Abstract
A limited decrease in mitochondrial membrane potential can be beneficial for cells, especially under some pathological conditions, suggesting that mild uncouplers (protonophores) causing such an effect are promising candidates for therapeutic uses. The great majority of protonophores are weak acids capable of permeating across membranes in their neutral and anionic forms. In the present study, protonophorous activity of a series of derivatives of cationic rhodamine 19, including dodecylrhodamine (C12R1) and its conjugate with plastoquinone (SkQR1), was revealed using a variety of assays. Derivatives of rhodamine B, lacking dissociable protons, showed no protonophorous properties. In planar bilayer lipid membranes, separating two compartments differing in pH, diffusion potential of H+ ions was generated in the presence of C12R1 and SkQR1. These compounds induced pH equilibration in liposomes loaded with the pH probe pyranine. C12R1 and SkQR1 partially stimulated respiration of rat liver mitochondria in State 4 and decreased their membrane potential. Also, C12R1 partially stimulated respiration of yeast cells but, unlike the anionic protonophore FCCP, did not suppress their growth. Loss of function of mitochondrial DNA in yeast (grande-petite transformation) is known to cause a major decrease in the mitochondrial membrane potential. We found that petite yeast cells are relatively more sensitive to the anionic uncouplers than to C12R1 compared with grande cells. Together, our data suggest that rhodamine 19-based cationic protonophores are self-limiting; their uncoupling activity is maximal at high membrane potential, but the activity decreases membrane potentials, which causes partial efflux of the uncouplers from mitochondria and, hence, prevents further membrane potential decrease.
Keywords: Antioxidant, Liposomes, Membrane Bilayer, Membrane Energetics, Mitochondria, Mitochondrial Transport, Quinones, Yeast, Penetrating Cation, Protonophore
Introduction
Transport of electrons along the mitochondrial respiratory chain is accompanied by the formation of an electrochemical gradient of hydrogen ions (ΔμH+)3 at the inner mitochondrial membrane. ΔμH+ is used for ATP production and other energy-consuming processes. However, high values of ΔμH+ can increase the production of dangerous reactive oxygen species (ROS) (1, 2). Although mitochondria are able to control ΔμH+ by adjusting the activity of natural uncoupling mechanisms (i.e. free fatty acids, anion carriers, and uncoupling proteins) (3), there is considerable interest in finding pharmacological agents to increase mitochondrial proton leak and, as a consequence, to prevent obesity and to decrease ROS production (4–7).
Uncouplers, or protonophores, are small organic compounds capable of carrying hydrogen ions across artificial and biological membranes. The strategy of “mild uncoupling” (2) relies on the fact that partial decrease in ΔμH+ can be beneficial for cells especially under some pathological conditions, suggesting that uncouplers are good candidates for therapeutic uses. Apparently, such applications are hindered by high toxicity, as in the case of 2,4-dinitrophenol (DNP), which was temporarily used at the beginning of the 20th century as an obesity-preventing drug (5). Therefore, there is a practical necessity for designing an uncoupler exhibiting a broad window between concentrations causing partial ΔμH+ decrease and toxic effects.
DNP and a majority of other protonophores are weak acids existing in neutral and anionic form, depending on the pH of the medium (8). The anionic form is characterized by substantial delocalization of its negative charge, facilitating permeation across membranes (9). Several cationic protonophores have been described, although their acting concentrations are in the submillimolar or even millimolar range (10–13). Murphy and co-workers (14) were the first who used conjugation with penetrating cations for addressed delivery of substances into mitochondria having Δψ negative inside (15, 16). Among other compounds, Murphy and co-workers (4) designed a conjugate of triphenylphosphonium with DNP to increase the protonophorous activity of DNP. It turned out, however, that the conjugate was substantially less active than the parent DNP. Another conjugate of triphenylphosphonium, with butylated hydroxytoluene, exhibited protonophorous activity within a wide range of concentrations because of its interaction with adenine nucleotide translocase (17). It has recently been shown that mitochondria-targeted antioxidants (SkQ1 and 10-(6′-ubiquinonyl) decyltriphenylphosphonium) as well as SkQ1 lacking quinone (dodecyltriphenylphosphonium) exhibit protonophorous action when added together with free fatty acids (7). Because a certain amount of free fatty acids is usually present in living cells, SkQ1 may be considered as a mitochondria-targeted protonophore.
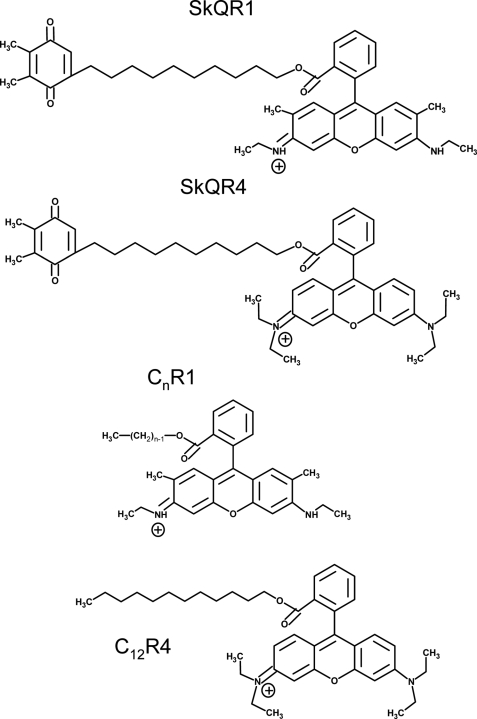
It was shown in our laboratory that rhodamine 19 can effectively substitute for cationic triphenylphosphonium in conjugates, ensuring delivery to mitochondria (18). Fig. 1 depicts a structure of a conjugate of plastoquinone and rhodamine 19 (SkQR1) as well as other derivatives of rhodamine 19 and rhodamine B used in the present work. Rhodamine B differs from rhodamine 19 by two additional ethyl groups on two nitrogen atoms of the rhodamine moiety, which hinder protonation of these nitrogens. According to our previous data (19), SkQR1 has better membrane permeability than triphenylphosphonium-based SkQ1. The present paper reveals that certain derivatives of rhodamine 19 show a protonophorous uncoupling effect even in the absence of fatty acids. Because these compounds can accumulate in mitochondria in a Δψ-dependent fashion, they can be considered as mitochondria-targeted cationic uncouplers. Due to the very fact that the accumulation results in uncoupling, which, in turn, lowers Δψ, novel uncouplers represent uncouplers of self-limited activity (i.e. mild uncouplers). This assumption was confirmed by experiments on isolated mitochondria and yeast cells.
FIGURE 1.
Chemical structures of rhodamine 19 derivatives (SkQR1, CnR1) and rhodamine B derivatives (SkQR4, C12R4) used in the present work.
MATERIALS AND METHODS
Earlier a series of mitochondria-targeted antioxidants having substituted 1,4-benzoquinone rings conjugated to hydrophobic triphenylphosphonium or the rhodamine cation through the decyl linker were synthesized (14, 18, 20). Structures of SkQR1 and other compounds used in this work are shown in Fig. 1. Rhodamine was modified by alkyl substitutes by esterification of its free carboxylic group by methods described in this group (20). An octyl ester of rhodamine 19 (C8R1) was prepared as described earlier (20).
