Abstract
Gram-negative bacteria expel diverse toxic chemicals through the tripartite efflux pumps spanning both the inner and outer membranes. The Escherichia coli AcrAB-TolC pump is the principal multidrug exporter that confers intrinsic drug tolerance to the bacteria. The inner membrane transporter AcrB requires the outer membrane factor TolC and the periplasmic adapter protein AcrA. However, it remains ambiguous how the three proteins are assembled. In this study, a hexameric model of the adapter protein was generated based on the propensity for trimerization of a dimeric unit, and this model was further validated by presenting its channel-forming property that determines the substrate specificity. Genetic, in vitro complementation, and electron microscopic studies provided evidence for the binding of the hexameric adapter protein to the outer membrane factor in an intermeshing cogwheel manner. Structural analyses suggested that the adapter covers the periplasmic region of the inner membrane transporter. Taken together, we propose an adapter bridging model for the assembly of the tripartite pump, where the adapter protein provides a bridging channel and induces the channel opening of the outer membrane factor in the intermeshing tip-to-tip manner.
Keywords: Antibiotics, Bacteria, Bacterial Genetics, Multidrug Transporters, Protein Assembly, TolC, Electron Microscopy
Introduction
The Escherichia coli AcrAB-TolC efflux pump has broad substrate specificity and expels a wide range of chemotherapeutic agents (1, 2). This efflux pump is mainly responsible for the intrinsic resistance of E. coli to most lipophilic antibiotics, detergents, and dyes (2). The AcrAB-TolC pump consists of a resistance-nodulation cell division-type transporter, AcrB, a multifunctional outer membrane factor, TolC, and a periplasmic membrane fusion protein, AcrA (3–6). The inner membrane transporter AcrB transports substrates by utilizing the proton gradient across the inner membrane (7), and TolC connects AcrB in the periplasm to the external medium by providing a continuous conduit (7, 8). AcrA forms a direct interaction between AcrB and TolC in the periplasm (9–11). AcrA contains a membrane proximal (MP)5 domain, a β-barrel domain, a lipoyl domain, and an α-hairpin domain, which are linearly arranged (12–16). In particular, the α-hairpin domain of AcrA is responsible for binding to TolC, whereas the other domains bind to AcrB (10, 17–20).
AcrB and TolC exist as homotrimers (8, 21). However, the oligomeric state of AcrA remains ambiguous despite the presence of the crystal structures of AcrA and its homologues (7, 12, 13, 17, 22–25). Several models for the assembly of the tripartite pump have been proposed and have focused on the direct interaction between AcrB and TolC, which was detected only when cross-linking was induced (12, 17, 26).
Recently, our research group has shown an alternative model (called MacA-bridging model) for the macrolide-specific efflux pump MacAB-TolC, based on functional and structural analyses of MacA, which is the functional counterpart of AcrA (7, 13, 20, 22, 27). In the MacA-bridging model, TolC make a tip-to-tip interaction with MacA but not with the cognate inner membrane transporter MacB. Crystal structures of the MacA protein showed a funnel-like structure in a hexameric assembly (13, 22, 28). The hexameric funnel-like structure of MacA is physiologically relevant, and the tip region of the α-hairpin is functionally important and is related to TolC binding (13, 20). Electron microscopic study revealed that the TolC aperture tip region makes intermeshing cogwheel-to-cogwheel interaction with the MacA hexamer (27). Furthermore, we demonstrated that the binding mode of AcrA to TolC is basically the same as that of MacA to TolC by using a chimeric AcrA in which the α-helical domain is substituted with that of MacA (19). Nevertheless, it remains under debate whether AcrA acts as a hexamer-like MacA in the functional state, because the hexameric model of AcrA is not compatible with the direct interaction between AcrB and TolC.
In this study, we provide further experimental verification showing that AcrA exhibits hexameric assembly similar to MacA and also that the MacA-bridging model is applicable to the AcrAB-TolC pump. Our findings are consistent with previous experimental results, except for some of the cross-linking results that have been used to build the currently prevailing models for the assembly of AcrAB-TolC pump (17, 26), which we discuss.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Expression Plasmids for Genetic Studies
Details are found in the supplemental material.
Construction and Expression of AcrA Dimer
The AcrA dimer was constructed in two steps. The first DNA fragment encoding AcrA (residues 26–397 without stop codon) was ligated into NcoI and NotI sites of the pPROEX-HTA vector (Invitrogen). The second DNA fragment encoding AcrA (residues 26–397 with stop codon) was inserted into NotI and XhoI sites of the resulting vector, generating pPRO-AcrA dimer. Expression and purification of the AcrA-linked dimer were performed using the same procedure for AcrA (19). Primers used in this study are listed in supplemental Table S4.
Isolation of TolC Variants That Complement AcrA-L132C Mutant
To introduce random mutations into the TolC aperture tip regions from both hairpins (145–155 and 363–373), the previously described method was applied (19). Briefly, random mutations were introduced into the DNA segments encoding the TolC aperture tip regions from both hairpins (amino acid residues 145–155 and 363–373) and ligated into pTolC1, which can express the wild-type TolC protein tagged with a C-terminal hexa-histidine sequence (19). The resulting ligation products were transformed into acrA- and tolC-deleted E. coli cells (BW25113 ΔacrA ΔtolC210::Tn10) harboring pAcrA2-L132C. The transformants were selected for novobiocin resistance at a concentration (3 μg/ml) that did not allow survival of the parental E. coli cells expressing AcrA-L132C and wild-type TolC.
Measurement of Minimum Inhibitory Concentration (MIC)
The procedure for measurement of MICs has been described previously (19).
Construction of E. coli MacA-AcrAα Hybrid and Aa MacA-TolCα Hybrid Dimer
To construct the E. coli MacA-AcrAα hybrid for the in vivo experiment, DNA fragments encoding the regions of E. coli MacA (residues 1–92), E. coli AcrA (residues 97–174), and E. coli MacA (residues 185–372) were separately amplified, and the three resulting PCR fragments were recombined using an overlap extension PCR method. The DNA was digested with EcoRI/HpaI and ligated into pMacAB1 (13), generating pMacA-AcrAα. To purify the chimeric protein E. coli MacA-AcrAα hybrid, DNA fragments encoding the regions of E. coli MacA (residues 32–92), AcrA (residues 97–174), and MacA (residues 185–372) were amplified using pMacA-AcrAα as a template by PCR and were subsequently ligated into NcoI/XhoI sites of pPROEX-HTA vector (Invitrogen), generating pPRO-MacA-AcrAα hybrid. Construction of Aa MacA-TolCα hybrid dimer was described previously (27).
Electron Microscopy and Image Processing
The sample was applied to a freshly plasma cleaned carbon-coated copper grid and allowed to adhere for 60 s before being blotted. Immediately after blotting, 3 μl of a 1% solution of uranyl formate was applied onto the grid and blotted off directly. This was repeated three times. Data were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 kV, using a dose of ∼15 electrons/Å2 and a nominal underfocus ranging from 1.0 to 2.0 μm. Details for image processing are found in supplemental material.
In Vivo Cross-linking Assay
The procedure for in vivo cross-linking assay has been described previously (10).
