Abstract
Yeast Hsp104 is an AAA+ chaperone that rescues proteins from the aggregated state. Six protomers associate to form the functional hexamer. Each protomer contains two AAA+ modules, NBD1 and NBD2. Hsp104 converts energy provided by ATP into mechanical force used to thread polypeptides through its axial channel, thereby disrupting protein aggregates. But how the action of its 12 AAA+ domains is co-ordinated to catalyze disaggregation remained unexplained. Here, we identify a sophisticated allosteric network consisting of three distinct pathways that senses the nucleotide state of AAA+ modules and transmits this information across the Hsp104 hexamer. As a result of this communication, NBD1 and NBD2 each adopt two distinct conformations (relaxed and tense) that are reciprocally regulated. The key element in the network is the NBD1-ATP state that enables Hsp104 to switch from a barely active [RT] state to a highly active [TR] state. This concerted switch involves both cis and trans protomer interactions and provides Hsp104 with the mechanistic scaffold to catalyze disaggregation. It prepares the chaperone for polypeptide binding and activates NBD2 to generate the power strokes required to resolve protein aggregates. ATP hydrolysis in NBD1 resolves the high affinity [TR] state and switches the chaperone back into the low affinity [RT] state. Our model integrates previously unexplained observations and provides the first comprehensive map of nucleotide-related allosteric signals in a class-1 AAA+ protein.
Keywords: Allosteric Regulation, ATPases, Chaperone Chaperonin, Molecular Motors, Protein Folding, AAA+, Disaggregation
Introduction
AAA+ proteins (ATPases associated with various cellular activities) comprise a functionally diverse family of molecular machines that share a large degree of sequence homology and an evolutionary conserved mechanism (1). AAA+ proteins use chemical energy provided by ATP hydrolysis to catalyze force-induced structural rearrangements of client molecules. However, the details of this mechano-chemical coupling are not well understood. AAA+ proteins consist of one or two conserved ATP-binding modules (NBDs)2 linked to function-specific auxiliary domains. Each AAA+ module harbors canonical Walker A and Walker B motifs that are critical for nucleotide binding and hydrolysis, respectively. In many cases, AAA+ proteins assemble into ring-shaped oligomers (2).
Hsp104 from yeast and its bacterial homolog, ClpB, are AAA+ chaperones that provide thermotolerance to their hosts by disassembling protein aggregates after heat shock (3–5). Reactivation of these proteins seems to be crucial because the removal of protein aggregates by degradation was found to be insufficient for restoring cell viability (6, 7). To disrupt noncovalent interactions between aggregated polypeptides, Hsp104 and ClpB are believed to employ a common mechanism that involves co-operation with the Hsp70 chaperone system (8, 9): ATP binding and hydrolysis induce a sequence of domain movements in Hsp104 which in turn exert a mechanical force on bound polypeptide substrates (10–12). As a result, individual polypeptide chains are extracted from aggregates and threaded through the central channel of the disaggregase (6, 7, 13). This proposed mode of action is reminiscent of models established for other AAA+ proteins, such as the unfoldase unit of the proteasome or the bacterial unfoldases ClpA and ClpX, which unfold target polypeptides and feed them into the proteolytic chamber of an associated protease (14–17). Nucleotide-induced domain rearrangements have been observed for several AAA+ proteins (18, 19), but to what extent these movements occur in a co-ordinated fashion and how AAA+ machines couple chemical energy to mechanical work are largely unknown. For ClpX it has been suggested that ATP hydrolysis occurs randomly, i.e. without co-ordination among the six subunits, and facilitates progressive polypeptide processing (20). For other AAA+ proteins concerted ATP hydrolysis in all domains has been proposed (21, 22).
The question of allosteric communication has also been addressed for AAA+ proteins involved in disaggregation, although with inconclusive results. Both Hsp104 and ClpB are class-1 AAA+ proteins that contain two AAA+ modules per subunit, NBD1 and NBD2. In the functional hexamer, these modules form two rings that are stacked on top of each other (18, 23). In the case of ClpB from Thermus thermophilus incorporation of nonfunctional subunits into functional hexamers was found to impair ATPase activity and disaggregation (24). For Escherichia coli ClpB it was reported that mixing nonfunctional subunits with wild-type subunits unleashed unexpected protein remodeling and threading activity (25). It was further shown for both Hsp104 and ClpB that ATP binding to NBD1 greatly stimulates ATP turnover by NBD2 (12, 26). Although these studies suggest an allosteric signaling network within the hexamer, the mechanistic details and implications for the chaperone function of Hsp104/ClpB remained unclear.
Here, we examined the roles of individual NBDs by mixing active Hsp104 subunits with inactive subunits. We identified three distinct communication pathways that propagate allosteric signals within the hexamer and establish the mechanistic link between ATP hydrolysis and polypeptide translocation. To our knowledge, this is the first mechanistic description of an interdomain network that encompasses all 12 ATPase domain of a class-1 AAA+ protein. Our model provides a mechanistic understanding of how AAA+ proteins sense and respond to changes in nucleotide state and how this allosteric circuitry enables Hsp104 to disrupt protein aggregates.
EXPERIMENTAL PROCEDURES
Unless stated otherwise, experiments were carried out in 50 mm HEPES/KOH, 150 mm KCl, 20 mm MgCl2, pH 7.5 (HKM buffer) at 30 °C. Pyruvate kinase, l-lactate dehydrogenase, phosphoenolpyruvate, ATP, ADP, and ATPγS were from Roche Applied Science. Recombinant luciferase was from Promega. Luciferin, acetylcoenzyme A (CoA), NADH, dithiothreitol (DTT), and bovine serum albumin (BSA) were from Sigma-Aldrich. All other chemicals were from EMD Chemicals. We used previously described protocols to produce wild-type Hsp104 and variants, reduced carboxymethylated α-lactalbumin (RCMLa) and fluorescently labeled, reduced carboxymethylated α-lactalbumin (f-RCMLa) (27), LaEYFP (12), and human Hsc70 (28). Sis1/Hsp40 was expressed in E. coli BL21 containing a plasmid encoding a His6-tagged version of Sis1 and purified via Ni2+-chelating chromatography.