Cell or Mitochondria Respiration
Respiration of yeast cells or isolated mitochondria was measured using a standard polarographic technique with a Clark-type oxygen electrode (Oroboros Instruments, Innsbruck, Austria) at 25 °C using DATLAB software. The incubation medium for yeast cells contained 50 mm KH2PO4, pH 5.5, and 0.005% glucose. The incubation medium for rat liver mitochondria contained 150 mm KCl, 20 mm HEPES, 0.5 mm EDTA, 0.2 mg/ml BSA, pH 7.4 (Fig. 2A) or 300 mm mannitol, 2.5 mm HEPES, 0.5 mm EDTA, 5.5 mm MgCl2, 5 mm KH2PO4, 0.5 mg/ml BSA, pH 7.4 (Fig. 2B). Mitochondrial protein concentration was 0.4 mg/ml.
FIGURE 2.
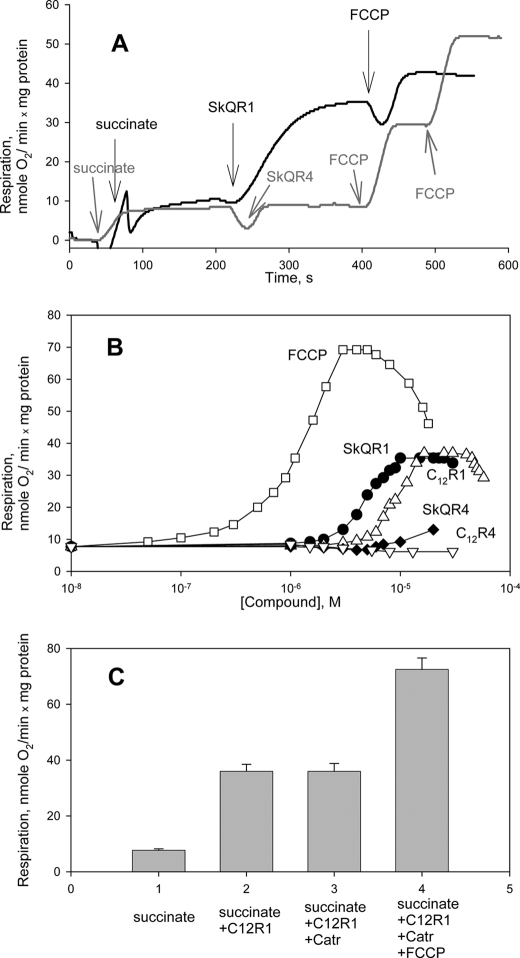
A, stimulation of rat liver mitochondria respiration by SkQR1. Shown are traces of the rate of respiration in the medium described under “Materials and Methods.” Additions were as follows: 5 mm succinate, 2 μm rotenone, 2 μm SkQR1, 2 μm SkQR4, 1 μm FCCP (trace with SkQR1), and two additions of FCCP (0.5 μm) in the trace with SkQR4. B, dose dependence of respiratory stimulation by SkQR1, SkQR4, C12R1, C12R4, and FCCP. C, the increase in the respiration rate caused by C12R1 was insensitive to the addition of carboxyatractilate (2 μm). Error bars, S.E.
Mitochondrial Membrane Potential Measurement
3,3′- Dipropylthiadicarbocyanine iodide (DiS-C3-(5)) was used as a membrane potential probe (21). Fluorescence intensity at 670 nm (excitation at 622 nm) was measured with a Panorama Fluorat 02 spectrofluorimeter (Lumex, St. Petersburg, Russia). The medium for measurements contained 150 mm KCl, 10 mm Tris, 0.5 mm EGTA, 5 mm succinate, 1 μm rotenone, and 1 μm DiS-C3-(5), pH 7.4. Mitochondrial protein concentration was 0.4 mg/ml.
Detection of Proton Transport in Pyranine-loaded Liposomes
The lumenal pH of the liposomes was assayed with pyranine by a slightly modified procedure of Ref. 22. To prepare pyranine-loaded liposomes, lipids (5 mg of egg phosphatidylcholine and 1 mg of cholesterol) in a chloroform suspension were dried in a round-bottom flask under a stream of nitrogen. The lipids were then resuspended in buffer (100 mm KCl, 20 mm MES, 20 mm MOPS, 20 mm Tricine titrated with KOH to pH 6.0) containing 0.5 mm pyranine. The suspension was vortexed and then freeze-thawed three times. Unilamellar liposomes were prepared by extrusion through 0.1-μm pore size Nucleopore polycarbonate membranes using an Avanti miniextruder. The unbound pyranine was then removed by passage through a Sephadex G-50 coarse column equilibrated with the same buffer solution. To measure the rate of pH dissipation in liposomes with lumenal pH 6.0, the liposomes were diluted in a solution buffered to pH 8 and supplemented with 10 mm p-xylene-bis-pyridinium bromide to suppress the fluorescence of leaked pyranine. The pH was estimated from the ratio I455/I410 of the intensities of fluorescence measured at 505 nm upon excitation at 455 nm (I455) and 410 nm (I410), respectively (23), as monitored with the Panorama Fluorat 02 spectrofluorimeter. At the end of each recording, 1 μm nigericin was added to dissipate the remaining pH gradient. To prevent the formation of H+ diffusion potential, the experiments were carried out in the presence of 10 nm valinomycin.
Planar Bilayers
Bilayer lipid membrane (BLM) was formed from 2% decane solution of diphytanoylphosphatidylcholine on a 0.6-mm aperture in a Teflon septum separating the experimental chamber into two compartments of equal size (3-ml volumes). Electrical parameters were measured with two AgCl electrodes placed into the solutions on the two sides of the BLM via agar bridges, using a Keithley 6517 amplifier (Cleveland, OH). The incubation mixture contained 50 mm Tris-HCl, 50 mm KCl, pH 7.0.
Isolation of Rat Liver Mitochondria
Rat liver mitochondria were isolated by differential centrifugation (24) in a medium containing 250 mm sucrose, 10 mm MOPS, 1 mm EGTA, and bovine serum albumin (0.1 mg/ml), pH 7.4. The final washing was performed in the medium of the same composition. Protein concentration was determined using bicinchoninic acid as described (25). Handling of animals and experimental procedures were conducted in accordance with international guidelines for animal care and use and were approved by the Institutional Ethics Committee of the A. N. Belozersky Institute of Physico-Chemical Biology at Moscow State University.
Strains and Cell Growth Conditions
Both yeast strains used in this study are of W303, Mat a genetic background. The petite strain was generated by growing the cells on plates supplemented with ethidium bromide (26). Liquid media contained yeast extract with peptone and were supplemented with 2% glucose, 2% glycerol, or 2% raffinose (27). The yeast was typically grown in liquid media to a density of 2 × 106 cells/ml. The growth rates were determined by plating culture aliquots on solid media and counting the colonies the next day or by counting them under the microscope. At least three experiments were performed for each treatment. Respiration rates were determined as described previously (28). Because multidrug resistance is hyperactivated in petite cells (29), the inhibitor of multidrug resistance verapamil (30) (100 μm) was added to all samples in the experiments comparing growth of the wild type (grande) and petite cells (Fig. 7C).
