Abstract
Estrogen receptor is a nuclear receptor superfamily member of transcriptional activators that regulate gene expression by recruiting diverese transcriptional coregulators. The Mediator complex is a central transcriptional coactivator complex that acts as a bridge between transcriptional activators and RNA polymerase II. MED1 (Mediator subunit 1) is the key Mediator subunit that directly interacts with estrogen receptor to mediate its functions both in vitro and in vivo. Interestingly, our previous biochemical analyses indicated that MED1 exists only in a subpopulation of the Mediator complex that is enriched with a number of distinct Mediator subunits and RNA polymerase II. Here, we report ARGLU1 as a MED1/Mediator-associated protein. We found that ARGLU1 (arginine and glutamate rich 1) not only colocalizes with MED1 in the nucleus, but also directly interacts with a far C-terminal region of MED1. Reporter assays indicate that ARGLU1 is able to cooperate with MED1 to regulate estrogen receptor-mediated gene transcription. Importantly, ARGLU1 is recruited, in a ligand-dependent manner, to endogenous estrogen receptor target gene promoters and is required for their expression. Furthermore, by ChIP-reChIP assay, we confirm that ARGLU1 and MED1 colocalize on the same estrogen receptor target gene promoter upon estrogen induction. Moreover, we found that depletion of ARGLU1 significantly impairs the growth, as well as anchorage-dependent and -independent colony formation of breast cancer cells. Taken together, these results establish ARGLU1 as a new MED1-interacting protein required for estrogen-dependent gene transcription and breast cancer cell growth.
Keywords: Breast Cancer, Estrogen, Gene Regulation, Nuclear Receptors, Transcription Coactivators
Introduction
Estrogen receptor belongs to a nuclear receptor superfamily of transcriptional activators that, in a ligand-dependent manner, regulates the expression of specific genes controlling the development, reproduction, homeostasis, and metabolism of an organism (1, 2). Like other nuclear receptors, estrogen receptor shares a common organization of functional domains: a highly variable N-terminal activation function-1 domain that mediates ligand-independent transcription, a highly conserved central DNA-binding domain that specifically binds to hormone-response elements in cognate promoters of target genes, and a moderately conserved C-terminal ligand-binding domain containing activation domain 2, which mediates ligand-dependent transcription (1, 2). Upon ligand binding, the ultimate action of these receptors on regulating target gene expression is to enhance the recruitment and/or function of the general transcriptional machinery, including RNA polymerase II and general transcription factors (3–6). In the past decades, increasingly diverse groups of transcriptional coregulators have been found to play key roles in this process (3, 4). Among them, Mediator has emerged as a key transcriptional coregulator complex that is responsible for communicating the signals from transcription activators to RNA polymerase II and the general transcription machinery (7–9).
Mediator is a large protein complex composed of about 30 distinct subunits and is conserved from yeast to humans (9–13). Recent studies have established the direct interactions between estrogen receptor and the MED1 (also known as TRAP220/PBP/DRIP205) subunit of the Mediator complex (14–17). Ectopic MED1 expression has been shown to markedly enhance estrogen receptor-dependent transcription, both in vitro and in cellular systems (15, 16, 18). Further studies indicated that MED1 is not only required for estrogen receptor-dependent reporter and endogenous gene expression, but is also required for estrogen-dependent breast cancer cell growth (15, 18–20). The estrogen receptor interacts in a ligand-dependent manner with two LXXLL motifs of MED1 through its activation function-2 domain (16, 19, 21–23). More recently, we have generated MED1 LXXLL motif-mutant knock-in mice and found that MED1 is required for estrogen receptor functions in pubertal mammary gland development and luminal cell differentiation (17). Surprisingly, these MED1 LXXLL motifs were apparently dispensable for mammary gland development during pregnancy and the development of other estrogen-responsive tissues such as the uterus (17).
Our biochemical analyses indicated that MED1 exists only in a subpopulation of Mediator complexes (20). Further characterization revealed that MED1-Mediator is enriched with RNA polymerase II and a number of additional Mediator subunits (20, 24). Interestingly, MED1 displays a relative functional specificity to its corresponding transcription factors. Thus, knockdown of MED1 expression abolishes the expression of estrogen receptor-dependent target genes but not that of genes controlled by other activators such as p53, which interacts with another subunit, MED17 (20). Importantly, our ChIP analysis revealed that the MED1-Mediator subpopulation is specifically recruited to estrogen receptor target gene promoters upon estrogen stimulation but not to the p53 target gene promoter by UV stimulation where a general population of Mediator is recruited by activated p53 (20). These results, together with our finding that MED1 is selectively expressed and required for luminal mammary epithelial cell differentiation, revealed additional functional specificity for this complex (17).
In this study, we report the novel finding that arginine and glutamate rich 1 (ARGLU1)2 is a MED1/Mediator-interacting protein by mass spectrometry and protein-protein interaction studies. Interestingly, we found that ARGLU1 not only colocalizes with MED1, but that it is also able to directly interact with MED1. Further deletion mapping experiments were carried out to dissect the region required for these interactions and establish ARGLU1 as the first protein reported to interact with a far C-terminal region of MED1. Moreover, the role of ARGLU1 in estrogen-mediated gene transcription and the requirement of ARGLU1 for estrogen-dependent cell growth and anchorage-dependent and -independent colony formation of breast cancer cells was investigated.