Formation of Hsp104 Hetero Complexes
A constant concentration of type A subunits (at least one NBD is wild type-like) was mixed with varying concentrations of type B subunits (none of the NBDs is wild type-like) to yield the indicated mixing ratios. Mixtures were incubated for 1 h at 30 °C and then diluted 10-fold into assay buffer. Mixing ratios are given as concentration of type B subunits over concentration of type A subunits. Experimental readouts, such as ATPase rate, were calculated based on the concentration of active subunits.
ATPase Activity
ATPase activity was monitored using a coupled enzymatic colorimetric assay (29). Kinetics were recorded in Cary50 spectrophotometer (Varian). ATP turnover/Hsp104 monomer was calculated from the slope of the reaction using a NADH extinction coefficient of 6200 m−1 cm−1.
ANS Fluorescence
5 μm Hsp104 was preincubated with 100 μm ANS (8-anilino-1-naphthalene sulfonic acid) in HKM buffer and then mixed with 500 μm nucleotides or buffer. For ATP measurements, residual ADP was converted to ATP using 10 mm phosphoenolpyruvate and pyruvate kinase prior to mixing with the protein. Fluorescence spectra were recorded with a QuantaMaster4 (Photon Technology International) at 20 °C. Excitation was at 370 nm using 0.8-nm slits. Emission spectra were collected from 380 nm to 650 nm using 2-nm slits. Spectra were buffer-corrected. Experiments were carried out as independent triplicates and averaged.
Polypeptide Binding
Binding of f-RCMLa was monitored by changes in fluorescence anisotropy as described (27). FITC fluorescence was excited at 488 nm, and polarized emission was detected at 515 nm in a QuantaMaster4 equipped with autopolarizers. The f-RCMLa fraction bound to Hsp104 was calculated from the observed change in anisotropy. Values were normalized with respect to the binding amplitude of Hsp104 [bb].
Luciferase Disaggregation
Firefly luciferase (FFL) disaggregation was carried out as described previously (12, 30). In short, FFL was denatured in HKM containing 8 m urea for at least 30 min and subsequently diluted 125-fold into refolding buffer (HKM, 2 units/ml pyruvate kinase, 20 mm phosphoenolpyruvate, 2 mm DTT, 0.24 mm CoA, 50 μg/ml BSA, 0.1 mm luciferin) containing Hsp104, Hsc70, and Sis1 (see also figure legends). The increase in FFL activity over time was monitored in 96-well plates using an Omega FluoroStar (BMG Labtech). The initial slope and the luminescence signal after 70 min were used as a measure of disaggregation activity.
Additional Methods are described in the supplemental Experimental Procedures. Target genes, primers, sequences, and templates are shown in supplemental Table 1.
RESULTS
Experimental Approach and Nomenclature
Allosteric communication in multimeric proteins is based on the ability to transmit changes in ligand occupation between protomers. Perturbation of this cross-talk, e.g. by mutating one of the protomers, will change the behavior of other (wild-type) protomers. Hsp104 is a dynamic hexamer, i.e. it rapidly dissociates and reassembles, and perturbations can be introduced by mixing mutant and wild-type forms of the protein. This yields a distribution of probabilistically assembled hetero hexamers, with the center of the distribution depending on the mixing ratio (Fig. 1) (24). The larger the mixing ratio of inactive over active protein, the more inactive subunits a hexamer will contain on average. Intersubunit communication can be detected by comparing properties of hetero complexes with those of the respective homo multimers (Fig. 1D). We established conditions for hetero complex formation of Hsp104 by mixing non-like variants and monitoring overall ATPase activity. Changes in activity with mixing time reflect the formation of hetero hexamers, which occurred with a half-life of ∼20 s under our conditions (supplemental Fig. S1), similar to the value reported for ClpB (24).
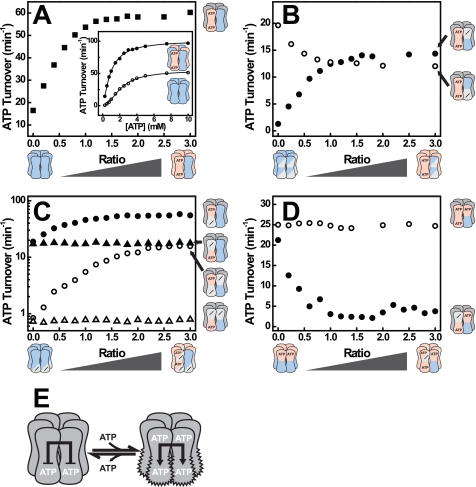
FIGURE 1.
Dissecting allosteric interactions using mixed hexamers. A, cryo-EM reconstruction of hexameric Hsp104ΔN (32) is shown. B, upon mixing, an active Hsp104 protein such as wild-type Hsp104 (blue) will form hetero hexamers with an inactive Hsp104 protein (red) by exchanging subunits. C, the resulting hetero hexamer distribution depends on the mixing ratio. The larger the mixing ratio, the more inactive subunits a hexamer contains on average. At a mixing ratio of 1, the most abundant hexamer contains three active and three inactive subunits. D, whether the presence of inactive subunits (red) changes the behavior of active subunits (blue) in a hexamer can be assessed by monitoring a property of the active subunits such as ATPase activity. These experiments were carried out by mixing a constant amount of an active hexamer with an increasing amount of an inactive hexamer. Three possible scenarios are depicted. Inactive subunits may not affect active subunits (●), they could stimulate active subunits (green circle) or they could inhibit active subunits (red circle). E, three types of NBD variants were used in this study. Wild-type NBDs (blue) bind and hydrolyze ATP. Variants with a mutated Walker B motif (red) no longer hydrolyze ATP, but still bind it. Variants with a mutated Walker A motif (white) are unable to bind nucleotide. In the text, the three types of NBDs are identified by the symbols [+], [b], and [−], respectively.
