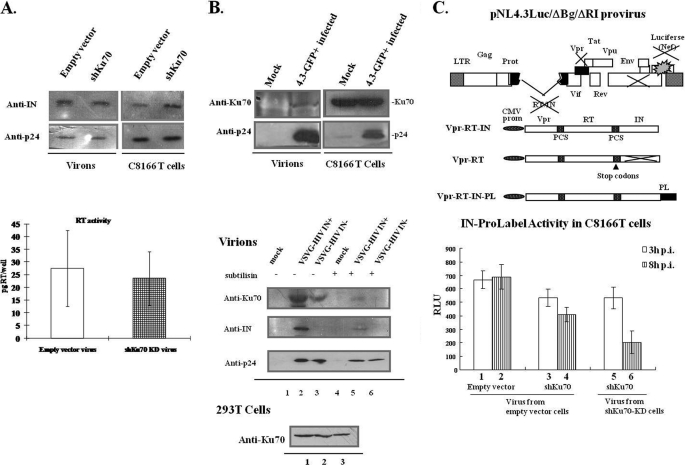
Because virus produced from Ku70-KD cells cannot infect C8166 cells efficiently, we first checked whether Ku70 knockdown may affect HIV-1 maturation. The progeny viruses produced from empty vector-transduced cells and Ku70-KD C8166 T cells were normalized by p24 value and loaded onto an SDS-polyacrylamide gel. Simultaneously, infected C8166 T cells were lysed and also subjected to the same analysis. Next, anti-p24 and anti-IN antibodies were used to check for the presence of p24 and IN in the virions and infected cells. We did not detect any difference in the p24/IN ratio in the E-virus- and Sh-virus-infected cells (Fig. 6A, top panel). This suggests that the virus Gag-pol processing remained unaffected in the progeny virus produced from Ku70-KD cells. To examine the RT activity in the virions produced in Ku70-KD cells, the same amounts of viruses (normalized by p24 ELISA) from either empty vector-transduced cells or Ku70-KD cells were lysed and subjected to an RT assay (Roche Applied Science). There was no significant difference in viral RT activity observed between normal and Ku70-KD cells (Fig. 6A, bottom panel). All of these data suggest that Ku70-KD does not affect the processing of Gag/Gag-Pol or virus maturation.
FIGURE 6.
Ku70 incorporation into HIV-1 particles and its effects on HIV-1 replication. A, knockdown of Ku70 does not affect the p24/IN profile in the virions or virus-producing cells. HIV pNL4.3-GFP virus produced from empty vector-infected and shKu70-KD C8166 T cells were pelleted through a 20% sucrose cushion and dissolved in RPMI 1640. Viral particles with the same amounts of p24 and equal amounts of infected cells were lysed, separated by SDS-PAGE, and immunoblotted with anti-IN and anti-p24 antibodies (top panel). The data shown represent two independent experiments. The same amount of pNL4.3-GFP virus (normalized by p24) produced from empty vector-transduced and shKu70-KD C8166 T cells was analyzed for HIV RT activity with a reverse transcriptase assay kit (Roche Applied Science). The data are shown as the amount of RT in each well, reported in pg/well, representing the means and S.D. values (error bars) from viruses produced in two independent experiments (bottom panel). B, top panel, Ku70 is present in the HIV-1 virion. C8166 T cells were infected with pNL4.3-GFP virus or uninfected. After 4 days of infection, supernatants were centrifuged through a 20% sucrose cushion at 35,000 rpm for 2 h at 4 °C. Pellets were dissolved in radioimmune precipitation assay buffer and subjected to a TCA precipitation assay. Protein contents in the viruses and the cells were analyzed by WB using anti-Ku70 and anti-p24 antibodies to determine the presence of various proteins. Bottom panel, Ku70 incorporation into HIV-1 virion is dependent on IN. The Vpr-RT-IN or Vpr-RT expressor was cotransfected with VSV-G and NL4.3lucΔBglΔRI to produce single cycle IN+ and IN− virus. The viruses were subjected to a subtilisin resistance assay as described under “Experimental Procedures.” Virus-associated Ku70, IN, and p24 were analyzed by WB. Endogenous Ku70 expressions in the transfected 293T cells were checked by blotting with anti-Ku70 antibody. C, top panel, schematic structure of RT/IN/Env-deleted HIV-1 provirus NL4.3lucΔBglΔRI and of the Vpr-RT-IN, Vpr-RT, and Vpr-RT-IN-PL fusion