EXPERIMENTAL PROCEDURES
Plasmids and Constructs
pERE-TK-Luc, pRL-CMV, and the MED1 expression construct pCDNA3.1-TRAP220 have been described previously (20). MED1 deletions and other Mediator subunits used for tnt assays were constructed by inserting the appropriate PCR-amplified cDNA coding regions into the BamHI/EcoRI sites of the pIRES-neo vector (20, 23, 25). The ARGLU1 expression plasmid pCDNA3.1-ARGLU1 was constructed by inserting the coding region of ARGLU1 into the BamHI/EcoRI sites of pCDNA3.1 after amplication using primers 5′-atggatccaccatgGGCCGGTCTCGGAGC-3′ and 5′-acgaattcTTAATCCTGGGTTTTTAATG-3′. The GST (glutathione S-transferase) fusion proteins of ARGLU1 were produced by subcloning the cDNA fragments encoding full-length human ARGLU1 (primers described above), N-terminal (amino acids (aa) 1–89, 5′-atggatccaccatgGGCCGGTCTCGGAGC-3′ and 5′-acgaattcCACCGTGCGCCCGAAGATGT-3′) and C-terminal (aa 83–273, 5′-atggatccaccatgGACATCTTCGGGCGCACGGT-3′ and 5′-acgaattcTTAATCCTGGGTTTTTAATG-3′) into the pGEX-4T1 vector.
Immunoprecipitation and GST Pull-down
HeLa nuclear extract was first cleared with protein A/G-agarose beads (Santa Cruz Biotechnology) for 1 h at 4 °C. For immunoprecipitation experiments, ∼10 μl of preimmune, IgG, anti-MED1 (20), or anti-ARGLU1 (H00055082-B01, Novus Biologicals) antibodies were mixed with 30 μl of protein A/G-beads and 2 mg of nuclear extract overnight at 4 °C. For GST pulldown assays, 3 μg of each immobilized GST fusion protein was incubated with 2 mg of nuclear extract for 4–6 h at 4 °C. For both experiments, the beads were then washed extensively with BC200 containing 0.1% Nonidet P-40, 1 mm DTT, and 0.25 mm PMSF, and subjected to SDS-PAGE and immunoblotting.
Immunofluorescent Staining
Immunofluorescent staining was carried out by first seeding MCF-7 cells on glass coverslips in 6-well plates. Cells were then grown to about 75% confluence and fixed for 15 min at room temperature in 2% (w/v) formaldehyde. After washing, cells were incubated overnight at 4 °C with control IgG, or primary anti-MED1 (20) and anti-ARGLU1 (H00055082-B01, Novus Biologicals) rabbit polyclonal antibodies, followed by Alexa 555- or Alexa 488-conjugated secondary goat anti-rabbit antibodies (Invitrogen). DAPI staining was then carried out for 15 min at RT before mounting of the coverslips. The images were visualized and captured using a LSM510 NLO two-photon confocal microscope (Zeiss).
Luciferase Reporter Assay
Cells were first grown in DMEM containing 10% charcoal-dextran-stripped fetal bovine serum for at least 3 days, and then seeded in 24-well plates at a density of 105 cells/well. The estrogen-responsive reporter plasmid pERE-TK-Luc was cotransfected with control plasmids or plasmids expressing ARGLU1 or MED1 by Oligofectamine (Invitrogen) according to the manufacturer's instructions. The pRL-CMV plasmids were also cotransfected to serve as an internal control for transfection efficiency. After 4 h, the transfection mixture was replaced with fresh medium containing 17β-estradiol (E2, 100 nm) or vehicle control (ethanol), and incubated for another 30 h. Luciferase assays were then performed using a Dual Luciferase Assay Kit (Promega, Madison, WI).
Real Time RT-PCR
Total RNAs were isolated with an RNeasy Mini Kit (Qiagen) and first-strand cDNA was synthesized by reverse transcription of mRNA using oligo(dT)21 primer and SuperScriptTM III Reverse Transcriptase (Invitrogen). Real time PCR was performed using the following primers: Myc, 5′-GAGCAGCAGAGAAAGGGAGA-3′ (forward) and 5′-CAGCCGAGCACTCTAGCTCT-3′ (reverse); pS2, 5′-TCCCAGTGTGCAAATAAG-3′ (forward) and 5′-ATTCACACTCCTCTTCTGG-3′ (reverse); 18S, 5′-CGATCCGAGGGCCTCACTA-3′ (forward) and 5′-TCTTCGGCTGGAGCGGGAGGAGTA-3′ (reverse). Experiments were repeated in triplicate, and the relative gene expression was analyzed using the 2−ΔΔCT method (26) by normalization to the 18S rRNA levels.
Chromatin Immunoprecipitation (ChIP) and Sequential Chromatin Immunoprecipitation (ChIP-reChIP) Assays
ChIP assays were performed as described previously (27). Briefly, MCF-7 cells were grown in 100-mm cell culture dishes with or without 17β-estradiol (E2) for 1 h, and fixed with 1% formaldehyde for 15 min. After PBS washing, cells were harvested and chromatin was sheared using a Bioruptor (Diagenode). Chromatin fractions were subjected to immunoprecipitation overnight with control, anti-MED1, or anti-ARGLU1 antibodies. The immunoprecipitated DNA was obtained by heating to reverse formaldehyde cross-linking followed by purification using a PCR purification kit (Qiagen). For ChIP-reChIP assays, cross-linked protein-DNA complexes were eluted from primary immunoprecipitates by incubation with 10 mm dithiothreitol (DTT) for 30 min at 37 °C. The eluates were diluted 1:50 in dilution buffer and then subjected to immunoprecipitation with the secondary antibodies. Real-time PCR using a 7900 HT Fast Real-time PCR System (Applied Biosystems) was performed with SYBR Green Master Mix (Roche Applied Science). The primers used were as follows: c-Myc, 5′-GAGCAGCAGAGAAAGGGAGA-3′ (forward) and 5′-CAGCCGAGCACTCTAGCTCT-3′ (reverse); pS2, 5′-TATGAATCACTTCTGCAGTGAG-3′ (forward) and 5′-GAGCGTTAGATAACATTTGCC-3′ (reverse); GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTA-3′ (forward) and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse). The relative amount of immunoprecipitated DNA was analyzed using the 2−ΔΔCT method (26) by normalization to input.