Hsp104 variants are denoted by either symbols (in the text) or colors (in figures) reflecting their properties (Table 1 and Fig. 1E): Wild-type NBDs [+] (blue) bind and hydrolyze ATP. The E285Q and E687Q mutations in the Walker B motif, which abolish ATP hydrolysis in the affected NBD while maintaining nucleotide binding, are designated by [b] (red). [−] (white) denotes the K218T and K620T mutations in the Walker A motif, which disrupt nucleotide binding to the respective NBD. Because each Hsp104 protomer contains two NBDs, it is represented by two of these symbols reflecting the properties of NBD1 (superscripted) and NBD2 (subscripted). As an example, wild-type Hsp104 is denoted by the symbol [++], whereas the symbol [bb] designates the Hsp104E285Q/E687Q double mutant (27). Mixing these proteins yields the hetero complex distribution [bb·++]. In our experiments, we mixed an inactive variant (none of the NBDs is wild type-like) with an active variant (at least one of the NBDs is wild type-like). Depending on the type of mutants used, a number of NBDs in the resulting hetero hexamers is either empty [−] or permanently occupied with ATP [b]. The properties of these hetero hexamers reflect how active subunits are affected by the nucleotide state of other protomers (Fig. 1D).
TABLE 1.
Hsp104 variants and their properties
| Symbol | NBD1 | NBD2 | Mutation |
|---|---|---|---|
| [++] | Wild type | Wild type | None |
| [−+] | Empty | Wild type | K218T |
| [+−] | Wild type | Empty | K620T |
| [b+] | ATP | Wild type | E285Q |
| [+b] | Wild type | ATP | E687Q |
| [bb] | ATP | ATP | E285Q/E687Q |
| [−b] | Empty | ATP | K218T/E687Q |
| [b−] | ATP | Empty | E285Q/K620T |
ATP-bound State of NBD1 Increases ATP Hydrolysis by NBD2 of an Adjacent Protomer
We first examined whether the ATPase activity of Hsp104 [++] subunits changes upon incorporation of inactive subunits, such as Hsp104 [bb], into the hexamer. As shown in Fig. 2A, the addition of increasing amounts of [bb] to a constant amount of [++] resulted in a hyperbolic increase in ATP turnover. Because [bb] subunits do not exhibit ATPase activity, this increase must originate from an enhanced hydrolysis of the [++] subunits. In other words, [bb] subunits exert a stimulatory effect on [++] subunits in [bb·++] hexamers. The increased ATPase activity appears to be the result of a decreased Km and an increased kcat (Fig. 2A, inset). To determine which of the two NBDs in [++] becomes stimulated, we mixed [bb] with the Walker A variants [−+] and [+−], respectively, to form [bb·−+] and [bb·+−] complexes. Addition of [bb] to [−+] led to a pronounced increase in ATP hydrolysis (Fig. 2B). In contrast, hydrolysis rates decreased when we mixed [bb] with [+−]. This implies that [bb] subunits enhance ATP hydrolysis in NBD2 [bb·−+] while inhibiting ATP hydrolysis in NBD1 [bb·+−], of a neighboring subunit (more about the inhibitory effect in the following section). We then determined which of the NBDs in [bb] is responsible for the stimulation of NBD2. To this end, we generated the Walker A/B double mutants Hsp104K218T/E687Q [−b] and Hsp104E285Q/K620T [b−]. In contrast to [bb], only the NBD carrying the Walker B mutation can be occupied by ATP, whereas the domain with the Walker A mutation remains nucleotide-free. Both mutants lacked ATPase activity (data not shown). When subjected to our mixed hexamer assay, only [b−] but not [−b] enhanced the ATPase activity of NBD2 in either [++] or [−+] (Fig. 2C). The Walker A mutation in [−b] not only disrupts ATP binding to NBD1 but also its ability to activate NBD2. These data demonstrate that NBD2 receives an allosteric signal from an adjacent ATP-bound NBD1 and responds with an increase in ATPase activity (Fig. 2E).
FIGURE 2.
ATP binding to NBD1 stimulates ATP hydrolysis in an adjacent NBD2. A, ATPase activity of [++] mixed with [bb] at various ratios (■). Inset, ATPase rate plot of [++] (○) and a [bb·++] mixture at a ratio of 1 (●). B, ATPase activities of [−+] (●) and [+−] (○) mixed with [bb] at the indicated mixing ratios. C, ATPase activities of [++] (filled symbols) and [−+] (open symbols) mixed with [−b] (triangles) or [b−] (circles) at the indicated ratios. D, ATPase activity of [b+] mixed with [bb] (○) or [−b] (●) at the indicated ratios. E, model of Hsp104 showing the stimulatory effect of ATP binding to NBD1 on ATP hydrolysis in NBD2. ATP hydrolysis rates were determined at 30 °C and 2 mm ATP. The concentration of active subunits was 1 μm, whereas the concentration of inactive subunits varied. Hetero complexes were formed by mixing both subunits in the absence of nucleotide and incubation for 1 h at 30 °C prior to measurement.