FIGURE 7.
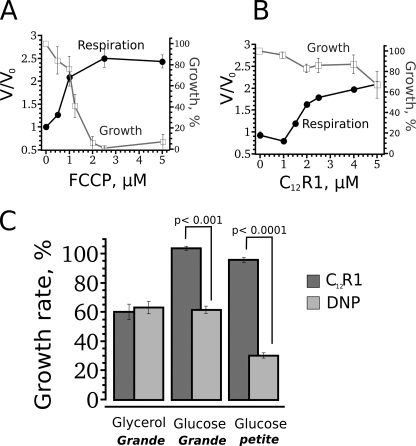
C12R1, unlike anionic uncouplers, affects yeast cells, depending on their mitochondrial membrane potential. Shown are the dependences of stimulation of yeast cell respiration rate (V/V0) and growth on the concentration of FCCP (A) and C12R1 (B). C, C12R1 (10 μm; dark gray) or DNP (500 μm; light gray) were added to yeast cells (grande or petite) growing on non-fermentable (glycerol) or fermentable (glucose) carbon sources. The growth rate without uncouplers corresponds to 100%. Error bars, S.E.
Molecular Dynamics
Two models for exploring the ion transfer thermodynamics within the membrane were constructed. One of those is a homogeneous lipid bilayer with dodecyltriphenylphosphonium cation (C12-TPP), which has been used extensively in previous studies (7) and could be considered as a reference for the transfer of SkQR1 ion that comprises the second model. A lipid vesicle was modeled by a bilayer consisting of 100 dipalmitoylphosphatidylcholine molecules (50 per leaflet) surrounded by 3204 water molecules. The system was large enough to ensure that ions in the middle of the membrane patch did not perturb the general membrane structure. Orthogonal periodic boundaries with x,y-translation lengths of ∼5.7 nm and average height of ∼6.8 nm were used. The negative net charge of the membrane was neutralized by a Cl− anion. The system consisted of 14,661 or 14,668 atoms in total for the C12-TPP and SkQR1 ions, respectively. Simulations were performed with the program Gromacs (31) using the G43A1-S3 force field (32) and SPC/E water model (33). The electrostatics were calculated using the particle mesh Ewald method (34), and all bonds were maintained with the LINCS routine (35). Molecular dynamics simulations were carried out at a constant number of particles (N), constant pressure (P = 1 atm), and constant temperature (T = 323 K) (NPT ensemble). Three-dimensional periodic boundary conditions were applied with the z axis lying along a direction normal to a bilayer. The pressure was controlled semi-isotropically, so that the x-y and z sizes of the simulation box were allowed to fluctuate independently from each other, keeping the total pressure constant. The averaged area per lipid was 0.65 nm2. The force field parameters for C12-TPP were taken from Ref. 7, and parameters for SkQR1 were developed using those for sulforhodamine (36). All molecular dynamics calculations were performed at the supercomputer SKIF “Chebyshev” in the Computation Center, Moscow State University.
The potential of mean force, W, is the thermodynamic work along a coordinate z, relative to some reference value z′,
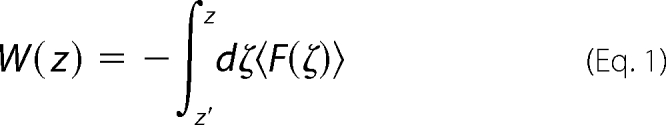 |
where F(ζ) is the instantaneous force acting on the transferred ion at a chosen coordinate, ζ. To assist in computing this integral, we employ umbrella sampling (37), whereby we add a biasing potential (Equation 2) to the system potential.
Here z is the z-coordinate of the charge center of transferred ions (phosphorus atom in the case of TPP cation and the center of the π-conjugated aromatic ring in the case of rhodamine, respectively), z0 is its initial coordinate, the biasing harmonic force constant K used was 2000 kJ mol−1 nm−2, and the pulling rate v was 2·10−6 nm/ps.
RESULTS
Mitochondrial Respiration and Membrane Potential
The rate of mitochondrial respiration is determined by the activity of respiratory electron transport chains, which, in turn, is limited by ΔμH+. Under the conditions of upper ΔμH+ limit, the respiration rate is minimal and depends on the intrinsic proton leaks across the inner mitochondrial membrane. The addition of an uncoupler, which diminishes the ΔμH+ value, stimulates respiration. Fig. 2A shows changes in the rate of respiration of rat liver mitochondria upon the addition of SkQR1 and SkQR4 (i.e. plastoquinone-containing derivatives of rhodamine 19 or rhodamine B, respectively). In contrast to SkQR4, SkQR1 stimulated respiration, suggesting uncoupling action of the rhodamine 19 derivative. The addition of FCCP after SkQR1 stimulated the rate of respiration, suggesting incomplete loss of Δψ in the presence of SkQR1 (Fig. 2A). The same result was observed in a wide range of SkQR1 concentrations (data not shown). Fig. 2B displays the dependences of the respiration rate on the concentrations of SkQR1 and SkQR4 and the corresponding compounds lacking quinone moieties (i.e. C12R1 and C12R4, as well as of a conventional uncoupler FCCP). SkQR1 stimulated respiration at concentrations exceeding those of FCCP by approximately 1 order of magnitude. Moreover, the maximal stimulating effect of SkQR1 on respiration was lower than that of FCCP. According to our data (not shown), the SkQR1 concentrations that induced maximal stimulation did not inhibit respiration in the presence of FCCP. Therefore, the relatively low stimulation of respiration is not due to the inhibition of the respiratory chain activity when [SkQR1] rises. It is also seen from Fig. 2B that the acting concentrations of C12R1 exceeded those of SkQR1 by a factor of 2. Importantly, both derivatives of rhodamine B, C12R4 and SkQR4, did not stimulate mitochondrial respiration (Fig. 2B).
It has been shown previously that ATP/ADP antiporter can enhance uncoupling of mitochondria, mediated by DNP and several other uncouplers (38–40). Fig. 2C shows that the increase in the respiration rate caused by C12R1 was insensitive to the addition of carboxyatractilate (an inhibitor of ATP/ADP antiporter). Control experiments showed that the selected concentration of carboxyatractilate blocked completely stimulation of respiration caused by the addition of ADP. It can be concluded that the uncoupling action of the derivatives of rhodamine 19 does not depend on the functioning of ATP/ADP antiporter. Similar experiments were carried out with other inhibitors or modulators of mitochondrial inner membrane carriers (i.e. mersalyl and N-ethylmaleimide (phosphate carrier), glutamate (aspartate/glutamate antiporter), and ruthenium red (calcium uniporter). As in the case of carboxyatractilate, the inhibitors did not affect the respiration stimulated by C12R1 (supplemental Fig. 1S).