MTT Assay
For cell proliferation assays, 2500 cells/well were seeded in a 96-well plate. MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) was added at a concentration of 0.5 mg/ml for 3 h at 37 °C. The medium was then removed and 0.2 ml/well of acidic isopropyl alcohol (0.04 m HCl in absolute isopropyl alcohol) was added. The absorbance of the converted dye was measured at 570 nm using a Synergy II spectrophotometer (Biotek).
Colony Formation and Soft Agar Assay
For colony formation assays, MCF7-scramble and MCF7-shARGLU1 cells were seeded at a density of 4000 cells/well in 6-well plates containing DMEM with 10% FBS and puromycin (5 μg/ml). The cells were allowed to grow for 2 weeks and then stained with 0.1% crystal violet. For anchorage-independent growth assays, we first established a bottom layer with 1% agarose in DMEM containing 10% FBS in a 24-well plate. The top layer containing 0.35% agarose mixed with 4000 of each of the above cell types was then added. Additional liquid medium was placed on top of the solidified agar in each well and replaced every 3 days. Each sample had three replicate wells. The colonies formed were stained by crystal violet after 3 to 4 weeks when the colonies were visible and counted.
RESULTS
ARGLU1 Associates with MED1-Mediator Complex
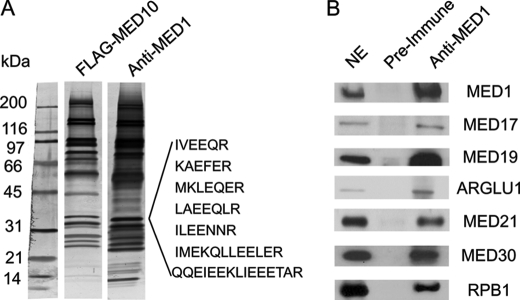
We have previously shown that MED1 exists predominantly in a Mediator subpopulation enriched with RNA polymerase II and several other proteins (20). Furthermore, mass spectrometry (MS) assays identified several peptide sequences that matched with ARGLU1, a previously uncharacterized protein named based on its high content of arginine and glutamate (Fig. 1A). To further confirm that ARGLU1 exists in the Mediator complex, we performed Western blot analyses using anti-ARGLU1 antibody. As expected, we observed ARGLU1 in the anti-MED1 preparations but not in the control preimmune preparations (Fig. 1B). In addition, we also found RNA polymerase II subunit RPB1, in the immunoprecipitates. This is consistent with our previous report that the MED1-Mediator complex is enriched with RNA polymerase II (20).
FIGURE 1.
ARGLU1 associates with MED1-Mediator complex. A, the MED1-Mediator complex was isolated from nuclear extract by using anti-MED1 antibody and compared with total Mediator (isolated by using anti-FLAG MED10). Proteins were visualized by silver stain. New or enriched bands were subjected to mass spectrometry analyses. B, anti-MED1 and control preimmune antibodies were employed for immunoprecipitation (IP) experiments using HeLa nuclear extracts. Immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blot using the indicated antibodies.
ARGLU1 Is Predominantly a Nuclear Protein That Colocalizes with MED1
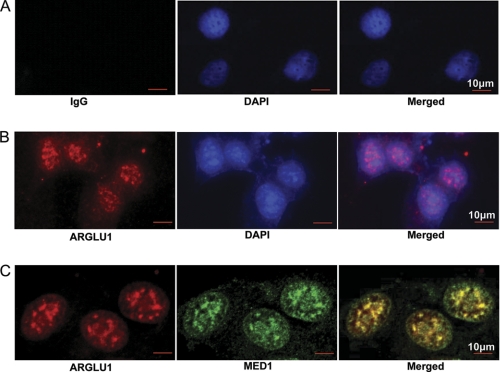
The studies described above established ARGLU1 as a novel interacting protein of the MED1-Mediator complex. Because it is a protein that has not previously been characterized except for its amino acid composition, we first examined its intracellular localization by immunostaining using anti-ARGLU1 antibody. As shown in Fig. 2, we found that the ARGLU1 protein is predominantly localized to the nucleus of MCF-7 cells. This is consistent with the prediction that ARGLU1 possesses several nuclear localization signals (WoLFPSORT). We further compared the cellular localization of ARGLU1 to that of MED1 using double labeling with antibodies against each of these proteins. Interestingly, we found that ARGLU1 clearly colocalized with MED1 in the nucleus (Fig. 2).
FIGURE 2.
ARGLU1 colocalizes with MED1 in the nucleus. Immunofluorescent staining to detect ARGLU1 (red) and MED1 (green) was carried out as described under “Experimental Procedures.” IgG was included to serve as a negative control, whereas DAPI staining (blue) was used to stain nuclei. The data indicate that ARGLU1 is a predominantly nuclear protein (upper panel) and that it colocalizes with MED1 (Merged, bottom panel).