proteins. NL4.3lucΔBglΔRI and Vpr-RT-IN were described earlier (34, 69), and Vpr-RT without IN expression has two stop codons TAGTGA after the last nucleotide of the RT sequence. Vpr-RT-IN-PL was obtained through removal of the IN stop codon and insertion of the ProLabel gene after IN of Vpr-RT-IN plasmid as described under “Experimental Procedures.” Bottom panel, NL4.3lucΔBglΔRI was cotransfected with Vpr-RT-IN-PL and VSV-G expressor into 293T cells to generate VSV-G-pseudotyped HIV-1 single cycle IN-PL virus. 2 × 106 shKu70-KD or empty vector-transduced C8166 T cells were infected with of IN-PL virus (2000 pg of p24) produced from shKu70-KD or normal empty vector-transduced 293T cells for 3 h. The cells were washed three times, and half of the cells were collected. The rest of cells were kept in RPMI medium and harvested at 8 h p.i. All of the cells were then lysed and measured for ProLabel activity by the POLARstar OPTIMA multidetection microplate reader. The results are representative of three experiments, shown as mean ± S.D. RLU, relative light units.
The observation that the Ku70-KD phenotype resulted in defective progeny virus (Fig. 5) implies that Ku70 might be packaged into the progeny virus particles affecting HIV-1 replication. To test this hypothesis, we checked for the presence of Ku70 in the HIV virion. Briefly, the C8166 T cells were mock-infected or infected with HIV pNL4.3-GFP at an MOI of 1 for 2 h. Virions were isolated 4 days after infection on a 20% sucrose gradient by ultracentrifugation (35,000 rpm) for 1.5 h. Virions were lysed in radioimmune precipitation assay buffer and precipitated in 20% TCA to concentrate their protein contents before WB analysis. Simultaneously, the infected or uninfected C8166 T cells were lysed and analyzed by WB. Note that Ku70 was detected in the virions prepared from infected cells but not in the mock-infected cells (Fig. 6B, top panel). Additionally, the presence of p24 in the virions and the cells was detected by immunoblotting with anti-p24 antibody on the same membrane (Fig. 6B, top panel).
To verify that Ku70 incorporation into HIV-1 virion is mediated by IN, the VSV-G pseudotyped HIV-1 single cycle infection system was used. Vpr-RT-IN or Vpr-RT was transcomplemented with VSV-G and RT/IN/Env gene-deleted NLlucΔBglΔRI provirus into 293T cells to produce IN+ and IN− virus (Fig. 6C). The viruses were collected and ultracentrifuged through 20% sucrose at 35,000 rpm for 2 h. Because subtilisin treatment can effectively remove the proteins (either microvesicles or exosomes) outside the virions of purified HIV-1 virions (51), the IN+ and IN− virus were treated with 0.1 mg/ml subtilisin to remove potential contamination outside the virion. As shown in Fig. 6B (bottom panel), the presence of Ku70 is significantly higher in IN+ virus than IN− virus without subtilisin treatment (Fig. 6B (top panel), compare lane 2 with lane 3). However, when virions were treated with subtilisin, Ku70 was evidently detected in IN+ virus but not IN− virus (lanes 5 and 6, top panel). IN and p24 expression in both viruses were monitored by blotting with anti-IN and anti-p24 antibodies. Meanwhile, Ku70 expressions in mock-transfected or transfected cells were assessed (Fig. 6B, bottom panel). Taken together, these results suggest that, mediated by IN, Ku70 is incorporated into the HIV-1 particle.