RESULTS
Functional Oligomer of AcrA Is a Trimer of a Dimeric Unit
Several crystal structures of membrane fusion proteins have been determined, and a parallel side-by-side interaction between the two neighboring protomers was commonly found (12–16). In the crystal structure of AcrA and its homologue ZneB, two protomers make a parallel contact in the asymmetric unit of the crystals (14, 25, 29). The MexA protein, which is the AcrA homologue of the Pseudomonas aeruginosa MexAB-OprM pump, showed side-by-side interactions between six or seven protomers (15, 16). Furthermore, the crystal structure of MacA showed a funnel-like hexameric assembly by trimerization of two protomers in the side-by-side ring arrangement (13). It was demonstrated biochemically and genetically that this side-by-side interaction is crucial for MacA function. Indeed, when the side-by-side interaction of MacA was disrupted, the efflux function of MacAB-TolC pump was abolished (13).
To investigate whether the side-by-side interaction between two protomers of AcrA is important for its function, we employed an experimental approach similar to that used to characterize the multimeric status of functional AcrB (30). We constructed an AcrA dimer that is covalently linked in a tandem manner (Fig. 1A). In the AcrA dimer, the first AcrA is full-length, including the signal sequence plus the lipid modification moiety, although the second AcrA lacks the signal sequence and the lipid modification moiety to direct the protein to the right position in vivo (Fig. 1A). Because both the N and C termini of AcrA are anchored or located close to the membrane, it is likely that the AcrA dimer is arranged in the parallel side-by-side manner. Expression of the AcrA dimer rendered acrA-deleted E. coli cells resistant to novobiocin at degrees similar to those expressing wild-type AcrA (supplemental Table S1). This result implies that AcrA functions as a dimeric unit, suggesting that the parallel interaction observed in all membrane fusion protein structures is physiologically relevant. This result further suggests that the oligomerization number of AcrA is an even number but not an odd number.
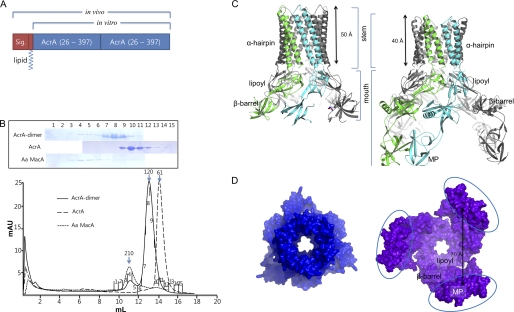
FIGURE 1.
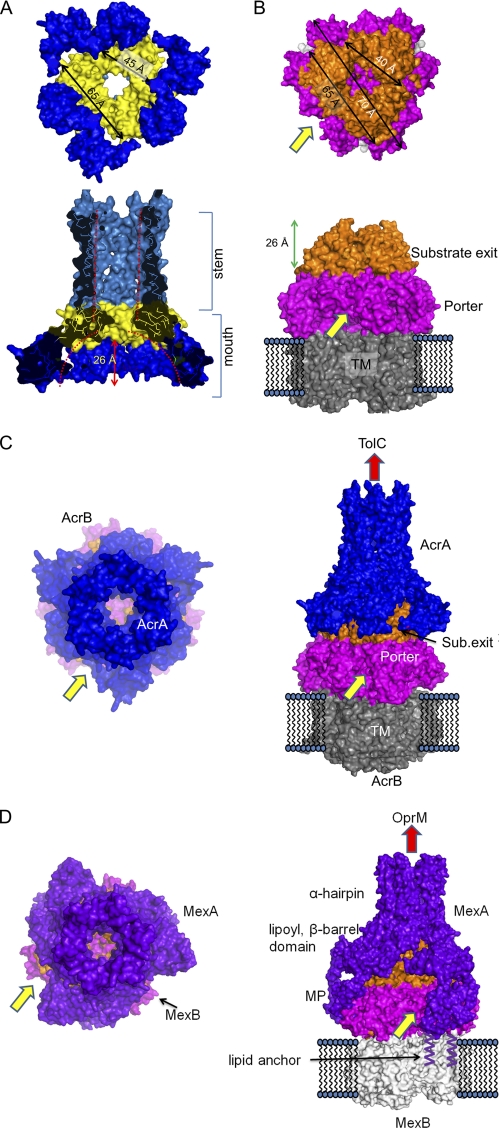
AcrA hexameric structure. A, bar diagram for AcrA dimer constructs. The first AcrA unit contains the signal peptide and the lipid modification moiety to properly locate the protein at the inner membrane in the periplasm for the in vivo genetic study. For the in vitro binding assay, the signal peptide and the lipid modification moiety were removed. B, elution profiles of E. coli AcrA dimer, E. coli AcrA, and A. actinomycetemcomitans (Aa) MacA from a size-exclusion chromatographic column (Superdex 200 HR 10/30). The SDS-PAGE analyses of the fractions are shown. The numbers correspond to the fractions indicated on each chromatogram. The peaks indicated by 210, 120, and 61 (kDa) arrows are estimated as a hexamer, dimer, and monomer of AcrA or MacA protomer, respectively. C, side views of the E. coli AcrA hexamer (left) and the P. aeruginosa MexA hexamer models (right). The dimeric unit of AcrA structure (A and B chains of PDB code 2F1M), devoid of the MP domain, was used as the initial model (left). Two neighboring MexA protomers (L and M chains of PDB code 2V4D (12)), which are paired by the interaction between the MP domains, contain all four domains. The funnel stem comprising α-hairpins and the funnel mouth comprising lipoyl, β-barrel, and MP domains are indicated. D, top view of the E. coli AcrA hexameric model (left) and bottom view of the P. aeruginosa MexA hexameric model (right). The distance between MexA MP domains is indicated. Note the triangular cone-shaped internal hollows and the central pores in the modeled structures.
We next characterized the multimeric status of the purified AcrA dimer protein that does not contain the N-terminal lipid modification moiety. As shown in Fig. 1B, some AcrA dimer proteins formed a complex whose molecular size was similar to that of the MacA hexamer in a size-exclusion chromatographic column, whereas the AcrA protein exclusively eluted as a monomer. This result indicates that AcrA dimer protein has a propensity to form a trimer, resulting in an oligomer composed of six AcrA protomers. In addition, the dynamic light scattering experiment suggested that the AcrA monomer protein also has a propensity to form a dimer when the protein was concentrated (supplemental Fig. S2).
Molecular Modeling of the AcrA and MexA Hexamers
Given the trimeric assembly that was composed of the dimeric units of AcrA, we constructed an AcrA hexamer model based on the known features of the AcrA and the MacA hexamer structures. We separately modeled the α-hairpin domain and the other domains of AcrA.
Part 1. α-Hairpin Domain
The hexameric assembly of the α-hairpin regions of AcrA was modeled based on the known MacA hexamers because binding sites and modes of the α-hairpin domains of AcrA and MacA to TolC are likely to be the same, as was revealed by functional analyses of chimeric AcrA proteins containing α-hairpin domains of MacA (19). The α-hairpin from each AcrA protomer was superposed on one of the α-hairpins from six MacA protomers using the start and the end regions of the α-hairpin as references (supplemental Fig. S3). From this superposition, we modeled the α-barrel structure of AcrA, consisting of six α-hairpins, which exhibits the same inclination of the α-helix as the MacA hexamer and the periplasmic α-barrel of TolC (8) (supplemental Fig. S3). Because the length of the α-hairpin of AcrA is just one heptad (7 amino acids) shorter than that of MacA, the cogwheel structure of the α-barrel consisting of the six α-hairpin tip region is exhibited. Despite the difference in the length of α-barrels of AcrA and MacA, the cogwheel structure of the AcrA hexamer is similar to that of the α-barrel of the MacA hexamer because the heptad rule is also kept in AcrA. This structural resemblance of the AcrA and MacA α-barrels is consistent with previous experimental results that showed the importance of the α-hairpin tip region of AcrA and MacA and the flexibility of the length of AcrA α-hairpin for the function of TolC-mediated efflux pumps (19, 20).