ARGLU1 Interacts Directly with MED1
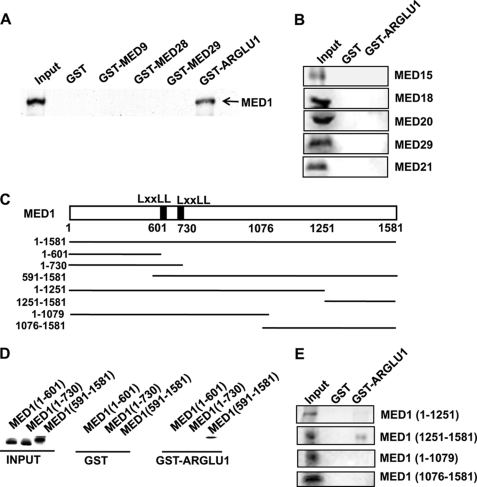
To further investigate a potential physical interaction between ARGLU1 and MED1, we performed GST pulldown assays. Control GST or GST fused to ARGLU1 was expressed, purified, and incubated with in vitro transcribed 35S-labeled full-length MED1 (tnt, Promega). After extensive washing, bound proteins were eluted and subjected to SDS-PAGE. The gel was then exposed to a phosphorscreen and the signals were detected by a Typhoon PhosphorImager (GE Healthcare). We found that GST-ARGLU1, but not control GST, could interact with MED1 (Fig. 3A). Interestingly, GST fusion proteins with several other proteins (FLJ10193/MED9, FSKG20/MED28, and Hintersex/MED29) that were also enriched in MED1-Mediator complex (20) failed to bind to MED1 (Fig. 3A; see input for GST fusion proteins under supplemental Fig. S1). Conversely, we tested a number of other known Mediator subunits and found that they also could not interact directly with GST-ARGLU1 (Fig. 3B). These results strongly support a specific direct interaction between ARGLU1 and MED1.
FIGURE 3.
ARGLU1 directly interacts with MED1. A and B, bacterially produced control GST and GST fused to full-length ARGLU1 were incubated with 35S-labeled in vitro transcribed MED1. GST fused to several other MED1-Mediator-enriched proteins (A) and five 35S-labeled Mediator subunits (B) were included as controls. C, the diagram shows the 35S-labeled MED1 fragments generated for the studies in D and E. The MED1 fragments shown in C were incubated with either GST or GST-ARGLU1 and the bound proteins were eluted, separated by SDS-PAGE, and visualized by using a phosphorimager.
MED1 is composed of 1581 amino acids and can be largely divided into three functional domains (Fig. 3C). The N-terminal domain of MED1 consists of about 600 amino acids and is required for the assembly of MED1 into Mediator complex (22, 23, 25). Immediately following this N-terminal domain are two classical LXXLL motifs that are required for ligand-dependent interactions with a number of nuclear receptors and the large C terminus that may play some regulatory roles (28, 29). To further map the region(s) of MED1 responsible for its interactions with ARGLU1, we generated the indicated MED1 deletion fragments for GST pulldown assays. Interestingly, we found that neither MED1(1–601) nor this N-terminal domain plus the LXXLL motifs (1–730) could bind to ARGLU1. Instead, the C-terminal region (591–1581) of MED1 exhibited a strong interaction with ARGLU1 (Fig. 3D). To further define the C-terminal region of MED1 that interacts with ARGLU1, four additional fragments (1–1251, 1251–1581, 1–1079, and 1076–1581) were in vitro translated and assayed for their interactions with ARGLU1. As shown in Fig. 3E, only the MED1(1251–1581) fragment was able to bind GST-ARGLU1. Interestingly, MED1(1076–1581), which includes the binding fragment, could not bind to ARGLU1, suggesting a potential inhibitory role of amino acids 1076 to 1251 with respect to this interaction.
Mapping the ARGLU1 Region That Interacts with MED1
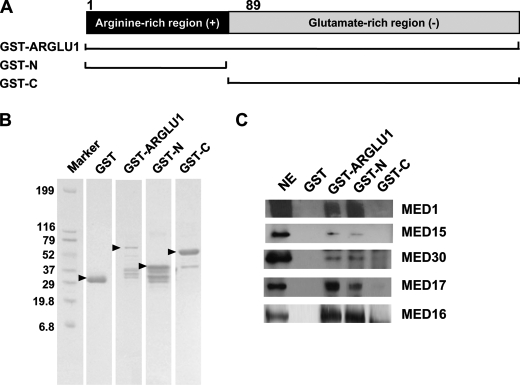
Based on an analysis of amino acid composition, ARGLU1 can be largely divided into two regions: an N-terminal, positively charged region enriched with arginine, and a C-terminal, negatively charged region enriched with glutamate (Fig. 4A). Here, we decided to further determine which of these regions is responsible for interaction with the Mediator complex. GST fused to either the N terminus (aa 1–89) or the C terminus (aa 83–273) of ARGLU1 were first expressed and purified from bacteria (Fig. 4B). GST pulldown assays were then carried out using HeLa nuclear extract as described above. As expected, GST-ARGLU1 (full-length) but not control GST could pull down MED1 and other Mediator subunits. Interestingly, GST fused to the N terminus but not the C terminus of ARGLU1 could pull down MED1-Mediator (Fig. 4C). These results indicate an essential role for the N terminus of ARGLU1 in interacting with MED1.
FIGURE 4.
ARGLU1 interacts with MED1-Mediator through its N-terminal arginine-rich region. A, a schematic diagram of the ARGLU1 protein shows that it can be largely divided into a positively charged arginine-rich region and a negatively charged glutamate-rich region. B, purified GST alone and the indicated GST fusion proteins were stained with Coomassie Brilliant Blue R-250 following SDS-PAGE. C, GST pulldown assays were performed on HeLa nuclear extracts. Control GST and GST fused to ARGLU1 fragments were used. The bound proteins were eluted and analyzed by Western blot using the indicated antibodies.