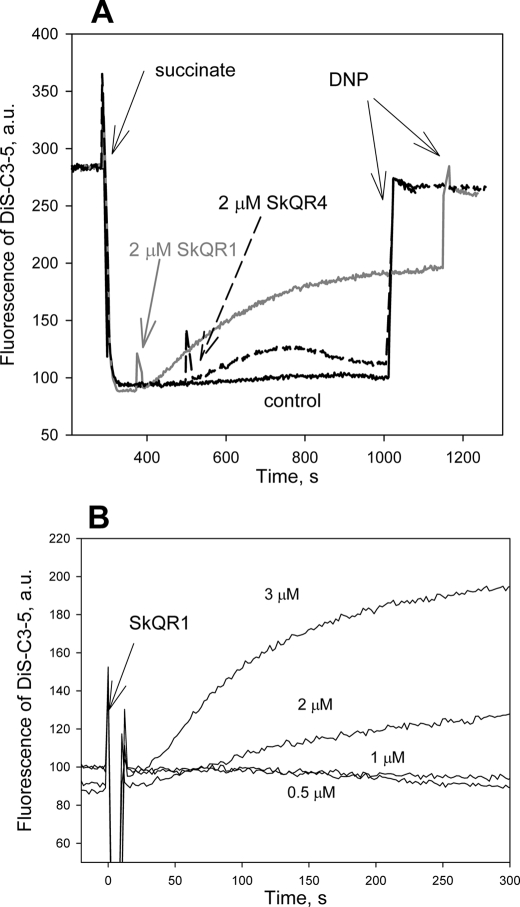
Fig. 3 shows Δψ measurements by means of the potential-sensitive dye DiS-C3-(5). In contrast to widely used safranine O, the spectral range of DiS-C3-(5) differs considerably from that of rhodamine (i.e. excitation 622 nm, emission 670 nm). According to Ref. 21, energization of mitochondria leads to quenching of DiS-C3-(5) fluorescence because of dye aggregation in the matrix. Fig. 3A demonstrates typical records of the action of 2 μm SkQR1 and 2 μm SkQR4 on the fluorescence of DiS-C3-(5) in mitochondria. At the end of the recordings, 100 μm DNP was added to cause complete deenergization of the mitochondria. As can be expected from the previous data, the addition of SkQR1 induced a steady decrease in the membrane potential. This confirms its uncoupling activity. As to SkQR4, it caused a small and transient Δψ decrease, which can be explained by electrophoretic import of this cation by mitochondria. Fig. 3B shows several recordings with different concentrations of SkQR1. As in the experiments with mitochondrial respiration, the acting concentrations were in the micromolar range. The time course of the SkQR1-induced Δψ decrease appeared to be slow, with the characteristic time constant of about 3 min. By contrast, the effect of low concentrations of DNP on Δψ was fast and limited by the instrument response time (data not shown). Apparently, the slow Δψ response to the addition of cationic SkQR1 was due to slow accumulation of SkQR1 in the matrix.
FIGURE 3.
A, effect of SkQR1 and SkQR4 on the membrane potential of rat liver mitochondria measured by DiS-C3-(5). Shown are traces of fluorescence in the medium described under “Materials and Methods.” In all traces, 5 mm succinate and 1 μm rotenone were added at 300 s, and 100 μm DNP was added at the end of each trace. Control, no other additions. Gray trace, 2 μm SkQR1 was added at 380 s. Dashed trace, 2 μm SkQR4 was added at 500 s. B, dose dependence of the effect of SkQR1 on the potential of mitochondria. a.u., arbitrary units.
Liposomes Loaded with Pyranine
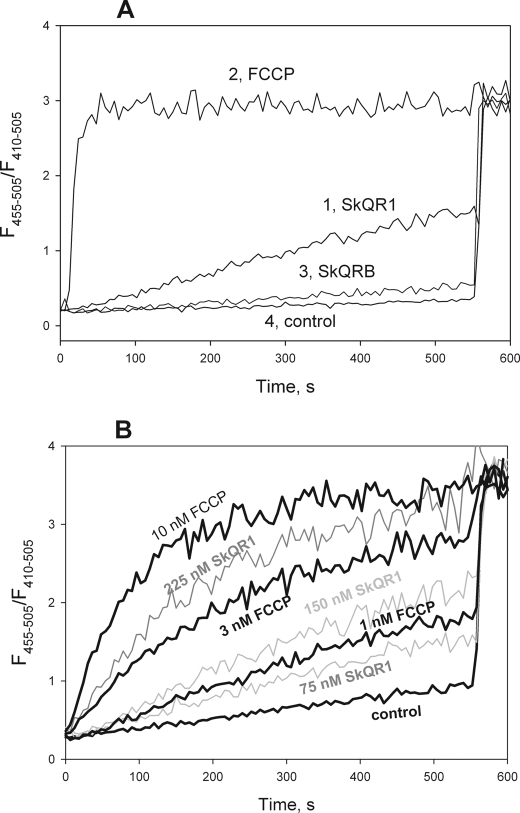
The uncoupling action of SkQR1 and C12R1 can be a result of either direct protonophorous activity or indirect action leading to the induction of a proton leak due to, for example, some damage to the inner membrane of mitochondria. To address this point, we conducted experiments on artificial lipid membranes (i.e. liposomes and planar bilayers). According to Refs. 22 and 41, liposomes loaded with the pH-sensitive probe pyranine can serve as a useful system to study the action of protonophores. Fig. 4A shows the kinetics of dissipation of a preformed pH gradient on membranes of liposomes after the addition of 0.1 μm FCCP, SkQR1, or SkQR4. The addition of SkQR1 led to significant changes in liposomal pH within minutes, suggesting specific protonophorous activity (Fig. 4A, curve 1). FCCP displayed higher activity, leading to pH equilibration within less than 1 min (Fig. 4A, curve 2). Remarkably, SkQR4 was completely inactive in this system (Fig. 4A, curve 3). The measurements were carried out in the presence of valinomycin, a powerful K+-ionophore. No pH changes were detected in the absence of valinomycin, as shown with FCCP in Fig. 4A, curve 4. According to Ref. 22, this effect is associated with formation of Δψ on membranes of liposomes, which blocks proton transport without valinomycin. Valinomycin per se did not induce pH changes under our conditions (data not shown).
FIGURE 4.
A, dissipation of pH gradient on membranes of pyranine-loaded liposomes by SkQR1, SkQR4, and FCCP. Inner liposome pH was estimated from the ratio (I455/I410) of pyranine fluorescence intensities measured at 505 nm upon excitation at 455 and 410 nm, respectively. Nigericin (1 μm) was added at 550 s to equilibrate the pH. The control curve is with FCCP but without valinomycin. Concentrations were as follows: SkQR1 (100 nm), SkQR4 (100 nm), FCCP (100 nm), valinomycin (10 nm), and lipid (75 μm). B, dose dependence of the effect of SkQR1 and FCCP. The control curve corresponded to 10 nm valinomycin without protonophores.
Fig. 4B illustrates the dose dependences of the effect of SkQR1 (75–225 nm) and FCCP (1–10 nm) on liposomal pH. The acting concentrations of SkQR1 in this system exceeded those of FCCP by about 1 order of magnitude, as it occurred in mitochondria. Therefore, the uncoupling action of SkQR1 on mitochondria can be accounted for by the direct induction of proton fluxes in the lipid part of the mitochondrial membrane.
