Abstract
Bacterial polysaccharides are known to induce the immune response in macrophages. Here we isolated a novel extracellular polysaccharide from the biofilm of Thermus aquaticus YT-1 and evaluated its structure and immunomodulatory effects. The size of this polysaccharide, TA-1, was deduced by size-exclusion chromatography as 500 kDa. GC-MS, high performance anion-exchange chromatography with pulsed amperometric detection, electrospray ionization-MS/MS, and NMR revealed the novel structure of TA-1. The polysaccharide is composed of tetrasaccharide-repeating units of galactofuranose, galactopyranose, and N-acetylgalactosamine (1:1:2) and lacked acidic sugars. TA-1 stimulated macrophage cells to produce the cytokines TNF-α and IL-6. Screening of Toll-like receptors and antibody-blocking experiments indicated that the natural receptor of TA-1 in its immunoactivity is TLR2. Recognition of TA-1 by TLR2 was confirmed by TA-1 induction of IL-6 production in peritoneal macrophages from wild-type mice but not from TLR2−/− mice. TA-1, as a TLR2 agonist, could possibly be used as an adjuvant and could enhance cytokine release, which increases the immune response. Furthermore, TA-1 induced cytokine release is dependent on MyD88/TIRAP.
Keywords: Bacteria, Cytokines Induction, Immunology, Polysaccharide, Toll-like Receptors (TLR), Biofilm, Immunomodulatory
Introduction
The excessive use of antibiotics poses tremendous selection pressure on microorganisms to develop drug resistance, which eventually leads to incurable diseases. Recent alternatives to antibiotics are immunomodulators. Instead of combating pathological microbes directly, the immunomodulators act to enhance the host defense responses without the development of drug resistance.
Natural products extracted from microorganisms, mushrooms, algae, lichens, and higher plants were known to induce positive immunological effects and have been frequently used in the ancient practice of Chinese medicine (1). The mechanisms of action of these substances are often unknown, but polysaccharides in these extracts have been found to be the primary factor for macrophage stimulation through induction of the immune system of Toll-like receptors (2, 3). These biopolymers often show advantages over the polysaccharides that are currently in use, especially in combating microbial infections (4). Consequently, modulation of the innate immune system significantly improves the host ability to respond to different pathogens and diseases.
The innate immune system is the first line of host defense. It protects the host from a variety of pathogens based on a limited repertoire of germ line-encoded receptors called pattern recognition receptors (PRRs).4 PRRs include members of the Toll-like receptor (TLR) family and nucleotide binding oligomerization domain-like receptors and retinoic acid-inducible gene-I-like receptors, which recognize pathogen-associated molecules, such as microbial components, and then trigger the release of inflammatory cytokine and type I interferons for host defense (5–8). These PRRs are localized in distinct cellular compartments. TLR1, TLR2, TLR4, TLR5, and TLR6 are expressed on the cell surface, whereas TLR3, TLR7, TLR8, and TLR9, nucleotide binding oligomerization domain-like receptors, and retinoic acid-inducible gene-I-like receptors are found in the cytoplasm (7, 8). In the past decade, a series of microbial ligands has been identified that are sensed by the different TLR complexes, including bacterial and viral DNA/RNA (TLR3, TLR7, TLR8, and TLR9), lipopeptides (TLR1/TLR2, TLR6/TLR2), bacterial glycolipids (lipopolysaccharide, LPS; TLR4/MD-2), and bacterial flagellin (TLR5) (9). From a microbiological point of view, the generalization of the aforementioned compounds as TLR ligands is a gross simplification and ignores the tremendous variation in the various types of molecules found in different bacterial species or even in a single bacterial strain. Once TLRs recognize their cognate ligands, they dimerize and initiate a signaling cascade that results in NF-κB activation followed by inflammatory cytokine (TNF-α and IL-6, etc.) release.
Bacteria produce large quantities of extracellular polysaccharide (EPS) when they form biofilm, providing a natural resource for novel polysaccharides (10). Biofilm is a consortium of microorganisms immobilized and penned within EPS that can restrict the diffusion of substances and antimicrobial agents (11). The EPSs synthesized by microbial cells vary greatly in their composition and, hence, in their chemical and physical properties. Bacterial EPSs are highly heterogeneous polymers containing a number of distinct monosaccharides and non-carbohydrate substituents that are species-specific. Polysaccharide chains are usually formed by using an oligosaccharide as a repeating unit. The oligosaccharide may also be both functional and species specific and can vary in size depending on the degree of polymerization.
Thermus aquaticus YT-1 (ATCC 25104) is a Gram-negative, rod-shaped bacterium that forms biofilm under certain conditions (12). Unlike typical Gram-negative bacteria, it does not contain lipopolysaccharide in the outer membrane (13). It is widespread in natural and thermal habitats and grows from 50 to 85 °C (14). T. aquaticus and other thermophiles may be protected from environmental stress, such as high temperature, by biofilm, unusual glycolipids, and high DNA GC contents (13–15). Although many thermophiles have been isolated from hot springs, there are few studies of their biofilm EPS. Here we characterized the primary structure of TA-1, the major EPS secreted by T. aquaticus YT-1. We show that TA-1 possesses immunological activity, which can possibly be medically applied in the future.
EXPERIMENTAL PROCEDURES
Materials
Pam3csk4, poly(I:C), LPS, flagellin, and R848 were purchased from InvivoGen (San Diego, CA). Phosphorothioate-modified ODN1668 were synthesized by MEG Biotech (Ebersberg, Germany). siRNA pools against MyD88 and TIRAP genes, and scrambled siRNA were all obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-α-tubulin and anti-TIRAP antibodies were purchased from Epiomics (Epiomics Inc., CA), and anti-MyD88 antibody was from Abcam (Abcam, Cambridge, UK).
Bacterial Culture and EPS Purification
The crude EPS of biofilm was isolated as described in Wozniak (16) and Yildiz and Schoolnik (17). Briefly, T. aquaticus YT-1 (ATCC 25104), purchased from Bioresource Collection and Research Center, Taiwan, was cultured on Thermus agar plates covered with cellophane for 36 h at 60 °C (18). The cellophane membrane was then placed in 0.9% NaCl and centrifuged at 8000 × g for 15 min to pellet bacterial cells. The supernatant was mixed with 3 volumes of 95% alcohol and incubated at 4 °C overnight. The crude EPS was precipitated after centrifugation at 2500 × g at 0 °C for 15 min, then dissolved in double distilled H2O and lyophilized. Crude EPS (4 mg) was dissolved in 1 ml of double distilled H2O and centrifuged at 3000 × g for 3 min. The solution was filtered on 0.45-μm filters. The filtrate was loaded onto an HW-65F column (130 cm × 16 mm) and eluted with H2O at 0.5 ml min−1. Dextrans with sizes of 10, 70, and 500 kDa were used as standards.