The above results (Figs. 1 and 6B) suggest that virus-associated Ku70 might stabilize IN or protect it from degradation in the infected cells. To investigate the protective effects of Ku70 on IN during viral infection, we assessed IN expression in the presence or absence of Ku70 during VSV-G pseudotyped HIV-1 single cycle infection. To do so, we have constructed Vpr-RT-IN-PL plasmid with a ProLabel tag in the C terminus of IN, which enables us to quantify IN expression by measuring ProLabel activity (Fig. 6C). Vpr-RT-IN-PL is cotransfected with VSV-G and NLlucΔBglΔRI provirus into 293T cells to produce single cycle IN-PL virus (Fig. 6C). C8166 T cells were first infected with the same amount of normal IN-PL virus or virus produced from shKu70-KD 293T cells (normalized by p24 ELISA) and washed thoroughly at 3 h p.i., and half amounts of the cells were collected. The rest of the cells were harvested at 8 h p.i. All of the cells were lysed and measured for IN-ProLabel activity. The result showed that when C8166 T cells were infected with virus produced from normal 293T cells, the IN level remains unchanged after 8 h p.i., as compared with that at 3 h p.i. (Fig. 6C, bottom panel, compare bar 2 with bar 1). When Ku70-KD C8166 T cells were infected with virus produced from normal 293T cells, the IN level remained 77% after 8 h p.i., as compared with that at 3 h p.i. (Fig. 6C, bottom panel, compare bar 4 with bar 3). Remarkably, when Ku70-KD C8166 T cells were infected with virus produced from Ku70-KD 293T cells, at 8 h p.i., the IN level was reduced to ∼34%, as compared with that at 3 h p.i. (Fig. 6C, compare bar 6 with bar 5). All of these results together suggest that Ku70 present in the progeny virus and in the target cells contributes to stabilizing IN in the early stage of HIV-1 replication.
DISCUSSION
We studied the interaction between the cellular DNA repair protein Ku70 and HIV-1 IN and the potential roles of Ku70 in HIV-1 replication. By using a cell-based co-IP assay, we demonstrated that Ku70 interacts with HIV-1 IN in 293T cells and HIV-1-infected CD4+ T cells. Deletion analyses on IN and Ku70 indicated that an IN region encompassing aa 1–230 was unable to bind to Ku70, whereas the N-terminal region of Ku70 (aa 1–430) still retained the IN binding ability, and that their interaction is independent of Ku70/80 heterodimerization. We further discovered a dual mechanism for Ku70 in protecting IN from proteasomal degradation (i.e. by reducing overall protein ubiquitination levels within the host cells and by specifically reducing the ubiquitination of IN via their binding interaction). Finally, the knockdown of Ku70 expression in both virus-targeting and virus-producing CD4+ T cells significantly impaired HIV-1 replication. More specifically, Ku70-KD resulted in undetectable 2-LTR and integration levels in the early stage of viral replication. Taken together, our current study suggests that Ku70 is required for both the early and late stages of the HIV life cycle.
A recent study has found that Ku70 is able to reduce the ubiquitination of Bax in the regulation of apoptosis (30). This study indicated that the presence of Ku70 is able to specifically deubiquitinate Bax; however, whether Ku70 could affect total ubiquitination level in the host cells remained unknown. Presently, we discovered that, besides reducing the ubiquitination of Bax, Ku70 is able to universally down-regulate the ubiquitination of the entire complement of cellular proteins. The mechanism underlying this down-regulation of protein ubiquitination levels by Ku70 is unclear. A prior in vitro study revealed that Ku70, defined as a novel deubiquitination enzyme, was able to hydrolyze polyubiquitin chains into monoubiquitin units (30). In the cascade of the ubiquitin-proteasome pathway, deubiquitination enzymes remove ubiquitin chains from protein substrates and recycle the polyubiquitin chain into free ubiquitin, before or after the polyubiquitination chain is recognized by the 19 S cap of the 26 S proteasome (52). Thus, if Ku70 does exert an overall deubiquitinating activity on cellular protein, an increased free ubiquitin level in the cells would be expected. Unfortunately, under our experimental conditions, we were not able to detect an increased monoubiquitin form of ubiquitin when Ku70 was overexpressed. It appears that the total pool of monoubiquitin in the cells was reduced when Ku70 was overexpressed. Thus, how Ku70 interacts with the host ubiquitin-proteasome system to regulate the total pool of monoubiquitin in the cells and down-regulate the polyubiquitination of proteins remains an open question.