Part 2. Lipoyl and β-Barrel Domains of AcrA
Crystal structures of AcrA and MexA revealed that the lipoyl and β-barrel domains of AcrA are present as a single body, whereas the AcrA α-hairpin domain is flexible with respect to the lipoyl and β-barrel domains (14–16). The MP domain is known to exhibit extremely high mobility and forms a pair with the MP domain of the adjacent protomer, as was revealed in the crystal structures of the AcrA homologues, MexA and CusB (12, 25, 31). However, the structures of the MP domains of AcrA and MacA are not known (13, 14). Because the dimer of AcrA in the parallel arrangement is a fundamental unit, we selected a pair of the lipoyl and β-barrel domains from the asymmetric protomer of AcrA in the crystal (14). Three pairs of lipoyl and β-barrel domains of AcrA (residues 64–97 and 174–208) were superposed onto the corresponding regions of E. coli MacA hexamer (PDB code 3FPP; residues 60–92 and 184–219) as references (root mean square deviation of 2.128 Å between 120 Cα atoms) (supplemental Fig. S3). The model was composed of three pairs of lipoyl and β-barrel domains of AcrA that formed a triangular dome structure (Fig. 1, C and D).
When an AcrA protomer in the AcrA hexamer is superposed onto the crystal structure of AcrA protomer, we found that the AcrA α-hairpin domains had to be rotated by about 10° with respect to its lipoyl domain for the structural superposition. This rotation could be justified by the natural conformational flexibility of the α-hairpin domains (supplemental Fig. S4). Likewise, a MexA hexamer model was also built using a pair of MexA protomers whose MP domains were coupled as an initial model (Fig. 1, C and D) (12). The coordinates for the AcrA and MexA hexameric models are very similar (root mean square deviation of 1.847 Å between 1148 Cα atoms).
Part 3. Overall Structures of Models for AcrA and MexA Hexamers
The overall structures of the AcrA and MexA hexamer models resemble the funnel-like MacA hexamer structure, showing 3-fold symmetry (or pseudo 6-fold symmetry) along the central axis. The top or bottom view of the AcrA hexamer shows a ring-like structure and is strikingly similar to the electron microscopic picture from the AcrA two-dimensional crystals (32), rendering the model more feasible. The funnel stems of AcrA and MexA hexamers, comprising six α-hairpins, are very similar to that of the MacA funnel, especially in the tip region, although the lengths of the α-hairpins are different from each other (Fig. 1C). The α-barrel made by the six α-hairpins forms a channel that is ∼50 Å long and ∼30 Å wide with a cogwheel structure at the end of the α-hairpins that contains invariant residues (Fig. 1C). The inner and outer diameters of the funnel stem of the AcrA and MexA hexamers are virtually identical to that of MacA (Fig. 1C) and the outer membrane proximal region of the TolC α-helical barrel.
A central pore connecting the funnel stem to the funnel mouth region is found at the center of the AcrA and MexA funnel structures, the widths of which are close to that of the substrate exit funnel of AcrB and wider than that of the MacA hexamer (Fig. 1D). The AcrA funnel mouth, composed of three sets of a pair of lipoyl and β-barrel domains, forms a triangular cone-shaped hollow that is wider than the MacA funnel mouth (Fig. 1). In the case of the MexA hexamer model that contains the MP domains, three packs of the six MP domains are flared at the rim of the triangular cone-shaped funnel mouth (Fig. 1, C and D). In the MexA hexamer model, the length of the funnel stem is ∼40 Å because the α-hairpin of MexA is shorter by one heptad than that of AcrA (Fig. 1C left).
Validation of the Hexameric Model of AcrA and Its Binding Mode with TolC
Part I. Mutations at the Putative Pore Region of the AcrA Hexamer Change the Substrate Specificity of the AcrAB-TolC Pump
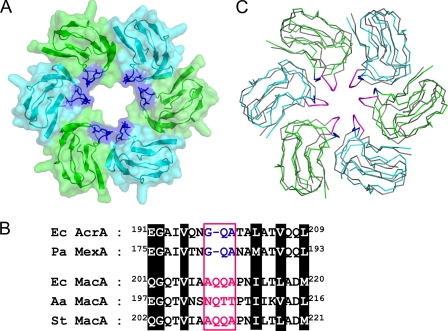
Given the funnel-like structure of the AcrA hexamer, a central pore is found at the lipoyl domain (Figs. 1D and 2A). Compared with the MacA hexamer, the pore in the AcrA hexamer is wider and is lined with a loop that is one amino acid shorter than that of MacA (Fig. 2B). To determine whether AcrA contains a pore region that functions as a channel for the passage of substrates in vivo, as proposed from the AcrA hexamer model, we selected the putative pore-lining loop for mutational analysis. We reasoned that the addition of a single amino acid to the pore-lining loop would limit the substrate of the pump because the diameter of the pore would be reduced by the elongated loop.
FIGURE 2.
Putative pore region of the AcrA hexameric model. A, central pore in the lipoyl domains from the AcrA hexamer. Only the lipoyl domains are shown for clarity. Each protomer is colored green or cyan with transparent surface representations. The Gly-Gln-Ala sequence conserved between E. coli AcrA and P. aeruginosa MexA is shown in blue. B, alignment of the sequences around the pore-lining loops from E. coli AcrA (Ec AcrA), P. aeruginosa MexA (Pa MexA), E. coli MacA (Ec MacA), A. actinomycetemcomitans MacA (Aa MacA), and Salmonella typhimurium MacA (St MacA). The pore-lining sequences are colored in blue or magenta in a box. The conserved residues are highlighted. C, superposition of the lipoyl domains from E. coli MacA and E. coli AcrA hexamers, displayed in the Cα tracing representations. The E. coli MacA protomers are colored in gray, although E. coli AcrA protomers are in green or cyan. The pore-lining residues of MacA are in magenta, and those of AcrA are in blue.
When AcrA was substituted with the longer MacA loop (198AQQA201) or its derivative (198AKEA201), E. coli cells expressing the mutant AcrA exhibited a marked change in substrate specificity compared with E. coli cells expressing the wild-type AcrA (Table 1). Expression of the longer MacA loop-like mutations (198AQQA201) did not affect the degree of resistance of E. coli cells to erythromycin, chloramphenicol, SDS, and tetracycline but had a notable effect on the degree of resistance to ethidium bromide and novobiocin (Table 1). Expression of the other AcrA mutant (198AKEA201) resulted in a more dramatic change in substrate specificity. Because the mutations did not alter the wild-type function of AcrAB-TolC pump on some substrates, we concluded that the mutant AcrA proteins are capable of forming a functional assembly with AcrB and TolC in vivo. The altered substrate specificity by the mutations at the putative pore region of AcrA strongly suggests that the loop region of AcrA is lining the pore in the functional assembly of the pump, which provides a passing channel for the substrate.
TABLE 1.