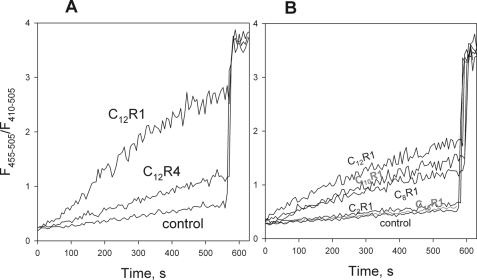
The experiments on mitochondrial respiration revealed that the rhodamine derivative lacking the quinone moiety retained uncoupling activity (Fig. 2B). Fig. 5A shows the effects of C12R1 and C12R4 on the liposome system. Consistent with our expectations, C12R1 exhibited protonophorous activity, in contrast to C12R4. Fig. 5B depicts the dependence of the protonophorous activity of alkyl rhodamines on the length of the alkyl chain. The activity of C8-, C10-, and C12-R1 was similar, whereas hexadecylrhodamine (C16R1) was ineffective (Fig. 5B). Rhodamine 19 ethyl ester (i.e. C2R1, also known as rhodamine 6G) was also inactive. It can be noted that nonlinear dependence of the activity on the alkyl chain length was also found for the ability of these rhodamines to transport organic anions across model membranes (20).
FIGURE 5.
A, dissipation of pH gradient on membranes of pyranine-loaded liposomes by C12R1 and C12R4 (both 150 nm). Inner liposome pH was estimated from the ratio (I455/I410) of pyranine fluorescence intensities measured at 505 nm upon excitation at 455 nm and 410 nm, respectively. Nigericin (1 μm) was added at 550 s to equilibrate the pH. Other conditions were as described in the legend to Fig. 2. B, protonophorous action of rhodamine 19 alkyl esters (CnR1, 150 nm). Lipid concentration was 200 μm.
Diffusion Potentials on Planar Bilayer Lipid Membranes
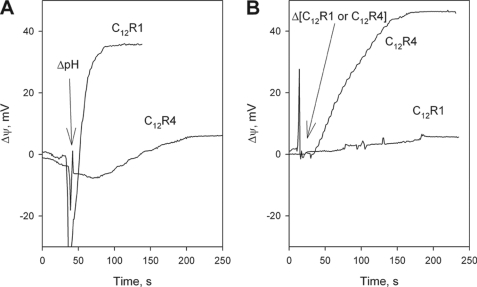
To measure the protonophorous effect of studied compounds, we also used a planar BLM separating two solutions differing in pH values. In such a system, the addition of a protonophore (FCCP) resulted in a transmembrane H+ flux generating an electrical potential difference across the BLM, the more acidic compartment being negatively charged. It was found (Fig. 6A) that C12R1, but not C12R4, can effectively substitute for FCCP. The magnitude of the generated Δψ proved to be slightly lower than the theoretical (Nernstian) limit (i.e. about 40 mV instead of 59 mV at ΔpH = 1). The effect of SkQR1 was similar to that of C12R1 (data not shown). Interestingly, C12R4 generated Δψ (negative at the side of the addition) when added into one of the membrane-separated compartments (Fig. 6B), suggesting that C12R4 can permeate through BLM in the cationic form (in this experiment, pH values were equal in both compartments). In contrast to C12R4, the addition of C12R1 at one side of the BLM did not generate the electrical potential (Fig. 6B), which was probably a result of the induction of H+ cycling and effective shunting of the diffusion potential generated by the flux of the cationic form of C12R1. Similar action of other protonophores on the diffusion potential of planar bilayers was described previously for CCCP (42), TTFB (43), and other anionic uncouplers (8).
FIGURE 6.
A, C12R1 but not C12R4 induced formation of H+ diffusion potential on planar bilayer lipid membrane made from diphytanoylphosphatidylcholine. Concentrations of C12R1 and C12R4 were 0.5 μm. pHcis = 7, and pHtrans = 8. B, formation of diffusion potential of penetrating cations C12R4 and C12R1 (both 1 μm at one side and 0.1 μm at the other) added at the cis side of the planar membrane at symmetrical pH 7.
pK of Rhodamine Derivatives
Conventional protonophores are weak acids with pK in the physiological range. All of them are capable of permeating lipid membranes in neutral as well as anionic forms. The ACD Labs data base predicts two pK values for the rhodamine moiety of SkQR1 (7.1 and 4.2), which correspond to protonation of one or two nitrogen atoms in the rhodamine core of the molecule (Fig. 1). In the case of SkQR4, the data base suggests a single pK (5.7) because double protonation of the rhodamine B molecule is impossible, implying the presence of one positive charge at pK <5.7 and two positive charges at pH >5.7. Although the pK of rhodamine derivatives is usually estimated spectrophotometrically (44), the absorption and fluorescence spectra of SkQR1 and SkQR4 were pH-independent in the range of pH from 3 to 10 (data not shown). To estimate the pK of rhodamine derivatives, we measured the pH dependence of self-aggregation of short-chain C4R1 and C4R4 by fluorescence correlation spectroscopy (supplemental Fig. 2S). The estimated pK of C4R1 was 7.3, and pK of C4R4 was 5.3. Apparently, the estimated pK of C4R1 corresponded to the first protonation of the neutral form of rhodamine 19, whereas the second protonation of C4R1 did not affect the fluorescence correlation spectroscopy data. The inability of derivatives of rhodamine B to transport protons across membranes can be attributed to the absence of a neutral form of the rhodamine B moiety, in contrast to rhodamine 19. We will further address this point when discussing the mechanism of the protonophorous action of alkyl rhodamines.
Respiration and Growth of Saccharomyces cerevisiae Cells
In the last series of experiments, we studied the effects of our compounds on intact cells of yeast S. cerevisiae. First, we checked the intracellular localization of C12R1 and C12R4. As expected, both compounds co-localized with mitochondria (supplemental Fig. 3SA), and the localization was sensitive to the addition of FCCP (supplemental Fig. 3SB). Next, we tested the effects of the compounds on respiration of the cells. Fig. 7 (closed circles) shows the dependence of the rate of respiration on the concentration of FCCP (A) and C12R1 (B). As in the case of isolated mitochondria, the acting concentrations of C12R1 were higher compared with those of FCCP, being in the micromolar range. Besides, the maximal level of the stimulation by C12R1 was lower than that by FCCP. Fig. 7, A and B (open squares), presents also the dependences of the cell growth rate on the concentrations of these two uncouplers. In contrast to FCCP, C12R1 did not suppress the growth at the concentrations studied, presumably because C12R1 does not reduce the membrane potential below the level critical for growth. We applied electron microscopy to monitor the effect of high concentrations of C12R1 and C12R4 on mitochondrial ultrastructure. Representative microphotographs show that whereas C12R4 induced massive swelling and dramatic changes in mitochondrial ultrastructure, the changes caused by the same level of C12R1 were much less pronounced (supplemental Fig. 4S).