Sugar Composition Analysis
The carbohydrate composition was analyzed by high performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) and GC-MS as follows.
For HPAEC-PAD analysis, TA-1 was hydrolyzed by 2 m trifluoroacetic acid at 120 °C for 2 h. The monosaccharides obtained were loaded onto a Carbopac PA10 analytical column (4 × 250 mm) with a Carbopac PA10 Guard column (4 × 50 mm) and eluted at a flow rate of 1 ml min−1 at 30 °C. The detector pulse potentials and durations were E1 = 0.05 V (0.4 ms), E2 = 0.75 V (0.2 ms), and E3 = −0.15 V (0.4 ms). The integration was recorded from 0.2 to 0.4 ms during the E1 application. Arabitol was used as an internal standard.
For GC-MS analysis, purified TA-1 was first treated with 0.5 m methanolic-HCl at 80 °C for 16 h, re-N-acetylated with methanol/pyridine/acetic anhydride (50:1:10, by volume) at room temperature, and then trimethylsilylated with Sylon HTP trimethylsilylation reagent (Supelco). The final trimethylsilylated products were dissolved in n-hexane for GC-MS on a Hewlett Packard gas chromatography HP6890 system with an HP5973 mass selective detector.
Uronic Acid Assay
The amount of uronic acid in the EPS supernatant fraction of TA-1 dissolved in double distilled H2O was determined using a modified carbazole assay with glucuronic acid as a standard.
Linkage Analysis
Sugar linkages were analyzed using Hakomori methylation (19). Briefly, the polysaccharides were permethylated in CH3I/DMSO under alkaline conditions and subsequently hydrolyzed by 2 m trifluoroacetic acid (0.2 ml) at 120 °C for 2 h. The sample was dried under a stream of nitrogen and then reduced in 200 μl of 1 m NH4OH/5 mg of NaBD4/ethanol for 2 h at room temperature. Acetic acid (100%) was added to terminate the reaction. After evaporation under a stream of nitrogen, 200 μl of 10% acetic acid in methanol was added and then evaporated; this procedure was repeated at least 5 times to yield crystals of the sugar derivatives. The crystallized derivatives were further acetylated in 200 μl of acetic anhydride at 100 °C for 1 h and then cooled to room temperature. The partially methylated aditol acetate derivatives were extracted three times with chloroform/double distilled H2O. The chloroform was evaporated, and the residue was suspended in n-hexane for analysis of the sugar linkages by GC-MS.
Endo-β-1,4-d-galactanase Digestion
Native TA-1 was digested by endo-β-1,4-d-galactanase to form small fragments. Endo-β-1,4-d-galactanase was added to dissolved in 20 mm ammonium acetate buffer (pH 4.5) for 1 h at 50 °C. The mixture was incubated for 10 min at 95 °C. The mixture was then centrifuged at 8000 × g, and the supernatant was lyophilized. The depolymerized TA-1 EPS was examined by NMR, electrospray ionization-MS and -MS/MS.
Nuclear Magnetic Resonance Spectroscopy
NMR spectra of in D2O were recorded on Bruker Avance 500 and 600 spectrometers (equipped with the cryoprobe) at 350 K. One- and two-dimensional total correlation spectroscopy spectra were recorded with mixing times of 80 to 250 ms, which allows proton chemical shifts of carbohydrates to be assigned. Two-dimensional 13C,1H HMBC spectra were recorded with 2J or 3J H-C coupling constants at 8 and 5 Hz. Two-dimensional 13C,1H HMQC spectra were recorded with 1J H-C coupling constants at 145 Hz. Two-dimensional nuclear Overhauser effect (NOE) spectra were recorded with mixing times of 200 to 500 ms.
Cell Culture
Human embryonic kidney (HEK) fibroblast cells (HEK293T) and murine RAW264.7 macrophages were obtained from the American Type Culture Collection (Manassas, VA). Human monocyte cell lines, THP-1, and MyD88-deficient THP-1 (THP-1 MyD88−/−) were purchased from InvivoGen). HeNC2 (TLR4+/+) and GG2EE (TLR4−/−) cells, kindly provided by Dr. Danuta Radzioch (McGill University, Montreal, Canada), and THP-1 cells were propagated in RPMI 1640 medium supplemented with 10% heated- inactivated fetal bovine serum (HyClone, Logan, UT) and 2 mm l-glutamine (Invitrogen) and cultured at 37 °C under a 5% CO2 atmosphere. HEK293T and RAW264.7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 50 μg ml−1 penicillin, 50 μg ml−1 streptomycin sulfate, and 100 μg ml−1 neomycin sulfate (Invitrogen) under a humidified atmosphere of 5% CO2 at 37 °C.
Nuclear Factor κ Light-chain Enhancer of Activated B Cells (NF-κB) Reporter Gene Assay
HEK293T cells (2.5 × 104 cells per well) were seeded onto a 96-well plate in DMEM medium and incubated overnight. The cells were transfected using Lipofectamine2000 (Invitrogen) plus 0.01 μg of TLR-expressing plasmid, 0.07 μg of p5xNF-κB-luc plasmid (Stratagene, TX), and 0.02 μg of pcDNA3.1-βgal according to the manufacturer's instructions. The cells were incubated with TLR ligands for 6 h, washed twice with PBS, and then lysed. NF-κB luciferase activities were measured using the luciferase assay system (Promega) according to the manufacturer's instructions. Levels of firefly luciferase expression were normalized against β-galactosidase activity as a control for transfection efficiency and expressed as -fold stimulation over the unstimulated pcDNA3.1 empty vector control.
ELISA
RAW264.7 cells (2 × 105 cells per well) were seeded onto a 24-well plate in EMEM medium and incubated overnight. The cells were treated with polysaccharide samples for 24 h. The cytokine concentration in the medium of each well was determined by ELISA. The ELISA reagents and protocol used are commercially available from R&D System. The final cytokine concentrations were measured at 550 nm on an MRX microplate reader.