The in vivo effect of the mutations at the putative pore region of AcrA
The in vivo effect of the mutations (198AQQA201 and 198AKEA201) on AcrA function was determined by the degree of resistance of E. coli strain BW25113ΔacrAB to several substrates of AcrAB-TolC pump. The level of AcrA protein expressed in the E. coli strain is shown in supplemental Fig. S6.
| Plasmid | MIC (μg/ml)a |
|||||
|---|---|---|---|---|---|---|
| CHL (323.132)b | EM (733.93) | EtBr (394.294) | NOV (612.624) | SDS (288.38) | TET (444.435) | |
| pKAN6B | 1.00 | 2.50 | 7.80 | 3.00 | 31.20 | 0.50 |
| pMacAB1 | 1.00 | 40.00 | 7.80 | 3.00 | 31.20 | 0.50 |
| pAcrAB1 | 8.00 | 200.00 | 1000.00 | 800.00 | 1000.00 | 4.00 |
| pAcrA-AQQA | 8.00 | 200.00 | 500.00 | 600.00 | 1000.00 | 4.00 |
| pAcrA-AKEA | 4.00 | 140.00 | 250.00 | 400.00 | 1000.00 | 2.00 |
a All MIC measurements were independently performed in triplicate. The abbreviations used are as follows: CHL, chloramphenicol; EM, erythromycin; EtBr, ethidium bromide; NOV, novobiocin; TET, tetracycline. Chloramphenicol and tetracycline concentrations used to measure MICs were 0, 0.50, 1.00, 2.00, 4.00, 8.00, 16.00, and 32.00 μg/ml. Erythromycin concentrations used to measure MICs were 0, 1.25, 2.50, 5.00, 10.00, 20.00, 40.00, 80.00, 100.00, 120.00, 140.00, 160.00, 180.00, and 200.00 μg/ml. EtBr and SDS concentrations used to measure MICs were 0, 3.91, 7.81, 15.63, 31.25, 62.50, 125.00, 250.00, 500.00, and 1000.00 μg/ml. Novobiocin concentrations used to measure MICs were 0, 1.50, 3.00, 6.00, 12.00, 25.00, 50.00, 100.00, 200.00, 400.00, 600.00, and 800.00 μg/ml.
b The molar mass (g/mol) of substrates is shown.
Part II. Functional and Physical Interactions between the AcrA α-Hairpin Tip Region and the TolC Aperture Tip Region
We previously showed that the TolC aperture tip region is functionally related to the AcrA α-hairpin tip region using genetic complementation experiments (19). To further examine the functional interactions between the AcrA α-hairpin tip region and the TolC aperture tip region, we designed experiments that utilized genetic complementation and in vivo chemical cross-linking. First, we constructed a nonfunctional AcrA mutant bearing a cysteine substitution at a strictly conserved residue, Leu-132, in the tip region of AcrA. This mutation abolished the resistance of E. coli cells to antibiotics (Table 2), indicating the functional importance of Leu-132 for the formation of efflux-ready AcrAB-TolC pump. Next, TolC variants containing random mutations at the TolC aperture tip region were constructed as described previously (19). This pool of TolC variants was used to select for clones that could complement AcrA (L132C). We obtained seven of about 104 transformants that showed a gain-of-function phenotype (Table 2). The isolated tolC clones greatly restored the novobiocin resistance of the cells expressing the nonfunctional AcrA (L132C) (MIC values from 1.56 to 6.25–12.50 μg/ml). These TolC mutants did not appear to form functional AcrAB-TolC pumps with wild-type AcrA, as indicated by lower levels of antibiotic resistance of E. coli cells expressing wild-type AcrA and the isolated TolC mutants compared with those expressing AcrA (L132C) and the complementing TolC mutants.
TABLE 2.
TolC variants complementing the AcrA (L132C) mutant
E. coli strain BW25113 ΔacrA Δtolc210::Tn10 was used to select TolC variants that were capable of complementing the nonfunctional AcrA (L132C) variant. Clones resistant to novobiocin at 3 μg/ml were selected. The levels of wild-type AcrA and TolC variant protein expressed are shown in supplemental Fig. S7.
| TolC proteina | MIC (μg of novobiocin/ml)b, AcrA proteinc |
|
|---|---|---|
| AcrA (WT) | AcrA (L132C) | |
| TolC−d | 0.78 | 0.78 |
| TolC (WT) | 25.00 | 1.56 |
| TolC (N145K/D153E/A159V/T366K) | 3.13 | 12.50 |
| TolC (T152S/D153N) | 3.13 | 6.25 |
| TolC (D153E/V370G) | 1.56 | 6.25 |
| TolC (D153Y/Q155H/V364I) | 0.78 | 6.25 |
| TolC (I369F/V370G) | 1.56 | 6.25 |
| TolC (L373R) | 1.56 | 6.25 |
| TolC (L373V) | 3.13 | 6.25 |
a TolC (WT) and TolC variants were expressed from pTolC1.
b 0, 0.78, 1.56, 3.13, 6.25, 12.50, and 25.00 μg of novobiocin /ml were used to measure MICs.
c AcrA (WT) and AcrA (L132C) were expressed from pAcrA2.
d An empty vector, pKAN6B, was used.
We further tested whether AcrA (L132C) makes a direct contact with the complementing TolC variant using the in vivo site-specific cross-linking method that has been used to identify interactions between AcrA and TolC (10). Exponential cultures of the acrAB- and tolC-deleted E. coli strain that expressed AcrA (L132C) and wild-type TolC or TolC (N145K/D153E/A159V/T366K) were subjected to two different membrane-permeable hetero-bifunctional cross-linkers, N-succinimidyl 3-(2-pyridyldithio)propionate and sulfosuccinimidyl 6-[3′(2-pyridyldithio)-propionamido] hexanoate. These cross-linkers are reactive toward sulfhydryl and primary amine groups at either end. TolC (N145K/D153E/A159V/T366K) was chosen because it best complemented AcrA (L132C). Hexahistidine-tagged TolC and cross-linked AcrA proteins were affinity-purified from the E. coli lysate using the nickel-nitrilotriacetic acid column and were analyzed by Western blot. AcrA (L132C) was cross-linked to TolC (N145K/D153E/A159V/T366K), whereas AcrA (L132C) was not cross-linked to wild-type TolC (Fig. 3A), indicating that AcrA (L132C) makes a specific contact with the TolC aperture tip region bearing the complementing mutations. In a positive control, co-expression of AcrA (D111C) and wild-type TolC resulted in positive cross-links as reported previously (10). These results showed that the AcrA α-hairpin tip region is not only functionally related to, but also interacts with, the TolC aperture tip region.
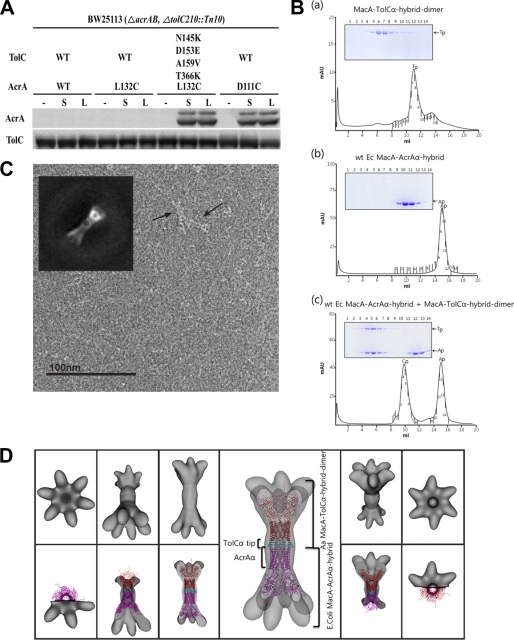
FIGURE 3.