The postulated NBD1→NBD2 pathway explains why a variant carrying a Walker B mutation in NBD1, Hsp104 [b+], hydrolyzes ATP faster than the wild-type protein (12). The hyperactivity of [b+] reflects that NBD1 is permanently occupied with ATP and exerts a (permanent) stimulation of ATP hydrolysis in NBD2. Our novel finding is that this stimulation is a trans rather than a cis effect, i.e. it is caused by the NBD1-ATP state in an adjacent protomer and not in the same protomer. The trans model predicts that mixing [b+] with a variant deficient in nucleotide binding to NBD1 should decrease its ATPase activity. As shown in Fig. 2D, the [−b] mutant exerts indeed a strong inhibitory effect on [b+], demonstrating the critical role of the NBD1-ATP state in adjacent protomers. The trans model also predicts that addition of [bb] to [b+] does not decrease the hydrolysis rate, in agreement with our experimental findings (Fig. 2D).
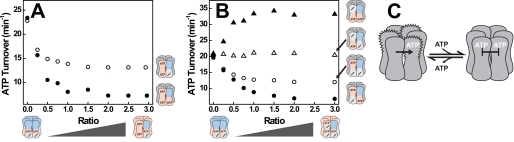
ATP-bound State of NBD1 Decreases ATP Turnover in Adjacent NBD1s
Next, we examined the interaction between adjacent NBD1 domains. The Walker A variant [+−] and the Walker B variant [+b] both carry an inactive NBD2 and hydrolyze ATP only in NBD1. As shown in Fig. 3A, incorporation of [bb] subunits decreased the activity of both proteins. To determine the origin of this allosteric signal, we again employed the Walker A/B double mutants. Only [b−], but not [−b], was able to inhibit hydrolysis of either [+−] or [+b], showing that an ATP-bound NBD1 decreases the ATPase activity of an adjacent NBD1 (Fig. 3B and supplemental Fig. S2). In fact, turnover of [+b] increased when it was mixed with [−b]. Presumably, NBD1 in the [+b] homo hexamer is down-regulated in its capability to hydrolyze ATP by the trans NBD1↔NBD1 pathway. This inhibition is attenuated when an ATP-bound NBD1 is replaced by the nucleotide-free NBD1 of [−b] (Fig. 3C).
FIGURE 3.
ATP binding to NBD1 inhibits ATP hydrolysis in an adjacent NBD1. A, ATPase activities of [+−] (○) and [+b] (●) mixed with [bb] at the indicated ratios. B, ATPase activities of [+b] (filled symbols) and [+−] (open symbols) after mixing with [−b] (triangles) or [b−] (circles) at the indicated ratio. Experiments were carried out as described in the legend for Fig. 2. C, model of Hsp104 showing the inhibitory effect of ATP binding to NBD1 on ATP hydrolysis in NBD1.
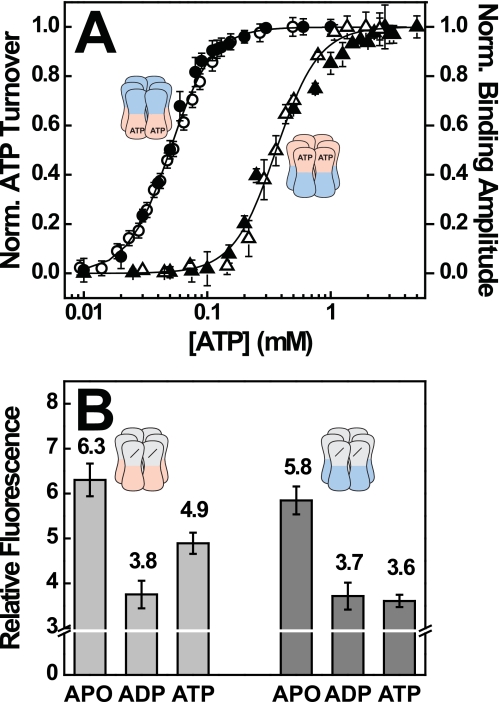
Nucleotide State of NBD2 Modulates the Affinity of NBD1 for ATP
Our data indicate that changes in the nucleotide state of NBD1 are transmitted to NBD2 but show no evidence for a corresponding pathway from NBD2 to NBD1. However, we noticed that the variant [+−] had a significantly higher Km for ATP compared with [+b] (supplemental Fig. S2). We initially attributed this difference to the known oligomerization defect of [+−] (3) (supplemental Fig. S3). But even at high protein concentrations, Km of [+−] was ∼10 times higher than of [+b] (1 mm versus 0.1 mm; supplemental Fig. S4), arguing against an artifact caused by poor oligomerization.
To obtain reliable evidence for a regulatory effect of NBD2 on NBD1, we exploited that polypeptide binding to Hsp104 requires NBD1 to be occupied with either ATP or the slowly hydrolyzed analog ATPγS (27). In other words, ATP occupancy of NBD1 can be probed by measuring binding of Hsp104 to a model polypeptide such as f-RCMLa. As shown in Fig. 4A, [+b] interacts with f-RCMLa already at low ATP concentrations, and the binding amplitude reaches saturation at ∼0.1 mm ATP. We also determined the ATPase activity of [+b] in the presence of polypeptide and included the normalized hydrolysis rates in the plot (Fig. 4A, filled circles). The two datasets superimpose, demonstrating that both, ATP hydrolysis and substrate binding are governed by the same event, ATP binding to NBD1. We then performed an analogous set of experiments using the [b+] mutant. As pointed out above, the ATPase activity of [b+] critically depends on the activation of NBD2 by ATP binding to NBD1. The increase in ATPase activity with increasing ATP concentration, as depicted in Fig. 4A, thus reflects an increase in ATP occupancy of the regulatory NBD1, and not of the hydrolytically active NBD2. As in the case of [+b], the datasets for substrate binding and ATP hydrolysis of [b+] were superimposable, consistent with the notion that both activation of NBD2 in [b+] and polypeptide binding depend on ATP binding to NBD1. But occupation of NBD1 in [b+] occurred at ∼10 times higher ATP concentrations (Fig. 4A), demonstrating that NBD1 has a lower affinity toward ATP in [b+] than in [+b]. We conclude that during steady-state ATP hydrolysis, NBD2 of [b+] is not occupied with ATP, but with ADP. Otherwise, this mutant should show an ATP dependence similar to [+b]. This suggests that the hydrolysis step in NBD2 is fast and the overall reaction rate is determined by the release of ADP and/or Pi.