The aforementioned logic predicts that the effects of C12R1 should be less pronounced on cells with poorly energized mitochondria compared with the action of anionic uncouplers. To address this point, we compared the inhibition of S. cerevisiae growth on fermentable and non-fermentable carbon sources. It is known that yeast cells growing in the presence of glucose exhibit suppressed mitochondria functioning and have lower mitochondrial membrane potential than yeast growing on glycerol (reviewed in Ref. 45). First, we determined concentrations of cationic (C12R1) and anionic (DNP) uncouplers that induced 2-fold suppression of growth on the non-fermentable carbon source (glycerol). Second, the same concentrations of the uncouplers were tested on yeast grown on the fermentable carbon source. As expected, C12R1 did not inhibit the growth, in contrast to DNP (Fig. 7C). We also used petite cells in this series of experiments. These cells lack a functional respiratory chain and, therefore, are not capable of growing on the non-fermentable carbon source. It is known that the mitochondrial membrane potential is much lower in petite cells compared with the normal (grande) cells (46). Again, C12R1 did not slow the growth of the petite strain, whereas the growth was strongly inhibited by DNP (Fig. 7C).
Molecular Dynamics Simulations
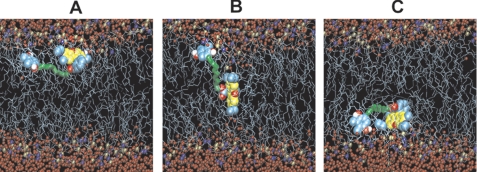
The process of permeation of RH+ across the membrane was studied by molecular dynamics simulations. Fig. 8 shows snapshots of SkQR1 residing at one side (A), in the middle (B), and at the other side (C) of a dipalmitoylphosphatidylcholine bilayer. The movement was induced by applying bias voltage (Equation 2), starting form the ion equilibrium position at z0. When entering the lipid bilayer, the π-conjugated aromatic ring of rhodamine appeared to lie parallel to the membrane surface, and the hydrocarbon linker in SkQR1 had sufficient freedom to adopt arbitrary conformations in the membrane. Upon immersion into the bilayer with the distance from the membrane center being larger than 0.5 nm, the aromatic ring of rhodamine in SkQR1 retained a parallel orientation with respect to the membrane surface. However, when the ions penetrated deeper into the membrane, the rhodamine ring orientation changed and became orthogonal to the membrane surface. A gradual change of the ring orientation was observed in the distance range from +0.5 to −0.5 nm from the membrane center. Remarkably, when located in the middle of the bilayer, the rhodamine ring spanned the nonpolar slab of the membrane, so that one of the nitrogen atoms of the rhodamine ring interacted with the water “finger” at one side of the membrane, and the second nitrogen atom interacted with the symmetrical water finger at the other side of the membrane. The entire process of SkQR1 permeation can be viewed in a movie presented in the supplemental material along with a free energy profile (supplemental Fig. 5S) and other details of the molecular dynamics simulations.
FIGURE 8.
Snapshots of SkQR1 permeation through a bilayer lipid membrane driven by the biasing potential (umbrella sampling method in molecular dynamics simulations). Shown are the lipid carbon tails (cyan thin sticks), nitrogen atoms of choline and phosphorus atoms of phosphate groups (blue and brown spheres), water molecules (small red spheres), and SkQR1 (aromatic rings (yellow), dodecyl residue (green), carbon (cyan), oxygen (red), and hydrogen (white)). A, equilibrium structure obtained after 100 ns of unperturbed molecular dynamics simulation (parallel position of rhodamine ring relative to the membrane surface). B, structure after 590 ns of umbrella sampling simulation (orthogonal conformation). C, structure after 940 ns of simulation (parallel conformation).
DISCUSSION
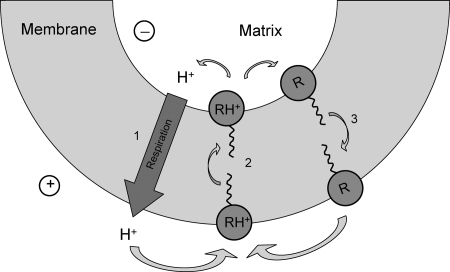
A scheme of functioning of the cationic protonophores SkQR1 and C12R1 is displayed in Fig. 9. The scheme is similar to that for conventional anionic protonophore except for the sign of the charged form moving across the membrane. A similar mechanism was proposed for the protonophorous action of the cationic antitumor alkaloid ellipticine (11) and pyridine AU-1421 (13). Our scheme differs from that for ellipticine by the intramembrane location of the carrier due to high affinity of SkQR1 and C12R1 with membranes (20). The scheme suggests a transmembrane movement for two forms of our H+ carriers: a deprotonated neutral form R and a monoprotonated cationic form RH+. We also assume that a doubly protonated form, RH22+, has considerably lower membrane permeability because dimers of alkyltriphenylphosphonium are known to be substantially less permeable than tetraphenylphosphonium itself (47, 48). Apparently, the neutral form of the carrier R should have much higher permeability than the charged form RH+. In the case of the protonophore CCCP, the ratio of permeabilities of neutral and charged forms exceeds 3 orders of magnitude (8, 42).
FIGURE 9.
Scheme of functioning of the cationic protonophores SkQR1 and C12R1. A deprotonated neutral form (R) and a singly protonated cationic form (RH+) can cross the membrane, whereas a doubly protonated form (RH22+) cannot. The scheme shows the following three steps of the carrier cycling: a voltage-dependent stage of RH+ translocation from cytoplasmic to the matrix side (step 2); RH+ deprotonation on the matrix side and movement of the neutral form R across the membrane along its concentration gradient (step 3); and protonation of R on the outer side of the membrane. Proton pumping by respiratory chains is designated by step 1.
The rhodamine B derivatives C12R4 and SkQR4 do not transport protons because they cannot exist in a neutral form R. Both R+ and RH2+ forms of C12R4 might, in principle, transverse a membrane, but both should move into the matrix driven by Δψ at the inner mitochondrial membrane. Besides, low permeability of RH2+ should prevent the effective functioning of a proton cycle even in the absence of transmembrane potential. In fact, C12R4 was unable to form a diffusion potential of hydrogen ions on planar bilayer lipid membrane under the conditions of a pH gradient (Fig. 5). Interestingly, C12R4 decelerated mitochondrial ATP production, suggesting its direct interaction with ATP synthase (supplemental Fig. 6S).
Cationic versus anionic character in permeating forms of the carriers leads to substantial differences in their transmembrane mobility because of the existence of oriented dipoles of phospholipid headgroups and dipoles of immobilized water molecules at the membrane-solution interface (49, 50). The difference in permeability of similar species carrying charges of opposite sign can reach several orders of magnitude, favoring anion permeation (51, 52). This property of lipid membranes suggests that anionic protonophores must be more effective, which agrees with their lower acting concentrations. However, the existence of Δψ on the inner mitochondrial membrane being negative inside makes cationic compounds capable of being accumulated inside mitochondria, which should increase their efficiency. It has been shown that SkQR1 accumulates effectively in mitochondria (experiments on isolated mitochondria, mammalian cell cultures, yeast cells, and intact organisms (7, 53, 54).