Tip-to-tip interaction between TolC and AcrA. A, in vivo interaction between AcrA (L132C) and TolC (N145K/D153E/A159V/T366K), detected using chemical cross-linking agents (N-succinimidyl 3-(2-pyridyldithio)propionate and sulfosuccinimidyl 6-[3′(2-pyridyldithio)-propionamido] hexanoate. E. coli BW25113 ΔacrAB ΔtolC210::Tn10 cultures that co-expressed hexa-His-tagged wild-type TolC (WT) or TolC (N145K/D153E/A159V/T366K) and wild-type AcrA (WT) or one of the AcrA cysteine variants (AcrA (D111C) or AcrA (L132C)) are shown. All cultures were treated with N-succinimidyl 3-(2-pyridyldithio)propionate (S), sulfosuccinimidyl 6-[3′(2-pyridyldithio)-propionamido] hexanoate (L), or none (−). Affinity-purified TolC and cross-linked AcrA proteins were separated by SDS-PAGE and immunoblotted as indicated. B, physical interaction between the TolC tip and the membrane fusion protein tip regions, revealed by the size-exclusion chromatography with the SDS-PAGE analysis. Panel a, MacA-TolCα hybrid dimer. The result indicates that this protein forms the funnel-like structure because the elution volume is identical to the hexameric Aa MacA (see Fig. 1B). Panel b, wild-type E. coli MacA-AcrAα hybrid. E. coli MacA-AcrAα hybrid, which has AcrA α-hairpin, is eluted as a monomer. Panel c, mixture of E. coli MacA-AcrAα hybrid and MacA-TolCα hybrid dimer. The complex peak (Ap-Tp) and the E. coli MacA-AcrAα hybrid peak (Ap) are indicated in the chromatogram. Ap and Tp stand for MacA-AcrAα hybrid and MacA-AcrAα hybrid dimer, respectively, and Ap-Tp indicates complex between them. See supplemental Fig. S8 for the interaction between E. coli MacA-AcrAα hybrid mutants and MacA-TolCα hybrid dimer. C, representative electron microscopic image of a complex consisting of E. coli MacA-AcrAα hybrid and Aa MacA-TolCα hybrid dimer, which was purified using the size-exclusion chromatography as in Fig. 3B, panel b. The sample was preserved in negative stain (uranyl formate) and imaged at ×80,000. A reference-free class formed from an average of the side views is shown as an inset and can be compared with a projection of the reconstructed density map. D, surface representation of the reconstructed density map displayed in top view, tilted, and side view (top row). The complex model of E. coli MacA-AcrAα hybrid (magenta) and Aa MacA-TolCα hybrid dimer (cyan for the TolC α-barrel tip, and red for the Aa MacA part) is docked into the density map (bottom row).
Part III. Direct Interaction between the AcrA α-Hairpin Tip Region and the TolC Aperture Tip Region
It was recently reported that E. coli MacA makes intermeshing cogwheel-to-cogwheel interaction with the TolC aperture tip region using a chimeric Actinobacillus actinomycetemcomitans MacA protein containing the TolC tip regions, which was called “Aa MacA-TolCα hybrid dimer.” The Aa MacA-TolCα hybrid dimer protein is an A. actinomycetemcomitans MacA (Aa MacA)-linked dimer whose α-helical tip region was replaced with the first or second aperture tip region of TolC. This chimeric protein exhibited a funnel-like hexameric structure like Aa MacA (13), which enabled us to use this protein as a structural imitator of the TolC α-barrel (27). To further determine whether AcrA shares a binding mode to TolC with MacA, we carried out the in vitro binding experiment with E. coli AcrA and Aa MacA-TolCα hybrid dimer on a size-exclusion chromatographic column. However, the result was not conclusive presumably due to the low propensity of AcrA to oligomerize.
To overcome the low oligomerization propensity of AcrA, we generated a chimeric E. coli MacA bearing a substitution with the AcrA α-hairpin domain (hereafter referred to as E. coli MacA-AcrAα hybrid), because E. coli MacA has greater propensity to oligomerize (33). First, we confirmed that the E. coli MacA-AcrAα hybrid was able to substitute for MacA in vivo (supplemental Table S2). We next observed the physical binding between the E. coli MacA-AcrAα hybrid protein and the Aa MacA-TolCα hybrid dimer protein (see below) (27) through size-exclusion chromatography, which indicated 2:1 binding stoichiometry between Aa MacA-TolCα hybrid dimer and the E. coli MacA-AcrAα hybrid (Fig. 3B), which was described to make a tip-to-tip contact with E. coli MacA hexamer in our previous work (27).
The mutation at the conserved residues of MacA or the AcrA tip region in MacA-AcrAα hybrid abolished the binding to the protein containing the TolC aperture tip region, as well as the formation of a functional efflux pump in vivo (supplemental Fig. S8 and supplemental Table S2). Based on these results, we concluded that the hexameric assembly of AcrA is required to bind to the TolC trimer, where the TolC aperture end regions are sufficient to serve as a binding motif.
Part IV. Electron Microscopy Visualizes the Tip-to-Tip Binding between TolC and AcrA
To visualize how AcrA interacts with the TolC α-barrel tip region, the complex composed of E. coli MacA-AcrAα hybrid and Aa MacA-TolCα hybrid dimer was applied to electron microscopy and single particle analysis. As mentioned above, E. coli MacA-AcrAα hybrid contains E. coli AcrA α-hairpin, and Aa MacA-TolCα hybrid dimer contains E. coli TolC α-barrel tip region, which are both oligomerized by help of the lipoyl, β-barrel, and MP domains of Aa MacA.
A projection matching approach using negatively stained single particles of the complex resulted in a three-dimensional density map at 37 Å resolution (Fourier shell correlation = 0.5). We constructed three starting models with different symmetries (C3, C6, and D6) by directly reconstructing a single selected side view class average using the applied symmetry. Additionally, a low pass filtered density map of the generated PDB model (also used for docking, see below) was used for comparison. We refined the C3 and C6 initial models with their respective symmetries, the D6 starting model using C6 symmetry and the simulated density map using C1 symmetry. After several iterations, all three-dimensional models converged to C6 symmetry. Despite the high similarity of the two protomers, an additional symmetry axis orthogonal to the 6-fold (D6-symmetry) could not be identified at this resolution.
The map exhibits a characteristic dumb-bell-like (bridge symbol) side view with flared ends at both sides, a motif that can also easily be identified in the raw images (Fig. 3C). To dock into the density map, we first generated a complex model composed of the two chimeric proteins (E. coli MacA-AcrAα hybrid and Aa MacA-TolCα hybrid dimer) by recombination of the structural domains available. For the coordinates of Aa MacA, we used a partly refined model of Aa MacA, which was partly built in the low resolution electron density map generated by the multiple wavelength anomalous diffraction method using selenomethionine-substituted protein (Fig. 3D) (13). However, the MP domains were not able to be built because of the lack of the available structure. The relative position between the proteins was determined by superposition with the two hexameric units of Aa MacA in the crystal, which makes the intermeshing cogwheel interaction (13). Docking of the complex structure into the experimental map shows an excellent correlation using the program Chimera (Fig. 3D) (34). The overall dimensions of the intermeshing α-barrels are exactly matched to the central long cylinder. Although the lipoyl and the β-barrel domains were fitted into the density map adjacent to the long cylinder, the density at both the ends was not docked by the model because the model does not contain the MP domains. Considering the overall size of the MP domain from the P. aeruginosa MexA (12), the undocked electron density map is consistent with the missing MP domains (data not shown). Combined with the genetic and biochemical studies, the results from the electron microscopy reveal that the TolC α-barrel makes a direct interaction with the α-barrel of the hexameric AcrA in the intermeshing cogwheel-like manner.