Nitric Oxide Detection
NO production in culture medium was determined by measuring nitrite (NO2−), a stable breakdown product of NO, using the Griess reagent (Sigma). Griess reagent (50 μl) was added to 50 μl of culture medium. After 15 min of incubation at room temperature, the nitrite concentration was measured at 540 nm on a microtiter plate reader. Nitrite concentrations were calculated by comparison with a standard curve of sodium nitrite.
TLR Antibody Blocking
RAW264.7 or THP-1 cells were treated with 10 μg ml−1 mouse monoclonal anti-TLR2 (eBioscience) or 10 μg ml−1 mouse isotype control IgG (Pierce) for 1 h before exopolysaccharide treatment. Cytokine production and nitric oxide production in the culture medium were measured 24 h later using ELISA and Griess reagent, respectively.
Isolation of Peritoneal Macrophages
Wild-type and TLR2−/− mice with a B6.129 background were purchased from The Jackson Laboratory (Bar Harbor, ME). Macrophages were isolated from peritoneal exudates of mice 3 days after intraperitoneal injection of 2 ml of 3% w/v thioglycolate solution.
Transfection
Raw 264.7 cells (2 × 106) were transfected with 40 nm siRNA by using the Amaxa Nucleofector (program D-032). All subsequent assays were performed after 48 h of transfection.
RESULTS
Structural Characterization of TA-1
Crude EPS from T. aquaticus YT-1 was fractionated by gel filtration on an HW-65F column. Four fractions with EPS were collected (supplemental Fig. S1); the EPS in the first fraction was named TA-1 and was selected for further structural characterization and immunomodulatory analysis.
The chemical structure of TA-1 was determined by sugar analyses, mass spectrometry, and NMR spectroscopy. The sugars of TA-1 consisted of galactose and N-acetylgalactosamine at a molar ratio of 1:1, as determined by GC-MS and HPAEC-PAD (not shown). The uronic acid assay indicated that TA-1 lacked acidic sugars (not shown), which is unusual for in bacterial biofilm EPS. Linkage analysis revealed two types of galactose derivatives (terminal Galf (D) and 3-substituted Galp (B)) and two types of N-acetylgalactosamines (3-substituted GalNAc (C) and 3,4-substituted GalNAc (A)); see Fig. 1F, for residue designations.
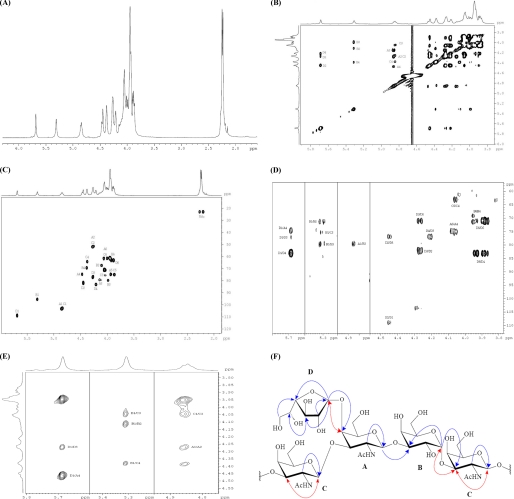
FIGURE 1.
Serious spectra of NMR analysis of the EPS TA-1 from T. aquaticus YT-1 (A–F). A, shown is an 1H NMR (600 MHz, D2O, 350 K) spectrum. B, two-dimensional total correlation spectroscopy (600 MHz, D2O, 350 K, 150 ms) spectrum is shown. C, HMQC (600 MHz, D2O, 350 K) is shown. D, HMBC (600 MHz, D2O, 350 K) is shown. E, two-dimensional NOE spectroscopy (600 MHz, D2O, 500 ms) is shown. F, shown is the structure of TA-1. In all figure parts, the residues are as follows: A = 3,4-substituted N-acetyl-galactosamine, B = 3-substituted galactopyranose, C = 3-substituted N-acetyl-galactosamine, and D = terminal galactofuranose.
The 1H NMR spectrum (Fig. 1A) revealed the anomeric protons δ 4.86 (residue A), δ 5.31 (residue B), δ 4.84 (residue C), and δ 5.69 (residue D) of the four residues deduced from linkage analysis. In two-dimensional total correlation spectroscopy (Fig. 1B), HMQC (Fig. 1C), and HMBC (Fig. 1D), residue D was identified as a non-reducing terminal galactofuranose. The signal characteristics for the downfield signals of C-4 (d 82.8) and H-1 also confirmed the assignment. The anomeric proton of terminal galactofuranose (residue D) was confirmed by the presence of a cross-peak with C-4 of residue A in HMBC (Fig. 1D). This result together with the NOE correlation (Fig. 1E) between H-1 of the terminal galactofuranose and H-4 of residue A suggested that the terminal galactofuranose is linked to the C-4 of residue A (Fig. 1F). Residue A was identified as 1,3,4-linked N-acetylgalactosamine according to the downfield chemical shifts of C-3 and C-4 (d 78.9 and d 74.2).
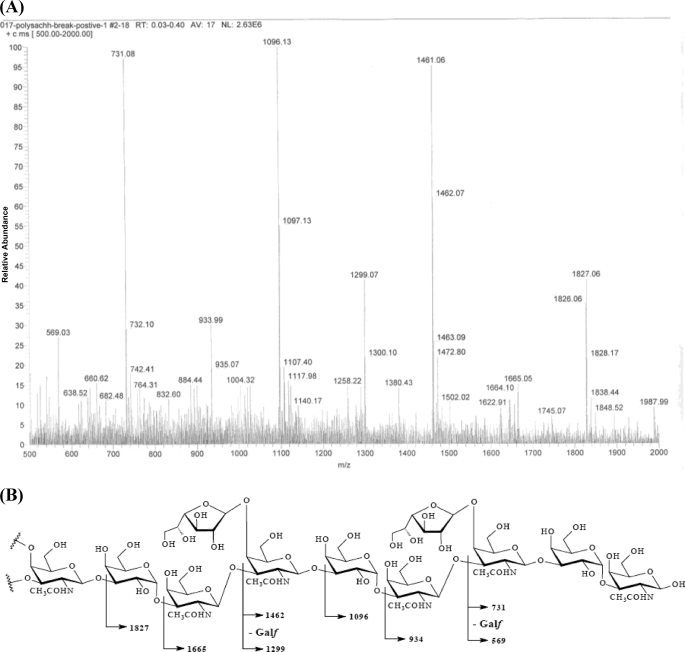
The similar features of residue B (dC-3 79.3) and residue C (dC-3 75.0) indicated that these residues, B and C, are 1,3-linked galactopyranose and N-acetyl-galactosamine, respectively. The HMBC correlations (Fig. 1D) between H-1 of residue A and C-3 of residue B and H-1 of residue B and C-3 of residue C together with the NOE cross-peak (Fig. 1E) between H-1 of residue B and H-3 of residue C revealed the sequence of the glycosyl moieties of TA-1: D-A-B-C (Fig. 1F). Although there was no long-range 13C,1H correlation and the NOE cross-peak indicated that residue C was linked to residue A, the electrospray ionization-MS/MS fragmentation of depolymerized TA-1 (Fig. 2A) clearly coincided with the NMR results and unambiguously confirmed the structure and order of the glycosyl moieties (Fig. 2B). Based on these studies, TA-1 was shown to be a novel polysaccharide structure.