ARGLU1 Is Required for ER-mediated Gene Transcription
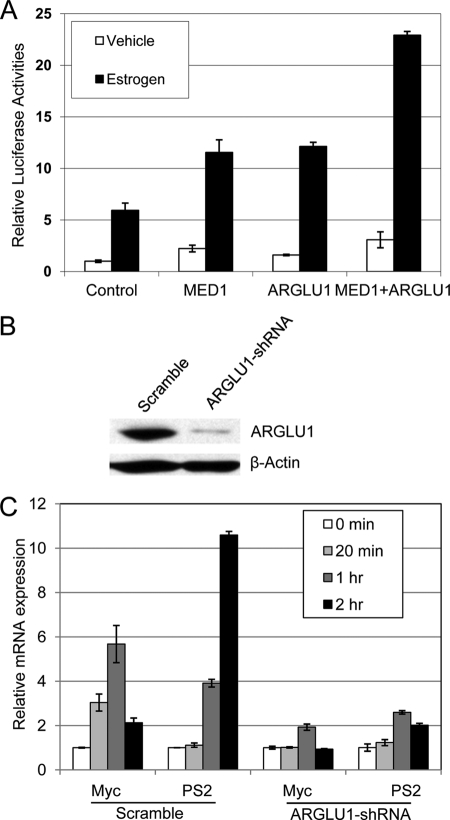
We and others have previously shown that MED1 plays critical roles in ER-mediated transcription (15–18, 20). To determine whether ARGLU1 also plays an important role in ER-mediated transcription, we first carried out transient transfection reporter assays. As expected, estrogen treatment alone significantly increased the expression of an ERE-TK-LUC reporter gene, and overexpression of MED1 further enhanced the luciferase reporter activity (Fig. 5A). Interestingly, ectopic expression of ARGLU1 itself led to increased estrogen-dependent reporter activity. Furthermore, we found that introduction of both MED1 and ARGLU1 further increased estrogen-dependent reporter activity, as compared with each of them alone (Fig. 5A). These data support a role for ARGLU1 in estrogen receptor-mediated transcription in conjunction with MED1.
FIGURE 5.
ARGLU1 is required for the expression of ER target genes. A, MCF-7 cells were first grown in estrogen-depleted medium and transfected with plasmids containing ERE-TK-LUC and pRL-CMV (control), along with plasmids expressing MED1, ARGLU1, or both. After treatment with vehicle or estrogen for 30 h, cells were harvested and analyzed for luciferase activity. B, MCF-7 cells were infected with lentivirus expressing scramble or ARGLU1 shRNA and subjected to Western blot analyses. C, cells in B were treated with estrogen for the indicated time and subjected to real time RT-PCR assays. The expression of indicated estrogen target genes was normalized to that of 18S rRNA. All experiments were repeated at least three times. Value = mean ± S.D.
To determine whether ARGLU1 is required for the expression of endogenous estrogen receptor target genes, lentiviruses expressing control scramble shRNA or shRNA against ARGLU1 were generated and introduced into MCF-7 cells. Western blot analyses were carried out to confirm the successful knockdown of the ARGLU1 protein in these cells (Fig. 5B). The cells were treated with estrogen for the indicated time and processed for real time RT-PCR analyses. We found that the estrogen-dependent induction of well known estrogen receptor target genes (c-Myc and pS2), but not that of the p53 target gene p21 (supplemental Fig. S2), was significantly impaired by treatment with ARGLU1 shRNA but not by control scramble shRNA (Fig. 5C). These data further indicate a key role for ARGLU1 in estrogen receptor-mediated gene transcription.
ARGLU1 Is Recruited to Endogenous ER Target Gene Promoters
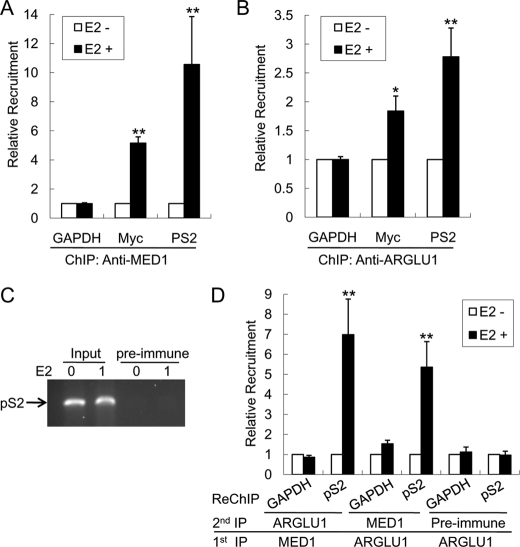
It is known that MED1-Mediator can be specifically recruited to ER target gene promoters in response to estrogen stimulation (20). To further examine the role of ARGLU1 in ER-mediated transcription, we determined whether ARGLU1 can also be recruited to the promoters of ER-target genes. Consistent with previous reports (20, 27), MED1 was recruited to the promoters of these ER target genes (pS2 and c-Myc) but not to the promoter of the control GAPDH gene upon estrogen stimulation (Fig. 6A). Interestingly, ARGLU1 was also significantly recruited to these endogenous ER target gene promoters upon estrogen treatment (Fig. 6B). As another negative control, we used preimmune serum and found that it failed to pull down the promoter fragment of these target genes (Fig. 6C). To further investigate whether MED1 and ARGLU1 can occupy the same ER target gene promoter together as part of a complex, we performed a sequential chromatin immunoprecipitation (ChIP-reChIP) assay. Our results clearly indicate that these two proteins do exist as a complex on the pS2 promoter (Fig. 6D). Taken together, these results support a direct role for ARGLU1 in ER-mediated transcription on its target gene promoters.
FIGURE 6.
ARGLU1 is recruited to the promoter of ER target genes in response to estrogen stimulation. A–C, MCF-7 cells were treated with vehicle or estrogen for 1 h and harvested for ChIP assay using anti-MED1 (A), anti-ARGLU1 (B), or control preimmune (C) antibodies. ChIP-reChIP assay (D) were carried out as described under “Experimental Procedures” under the same condition. The immunoprecipitated DNA fragments were analyzed by real time PCR with primers spaning the promoter regions of the control non-estrogen-responsive gene (GAPDH) and ER-target genes (pS2 and Myc). The relative amount of immunoprecipitated promoter fragments after normalizing to their respective levels in the input are shown. Data are presented as mean ± S.D. of three separate experiments. *, p < 0.05; **, p < 0.01.