During the HIV life cycle, the host ubiquitin-proteasome system is repeatedly used by HIV-1 viral proteins, such as Vif, Vpr, Vpu, and IN, to ensure viral replication, either by targeting host restriction factors for degradation, such as Vif/APOBEC3G or Vpu/BST2, or by protection from proteasomal degradation by host cellular cofactors (e.g. IN is protected by LEDGF/p75, and Vpr is protected by Cul4A-DDB1DCAF1 ubiquitin ligase) (53–58). Here, we provide another example of a host cellular cofactor of IN, the DNA repair protein Ku70, which protects IN from host proteasomal degradation in the overexpression system and under HIV-1 infection (Figs. 1, 4B (top panel), and 6C (bottom panel)). Interestingly, our results showed that, in addition to down-regulating the ubiquitin pool within the cells, Ku70 is also able to specifically reduce the ubiquitination level of IN and its associated proteins through IN/Ku70(1–430) binding. Indeed, the IN-binding peptide Ku70(1–430) was able to increase IN expression and reduce the ubiquitination of IN (and its associated proteins) although it does not have any deubiquitinating activity (Fig. 4B, first and second panels, compare lane 4 with lane 2). This suggests that Ku70 specifically binds IN and possibly masks ubiquitin attachment site(s) in IN to reduce the ubiquitination of IN, thus protecting IN from degradation (Fig. 4B, top panel). This scenario seems very likely because the attachment of ubiquitin to protein substrates, conducted by ubiquitin ligases E3 during the last step, is via the covalent binding of the C-terminal Gly76 of ubiquitin to the ϵ-amino group of an internal lysine residue in the substrate protein (59), and the C terminus of IN(230–288) is known to be enriched in lysine residues (e.g. Lys240, Lys244, and Lys264). Thus, in a future study, it would be intriguing to investigate which lysine residue(s) in IN is specifically recognized by ubiquitin and subjected to proteasomal degradation.
We also provided solid evidence that HIV-1 IN directly interacts with Ku70 in Ku70/IN-overexpressing mammalian cells and in T-lymphocytes during HIV-1 infection (Fig. 2). Indeed, this finding is not surprising, considering that previous studies using MMLV IN as bait in a yeast two-hybrid system were able to fish out Ku70 (5), that Ku70 is present in MMLV PICs (27), and that Ku70 associates with the IN of Ty elements in Saccharomyces cerevisiae, yeast retrotransposons with life cycles similar to retroviruses (60). We then extended our study to delineate the mutual binding interface of IN and Ku70 and found that the C terminus of IN (aa 230–288) appears to be involved in the interaction with Ku70. One interesting observation here is that the Ku70 deletion mutant 1–430, which was able to bind IN, cannot form the Ku70/80 heterodimer (Fig. 3). This result is consistent with a previous finding from a two-hybrid analysis that the C-terminal 20 kDa of Ku70 (aa 430–609) is essential for Ku70/80 heterodimerization (44) and suggests that the Ku70/IN interaction may be independent of Ku80. It has been shown that Ku70 has unique functions independent of Ku80, although many cellular functions in which Ku70 participates do require Ku70/80 heterodimerization. For example, the antiapoptotic activity of Ku70 by inhibiting Bax-mediated apoptosis is independent of Ku80 (61). One concern here is that, despite the fact that the Ku70 N terminus (aa 1–430) can still mediate IN binding, we cannot rule out the possibility that IN is able to bind the Ku70/80 heterodimer or that IN binding to Ku70 may be enhanced by Ku70/80 association in mammalian cells. Indeed, several cellular functions of Ku70 require association with Ku80, and this heterodimerization formation enhances the stability of each subunit (62, 63).