FIGURE 2.
Electrospray ionization-MS/MS fragmentation spectra of depolymerized TA-1 (A) and primary structure of TA-1 (B). A, the MS/MS fragments revealed the TA-1 primary sequence unambiguously. B, the labeled molecular weights corresponded to the MS/MS fragments in A.
TA-1 Stimulated Macrophages to Secrete Cytokines and NO
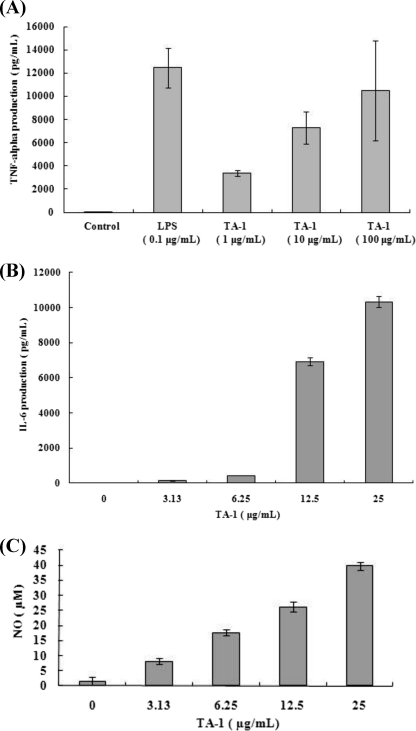
Many bacterial components are immunoactive, e.g. lipopeptides and polysaccharides. Therefore, we tested the ability of TA-1 to stimulate macrophages to secrete cytokines involved in nonspecific primary defense against infectious agents. Treatment of macrophage cell line RAW264.7 with TA-1 resulted in a marked increase in TNF-α (Fig. 3A) and IL-6 (Fig. 3B) production in a dose-dependent manner.
FIGURE 3.
Stimulation of macrophages by TA-1 (A–C). RAW264.7 cells were treated with TA-1 at the dosages indicated. TNF-α (A), IL-6 (B), and NO (C) in culture medium were measured using ELISA (A and B) or Griess reagent (C). Data represent the mean ± S.D. of triplicates from one of at least three independent experiments.
Because such induced immunological events accompany oxidative bursts when cells are exposed to foreign substances, we tested whether TA-1 stimulates macrophages to release NO. Indeed, TA-1 induced RAW264.7 macrophages to release NO (Fig. 3C).
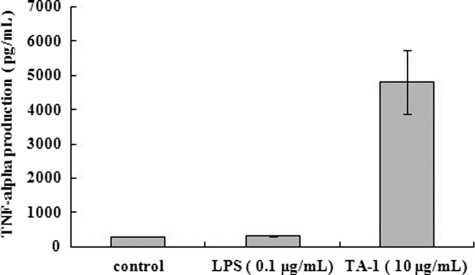
LPS also induces an immunological response in murein macrophages (20). Although T. aquaticus does not possess LPS (12, 13), we tested whether our assays were contaminated with LPS by pretreating the murine macrophages with polymyxin B, which binds to LPS. After pretreatment of macrophages with polymyxin B, TA-1 remained immunologically active, but LPS lost its immunological effect as expected (Fig. 4). Our results indicated that TA-1, not contaminating LPS, induced macrophages to produce TNF-α, IL-6, and NO.
FIGURE 4.
Pretreatment of murine macrophages with polymyxin B to exclude LPS contamination of assays. Effect of LPS and TA-1 on TNF-α secretion by Raw 264.7 macrophages after pretreatment with polymyxin B is shown. The control was pretreated with polymyxin B and not treated with LPS or TA-1.
TLR2 Was the Natural Receptor of TA-1 in Macrophages
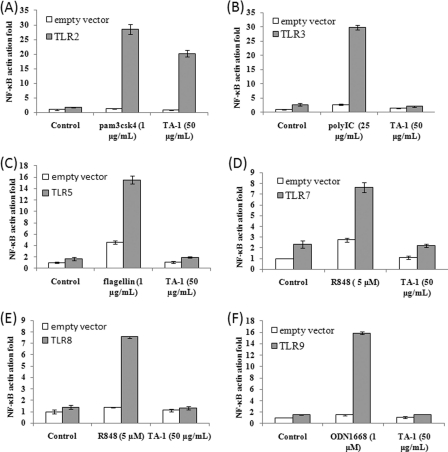
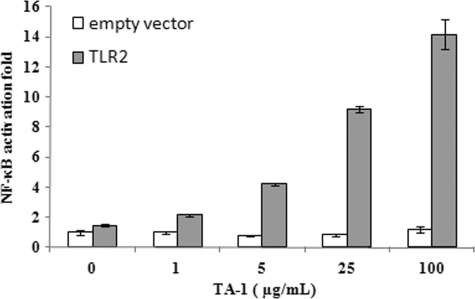
PRRs, such as TLRs, are responsible for initiating the diverse immune responses (21). We used reporter assays to identify the key TLR for TA-1. HEK293T cells, which do not normally express any TLRs, were transiently transfected with specific TLR-expressing plasmids and co-transfected with the NF-κB reporter plasmid to evaluate which TLRs participated in the response to TA-1. TA-1 activated NF-κB through TLR2 but not through TLRs 3, 5, 7, 8, or 9 (Fig. 5, A–F). Also, the TLR2 activation induced by TA-1 was dose-dependent (Fig. 6).