ARGLU1 Is Required for the Growth of MCF-7 Cells
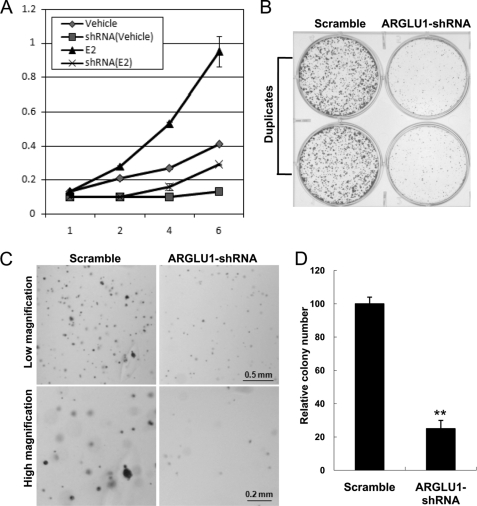
Estrogen, through its receptor, plays prominent roles in promoting breast cancer cell growth. Because our above data indicated a key role for ARGLU1 in ER-mediated transcription, we further assessed the role of ARGLU1 in the growth of ER-positive human breast cancer MCF-7 cells. We first performed cell proliferation assays on cells infected with lentivirus expressing shRNA against a scramble sequence or ARGLU1. Obviously, and as expected, estrogen treatment strongly promoted the proliferation of MCF-7 cells. However, knockdown of endogenous ARGLU1 significantly diminished the growth of MCF-7 cells (Fig. 7A). As a control, the GFP-shRNA showed no effect on the growth of MCF-7 cells (data not shown). Next, we performed colony formation assays and the results were consistent with the above cell proliferation assays (Fig. 7B). Finally, we determined the role of ARGLU1 in anchorage-independent colony formation using a soft agar assay. We found that knockdown of ARGLU1 significantly reduced the number and size of the colonies formed, as compared with that of the control group treated with scramble shRNA (Fig. 7, C and D). To further confirm the specificity of the above ARGLU1 shRNA knockdown experiments, we performed rescue experiments using lentivirus expressing shRNA-resistant ARGLU1 created by introducing two nonsense mutations within the shRNA targeting site (supplemental Fig. S3A). The ARGLU1 shRNA-treated MCF-7 cells were first infected with lentivirus containing vector control or shRNA-resistant ARGLU1. Cell proliferation and colony formation assays were then carried out as described above. We found that shRNA-resistant ARGLU1 can rescue protein expression of ARGLU1, as well as the growth defect of ARGLU1 shRNA knockdown cells in both MTT and colony formation assays (supplemental Fig. S3, B–D). Taken together, these data indicate that ARGLU1 is required for both anchorage-dependent and -independent growth of breast cancer cells.
FIGURE 7.
ARGLU1 is required for the growth of MCF-7 cells. A, MCF-7 cells were infected with lentivirus expressing scrambled or ARGLU1 shRNA and grown in estrogen-depleted medium supplemented with vehicle or estrogen. Cell proliferation was monitored every 2 days by MTT assay. B, control and ARGLU1 shRNA knockdown cells were cultured in 6-well plates, and the colonies that formed after 2 weeks were detected by crystal violet staining. C and D, 4000 cells/well of the above described cells were mixed with 0.35% agarose and plated in a 24-well plate. The resulting colonies were stained (C) with crystal violet and photographed after 3 weeks incubation and quantified (D). Data are presented as mean ± S.D. of three separate experiments. **, p < 0.01.
DISCUSSION
Recent studies revealed unexpected compositional heterogeneity and functional specificity of the Mediator complex (7, 10, 12, 13). We previously showed that MED1 exists only in a subpopulation of the Mediator complex with enriched RNA polymerase II and several additional Mediator subunits (20). In this study, we further identify ARGLU1 as a new MED1/Mediator-associated protein. We found that ARGLU1 colocalizes with MED1 in the cell nucleus and is able to interact with MED1-Mediator both in vitro and in vivo. One question arising is whether ARGLU1 is a bona fide Mediator subunit, or instead merely a MED1-interacting protein that is associated with the MED1-Mediator complex. We prefer the latter possibility because our sequence analysis of ARGLU1 shows that it has no homology to any known Mediator subunits from yeast to human. In fact, although ARGLU1 is conserved from Drosophila to mice and human, it cannot been found in yeast. Furthermore, previous studies of the Mediator complex (10, 11, 14), including one most extensive investigation that analyzed six independent Mediator preparations (immunopurified through MED10, MED9, MED29, MED19, MED28, and MED26 subunits) by multidimensional protein identification technology (MudPIT) (30), did not identify ARGLU1. Therefore, we believe it is more likely that ARGLU1 is not a subunit of the Mediator complex but rather an interacting protein that may play some regulatory roles through its interactions with MED1-Mediator.
Interestingly, we found that ARGLU1 is able to directly interact with MED1. Further deletion analyses narrowed down this interaction region to the far C terminus (1251–1581 aa) of MED1, a region with which no other protein has been reported to interact. Previous structural and functional analyses of MED1 has divided MED1 largely into three regions: the N-terminal region (1–501 aa) essential for incorporation of MED1 into Mediator complex (23), the central region (501–738 aa) containing two classical LXXLL motifs that interact with a number of nuclear receptors (10, 17, 19, 21–23, 31), and the large C-terminal region that may play some regulatory roles through phosphorylation by the MAPK pathway (24, 28, 29). In addition to interactions with the above mentioned nuclear receptors through its LXXLL motifs, MED1 has also been reported to interact with some other transcription factors and transcriptional coactivators. Interestingly, most of these interaction sites for MED1 are also restricted to either its N terminus or the region containing LXXLL motifs. MED1 has been reported to interact with GATA family transcriptional factors through several regions located at the N terminus that include amino acid residues 1–230, 1–327, 230–440, or 622–701 (32–34). Furthermore, MED1 was found to interact with another transcriptional coactivator peroxisome proliferator-activated receptor coactivator 1α through the region spanning the LXXLL motifs (506–753 aa) (35). Thus, our finding that ARGLU1 interacts with the far C-terminal region (1251–1581 aa) of MED1 represents the first case for a MED1 C terminus interacting protein. As mentioned above, MED1 can be phosphorylated at threonine 1032 and 1457 by the MAPK pathway, which in turn enhances its transcriptional coactivator activities through stabilizing MED1 and facilitating its subsequent assembly into Mediator complex (24, 28, 29). Interestingly, one of these phosphorylation sites (Thr-1457) is located right within this ARGLU1-interacting region of MED1. Therefore, in future studies it could be particularly interesting to examine whether a modification at this site could affect the interactions between MED1 and ARGLU1 and contribute to regulation of the MED1 functions.