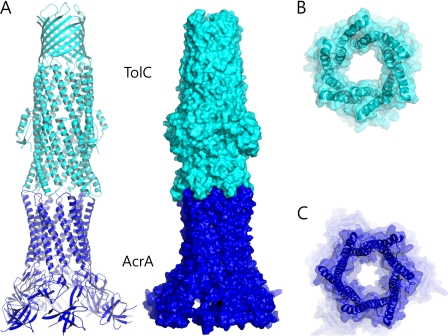
Given the evidence supporting the intermeshing cogwheel interaction between the TolC trimer and the AcrA hexamer, we built an assembly model for TolC and AcrA in the tip-to-tip intermeshing cogwheel-like interaction. We constructed a model of an open structure of TolC in which the TolC α-barrel exhibits the conformation of the α-barrel structure of the AcrA hexamer (Fig. 4A) because the TolC α-helical tip exhibiting the AcrA α-barrel conformation showed a strong binding affinity to AcrA in our experiment (Fig. 3B). Because of the striking shape complementarity between the cogwheels of TolC and AcrA, we generated putative cogwheel-to-cogwheel binding conformations by bringing the AcrA hexamer close to TolC (Fig. 4). Because formation of the docked complex buries 1,246 Å2 of solvent-exposed surface, which is comparable with the modeling based on in vivo chemical cross-linking experiments by Lobedanz et al. (10), our docking model is feasible and stable to form the complex in vivo.
FIGURE 4.
A TolC and AcrA docking model based on the tip-to-tip interaction. A, ribbon representation (left) and the surface representation (right) of a putative model of the TolC (cyan) and AcrA (blue) complex. The open conformation of TolC was generated by adopting the AcrA α-barrel conformation to the TolC α-barrel end region. It was brought to the top of E. coli AcrA hexamer to establish the intermeshing cogwheel interactions. B, bottom view of TolC in the docked complex displayed in A. The semi-transparent surface representation is shown with the ribbon representation. The central channel is wide open. C, top view of the AcrA hexamer in the docked complex. Structural resemblance to the TolC open structure is shown.
Internal Hollow of the AcrA Hexamer Fits the Substrate Exit Domain of the AcrB Homotrimer
Our models for the hexameric AcrA and its binding mode with AcrB are distinct from the currently prevailing models. The current models propose the tip-to-tip AcrB-TolC interaction, and hairpin-wrapping AcrA-TolC interactions are heavily supported by in vivo cysteine-mediated cross-linking assays (10, 12, 17, 26). For this reason, we investigated mode of interactions between the hexameric model of AcrA with the AcrB homotrimer.
Given the notable structural complementarities between the AcrA funnel-mouth hollow and the AcrB substrate exit domains (Fig. 5), the AcrB substrate exit domains were docked onto the funnel mouth of the modeled AcrA hexamer by visual inspection (Fig. 5C). In the docking model, the AcrA triangular cone-shaped hollow fits the AcrB substrate exit with a complementary shape (Fig. 5, A and B). When the MexA hexamer was docked onto the cognate inner membrane transporter MexB, which is similar to AcrB (35), a similar molecular contact was observed in the lipoyl and β-barrel domains (Fig. 5D). However, each pair of MP domains, which are only present in the MexA hexamer model, contacted one protomer of the MexB porter domain and did not obstruct the substrate entrance sites (Fig. 5D).
FIGURE 5.
Structural complementarities between the AcrA hexamer model and the substrate exit domain of AcrB trimer and molecular docking of AcrA and TolC. A, surface representations of the AcrA hexamer model. Top, bottom view of the AcrA hexamer. From the distance, the approximate size of the triangular hollow can be deduced. Bottom, side view of the AcrA hexamer. The internal hollow at the funnel stem and the funnel mouth are indicated by red dotted lines. B, surface representations of the AcrB homotrimer using PDB code 2W1B (21). Substrate exit domain is in orange; porter domain is in magenta, and transmembrane (TM) domain is in gray. Top, top view of the AcrB trimer with approximate dimensions of the triangular “substrate exit domain.” The longest length of the porter domain is indicated by 70 Å, which matches the inter-MP domain distance shown in Fig. 1D. Bottom, side view of AcrB trimer. The height of substrate exit domain is indicated. The substrate entrance site is indicated by a yellow arrow and the substrate exit site by a red arrow. C, docking model for AcrA-AcrB complex is displayed by surface representations. The AcrA hexamer model is shown in blue, and the AcrB is colored as in B. Substrate entrance site and the putative substrate exit sites of the complex are indicated by yellow and red arrows, respectively. Top view is shown on the left panel, and the side view is shown on the right. D, docking model for MexA-MexB complex (PDB code 2V50 was used for the MexB structure (35)). Because the MP domain is built in the MexA hexamer model, the docked complex includes the MP domains, which contact the porter domain but do not seem to preclude substrate entrance. Lipid modifications at the N terminus of MexA, which is responsible for the membrane anchorage, are shown.
AcrA Mediates the Interaction of AcrB to the TolC α-Barrel Tip Region
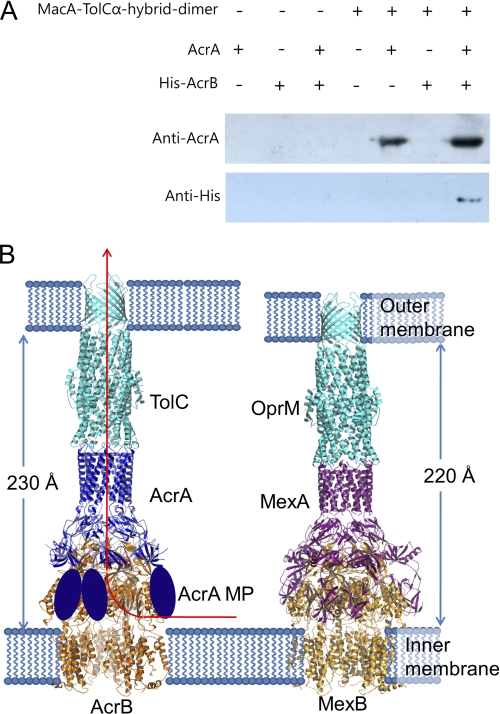
To investigate the interactions between AcrA, AcrB, and TolC in the absence of a reagent that induces cross-linking, the Aa MacA-TolCα hybrid dimer protein, which contains TolC α-barrel tip region (27), was covalently attached to resin and then incubated with the purified proteins of AcrA and/or a full-length AcrB. The Aa MacA-TolCα hybrid dimer protein was used instead of the full-length TolC because it provides TolC α-barrel tip region only, which was sufficient for interaction with the AcrA α-hairpin domain in vitro (Fig. 3B), without other regions of the TolC protein that may result in nonspecific interactions with other proteins. As shown in Fig. 6A, the AcrA protein was readily bound to the TolC α-barrel tip region-containing protein, whereas the AcrB protein did not show any affinity to the TolC α-barrel tip region. However, when AcrA and AcrB proteins were co-incubated with the TolC α-barrel tip region-coupled resin, the AcrB protein did bind to the resin with about 2-fold higher binding of AcrA than in the absence of AcrB (Fig. 6A). These results indicate that AcrA mediates and intervenes the interaction between TolC and AcrB and that AcrB enhances binding of AcrA to TolC.