In a finding of major relevance, we further determined that Ku70 is required for HIV-1 replication. HIV-1 infection was significantly impaired when Ku70 expression was knocked down in both producer and target cells (Fig. 5). Moreover, with a low MOI infection, Ku70-KD C8166 T cells were more significantly blocked in viral replication than for a high MOI infection (Fig. 5, A and B). These results indicate that Ku70 affects both the early and the late steps of the HIV-1 life cycle. In order to pinpoint the effect(s) of Ku70 on the early stage, we carried out real-time PCR to quantify late RT, 2-LTR circles, and integrated DNA under the same infection conditions as in Fig. 5C. Not surprisingly, 2-LTR circle formation in the Ku70-KD cells was undetectable, which is consistent with a previous report that NHEJ pathway is required for 2-LTR circle formation (Fig. 5D) (27, 50). In addition, integration of viral DNA was also abrogated by Ku70-KD, whereas reverse transcription measured by late RT products were reduced by around 50% in the Ku70-KD cells infected with shKu70 virus produced from Ku70-KD cells (Fig. 5D). Thus, Ku70 seems to be a multifaceted player in the early stage of viral infection. Ku70 is a component of the NHEJ pathway (15) that has been extensively investigated for its multiple functions during retroviral transduction or infection in previous studies. With respect to the effects of Ku70 on the early stage of HIV replication, one mechanism might be that Ku70 is actively involved in the gap repair of integration intermediates introduced by HIV-1 IN through the NHEJ pathway (20, 22–25, 27, 64). It explains an undetectable HIV integration event when Ku70 is absent during viral infection. The second possible mechanism, with a plausibility demonstrated in this study, is that Ku70 may protect IN from degradation before viral DNA integration by reducing overall cellular ubiquitination and/or by specifically interacting with IN in HIV-infected cells. This scenario was supported by studying the effect of Ku70 on IN-PL metabolism after viral entry in the early stage of viral replication (Fig. 6C). Depletion of Ku70 in the target cells had reduced IN expression to 77% at 8 h p.i. Remarkably, when Ku70-KD C8166 T cells were infected with virus produced from Ku70-KD 293T cells, the IN level at 8 h p.i., was reduced to ∼34%, as compared with that at 3 h p.i. (Fig. 6C, bottom panel, compare bar 6 with bar 5). These observations imply that the presence of Ku70 in both viruses and cells is able to protect IN to avoid host ubiquitin proteasomal degradation in the PIC. At this point, it should be noted that two other cellular proteins, hRad18 and LEDGF/p75, were also previously reported to protect IN from proteasomal degradation (58, 65). However, unlike these two IN cofactors, whose actions are mainly through their physiological binding to IN, Ku70 instead displays two different activities to protect IN from degradation. To date, it has not been clear how HIV-1 IN can coordinately recruit these cofactors to protect itself from the host proteasomal degradation machinery.