FIGURE 4.
The nucleotide state of NBD2 modulates the affinity of NBD1 toward ATP. A, correlation between ATPase activity and polypeptide binding. ATPase rates of [+b] (●) and [b+] (▴) were recorded in the presence of 3 μm RCMLa. Binding of f-RCMLa (160 nm) to [+b] (○) and [b+] (△) was monitored by fluorescence anisotropy at 515 nm. Datasets were recorded at 30 °C and a hexamer concentration of 80 nm and normalized. Error bars reflect the S.D. of three independent experiments. B, ANS fluorescence of [−b] and [−+] in the absence and presence of saturating concentrations of ADP and ATP. 5 μm Hsp104 was preincubated with 100 μm ANS and mixed with buffer or 500 μm nucleotide. Error bars reflect the S.D. of three independent experiments.
Because Hsp104 contains no tryptophan residues, the nucleotide state of NBD2 cannot be determined directly by fluorescence spectroscopy. We therefore tested whether nucleotide binding to NBD2 can measured indirectly using the interaction of Hsp104 with the hydrophobic dye ANS. We found that ANS fluorescence is not only a sensitive probe for nucleotide binding to NBD2, but is able to distinguish between ADP and ATP. This is illustrated by the inactive [−b] variant, which shows different ANS signals for the apo, ADP, and ATP states, respectively (Fig. 4B). When we carried out a corresponding experiment with [−+], which carries an active NBD2, the apo and ADP signals were very similar to values measured for [−b] (Fig. 4B). Remarkably though, ATP gave the same signal in the [−+] mutant as ADP, suggesting that its NBD2 is occupied with ADP even in the presence of ATP. This finding supports our previous conclusion that for NBD2, ADP release constitutes the rate-limiting step of ATP hydrolysis and results in the accumulation of the ADP state.
Communication between NBD1 Domains Determines How Hsp104 Interacts with Polypeptides
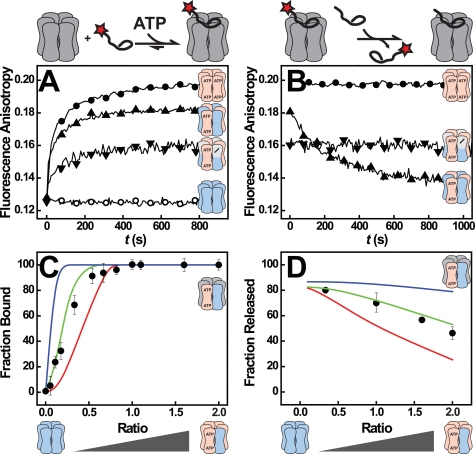
Our data imply that NBD1 serves as an allosteric center that controls ATP turnover in both NBD1 and NBD2. ATP binding to NBD1 also triggers binding of polypeptide substrates to Hsp104, whereas their release was shown to be induced by hydrolysis of ATP in either NBD1 or NBD2 (12). This immediately raised the question of whether the trans communication we have identified here is also involved in substrate binding.
In analogy to our ATPase experiments we tested whether the presence of [bb] subunits in a [++] hexamer affects the way Hsp104 interacts with polypeptide substrates such as f-RCMLa. As shown previously, wild-type Hsp104 [++] forms substrate complexes only when ATPγS is present, but not with ATP (supplemental Fig. S5B) (27). The [bb] mutant, on the other hand, binds polypeptides tightly in the presence of ATP but lacks the ATPase activity required for their release (Fig. 5, A and B) (27). Strikingly, [bb·++] exhibited properties of both parent homo hexamers. Similar to [bb] but unlike [++], [bb·++] interacted with f-RCMLa in the presence of ATP (Fig. 5A). And similar to [++] but in contrast to [bb], f-RCMLa could be displaced from [bb·++] complexes by nonfluorescent RCMLa (Fig. 5B). The dynamic substrate binding of [bb·++] hetero hexamers is not simply caused by a smaller number of binding competent subunits compared with [bb] homo hexamers, as [bb·−+] hexamers behaved very much trap-like, i.e. they bound f-RCMLa, albeit to a smaller extent, but were unable to release it even in the presence of a 20-fold excess of unlabeled RCMLa (Fig. 5, A and B). This leaves two other explanations: (i) The substrate is bound to [bb] subunits, and ATP hydrolysis in a neighboring NBD1 triggers its release. This is only possible in [bb·++], but not in [bb·−+] or [bb] hexamers. (ii) ATP-bound NBD1 of a [bb] subunit alters the properties of a neighboring NBD1 of a [++] subunit allowing it to adopt the high affinity state required for f-RCMLa binding even in the presence of ATP. Importantly, both scenarios require conformational coupling of NBD1 domains.
FIGURE 5.