Although no apparent known functional domains were found on ARGLU1, sequence analyses revealed that ARGLU1 is a highly polarized protein enriched in positively charged amino acids at the first 98 N-terminal amino acids (27 arginine and 8 lysine, or 36%) with the rest of the sequence highly enriched for negatively charged residues (49 Glutamate, or 28%). Further deletion mapping indicated that the N-terminal 98 aa of ARGLU1 are sufficient to interact with MED1 and associated Mediator subunits. Although more future work will be required to further narrow down the minimal region required and to determine whether the positive charge itself is the main factor responsible for this interaction, we did find that the ARGLU1-interacting C terminus of MED1 (1251–1581 aa) contain two stretches of sequence (1509–1521 aa, 1563–1575 aa) with a high percentage (54%) of negatively charged amino acids. As shown in Fig. 3, somewhat surprisingly, we found that one slightly longer MED1 C-terminal fragment (1076–1581 aa) failed to bind to ARGLU1. Although it remains possible that the MED1 (1076–1581 aa) fragment may misfold when expressed in bacteria, one other possibility is that the MED1 region (1076–1251 aa) could potentially form intra-molecular interactions with the MED1 C terminus to interfere with the interactions between ARGLU1 and MED1. Interestingly, the MED1 (1076–1251 aa) region contains 20 positively charged amino acids with only 2 negatively charged amino acids. Future studies to further dissect the molecular details of these interactions may provide deep insights into the regulation and functions of MED1.
Previously, we and others have shown that MED1 directly interacts with the ER and is required for ER-mediated gene transcription both in vitro and in breast cancer cells (15, 16, 18, 20, 36). Here, we reported that ARGLU1 could cooperate with MED1 in mediating ER transcription in reporter assays. Importantly, ARGLU1 could also be recruited to coexist with MED1 in the same complex on the endogenous ER target gene promoter upon estrogen stimulation, whereas depletion of ARGLU1 significantly impaired the expression of these genes. One significant question remaining is how ARGLU1 functions in concert with MED1 to regulate ER-mediated transcription. Because we have shown that the N-terminal ARGLU1 can directly interact with MED1, one possibility is that the remaining charged regions of ARGLU1 may function to stabilize the MED1-Mediator complex on the chromatin through interactions with DNA/chromatin, similar to high mobility group proteins, which are also highly charged (37). Furthermore, given the known role of MED1 and Mediator complex in affecting the recruitment and function of RNA polymerase II and general transcription factors (e.g. TFIID and TFIIB) (38–40), it is also possible that ARGLU1 could directly facilitate the assembly of RNA polymerase II and general transcription factors on the transcriptional initiation site. Further efforts to study the interplay of ARGLU1 and MED1 with DNA/chromatin and to identify additional interacting proteins for ARGLU1 or the MED1 C terminus may help us start to unravel the underlying molecular mechanisms.
We have shown that MED1 is not only required for estrogen-dependent transcription but also for estrogen-dependent growth of breast cancer cells (20). Interestingly, our most recent studies indicate that the disruption of MED1 in mice results in selective impairment of estrogen receptor functions in mammary gland development but not in other estrogen-responsive tissues like uterus, which suggests that MED1 could potentially be used as a tissue-specific target to selectively block the estrogen signaling pathway (17). Significantly, MED1 has been found to be overexpressed and amplified in a high percentage of human breast cancers, in both cell lines and primary tumors (41–44). Guided by these findings, we further examined the role of ARGLU1 in estrogen-dependent transcription and breast cancer cell growth. Our results clearly indicate that ARGLU1 not only plays a key role in ER-mediated transcription but that it is also required for the growth, as well as both the anchorage-dependent and -independent colony formation of human breast cancer cells. Therefore, further studies of the expression and function of ARGLU1 in breast cancer may provide potentially novel therapeutic targets for the treatment of breast cancer.
Supplementary Material
Acknowledgments
We thank Drs. Robert Roeder, Joan Conaway, Anders Naar, W. Scott Moye-Rowley, Sohaib Khan, and their laboratory members for plasmids, antibodies, advice, or critical reading of the manuscript. We also thank Laura Robbins, Qiuping Hu, Maryellen Daston, and Glenn Doerman for technical and editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK P30 DK078392 from the United States Public Health Service, University of Cincinnati Cancer Center Start-up and Pilot Grants, American Heart Association Beginning Grant-in-aid, and a Ride Cincinnati Award (to X. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ARGLU1
- arginine and glutamate rich 1
- MED1
- Mediator complex subunit 1
- ER
- estrogen receptor
- MTT
- 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
- aa
- amino acid(s).