With regard to the effect of Ku70 on the late stage of the viral life cycle, we originally hypothesized that Ku70-KD might affect the Gag/Gag-Pol ratio because Ku70 might protect IN as an early Gag-Pol polyprotein precursor during viral assembly. In fact, the results shown in Fig. 6A demonstrate that Ku70 does not affect Gag/Gag-Pol processing and maturation. Further analysis of virion composition revealed that the host cellular protein Ku70 is present in the HIV-1 particles themselves (Fig. 6B). This finding indicates that Ku70 is packaged into HIV-1 particles as early as its assembly stage and becomes part of the HIV-1 PICs after the virus enters target cells. Within the PIC and associated with IN, Ku70 might deploy two mechanisms to contribute to the early stage of HIV replication: 1) protecting IN from the host proteasomal degradation pathway and 2) assisting viral protein IN in a specific replication step(s), including 2-LTR formation and integration. Consistently, our data revealed that the progeny virus produced from Ku70-KD cells was profoundly defective even when it was used to infect normal cells (Fig. 5C, bar 3). However, we also cannot exclude the possibility that the presence of Ku70 is required for other events during HIV-1 morphogenesis. This notion is strengthened by the previous findings that, during infection, free ubiquitin is incorporated into viral particles of HIV-1, simian immunodeficiency virus, MMLV, and equine infectious anemia virus (66) and that pr55Gag is monoubiquitinated during assembly and budding (67). In addition, proteasome inhibitor treatment interfered with the assembly and budding of HIV progeny virus and efficiently inhibited HIV infectivity (68). Thus, further investigation is certainly needed to fully understand how Ku70 impacts HIV-1 replication, and a better understanding of the interplay between HIV-1 IN and Ku70 during viral infection will rationalize the design of specific inhibitors to target this viral/cellular protein interaction and consequently inhibit HIV-1 replication.
Acknowledgments
We sincerely thank Dr. Heinrich Gottlinger for kindly providing HA-ubiquitin plasmids and Dr. Shigemi Matsuyama for valuable comments.
This work was supported by Canadian Institutes of Health Research (CIHR) Grants HOP-81180 and HBF 103212 and the Leaders Opportunity Fund Award from the Canadian Foundation of Innovation (to X.-J. Y.).
- IN
- integrase
- PIC
- preintegration complex
- IP
- immunoprecipitation
- WB
- Western blot
- MMLV
- Moloney murine leukemia virus
- NHEJ
- nonhomologous end-joining
- Ub
- ubiquitin
- VSV-G
- vesicular stomatitis virus G
- p.i.
- postinfection
- PL
- ProLabel
- aa
- amino acids
- MOI
- multiplicity of infection.
REFERENCES
- 1. Wang G. P., Ciuffi A., Leipzig J., Berry C. C., Bushman F. D. (2007) Genome Res. 17, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J. R., Bushman F. (2005) Nat. Med. 11, 1287–1289 [DOI] [PubMed] [Google Scholar]
- 3. Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Maele B., Debyser Z. (2005) AIDS Rev. 7, 26–43 [PubMed] [Google Scholar]
- 5. Studamire B., Goff S. P. (2008) Retrovirology 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee M. S., Craigie R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9823–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farnet C. M., Bushman F. D. (1997) Cell 88, 483–492 [DOI] [PubMed] [Google Scholar]
- 8. Kalpana G. V., Marmon S., Wang W., Crabtree G. R., Goff S. P. (1994) Science 266, 2002–2006 [DOI] [PubMed] [Google Scholar]
- 9. Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempé D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Christ F., Rain J. C., Dargemont C., Debyser Z., Benarous R. (2005) J. Biol. Chem. 280, 25517–25523 [DOI] [PubMed] [Google Scholar]