Substrate binding and processing by Hsp104 hetero complexes. A, binding of 160 nm f-RCMLa to Hsp104 [++] (○), [bb] (●), [bb·++] (▴), and [bb·−+] (▾) (both at a mixing ratio of 1) in the presence of 1 mm ATP as monitored by fluorescence anisotropy at 515 nm. The Hsp104 concentrations were 160 nm for homo and 320 nm for hetero hexamers. B, release of bound f-RCMLa from Hsp104, induced by the addition of a 20-fold excess of unlabeled RCMLa. Symbols and conditions are as in A. C, quantitative analysis of f-RCMLa binding to [bb·++]. The lines in color represent the predicted amplitude for scenarios in which at least three (red), two (green), and one (blue) [bb] subunit(s) per hexamer are required for f-RCMLa binding. The observed amplitudes (●) are the means of three independent measurements. f-RCMLa (100 nm) and Hsp104 hexamer (160 nm) concentrations were kept constant. D, quantitative analysis of f-RCMLa release from [bb·++]. The lines in color represent the predicted amplitudes for scenarios in which at least three (red), two (green), and one (blue) [++] subunit(s) per hexamer are required for f-RCMLa release. The observed amplitudes are shown as (●).
Incorporation of [bb] subunits enables the Hsp104 [++] hexamer to interact with polypeptides in the presence of ATP. To determine how many [bb] subunits are needed for this gain of function, we formed [bb·++] complexes at various mixing ratios and examined to what extent these mixtures bound f-RCMLa (Fig. 5C). Based on the probability distribution in Fig. 1C, we calculated the concentrations of all possible hexamer types in the mixture at a given mixing ratio. Using a binding stoichiometry of one f-RCMLa molecule/hexamer (supplemental Fig. S5A), we then predicted the expected binding amplitude at each mixing ratio assuming that f-RCMLa binding requires one, two, three, etc. [bb] subunits/hexamer (Fig. 5C, colored lines). The observed increase in polypeptide binding with increasing amounts of [bb] subunits is best described by a model in which at least two [bb] subunits must be present to promote polypeptide binding (Fig. 5C, green curve). If a single [bb] protomer was sufficient, binding should already occur at much lower ratios (Fig. 5C, blue curve). In a corresponding experiment we determined the minimum number of [++] subunits required in a [bb·++] hexamer to allow release of a bound polypeptide (Fig. 5D). In striking homology with the binding requirement, we found that only [bb·++] complexes containing at least two [++] subunits release polypeptide.
During disaggregation Hsp104 extracts polypeptide chains from aggregates by threading polypeptides through its central channel. This translocation activity of Hsp104 requires that polypeptides are bound in dynamic fashion. Because our results indicate that only certain [bb·++] complexes bind and release f-RCMLa, we asked whether polypeptide translocation displayed a similar dependence on protomer composition. To this end, we employed a previously developed assay that monitors threading of polypeptides by Hsp104 in real time (12). As summarized in supplemental Fig. S6, these experiments confirm our observation that dynamic polypeptide binding to [bb·++] mixtures is restricted to hetero hexamers with a subunit composition of 4:2, 3:3, or 2:4. Complexes with more than four [bb] subunits no longer release bound polypeptides, whereas complexes containing more than four [++] protomers do not interact with polypeptides to begin with (supplemental Results).
Hsp104 Hetero-oligomers Can Reactivate Non-native Polypeptides in an Hsp70/40-independent Manner
To understand how Hsp104 function is controlled by its intrinsic allosteric properties, we also analyzed the impact of altered subunit communication on Hsp104-mediated disaggregation. The current model assigns Hsp70/40 an important role in this process because it either changes properties of the aggregate or of the disaggregation machine. This requirement for Hsp70/40 does not appear to be stringent because a fraction of the aggregated protein may be recovered by Hsp100 alone (31–33).
We assessed the ability of Hsp104 to reactivate aggregated FFL in the absence of Hsp70/40. In agreement with previous reports (12, 27), [++] alone did not disaggregate FFL, whereas [bb·++] hetero complexes were able to recover active FFL. When plotted against the mixing ratio, the rates and amplitudes of the reactivation reaction (Fig. 6AB and supplemental Fig. S7) strongly resembled the data we obtained for LaEYFP unfolding (cf. supplemental Fig. S6). It is noteworthy that also the Walker B variants [b+] and [+b] were able to disaggregate FFL in the absence of Hsp70/Hsp40 (supplemental Fig. S8). Apparently, Hsp70-independent disaggregation is tightly correlated with the ability of Hsp104 to bind and thread polypeptides, explaining why [bb·++] mixtures were active in FFL reactivation, but not [++].
FIGURE 6.
Reactivation of chemically denatured luciferase by Hsp104. Disaggregation of 55 nm FFL by [bb·++] mixtures (●) is shown at the indicated ratios. The concentration of [++] subunits was kept constant at 1 μm, whereas the concentration of [bb] subunits varied. Disaggregation was carried out at 3 mm ATP and 25 °C in the absence of Hsp70/40. Activity of renatured FFL was monitored continuously. The initial slope of FFL activity versus time was used as a measure of disaggregation velocity in FFL activity after 70 min (A) was used to assess the yield of the disaggregation reaction in B.
DISCUSSION
Protein unfolding and aggregation pose severe threats to living cells. Yeast Hsp104 and bacterial ClpB are class-1 AAA+ chaperones that rescue proteins from the aggregated state (11, 34, 35). To this end, Hsp104 converts energy provided by ATP hydrolysis into a mechanical force used to disrupt protein aggregates. Previous studies indicated that the ATPase of Hsp104 is subject to allosteric regulation (12, 36, 37). Yet it was not clear (i) which of the AAA+ modules in Hsp104 can communicate with each other and to what effect, (ii) whether allosteric cross-talk occurs within and/or between subunits (cis or trans), and (iii) what significance this allosteric communication has for the chaperone cycle. Using Hsp104 hexamers consisting of subunits with differing ATP binding and hydrolysis properties, we were able to map a sophisticated communication network that senses nucleotide states of AAA+ modules and transmits this information across the Hsp104 hexamer (Fig. 7). Three allosteric relations can be distinguished: (i) ATP in NBD1 inhibits ATP hydrolysis in other NBD1 domains, (ii) ATP in NBD1 stimulates ATP turnover in NBD2, and (iii) ATP in NBD2 increases the affinity of NBD1 for ATP.