REFERENCES
- 1. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsai M. J., O'Malley B. W. (1994) Annu. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 3. McKenna N. J., Lanz R. B., O'Malley B. W. (1999) Endocr. Rev. 20, 321–344 [DOI] [PubMed] [Google Scholar]
- 4. Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 5. Roeder R. G. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 201–218 [DOI] [PubMed] [Google Scholar]
- 6. Roeder R. G. (2003) Nat. Med. 9, 1239–1244 [DOI] [PubMed] [Google Scholar]
- 7. Blazek E., Mittler G., Meisterernst M. (2005) Chromosoma 113, 399–408 [DOI] [PubMed] [Google Scholar]
- 8. Malik S., Roeder R. G. (2005) Trends Biochem. Sci. 30, 256–263 [DOI] [PubMed] [Google Scholar]
- 9. Kornberg R. D. (2005) Trends Biochem. Sci. 30, 235–239 [DOI] [PubMed] [Google Scholar]
- 10. Malik S., Roeder R. G. (2000) Trends Biochem. Sci. 25, 277–283 [DOI] [PubMed] [Google Scholar]
- 11. Boube M., Joulia L., Cribbs D. L., Bourbon H. M. (2002) Cell 110, 143–151 [DOI] [PubMed] [Google Scholar]
- 12. Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W. (2005) Trends Biochem. Sci. 30, 250–255 [DOI] [PubMed] [Google Scholar]
- 13. Taatjes D. J. (2010) Trends Biochem. Sci. 35, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fondell J. D., Ge H., Roeder R. G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burakov D., Wong C. W., Rachez C., Cheskis B. J., Freedman L. P. (2000) J. Biol. Chem. 275, 20928–20934 [DOI] [PubMed] [Google Scholar]
- 16. Kang Y. K., Guermah M., Yuan C. X., Roeder R. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang P., Hu Q., Ito M., Meyer S., Waltz S., Khan S., Roeder R. G., Zhang X. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wärnmark A., Almlöf T., Leers J., Gustafsson J. A., Treuter E. (2001) J. Biol. Chem. 276, 23397–23404 [DOI] [PubMed] [Google Scholar]
- 19. Acevedo M. L., Lee K. C., Stender J. D., Katzenellenbogen B. S., Kraus W. L. (2004) Mol. Cell 13, 725–738 [DOI] [PubMed] [Google Scholar]
- 20. Zhang X., Krutchinsky A., Fukuda A., Chen W., Yamamura S., Chait B. T., Roeder R. G. (2005) Mol. Cell 19, 89–100 [DOI] [PubMed] [Google Scholar]
- 21. Yuan C. X., Ito M., Fondell J. D., Fu Z. Y., Roeder R. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7939–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ren Y., Behre E., Ren Z., Zhang J., Wang Q., Fondell J. D. (2000) Mol. Cell. Biol. 20, 5433–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malik S., Guermah M., Yuan C. X., Wu W., Yamamura S., Roeder R. G. (2004) Mol. Cell. Biol. 24, 8244–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Belakavadi M., Pandey P. K., Vijayvargia R., Fondell J. D. (2008) Mol. Cell. Biol. 28, 3932–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ge K., Cho Y. W., Guo H., Hong T. B., Guermah M., Ito M., Yu H., Kalkum M., Roeder R. G. (2008) Mol. Cell. Biol. 28, 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 27. Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 28. Misra P., Owuor E. D., Li W., Yu S., Qi C., Meyer K., Zhu Y. J., Rao M. S., Kong A. N., Reddy J. K. (2002) J. Biol. Chem. 277, 48745–48754 [DOI] [PubMed] [Google Scholar]
- 29. Pandey P. K., Udayakumar T. S., Lin X., Sharma D., Shapiro P. S., Fondell J. D. (2005) Mol. Cell. Biol. 25, 10695–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., Conaway R. C. (2004) Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 31. Savkur R. S., Burris T. P. (2004) J. Pept. Res. 63, 207–212 [DOI] [PubMed] [Google Scholar]
- 32. Crawford S. E., Qi C., Misra P., Stellmach V., Rao M. S., Engel J. D., Zhu Y., Reddy J. K. (2002) J. Biol. Chem. 277, 3585–3592 [DOI] [PubMed] [Google Scholar]
- 33. Stumpf M., Waskow C., Krötschel M., van Essen D., Rodriguez P., Zhang X., Guyot B., Roeder R. G., Borggrefe T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18504–18509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gordon D. F., Tucker E. A., Tundwal K., Hall H., Wood W. M., Ridgway E. C. (2006) Mol. Endocrinol. 20, 1073–1089 [DOI] [PubMed] [Google Scholar]
- 35. Wallberg A. E., Yamamura S., Malik S., Spiegelman B. M., Roeder R. G. (2003) Mol. Cell 12, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 36. Acevedo M. L., Kraus W. L. (2003) Mol. Cell. Biol. 23, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hock R., Furusawa T., Ueda T., Bustin M. (2007) Trends Cell Biol. 17, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baek H. J., Malik S., Qin J., Roeder R. G. (2002) Mol. Cell. Biol. 22, 2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson K. M., Wang J., Smallwood A., Arayata C., Carey M. (2002) Genes Dev. 16, 1852–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baek H. J., Kang Y. K., Roeder R. G. (2006) J. Biol. Chem. 281, 15172–15181 [DOI] [PubMed] [Google Scholar]
- 41. Zhu Y., Qi C., Jain S., Le Beau M. M., Espinosa R., 3rd, Atkins G. B., Lazar M. A., Yeldandi A. V., Rao M. S., Reddy J. K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luoh S. W. (2002) Cancer Genet. Cytogenet. 136, 43–47 [DOI] [PubMed] [Google Scholar]
- 43. Miller L. D., Smeds J., George J., Vega V. B., Vergara L., Ploner A., Pawitan Y., Hall P., Klaar S., Liu E. T., Bergh J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13550–13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ivshina A. V., George J., Senko O., Mow B., Putti T. C., Smeds J., Lindahl T., Pawitan Y., Hall P., Nordgren H., Wong J. E., Liu E. T., Bergh J., Kuznetsov V. A., Miller L. D. (2006) Cancer Res. 66, 10292–10301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.