FIGURE 6.
Assembly model for tripartite efflux pumps. A, interaction of TolC α-barrel tip, AcrA, and AcrB. MacA-TolCα hybrid dimer-coupled resin was prepared using CNBr-activated resin (50 μg; GE Healthcare) and MacA-TolCα hybrid dimer protein (1 mg) according to the manufacturer's instruction. AcrA (residues 26–397; 2 mg/ml) and/or the full-length AcrB with C-terminally hexahistidine tag (His-AcrB; 2 mg/ml) were incubated with the coupled resin (20 μl) for 2 h at 4 °C, and then the resin was thoroughly washed with PBS and applied to SDS-PAGE, followed by Western blotting. To detect AcrA and AcrB, anti-AcrA and anti-His antibodies were used, respectively. B, left, the ribbon representation of the TolC3-AcrA6-AcrB3 complex is colored by its components. The TolC trimer (cyan) contacts the funnel stem of the AcrA hexamer (blue), and the AcrB trimer (orange) contacts the funnel mouth of the AcrA hexamer. In particular, the flexible AcrA MP domains (blue ovoid) make a pair and interact with the porter domain of AcrB, accommodating the dynamic structural movement coupled with the proton translocation and the substrate transport. The substrate moving passage is indicated by a red arrow. The complex spans the entire periplasmic space, inner membrane, and outer membrane. The periplasmic part of the ternary complex is 230 Å long. Right, the ribbon representation of the OprM3-MexA6-MexB3 complex. The OprM trimer (cyan), MexA hexamer (violet), and MexB trimer (orange) make a long complex in the same arrangement with the TolC-AcrA-AcrB complex. The MP domains of MexA are shown. The periplasmic part of the complex is 10 Å shorter because the α-hairpin domain of MexA is shorter than that of AcrA.
DISCUSSION
Assembly Model for AcrAB-TolC Pump
In our previous reports, we provided evidence that MacA functions as a funnel-like hexamer and proposed the “adapter bridging model” for the MacAB-TolC pump (13, 20, 27). In this study, genetic complementation and in vivo cross-linking experiments showed that the AcrA hairpin tip region functionally and physically interacts with the TolC aperture tip region (Fig. 3 and Table 2) and that AcrA has a binding mode similar to that of MacA to TolC (19). Using the hexameric arrangement of MacA as a backbone, AcrA and MexA hexameric models were built based on the functional AcrA dimer, which has the propensity for trimerization of the dimeric units in vitro. The AcrA hexameric model was evaluated by mutational and functional analyses of the putative pore region (Fig. 2 and Table 1), and chimeric analysis was designed based on the Aa MacA packing interaction (Fig. 3). In particular, the electron microscopic study visualized the intermeshing cogwheel-like interaction between the TolC α-barrel tip region and the AcrA α-barrel, which is essentially same as the interaction between TolC and MacA (27). The hexameric models for AcrA and MexA showed striking molecular complementarities in the AcrB substrate exit domain, illustrating the plausibility of the docking models of the hexameric AcrA/MexA and the trimeric AcrB/MexB (Fig. 5, C and D). We further presented experimental evidence for the bridging role of AcrA between TolC and AcrB (Fig. 6A).
Taken together, we propose that the adapter bridging model is a general assembly mechanism for tripartite drug efflux pumps (Fig. 6B). The α-barrel of the AcrA hexamer interacts with the α-barrel of the TolC trimer in a tip-to-tip manner like the intermeshing cogwheels, as was proposed in the MacA and TolC interaction (13, 20). The triangular hollow formed by lipoyl and β-barrel domains covers the substrate exit domain of AcrB with the structural complementarities in our model. A pair of MP domains in AcrA interacts with the porter domain of AcrB without blocking the substrate entrance site. It seems quite reasonable that the flexible MP domain interacts with the porter domain of AcrB, whose motion is driven by the proton gradient.
Our assembly model explains the tomographic map of the MexA-OprM complex produced by cryo-electron microscopy (36). When we accounted for the diameter and the length, the central long cylinder of the tomogram would reflect the α-barrels of MexA and OprM connected by the tip-to-tip binding. Furthermore, the 21-nm intermembrane distance of the MexA-OprM complex in the tomogram can be explained by our tripartite MexAB-TolC pump model, which shows a similar distance (22 nm) between the two lipid membranes (Fig. 6B).
Comparison of the Adapter Bridging Model with the Current Model
Our models for the hexameric AcrA and the assembly of the tripartite pumps are distinct from the currently prevailing models. The current models are heavily supported by in vivo cysteine-mediated cross-linking assays (10, 12, 17, 26) that detect and map the tip-to-tip AcrB-TolC interaction and hairpin-wrapping AcrA-TolC interaction (10, 17, 26). However, the fidelity of the results from the in vivo cross-linking experiments has begun to be re-evaluated (19, 37). AcrA variants harboring various cysteine mutations, which resulted in positive cross-links between AcrA and TolC (10), did not form an efflux-ready AcrAB-TolC pump as functional as the wild type, as indicated by the dramatically increased sensitivity to antibiotics of acrA-deleted E. coli cells that expressed these variant proteins (19). For this reason, we constructed a cysteine-less AcrB variant (AcrB-Δcys) that has been previously used to detect interactions between the components of the AcrAB-TolC pump by in vivo cross-linking (12). However, unlike what has been reported (12), AcrB-Δcys exhibited a severe loss-of-function phenotype, and therefore, we were unable to use this cysteine-less AcrB variant for in vivo cross-linking analyses of the assembly of AcrAB-TolC pump to confirm our results. This cysteine-less AcrB variant failed to form a functional tripartite pump as indicated by the loss of resistance to novobiocin (MIC = 200 μg/ml) of the acrAB-deleted E. coli strain co-expressing AcrB-Δcys and wild-type AcrA, whereas co-expression of wild-type AcrA and AcrB fully restored the resistance of acrAB-deleted E. coli cells to novobiocin (MIC = 800 μg/ml) (supplemental Table S3). Co-expression of an AcrA variant bearing a cysteine substitution at position 58 or 196 along with AcrB-Δcys that resulted in positive cross-links (12) further abolished the antibiotic resistance of E. coli cells to novobiocin (MIC = 100 μg/ml).
For the reasons listed below, we believe that in vivo cross-linking data using Cys variants of AcrA or AcrB that do not form a functional AcrAB-TolC complex need to be re-evaluated when such variants are used for the construction of the assembly model of the pump. First, it is hard to draw a meaningful conclusion from chemical cross-linking experiments using nonfunctional AcrA or AcrB variants because the cross-linked complex in vivo would mostly be derived from a nonfunctional assembly state of the pump. Even though Cys variants are partially functional, it is still not easy to interpret the data because the cross-linked complex in vivo could be derived from either a functional efflux-ready state or a nonfunctional assembly state. Compared with previous cross-linking data, our cross-linking data (Fig. 3A) better explain the functional interaction between AcrA and TolC because a loss-of-function, as well as interaction of AcrA with wild-type TolC by substitution of the conserved residue at the α-hairpin tip region of AcrA (L132C), were greatly restored by the complementing mutations at the aperture region of TolC, indicating that cross-links between AcrA-L132C and the complementing TolC variant were derived from a functional efflux-ready state of the pump. In addition to our data, Weeks et al. (37) also reported that AcrB-TolC binding is much weaker than AcrA-TolC binding in the presence of a cross-linker.