- 10. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 11. Lin C. W., Engelman A. (2003) J. Virol. 77, 5030–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aravind L., Koonin E. V. (2001) Genome Res. 11, 1365–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Downs J. A., Jackson S. P. (2004) Nat. Rev. Mol. Cell Biol. 5, 367–378 [DOI] [PubMed] [Google Scholar]
- 14. Doherty A. J., Jackson S. P., Weller G. R. (2001) FEBS Lett. 500, 186–188 [DOI] [PubMed] [Google Scholar]
- 15. Tuteja R., Tuteja N. (2000) Crit. Rev. Biochem. Mol. Biol. 35, 1–33 [DOI] [PubMed] [Google Scholar]
- 16. Giffin W., Torrance H., Rodda D. J., Préfontaine G. G., Pope L., Hache R. J. (1996) Nature 380, 265–268 [DOI] [PubMed] [Google Scholar]
- 17. Wang J., Dong X., Myung K., Hendrickson E. A., Reeves W. H. (1998) J. Biol. Chem. 273, 842–848 [DOI] [PubMed] [Google Scholar]
- 18. Roe T., Chow S. A., Brown P. O. (1997) J. Virol. 71, 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lau A., Swinbank K. M., Ahmed P. S., Taylor D. L., Jackson S. P., Smith G. C., O'Connor M. J. (2005) Nat. Cell Biol. 7, 493–500 [DOI] [PubMed] [Google Scholar]
- 20. Daniel R., Pomerantz R. J. (2005) Nat. Cell Biol. 7, 452–453 [DOI] [PubMed] [Google Scholar]
- 21. Skalka A. M., Katz R. A. (2005) Cell Death Differ. 12, Suppl. 1, 971–978 [DOI] [PubMed] [Google Scholar]
- 22. Daniel R., Greger J. G., Katz R. A., Taganov K. D., Wu X., Kappes J. C., Skalka A. M. (2004) J. Virol. 78, 8573–8581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Daniel R., Katz R. A., Merkel G., Hittle J. C., Yen T. J., Skalka A. M. (2001) Mol. Cell. Biol. 21, 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeanson L., Subra F., Vaganay S., Hervy M., Marangoni E., Bourhis J., Mouscadet J. F. (2002) Virology 300, 100–108 [DOI] [PubMed] [Google Scholar]
- 25. Daniel R., Katz R. A., Skalka A. M. (1999) Science 284, 644–647 [DOI] [PubMed] [Google Scholar]
- 26. Greene W. C., Peterlin B. M. (2002) Nat. Med. 8, 673–680 [DOI] [PubMed] [Google Scholar]
- 27. Li L., Olvera J. M., Yoder K. E., Mitchell R. S., Butler S. L., Lieber M., Martin S. L., Bushman F. D. (2001) EMBO J. 20, 3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller M. D., Farnet C. M., Bushman F. D. (1997) J. Virol. 71, 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeanson L., Mouscadet J. F. (2002) J. Biol. Chem. 277, 4918–4924 [DOI] [PubMed] [Google Scholar]
- 30. Amsel A. D., Rathaus M., Kronman N., Cohen H. Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5117–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yao X. J., Subbramanian R. A., Rougeau N., Boisvert F., Bergeron D., Cohen E. A. (1995) J. Virol. 69, 7032–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ao Z., Danappa Jayappa K., Wang B., Zheng Y., Kung S., Rassart E., Depping R., Kohler M., Cohen E. A., Yao X. (2010) J. Virol. 84, 8650–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ao Z., Huang G., Yao H., Xu Z., Labine M., Cochrane A. W., Yao X. (2007) J. Biol. Chem. 282, 13456–13467 [DOI] [PubMed] [Google Scholar]
- 34. Ao Z., Fowke K. R., Cohen E. A., Yao X. (2005) Retrovirology 2, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strack B., Calistri A., Göttlinger H. G. (2002) J. Virol. 76, 5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ao Z., Yu Z., Wang L., Zheng Y., Yao X. (2008) PLoS One 3, e1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Z., Zheng Y., Ao Z., Clement M., Mouland A. J., Kalpana G. V., Belhumeur P., Cohen E. A., Yao X. (2008) Retrovirology 5, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng Y., Ao Z., Jayappa K. D., Yao X. (2010) Virol. J. 7, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mouland A. J., Mercier J., Luo M., Bernier L., DesGroseillers L., Cohen E. A. (2000) J. Virol. 74, 5441–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mulder L. C., Muesing M. A. (2000) J. Biol. Chem. 275, 29749–29753 [DOI] [PubMed] [Google Scholar]