FIGURE 7.
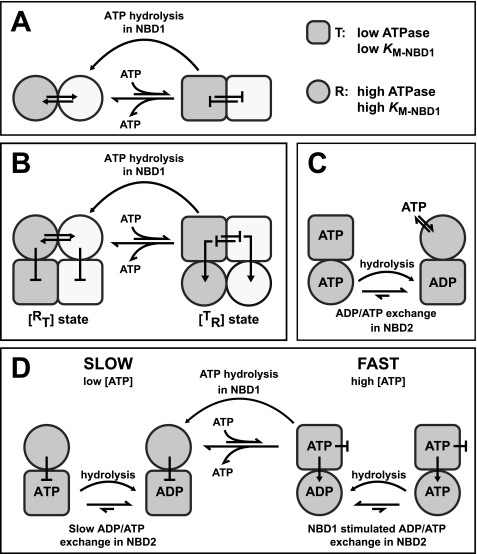
Model for the allosteric communication between AAA+ modules in Hsp104. A, conformational coupling between NBD1 domains is the mechanistic basis for the trans interaction of protomers and propagates nucleotide-induced structural rearrangements across the hexamer. NBD1 can adopt two states that differ in their ATPase activity: T (square, low activity) and R (circle, high activity). The transition between the two states is concerted, i.e. it occurs simultaneously in a number of NBD1s, two of which are depicted here in white and gray. ATP binding to NBD1 stabilizes the T state, whereas ATP hydrolysis induces the transition to the R state. B, the trans interaction of NBD1 co-ordinates the behavior of NBD2 in the Hsp104 hexamer. Depicted are two of six protomers in gray and white. Similar to NBD1 (top), NBD2 (bottom) can adopt T and R states with differing ATPase activity. However, the T↔ R transition in NBD2 is inversely linked to the T↔ R transition in NBD1. ATP binding to NBD1 induces a concerted transition of NBD1 to the T state. Adoption of the T state is propagated to NBD2 using a NBD1→NBD2 cis pathway. It triggers a conformational change of NBD2 to the R state and stimulates nucleotide exchange, thereby strongly activating ATP turnover in this domain. ATP hydrolysis in NBD1 triggers the co-operative transition of NBD1 to the R state and concomitantly causes NBD2 to revert to the T state. C, nucleotide occupancy in NBD2 modulates the affinity of NBD1 toward ATP. When NBD2 is occupied with ADP (or empty), NBD1 adopts an open conformation with a low affinity toward ATP (R state). Binding of ATP to NBD2 induces a conformation of NBD1 to which ATP binds more tightly (T state). ATP hydrolysis in NBD2 facilitates the transition of NBD1 to the R state. D, allosteric model for ATP hydrolysis in Hsp104 is shown. For details, see “Discussion.”
The first allosteric pathway interconnects the N-terminal AAA+ modules, NBD1. ATP hydrolysis in NBD1 was inhibited by incorporation of inactive, ATP-bound [b] NBD1 protomers. Conversely, the incorporation of nucleotide-free [−] NBD1s into [+b] hexamers stimulated ATPase activity in NBD1, demonstrating that the intrinsic ATPase of a given NBD1 depends on the nucleotide state of neighboring NBD1s. We conclude that NBD1 adopts two conformations, a T state with lower ATPase activity and an R state with higher ATPase activity (Fig. 7A). The R↔ T transition is concerted, i.e. it occurs simultaneously in a number (not necessarily all six) of protomers. ATP binding to NBD1 stabilizes the T state and slows down ATP hydrolysis in other NBD1s. The larger the number of subunits with ATP-bound NBD1s, the more likely the switch to the T state occurs. Conversely, ATP hydrolysis in NBD1 facilitates a concerted transition to the R state and increases the rate at which the other NBD1s hydrolyze ATP. Similar observations have been made for the hetero-oligomeric protease Yta10/12 (38), a class-2 AAA+ protein with only one NBD/protomer, suggesting a conserved mode of ATP hydrolysis in the NBD1 ring. We conclude that the intrinsic propensity of NBD1s to adopt the same conformational state is the mechanistic basis for interprotomer cross-talk and allows propagation of ATP-induced structural rearrangements across the hexamer.
The second allosteric signal generated by ATP binding to NBD1 is the stimulation of ATP turnover in NBD2, which has been noted previously but remained mechanistically unexplained (12, 26). In contrast to NBD1, which becomes inhibited, NBD2 responds with a pronounced increase in activity by ∼2 orders of magnitude. We propose that NBD2, like NBD1, adopts two states, T and R, with low and very high ATPase activity, respectively (Fig. 7B). This T↔ R transition and hence the ATPase activity of NBD2 is inversely linked to the T↔ R transition in NBD1: When NBD1 adopts the T state, NBD2 is in the R state and vice versa. Because of this inverse correlation, ATP binding to NBD1 (which stabilizes the T state in this domain) induces the R state in NBD2 and stimulates its ATPase activity. Although the NBD1→NBD2 pathway is intraprotomeric according to our model, the activation of NBD2 has nevertheless a strong trans component because the behavior of NBD1 is governed by the interprotomeric NBD1↔NBD1 interaction. NBD2 has low activity when the NBD1 ring is nucleotide free ([−+] or [−+·−b]) and gains maximum activity when all six NBD1s are occupied with ATP, as in [b+], [++·bb], and [b+·bb]. NBD2 activity is intermediate when the NBD1 ring is only partially occupied, although it appears to be irrelevant whether ATP is bound to the NBD1 of the stimulated NBD2 (cis), as in [b+·−b], or to an adjacent NBD1 (trans), as in [−+·bb]. Neither NBD1 ring configuration was able to activate NBD2 to its full extent. We conclude that rather than responding to ATP binding to a specific NBD1, NBD2 activation is coupled to the concerted T↔ R switch in NBD1. This linkage between the T↔ R switch in NBD1 and the conformational transition in NBD2 warrants that the activity of NBD2 is regulated in a concerted manner and responds to changes in the ATP occupancy of neighboring NBD1s. It also explains why NBD2 is sensitive to trans effects although cryo-EM data suggest that the structural interactions between adjacent NBD2 modules are very weak (39).