FIGURE 5.
Activation of NF-κB reporter plasmid by TA-1 TLRs (A–F). HEK293T cells were transfected with p5xNF-κB-luc, pcDNA3.1-β-galactosidase, and either the vector pcDNA3.1 (empty vector) or this vector expressing one of six TLR genes as indicated TLR2 (A), TLR3 (B), TLR5 (C), TLR7 (D), TLR8 (E), and TLR9 (F). Twenty-four hours after transfection, the cells were treated with TA-1 (50 μg ml−1) or left untreated for 6 h; specific TLR ligands were also added as positive controls. The cells were lysed, and the lysates were used for luciferase activity measurement. Data represent the mean ± S.D. of triplicates from one of at least two independent experiments.
FIGURE 6.
Activation of NF-κB by TA-1 via TLR2. HEK293T cells were transfected with p5xNF-κB-luc, pcDNA3.1-β-gal, and either pcDNA3.1 (empty vector) or pcDNA3.1 carrying the TLR2 gene. Twenty-four hours after transfection, the cells were treated with TA-1 at the concentrations indicated or left untreated for 6 h. The cells were lysed, and the lysates were used for luciferase activity measurement. Data represent the mean ± S.D. of triplicates from one of at least two independent experiments.
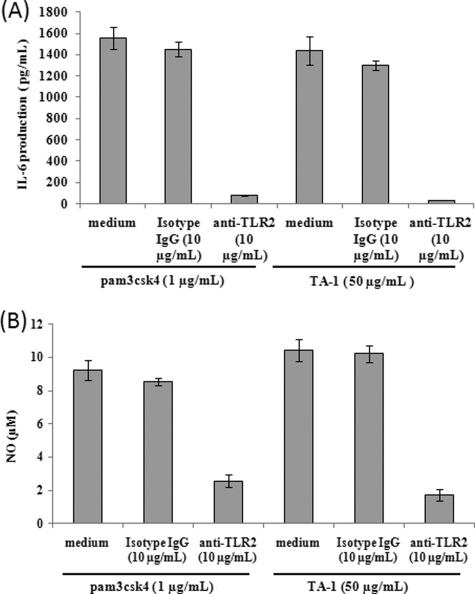
To examine whether TLR2 was involved in TA-1-induced cytokine or nitric oxide production in macrophages, RAW264.7 cells were pretreated with a specific anti-TLR2 antibody to block the extracellular domain of TLR2 on the cell membrane. Both IL-6 (Fig. 7A) and nitric oxide production (Fig. 7B) induced by TA-1 significantly decreased when cells were pretreated with TLR2-specific antibodies but not when cells were pretreated with the isotype control IgG. These results suggested that TA-1 induces cytokines and NO production mediated through TLR2 in macrophages.
FIGURE 7.
Blocking of TA-1 induced IL-6 and NO productions by TLR2-specific antibody (A and B). RAW264.7 cells were treated with TLR2 antibody, the isotype control IgG, or left untreated for 1 h followed by TA-1 and pamcsk4 treatments for 24 h. The pam3csk4 treatment was served as positive control. IL-6 (A) or NO (B) production by the macrophages in culture medium was measured by using ELISA or Griess reagent, respectively. Data represent the mean ± S.D. of triplicates from one of at least two independent experiments.
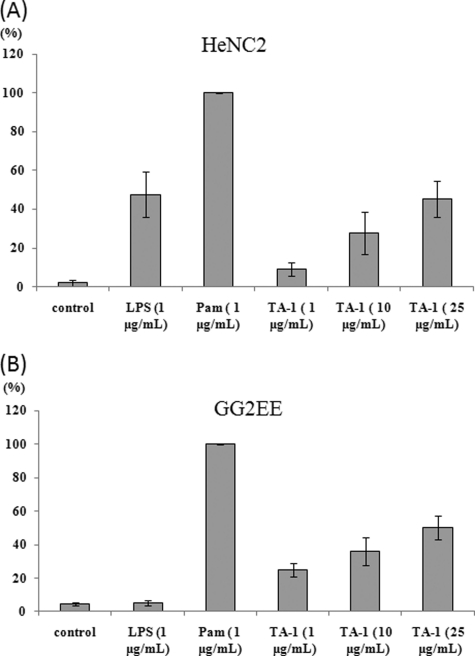
Because TLR4 has been reported to mediate cytokine expression by macrophages in response to several kinds of polysaccharides, such as polysaccharides from Ganoderma lucidum and Streptococcus pneumoniae (2, 22), we tested whether TLR4 takes part in the TA-1-induced immunoregulatory effects. In our assays, the murine macrophage cell lines HeNC2 (with functional TLR4; Fig. 8A) and GG2EE (lacking functional TLR4; Fig. 8B) produced TNF-α in a dosage-dependent manner with similar patterns upon stimulation by TA-1. In contrast, LPS treatment of GG2EE cells did not lead to significant TNF-α secretion (Fig. 8B). As a positive control, the TLR2 ligand Pam3csk4 was used in place of TA-1; after incubation with Pam3csk4, both cell lines secreted the same amount of TNF-α (Fig. 8). These results revealed that TLR2 and not TLR4 is the congenital receptor for TA-1 and is responsible for NF-κB activation to bring about the physiological cellular responses.
FIGURE 8.
Effect of TLR4 on the TA-1-induced immunoregulation (A and B). HeNC2 (A) and GG2EE (B) cells (1 × 106/ml) were incubated with TA-1 for 24 h. The TNF-α concentration was determined by ELISA. TNF-α secretion stimulated by Pam3csk4 (1 μg ml−1) served as positive control, and the response was considered as 100% to normalize differences in the two cell lines.