As pointed out previously (4), functioning of cationic protonophores should be strongly dependent on Δψ, exhibiting more effective uncoupling at high Δψ due to their accumulation in the matrix. The following results are in line with the self-limiting character of protonophores on the basis of rhodamine 19: 1) stimulation of respiration of mitochondria by C12R1 or SkQR1 did not reach its maximum despite the absence of inhibition of respiratory chain at the concentrations used (Fig. 2), and 2) no suppression of growth rate of yeast cells by C12R1 was observed over a wide range of concentrations (Fig. 7). In addition, the slow time course of the decrease in membrane potential by SkQR1 (Fig. 3) also points to dependence of its uncoupling efficiency upon accumulation in mitochondria, which presumes the self-limiting mode of action of this protonophore. It has been shown recently that a conjugate of triphenylphosphonium with butylated hydroxytoluene exhibited protonophorous action in mitochondria because of its interaction with adenine nucleotide translocase (17). Our experiments showed that the action of SkQR1 was not mediated by this and several other carriers operating in the inner mitochondrial membrane (Fig. 1C and supplemental Fig. 1S).
The negative feedback between protonophore concentration and Δψ suggests that Δψ decrease cannot be complete. This feature should be important in designing compounds aimed against obesity and several other disorders. Cationic protonophores are likely candidates for such drugs. Our results on yeast cells are in line with this reasoning. In contrast to FCCP, stimulation of respiration by C12R1 was not accompanied by growth suppression over a wide range of its concentrations (Fig. 7).
It has been shown recently in our laboratory that cationic mitochondria-targeted antioxidants (SkQ) and their dodecyltriphenylphosphonium derivatives lacking quinone exhibit protonophorous activity due to acceleration of flip-flop of free fatty acids (7). The protonophorous activity described in the present work can hardly correspond to this effect because cationic rhodamine B derivatives were inactive, in contrast to derivatives of rhodamine 19. Apparently, mitochondria used in the present work contained insufficient levels of endogenous fatty acids. Because different types of cells have varying levels of fatty acids, protonophorous action of SkQR1 or C12R1 can proceed by two different pathways (i.e. the direct mechanism H+ cycling or indirectly via formation of complexes with fatty acids).
It was shown that SkQR1 can effectively protect the brain and kidney from ischemia injuries that are accompanied by generation of ROS (54, 55). It has recently been shown that C12R1 can exhibit similar action although to a lesser extent (56). Further experiments with C12R4 may answer the question whether their effect is mediated by mild uncoupling, which should also suppress the level of ROS production. We are currently testing the involvement of this mechanism in the protective action of SkQR1 in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Elena Kotova and Dr. Lev Yaguzhinsky for valuable comments.
This work was supported in part by Russian Foundation for Basic Research Grant 09-04-00890; the Institute of Mitoengineering, Moscow State University; and the Rostok Group of A. V. Chikunov.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods,” “Results,” Table 1S, Figs. 1S–6S, and Movies 1 and 2.
- ΔμH+
- transmembrane difference in electrochemical potentials of hydrogen ions
- Δψ
- transmembrane electrical potential difference
- BLM
- bilayer lipid membrane
- C2R1
- rhodamine 19 ethyl ester (rhodamine 6G)
- C8R1
- rhodamine 19 octyl ester
- C10R1
- rhodamine 19 decyl ester
- C12R1
- rhodamine 19 dodecyl ester
- C16R1
- rhodamine 19 octadecyl ester
- C12R4
- rhodamine B dodecyl ester
- C12-TPP
- dodecyltriphenylphosphonium
- DiS-C3-(5)
- 3,3′-dipropylthiadicarbocyanine iodide
- DNP
- 2,4-dinitrophenol
- FCCP
- carbonyl cyanide-p-trifluoromethoxyphenylhydrazone
- ROS
- reactive oxygen species
- SkQ1
- 10-(6′-plastoquinonyl) decyltriphenylphosphonium
- SkQR1
- 10-(6′- plastoquinonyl) decylrhodamine 19
- SkQR4
- 10-(6′-plastoquinonyl) decylrhodamine B.
REFERENCES
- 1. Korshunov S. S., Skulachev V. P., Starkov A. A. (1997) FEBS Lett. 416, 15–18 [DOI] [PubMed] [Google Scholar]
- 2. Skulachev V. P. (1996) Q. Rev. Biophys. 29, 169–202 [DOI] [PubMed] [Google Scholar]
- 3. Nedergaard J., Cannon B. (2003) Exp. Physiol. 88, 65–84 [DOI] [PubMed] [Google Scholar]
- 4. Blaikie F. H., Brown S. E., Samuelsson L. M., Brand M. D., Smith R. A., Murphy M. P. (2006) Biosci. Rep. 26, 231–243 [DOI] [PubMed] [Google Scholar]
- 5. Harper J. A., Dickinson K., Brand M. D. (2001) Obes. Rev. 2, 255–265 [DOI] [PubMed] [Google Scholar]
- 6. Lanni A., Moreno M., Lombardi A., Goglia F. (2003) FEBS Lett. 543, 5–10 [DOI] [PubMed] [Google Scholar]
- 7. Severin F. F., Severina I. I., Antonenko Y. N., Rokitskaya T. I., Cherepanov D. A., Mokhova E. N., Vyssokikh M. Y., Pustovidko A. V., Markova O. V., Yaguzhinsky L. S., Korshunova G. A., Sumbatyan N. V., Skulachev M. V., Skulachev V. P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McLaughlin S. G., Dilger J. P. (1980) Physiol. Rev. 60, 825–863 [DOI] [PubMed] [Google Scholar]
- 9. Skulachev V. P., Sharaf A. A., Liberman E. A. (1967) Nature 216, 718–719 [DOI] [PubMed] [Google Scholar]
- 10. Sun X., Garlid K. D. (1992) J. Biol. Chem. 267, 19147–19154 [PubMed] [Google Scholar]
- 11. Schwaller M. A., Allard B., Lescot E., Moreau F. (1995) J. Biol. Chem. 270, 22709–22713 [DOI] [PubMed] [Google Scholar]
- 12. Blaustein R. O., Finkelstein A. (1988) Biochim. Biophys. Acta 946, 221–226 [DOI] [PubMed] [Google Scholar]
- 13. Nagamune H., Fukushima Y., Takada J., Yoshida K., Unami A., Shimooka T., Terada H. (1993) Biochim. Biophys. Acta 1141, 231–237 [DOI] [PubMed] [Google Scholar]
- 14. Kelso G. F., Porteous C. M., Coulter C. V., Hughes G., Porteous W. K., Ledgerwood E. C., Smith R. A., Murphy M. P. (2001) J. Biol. Chem. 276, 4588–4596 [DOI] [PubMed] [Google Scholar]