Recent studies of drug efflux pumps are also challenging the currently prevailing models (7, 36). The current models are not compatible with the tomogram averaging of MexA-OprM by cryo-electron tomography. When the current models were overlapped with the tomographic map, the lipoyl domain remains outside of the map (36). Furthermore, the current tripartite complex models gave an intermembrane distance of only 17 nm, although the distance was measured as 21 nm in the tomogram with only two components of the pump (36).
Conclusions
Given the hexameric structure of MacA (13), we studied the functional and structural parallels between MacA and AcrA (19). This study presents structural and functional evidence for an AcrA hexamer and the assembly model at the functional state, representing a new class of drug targets for multidrug-resistant bacteria. Further structural and functional studies on the binary and ternary complexes should help define the assembly and channel opening of tripartite efflux pumps.
Supplementary Material
Acknowledgments
We thank Erica Jacovetty for assistance with EM data collection. EM imaging was performed at the National Resource for Automated Molecular Microscopy, which is supported by National Institutes of Health Grant RR17573 through the NCRR P41 program.
This work was supported in part by National Research Foundation Grant NRF-2010-0016052 (to N.-C. H.) and Pioneer Research Center Program Grant 20100002201 from the National Research Program of Korea (to K. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S9, Tables S1–S4, and additional references.
- MP
- membrane proximal
- PDB
- Protein Data Bank
- MIC
- minimum inhibitory concentration.
REFERENCES
- 1. Nikaido H., Zgurskaya H. I. (1999) Curr. Opin. Infect. Dis. 12, 529–536 [DOI] [PubMed] [Google Scholar]
- 2. Nikaido H., Zgurskaya H. I. (2001) J. Mol. Microbiol. Biotechnol. 3, 215–218 [PubMed] [Google Scholar]
- 3. Koronakis V., Eswaran J., Hughes C. (2004) Annu. Rev. Biochem. 73, 467–489 [DOI] [PubMed] [Google Scholar]
- 4. Lewis K. (2000) Curr. Biol. 10, R678–R681 [DOI] [PubMed] [Google Scholar]
- 5. Zgurskaya H. I., Nikaido H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7190–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thanabalu T., Koronakis E., Hughes C., Koronakis V. (1998) EMBO J. 17, 6487–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Misra R., Bavro V. N. (2009) Biochim. Biophys. Acta 1794, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. (2000) Nature 405, 914–919 [DOI] [PubMed] [Google Scholar]
- 9. Husain F., Humbard M., Misra R. (2004) J. Bacteriol. 186, 8533–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lobedanz S., Bokma E., Symmons M. F., Koronakis E., Hughes C., Koronakis V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4612–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tikhonova E. B., Zgurskaya H. I. (2004) J. Biol. Chem. 279, 32116–32124 [DOI] [PubMed] [Google Scholar]
- 12. Symmons M. F., Bokma E., Koronakis E., Hughes C., Koronakis V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7173–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yum S., Xu Y., Piao S., Sim S. H., Kim H. M., Jo W. S., Kim K. J., Kweon H. S., Jeong M. H., Jeon H., Lee K., Ha N. C. (2009) J. Mol. Biol. 387, 1286–1297 [DOI] [PubMed] [Google Scholar]
- 14. Mikolosko J., Bobyk K., Zgurskaya H. I., Ghosh P. (2006) Structure 14, 577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins M. K., Bokma E., Koronakis E., Hughes C., Koronakis V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9994–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akama H., Matsuura T., Kashiwagi S., Yoneyama H., Narita S., Tsukihara T., Nakagawa A., Nakae T. (2004) J. Biol. Chem. 279, 25939–25942 [DOI] [PubMed] [Google Scholar]
- 17. Bavro V. N., Pietras Z., Furnham N., Pérez-Cano L., Fernández-Recio J., Pei X. Y., Misra R., Luisi B. (2008) Mol. Cell 30, 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerken H., Misra R. (2004) Mol. Microbiol. 54, 620–631 [DOI] [PubMed] [Google Scholar]
- 19. Kim H. M., Xu Y., Lee M., Piao S., Sim S. H., Ha N. C., Lee K. (2010) J. Bacteriol. 192, 4498–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu Y., Sim S. H., Song S., Piao S., Kim H. M., Jin X. L., Lee K., Ha N. C. (2010) Biochem. Biophys. Res. Commun. 394, 962–965 [DOI] [PubMed] [Google Scholar]
- 21. Murakami S., Nakashima R., Yamashita E., Yamaguchi A. (2002) Nature 419, 587–593 [DOI] [PubMed] [Google Scholar]
- 22. Xu Y., Sim S. H., Nam K. H., Jin X. L., Kim H. M., Hwang K. Y., Lee K., Ha N. C. (2009) Biochemistry 48, 5218–5225 [DOI] [PubMed] [Google Scholar]
- 23. Gristwood T., Fineran P. C., Everson L., Salmond G. P. (2008) Mol. Microbiol. 69, 418–435 [DOI] [PubMed] [Google Scholar]
- 24. Mima T., Joshi S., Gomez-Escalada M., Schweizer H. P. (2007) J. Bacteriol. 189, 7600–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. De Angelis F., Lee J. K., O'Connell J. D., 3rd, Miercke L. J., Verschueren K. H., Srinivasan V., Bauvois C., Govaerts C., Robbins R. A., Ruysschaert J. M., Stroud R. M., Vandenbussche G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11038–11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura N., Murakami S., Oyama Y., Ishiguro M., Yamaguchi A. (2005) Biochemistry 44, 11115–11121 [DOI] [PubMed] [Google Scholar]
- 27. Xu Y., Song S., Moeller A., Kim N., Piao S., Sim S. H., Kang M., Yu W., Cho H. S., Chang I., Lee K., Ha N. C. (2011) J. Biol. Chem. 286, 13541–13549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piao S., Xu Y., Ha N. C. (2008) Acta Crystallogr. F. Struct. Biol. Cryst. Commun. 64, 391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 30. Takatsuka Y., Nikaido H. (2009) J. Bacteriol. 191, 1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su C. C., Yang F., Long F., Reyon D., Routh M. D., Kuo D. W., Mokhtari A. K., Van Ornam J. D., Rabe K. L., Hoy J. A., Lee Y. J., Rajashankar K. R., Yu E. W. (2009) J. Mol. Biol. 393, 342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Avila-Sakar A. J., Misaghi S., Wilson-Kubalek E. M., Downing K. H., Zgurskaya H., Nikaido H., Nogales E. (2001) J. Struct. Biol. 136, 81–88 [DOI] [PubMed] [Google Scholar]
- 33. Tikhonova E. B., Dastidar V., Rybenkov V. V., Zgurskaya H. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16416–16421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goddard T. D., Huang C. C., Ferrin T. E. (2007) J. Struct. Biol. 157, 281–287 [DOI] [PubMed] [Google Scholar]
- 35. Sennhauser G., Bukowska M. A., Briand C., Grütter M. G. (2009) J. Mol. Biol. 389, 134–145 [DOI] [PubMed] [Google Scholar]
- 36. Trepout S., Taveau J. C., Benabdelhak H., Granier T., Ducruix A., Frangakis A. S., Lambert O. (2010) Biochim. Biophys. Acta 1798, 1953–1960 [DOI] [PubMed] [Google Scholar]
- 37. Weeks J. W., Celaya-Kolb T., Pecora S., Misra R. (2010) Mol. Microbiol. 75, 1468–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.