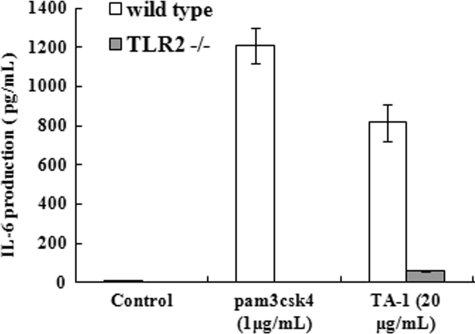
To verify that TA-1 is the agonist of TLR2, peritoneal macrophages isolated from wild-type or TLR2-knock-out mice were stimulated with TA-1, and the amount of IL-6 was measured. Peritoneal macrophages isolated from TLR2−/− mice did not respond to the synthetic TLR2 agonist pam3csk4, i.e. no IL-6 was produced (Fig. 9). Upon TA-1 stimulation, peritoneal macrophages isolated from wild-type mice produced much more IL-6 than those from TLR2−/− mice (Fig. 9). These results further confirmed that TA-1 is mainly recognized by TLR2.
FIGURE 9.
Production of IL-6 in peritoneal macrophages of wild-type and TLR2−/− mice induced by TA-1. Peritoneal macrophages were isolated from wild-type or TLR2−/− mice and treated with TA-1 or the TLR2 ligand pam3csk4 at the indicated concentrations for 24 h. IL-6 production in culture medium was measured using ELISA. Data represent the mean ± S.D. of triplicates from one of at least two independent experiments.
TA-1 Induced Cytokine Production through the TLR2/MyD88/TIRAP Signaling Pathway
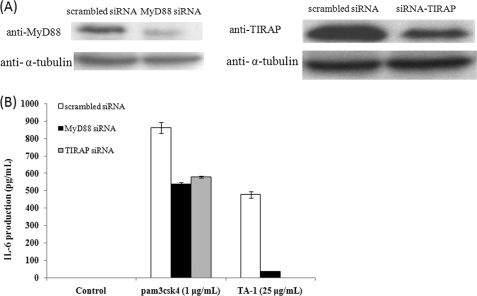
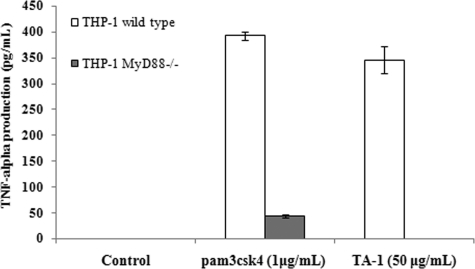
It is believed that ligand recognition by TLR2 leads to the recruitment of Toll/IL-1 receptor homology (TIR) domain-containing adaptors, MyD88 and TIRAP. This recruitment triggers the signaling cascade and then the activation of NF-κB, which ultimately leads to cytokine production (8, 23). To clarify whether TLR2 signaling mediated by TA-1 is dependent on MyD88/TIRAP or not, siRNA knockdown experiments were conducted. As shown in Fig. 10, IL-6 production was significantly reduced when MyD88 or TIRAP was knocked down. Furthermore, TA-1 stimulated wild-type THP-1 monocyte to secrete TNF-α but not MyD88-knock-out THP-1 monocyte (Fig. 11). Taken together, these results indicated that TA-1 was similar to other TLR2 ligands, in which function was in an MyD88- and TIRAP-dependent manner.
FIGURE 10.
Effect of MyD88 or TIRAP knockdown on IL-6 production in Raw264.7 macrophages. A, the protein expression level of scrambled siRNA, MyD88 siRNA, and TIRAP siRNA-transfected Raw264.7 cells was analyzed using Western blot with α-tubulin as the loading control. B, scrambled siRNA, MyD88 siRNA, and TIRAP siRNA-transfected Raw264.7 cells were treated with the TLR2 ligand, pam3csk4, and TA-1 polysaccharide for 24 h. The IL-6 level in the culture supernatants was measured by ELISA. Data represent the mean ± S.D. of triplicates from one of at least two independent experiments.
FIGURE 11.
MyD88 played a key role in TA-1 function. THP-1 and MyD88-deficient THP-1 cells were treated with TA-1 or pam3csk4 (1 μg ml−1) for 16 h, and the amount of TNF-α in the medium was measured by ELISA. Data represent the mean ± S.D. of triplicates from one of at least two independent experiments.
DISCUSSION
Biofilms are usually found on solid surfaces and are held together and protected by a matrix of excreted polymeric EPS. We purified the EPS TA-1 from the monospecies biofilm of T. aquaticus YT-1.
EPSs purified from biofilms to date vary in size from 500 to 2000 kDa, and most are polyanionic due to the presence of either uronic acids (most often d-glucuronic acid and occasionally d-galacturonic acid or d-mannuronic acids) or ketal-linked pyruvate (24, 25). TA-1 falls within the typical size range but lacks ionic sugars and has neutral sugars instead. Half of TA-1 is composed of N-acetylgalactosamines, which probably contributes to a regular and stable structure because the acetyl groups form random coils that tend to from helical aggregates at higher temperatures (24, 25). Besides, TA-1 contains d-galactofuranose residues, which are often found in important glycoconjugates on the surface of pathogenic bacteria, fungi, and protozoan parasites (26, 27) and are absent in the mammalian host. Thus, galactofuranose may be responsible for TA-1 immunoactivity, and the enzyme UDP-galactopyranose mutase, which converts UDP-galactopyranose to UDP-galactofuranose, may play a role in T. aquaticus YT-1 biofilm formation. The activity of this enzyme has been directly correlated to the synthesis of important galactofuranose-containing glycoconjugates in several microorganisms, including Escherichia coli (27) and Mycobacteria (28). The UDP-galactopyranose mutase activity may increase and may be involved in glycoconjugate formation in T. aquaticus YT-1 when it forms biofilm. More studies are required to test this hypothesis.
Our results indicated that the novel TA-1 of T. aquaticus YT-1 has immunoregulatory activity within macrophages. Macrophages are the first line of host defense against bacterial infection and tumor growth, and thus, they play an important role in the initiation of adaptive immune responses (4, 29, 30). In general, cells will produce both reactive oxygen species and reactive nitrogen species when they encounter microbes or foreign antigens. We showed that treatment of macrophages with various dosages of TA-1 induced NO production. Such an oxidative environment may not only facilitate bacterial killing and degradation but also enable antigen-presenting cells to process T-cell-dependent carbohydrate antigens for presentation (31). Also, TA-1 stimulated murine macrophages to secrete cytokines, which are produced mainly by activated macrophage and are important in various immune responses and in inflammatory reactions. We propose that TA-1 stimulates the immune system by activating macrophage functions, as shown for other polysaccharides from microbial and plant origins (32–35).