- 41. Berthoux L., Sebastian S., Muesing M. A., Luban J. (2007) Virology 364, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McKee C. J., Kessl J. J., Shkriabai N., Dar M. J., Engelman A., Kvaratskhelia M. (2008) J. Biol. Chem. 283, 31802–31812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jin S., Weaver D. T. (1997) EMBO J. 16, 6874–6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu X., Lieber M. R. (1996) Mol. Cell. Biol. 16, 5186–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koike M., Shiomi T., Koike A. (2001) J. Biol. Chem. 276, 11167–11173 [DOI] [PubMed] [Google Scholar]
- 46. Kiel C., Serrano L. (2006) J. Mol. Biol. 355, 821–844 [DOI] [PubMed] [Google Scholar]
- 47. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 48. Dubiel W., Gordon C. (1999) Curr. Biol. 9, R554–R557 [DOI] [PubMed] [Google Scholar]
- 49. Bukrinsky M. I., Sharova N., Dempsey M. P., Stanwick T. L., Bukrinskaya A. G., Haggerty S., Stevenson M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6580–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kilzer J. M., Stracker T., Beitzel B., Meek K., Weitzman M., Bushman F. D. (2003) Virology 314, 460–467 [DOI] [PubMed] [Google Scholar]
- 51. Ott D. E. (2009) Methods Mol. Biol. 485, 15–25 [DOI] [PubMed] [Google Scholar]
- 52. Klinger P. P., Schubert U. (2005) Expert Rev. Anti Infect. Ther. 3, 61–79 [DOI] [PubMed] [Google Scholar]
- 53. Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 54. Le Rouzic E., Morel M., Ayinde D., Belaïdouni N., Letienne J., Transy C., Margottin-Goguet F. (2008) J. Biol. Chem. 283, 21686–21692 [DOI] [PubMed] [Google Scholar]
- 55. Douglas J. L., Viswanathan K., McCarroll M. N., Gustin J. K., Früh K., Moses A. V. (2009) J. Virol. 83, 7931–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Conticello S. G., Harris R. S., Neuberger M. S. (2003) Curr. Biol. 13, 2009–2013 [DOI] [PubMed] [Google Scholar]
- 57. Marin M., Rose K. M., Kozak S. L., Kabat D. (2003) Nat. Med. 9, 1398–1403 [DOI] [PubMed] [Google Scholar]
- 58. Llano M., Delgado S., Vanegas M., Poeschla E. M. (2004) J. Biol. Chem. 279, 55570–55577 [DOI] [PubMed] [Google Scholar]
- 59. Sadowski M., Sarcevic B. (2010) Cell Div. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Downs J. A., Jackson S. P. (1999) Mol. Cell. Biol. 19, 6260–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sawada M., Sun W., Hayes P., Leskov K., Boothman D. A., Matsuyama S. (2003) Nat. Cell Biol. 5, 320–329 [DOI] [PubMed] [Google Scholar]
- 62. Gu Y., Jin S., Gao Y., Weaver D. T., Alt F. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8076–8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M. C., Li G. C. (1996) Nature 382, 551–555 [DOI] [PubMed] [Google Scholar]
- 64. Daniel R., Litwin S., Katz R. A., Skalka A. M. (2001) J. Virol. 75, 3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mulder L. C., Chakrabarti L. A., Muesing M. A. (2002) J. Biol. Chem. 277, 27489–27493 [DOI] [PubMed] [Google Scholar]
- 66. Ott D. E., Coren L. V., Copeland T. D., Kane B. P., Johnson D. G., Sowder R. C., 2nd, Yoshinaka Y., Oroszlan S., Arthur L. O., Henderson L. E. (1998) J. Virol. 72, 2962–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pornillos O., Garrus J. E., Sundquist W. I. (2002) Trends Cell Biol. 12, 569–579 [DOI] [PubMed] [Google Scholar]
- 68. Schubert U., Ott D. E., Chertova E. N., Welker R., Tessmer U., Princiotta M. F., Bennink J. R., Krausslich H. G., Yewdell J. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13057–13062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ao Z., Yao X., Cohen E. A. (2004) J. Virol. 78, 3170–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]