- 15. Murphy M. P., Smith R. A. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 629–656 [DOI] [PubMed] [Google Scholar]
- 16. Liberman E. A., Skulachev V. P. (1970) Biochim. Biophys. Acta 216, 30–42 [DOI] [PubMed] [Google Scholar]
- 17. Lou P. H., Hansen B. S., Olsen P. H., Tullin S., Murphy M. P., Brand M. D. (2007) Biochem. J. 407, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Antonenko Y. N., Avetisyan A. V., Bakeeva L. E., Chernyak B. V., Chertkov V. A., Domnina L. V., Ivanova O. Y., Izyumov D. S., Khailova L. S., Klishin S. S., Korshunova G. A., Lyamzaev K. G., Muntyan M. S., Nepryakhina O. K., Pashkovskaya A. A., Pletjushkina O. Y., Pustovidko A. V., Roginsky V. A., Rokitskaya T. I., Ruuge E. K., Saprunova V. B., Severina I. I., Simonyan R. A., Skulachev I. V., Skulachev M. V., Sumbatyan N. V., Sviryaeva I. V., Tashlitsky V. N., Vassiliev J. M., Vyssokikh M. Y., Yaguzhinsky L. S., Zamyatnin A. A., Jr., Skulachev V. P. (2008) Biochemistry 73, 1273–1287 [DOI] [PubMed] [Google Scholar]
- 19. Rokitskaya T. I., Klishin S. S., Severina I. I., Skulachev V. P., Antonenko Y. N. (2008) J. Membr. Biol. 224, 9–19 [DOI] [PubMed] [Google Scholar]
- 20. Rokitskaya T. I., Sumbatyan N. V., Tashlitsky V. N., Korshunova G. A., Antonenko Y. N., Skulachev V. P. (2010) Biochim. Biophys. Acta 1798, 1698–1706 [DOI] [PubMed] [Google Scholar]
- 21. Laris P. C., Bahr D. P., Chaffee R. R. (1975) Biochim. Biophys. Acta 376, 415–425 [DOI] [PubMed] [Google Scholar]
- 22. Chen Y., Schindler M., Simon S. M. (1999) J. Biol. Chem. 274, 18364–18373 [DOI] [PubMed] [Google Scholar]
- 23. Avnir Y., Barenholz Y. (2005) Anal. Biochem. 347, 34–41 [DOI] [PubMed] [Google Scholar]
- 24. Johnson D., Lardy H. (1967) Methods Enzymol. 10, 94–96 [Google Scholar]
- 25. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 26. Slonimski P. P., Perrodin G., Croft J. H. (1968) Biochem. Biophys. Res. Commun. 30, 232–239 [DOI] [PubMed] [Google Scholar]
- 27. Sherman F. (2002) Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 28. Pozniakovsky A. I., Knorre D. A., Markova O. V., Hyman A. A., Skulachev V. P., Severin F. F. (2005) J. Cell Biol. 168, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hallstrom T. C., Moye-Rowley W. S. (2000) J. Biol. Chem. 275, 37347–37356 [DOI] [PubMed] [Google Scholar]
- 30. Raymond M., Ruetz S., Thomas D. Y., Gros P. (1994) Mol. Cell. Biol. 14, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 32. Pandit S. A., Chiu S. W., Jakobsson E., Grama A., Scott H. L. (2008) Langmuir 24, 6858–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berendsen H. J., Grigera J. R., Straatsma T. P. (1987) Phys. Chem. 91, 6269–6271 [Google Scholar]
- 34. Darden T., Perera L., Li L., Pedersen L. (1999) Structure 7, R55–R60 [DOI] [PubMed] [Google Scholar]
- 35. Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. (1997) Comput. Chem. 18, 1463–1472 [Google Scholar]
- 36. Kyrychenko A. (2010) Chem. Phys. Lett. 485, 95–99 [Google Scholar]
- 37. Torrie G. M., Valleau J. P. (1977) Comput. Phys. 23, 187–193 [Google Scholar]
- 38. Andreyev A. Y., Bondareva T. O., Dedukhova V. I., Mokhova E. N., Skulachev V. P., Volkov N. I. (1988) FEBS Lett. 226, 265–269 [DOI] [PubMed] [Google Scholar]
- 39. Andreyev A. Y., Bondareva T. O., Dedukhova V. I., Mokhova E. N., Skulachev V. P., Tsofina L. M., Volkov N. I., Vygodina T. V. (1989) Eur. J. Biochem. 182, 585–592 [DOI] [PubMed] [Google Scholar]
- 40. Skulachev V. P. (1998) Biochim. Biophys. Acta 1363, 100–124 [DOI] [PubMed] [Google Scholar]
- 41. Kamp F., Hamilton J. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11367–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. LeBlanc O. H. (1971) J. Membr. Biol. 4, 227–251 [DOI] [PubMed] [Google Scholar]
- 43. Borisova M. P., Ermishkin L. N., Liberman E. A., Silberstein A. Y., Trofimov E. M. (1974) J. Membr. Biol. 18, 243–261 [DOI] [PubMed] [Google Scholar]
- 44. Duvvuri M., Gong Y., Chatterji D., Krise J. P. (2004) J. Biol. Chem. 279, 32367–32372 [DOI] [PubMed] [Google Scholar]
- 45. Gancedo J. M. (1998) Microbiol. Mol. Biol. Rev. 62, 334–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petit P., Glab N., Marie D., Kieffer H., Métézeau P. (1996) Cytometry 23, 28–38 [DOI] [PubMed] [Google Scholar]
- 47. Ross M. F., Da Ros T., Blaikie F. H., Prime T. A., Porteous C. M., Severina I. I., Skulachev V. P., Kjaergaard H. G., Smith R. A., Murphy M. P. (2006) Biochem. J. 400, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Severina I. I., Vyssokikh M. Y., Pustovidko A. V., Simonyan R. A., Rokitskaya T. I., Skulachev V. P. (2007) Biochim. Biophys. Acta 1767, 1164–1168 [DOI] [PubMed] [Google Scholar]
- 49. Flewelling R. F., Hubbell W. L. (1986) Biophys. J. 49, 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Flewelling R. F., Hubbell W. L. (1986) Biophys. J. 49, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liberman E. A., Topaly V. P. (1969) Biophysics Moscow 14, 477–487 [Google Scholar]
- 52. Pickar A. D., Benz R. (1978) J. Membr. Biol. 44, 353–376 [Google Scholar]
- 53. Fetisova E. K., Avetisyan A. V., Izyumov D. S., Korotetskaya M. V., Chernyak B. V., Skulachev V. P. (2010) FEBS Lett. 584, 562–566 [DOI] [PubMed] [Google Scholar]
- 54. Bakeeva L. E., Barskov I. V., Egorov M. V., Isaev N. K., Kapelko V. I., Kazachenko A. V., Kirpatovsky V. I., Kozlovsky S. V., Lakomkin V. L., Levina S. B., Pisarenko O. I., Plotnikov E. Y., Saprunova V. B., Serebryakova L. I., Skulachev M. V., Stelmashook E. V., Studneva I. M., Tskitishvili O. V., Vasilyeva A. K., Victorov I. V., Zorov D. B., Skulachev V. P. (2008) Biochemistry 73, 1288–1299 [DOI] [PubMed] [Google Scholar]
- 55. Plotnikov E. Y., Silachev D. N., Chupyrkina A. A., Danshina M. I., Jankauskas S. S., Morosanova M. A., Stelmashook E. V., Vasileva A. K., Goryacheva E. S., Pirogov Y. A., Isaev N. K., Zorov D. B. (2010) Biochemistry 75, 145–150 [DOI] [PubMed] [Google Scholar]
- 56. Plotnikov E. Y., Chupyrkina A. A., Jankauskas S. S., Pevzner I. B., Silachev D. N., Skulachev V. P., Zorov D. B. (2011) Biochim. Biophys. Acta 1812, 77–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.