TLRs serve as the recognition receptor to distinguish between self- and non-self-molecular patterns. TLRs receive external molecules and then trigger signal transduction to initiate immune responses, including cytokine release (36). We propose that TLR2 is the TA-1 cognate receptor and plays a critical role in initiating cytokine production; however, how TA-1 is recognized might be different from the recognition of other bacterial products, such as peptidoglycans and lipopeptides. Although many known TLR ligands contain carbohydrate moieties, it appears that the non-carbohydrate portion is usually critical for TLR recognition and activation. For example, the crystal structure of the TLR2-TLR1 complex shows that lipid chains of TLR2 ligands interact with the hydrophobic residues in TLR2 ectodomains and induce heterodimer formation (37). The constituents of TA-1, in contrast, are all the hydrophilic carbohydrates, and thus the TA-1 and TLR2 interaction probably differs from known interactions. To date, no crystal structure of a TLR-polysaccharide/oligosaccharide complex has been resolved, and it is still unclear whether these ligands adopt a distinct mode to trigger signal transduction through TLR2 homodimers or heterodimers.
Pure carbohydrates as ligands for TLR2 remain largely uninvestigated, and the known TLR2 polysaccharide ligands have almost a zwitterionic charge character (38). In contrast, the monosaccharides of TA-1 are neutral sugars without any charged groups. However, the zwitterionic charge alone is not sufficient for polysaccharide stimulation of TLR2 activity; for example, the zwitterionic Sp1 fails to stimulate cytokine production by RAW264.7 macrophages (31). Thus, these different TLR2 ligands assume similar three-dimensional structures and define a common scaffold to form a unique molecular pattern for reorganization. The tertiary structure of TA-1 may provide clues for analyzing the pattern recognition mechanism. Also, further studies of the specific structure/activity relationship of the carbohydrate-TLR2 interaction may provide more insights into the TLR-mediated recognition mechanism as well as the signal transduction system leading to cytokine production.
Our findings provide a step forward in the development of new adjuvants. The use of PRR agonists as vaccine adjuvants has been shown experimentally to enhance the vaccine immune response; PRRs determine the nature of the adaptive immune response through the emission of a “signal zero” that precedes T-cell receptor activation (signal 1) and T-helper cell antigen-presenting cell co-stimulation (signal 2) (39–41). In addition, PRR agonists are mainly used to trigger antigen-presenting cell activation in animal models (42); thus, the use of adjuvants is believed to increase the activation of both antigen-presenting cells and B cells. The interest in these molecules and in new adjuvants to improve the overall immune response is growing. Conjugating the TLR2 agonist to a vaccine antigen will increase both antibody and T-cell responses, and thus TLR2 ligands can serve as adjuvants (43). For example, the efficiency of vaccines against yellow fever (44) and against pneumococcus (45) is partly attributed to the association of TLR2 agonists. TA-1, as a TLR2 agonist, could be used as an adjuvant and could enhance cytokine release, which increases the immune response (40, 46, 47). However, the molecular basis of the TA-1 effect in cells remains to be deciphered.
In summary, we determined the structure of TA-1 and showed that TA-1 induces murine macrophage and human monocyte cell lines to secrete IL-6, TNF-α, and NO. TA-1 is a novel TLR2 agonist without charged groups, in contrast to zwitterionic polysaccharides found previously. TA-l had no bioactivity when TLR2 receptors were not present. Our findings contribute to our understanding of polysaccharide-TLR2 interactions.
Supplementary Material
Acknowledgment
We express our gratitude to Dr. Yu-Ju Chen of the Institute of Chemistry, Academia Sinica for mass spectrometry analyses.
This work was supported by the National Science Council and Academia Sinica, Taiwan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PRR
- pattern recognition receptor
- TLR
- Toll-like receptor
- EPS
- extracellular polysaccharide
- NF-κB
- nuclear factor κ light-chain enhancer of activated B cells
- LPS
- lipopolysaccharide
- HPAEC-PAD
- high performance anion-exchange chromatography with pulsed amperometric detection
- NOE
- nuclear Overhauser effect
- HMQC
- heteronuclear multiple quantum coherence
- HMBC
- heteronuclear multiple bond coherence
- TIR
- Toll/IL-1 receptor homology.
REFERENCES
- 1. Ooi V. E., Liu F. (2000) Curr. Med. Chem. 7, 715–729 [DOI] [PubMed] [Google Scholar]
- 2. Hsu H. Y., Hua K. F., Lin C. C., Lin C. H., Hsu J., Wong C. H. (2004) J. Immunol. 173, 5989–5999 [DOI] [PubMed] [Google Scholar]
- 3. Schepetkin I. A., Quinn M. T. (2006) Int. Immunopharmacol. 6, 317–333 [DOI] [PubMed] [Google Scholar]
- 4. Yim J. H., Son E., Pyo S., Lee H. K. (2005) Mar. Biotechnol. 7, 331–338 [DOI] [PubMed] [Google Scholar]
- 5. Akira S., Uematsu S., Takeuchi O. (2006) Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 6. Fritz J. H., Ferrero R. L., Philpott D. J., Girardin S. E. (2006) Nat. Immunol. 7, 1250–1257 [DOI] [PubMed] [Google Scholar]
- 7. Albiger B., Dahlberg S., Henriques-Normark B., Normark S. (2007) J. Intern. Med. 261, 511–528 [DOI] [PubMed] [Google Scholar]
- 8. Kumar H., Kawai T., Akira S. (2009) Biochem. Biophys. Res. Commun. 388, 621–625 [DOI] [PubMed] [Google Scholar]
- 9. van Putten J. P., Bouwman L. I., de Zoete M. R. (2010) Immunol. Lett. 128, 8–11 [DOI] [PubMed] [Google Scholar]
- 10. Watnick P., Kolter R. (2000) J. Bacteriol. 182, 2675–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mojica K., Elsey D., Cooney M. J. (2007) J. Microbiol. Methods 71, 61–65 [DOI] [PubMed] [Google Scholar]
- 12. Brock T. D., Edwards M. R. (1970) J. Bacteriol. 104, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leone S., Molinaro A., Lindner B., Romano I., Nicolaus B., Parrilli M., Lanzetta R., Holst O. (2006) Glycobiology 16, 766–775 [DOI] [PubMed] [Google Scholar]
- 14. Brock T. D., Freeze H. (1969) J. Bacteriol. 98, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chien A., Edgar D. B., Trela J. M. (1976) J. Bacteriol. 127, 1550–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wozniak D. J., Wyckoff T. J., Starkey M., Keyser R., Azadi P., O'Toole G. A., Parsek M. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7907–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yildiz F. H., Schoolnik G. K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu S. F., Shu H. Y., Lin L. C., Chen M. Y., Tsay S. S., Lin G. H. (2006) Plasmid 56, 46–52 [DOI] [PubMed] [Google Scholar]
- 19. Waeghe T. J., Darvill A. G., McNeil M., Albersheim P. (1983) Carbohydr. Res. 123, 281–304 [Google Scholar]
- 20. Alexander C., Rietschel E. T. (2001) J. Endotoxin Res. 7, 167–202 [PubMed] [Google Scholar]
- 21. Seong S. Y., Matzinger P. (2004) Nat. Rev. Immunol. 4, 469–478 [DOI] [PubMed] [Google Scholar]
- 22. Cohen N., Stolarsky-Bennun M., Amir-Kroll H., Margalit R., Nussbaum G., Cohen-Sfady M., Pevsner-Fischer M., Fridkin M., Bercovier H., Eisenbach L., Jung S., Cohen I. R. (2008) J. Immunol. 180, 2409–2418 [DOI] [PubMed] [Google Scholar]
- 23. Kumagai Y., Takeuchi O., Akira S. (2008) J. Infect. Chemother. 14, 86–92 [DOI] [PubMed] [Google Scholar]
- 24. Sutherland I. (2001) Microbiology 147, 3–9 [DOI] [PubMed] [Google Scholar]
- 25. Sutherland I. W. (2001) Water Sci. Technol. 43, 77–86 [PubMed] [Google Scholar]
- 26. Bakker H., Kleczka B., Gerardy-Schahn R., Routier F. H. (2005) Biol. Chem. 386, 657–661 [DOI] [PubMed] [Google Scholar]
- 27. Nassau P. M., Martin S. L., Brown R. E., Weston A., Monsey D., McNeil M. R., Duncan K. (1996) J. Bacteriol. 178, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weston A., Stern R. J., Lee R. E., Nassau P. M., Monsey D., Martin S. L., Scherman M. S., Besra G. S., Duncan K., McNeil M. R. (1997) Tuber. Lung Dis. 78, 123–131 [DOI] [PubMed] [Google Scholar]
- 29. Fidler I. J. (1988) Ciba Found Symp. 141, 211–222 [DOI] [PubMed] [Google Scholar]
- 30. Verstovsek S., Maccubbin D., Ehrke M. J., Mihich E. (1992) Cancer Res. 52, 3880–3885 [PubMed] [Google Scholar]
- 31. Wang Q., McLoughlin R. M., Cobb B. A., Charrel-Dennis M., Zaleski K. J., Golenbock D., Tzianabos A. O., Kasper D. L. (2006) J. Exp. Med. 203, 2853–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okazaki M., Adachi Y., Ohno N., Yadomae T. (1995) Biol. Pharm. Bull. 18, 1320–1327 [DOI] [PubMed] [Google Scholar]
- 33. Seljelid R. (1989) Scand J. Immunol. 29, 181–192 [DOI] [PubMed] [Google Scholar]
- 34. Otterlei M., Ostgaard K., Skjåk-Braek G., Smidsrød O., Soon-Shiong P., Espevik T. (1991) J. Immunother. 10, 286–291 [DOI] [PubMed] [Google Scholar]
- 35. Iwamoto M., Kurachi M., Nakashima T., Kim D., Yamaguchi K., Oda T., Iwamoto Y., Muramatsu T. (2005) FEBS Lett. 579, 4423–4429 [DOI] [PubMed] [Google Scholar]
- 36. Hirschfeld M., Ma Y., Weis J. H., Vogel S. N., Weis J. J. (2000) J. Immunol. 165, 618–622 [DOI] [PubMed] [Google Scholar]
- 37. Jin M. S., Kim S. E., Heo J. Y., Lee M. E., Kim H. M., Paik S. G., Lee H., Lee J. O. (2007) Cell 130, 1071–1082 [DOI] [PubMed] [Google Scholar]
- 38. Tzianabos A. O., Onderdonk A. B., Rosner B., Cisneros R. L., Kasper D. L. (1993) Science 262, 416–419 [DOI] [PubMed] [Google Scholar]
- 39. Guy B. (2007) Nat. Rev. Microbiol. 5, 505–517 [DOI] [PubMed] [Google Scholar]
- 40. Guy B. (2010) J. Comp. Pathol. 142, S133–S137 [DOI] [PubMed] [Google Scholar]
- 41. Maue A. C., Eaton S. M., Lanthier P. A., Sweet K. B., Blumerman S. L., Haynes L. (2009) J. Immunol. 182, 6129–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maletto B. A., Rópolo A. S., Liscovsky M. V., Alignani D. O., Glocker M., Pistoresi-Palencia M. C. (2005) Clin. Immunol. 117, 251–261 [DOI] [PubMed] [Google Scholar]
- 43. Jackson D. C., Lau Y. F., Le T., Suhrbier A., Deliyannis G., Cheers C., Smith C., Zeng W., Brown L. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15440–15445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Querec T., Bennouna S., Alkan S., Laouar Y., Gorden K., Flavell R., Akira S., Ahmed R., Pulendran B. (2006) J. Exp. Med. 203, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sen G., Khan A. Q., Chen Q., Snapper C. M. (2005) J. Immunol. 175, 3084–3091 [DOI] [PubMed] [Google Scholar]
- 46. Rosenberg S. A., Sportès C., Ahmadzadeh M., Fry T. J., Ngo L. T., Schwarz S. L., Stetler-Stevenson M., Morton K. E., Mavroukakis S. A., Morre M., Buffet R., Mackall C. L., Gress R. E. (2006) J. Immunother. 29, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sportès C., Hakim F. T., Memon S. A., Zhang H., Chua K. S., Brown M. R., Fleisher T. A., Krumlauf M. C., Babb R. R., Chow C. K., Fry T. J., Engels J., Buffet R., Morre M., Amato R. J., Venzon D. J., Korngold R., Pecora A., Gress R. E., Mackall C. L. (2008) J. Exp. Med. 205, 1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.