A third component of the allosteric network we have identified concerns the affinity of NBD1 toward ATP, which is modulated by the nucleotide state of NBD2. Our data show that ATP binding to NBD2 promotes ATP binding to NBD1, whereas ATP hydrolysis in NBD2 decreases the affinity of NBD1 toward ATP (Fig. 7C). In the context of our model, NBD2 facilitates concerted switching between T and R states of NBD1 by modulating its affinity for ATP.
What could a model for the ATPase cycle of Hsp104 look like that integrates the individual allosteric interactions? We suggest the mechanism outlined in Fig. 7D. At low ATP concentration, ATP binding occurs preferentially to NBD2, due to the low intrinsic Km of this domain (36). ATP is quickly converted to ADP, but the exchange of ADP against ATP is slow and hence rate-determining for the cycle. As a result, NBD2 is occupied predominantly with ADP, which inhibits ATP binding to NBD1 (Fig. 7C). NBD2 remains in the T state, and steady-state hydrolysis is slow (Fig. 7D, left cycle). With increasing ATP concentrations, NBD1 becomes increasingly occupied with ATP despite the destabilizing influence of NBD2. This induces a concerted allosteric transition in NBD1 (Fig. 7B), which in turn activates NBD2 by stimulating the nucleotide exchange reaction. Thus, ATP binding to NBD1 switches Hsp104 from the [RT] to the [TR] state (Fig. 7D, right cycle). Although the [b+] mutant can stay in the [TR] state with its highly active NBD2, this is not possible for wild-type Hsp104 due to its catalytically active NBD1, which forces the chaperone to revert to the [RT] state. However, the reader should be aware that the model in Fig. 7D is a simplification. Although ATP binding and hydrolysis in NBD1 allow Hsp104 to switch between [RT] and [TR] states, this is not an on/off switch in a mechanical sense. ATP binding and hydrolysis in NBD1 increase the probability that a given hexamer switches between the two states. Whether Hsp104 uses the slow, left branch of the ATPase cycle in Fig. 7D slow or fast branch on right is determined by the occupancy of NBD1 with ATP, which in turn depends on the levels of ATP and ADP as well as on the nucleotide state of NBD2. Under normal growth conditions, the ATP concentration in the yeast cytosol is in the low millimolar range whereas the ADP level is ∼5–10 times lower (40). The concentration of ADP is relevant because ADP is a competitive inhibitor (41) and thus increases the apparent Km for ATP. In view of these numbers, it is likely that the nucleotide state of NBD2 has a strong impact on the ATP occupancy of NBD1 and hence on the chaperone properties of Hsp104 in vivo. In particular, it opens the possibility that Hsp104 activity is regulated by intracellular ATP levels allowing its inactivation (i.e. switching to the slow branch) under conditions that are detrimental for protein refolding.
How does this model explain the polypeptide binding and threading properties of [bb], [++], and the respective hetero hexamers? Presumably, the NBD1 T state also represents the high affinity state for polypeptide binding. In [bb] homo hexamers, subunits readily adopt the T state in the presence of ATP, which leads to a tight binding of polypeptides. The switch to the low affinity R state is blocked (no ATP hydrolysis), and the bound polypeptide cannot be released. In [++], ATP hydrolysis in NBD1 antagonizes the population of the high affinity T state and prevents its interaction with polypeptides. Incorporation of [bb] subunits into a wild-type hexamer causes an overall increase in ATP turnover. The basis for this increase is complex. As pointed out above, protomers deficient in NBD1 hydrolysis stabilize the [TR] state. Thus, [bb·++] hetero hexamers display a decreased ATPase activity in NBD1 but an increased ATPase activity in NBD2. The latter effect must be dominating because the overall activity is increased. In essence, the incorporation of [bb] subunits shifts ATPase activity from NBD1 to NBD2. The reduced ATPase activity of NBD1 has a strong impact on polypeptide binding: It increases ATP occupancy of NBD1, which in turn activates the concerted transition to the T state in [bb·++] hetero hexamers. This is the molecular basis underlying the substrate binding and modeling activities that Wickner and co-workers observed for [bb·++] mixtures of E. coli ClpB (25). Our quantitative analysis suggests that in the case of Hsp104, at least two protomers must be in the NBD1·ATP state to establish polypeptide binding, whereas at least two protomers must remain wild type-like for effective polypeptide release and translocation. One explanation is that Hsp104 functions as a dimer of trimers, and polypeptide processing can only occur if each of the operating units contains both [++] and [bb] subunits. It so happens that [+b] hydrolyzes ATP with a Hill coefficient of ∼3, i.e. three ATP molecules in NBD1 are cleaved simultaneously (data not shown), suggesting that ATP hydrolysis is synchronized only among a subset of the six protomers. This would impose a structural asymmetry within the NBD1 ring, which has been proposed for a number of AAA+ proteins including Hsp104 (39, 42–44). Also, [b+] hydrolyzes ATP with a Hill coefficient of ∼3, in agreement with the linkage between the R↔ T transition in NBD1 and NBD2 activity. The reciprocal regulation of NDB1 and NBD2 implies that a similar asymmetry should also exist between the two AAA+ rings. Our model suggests that NBD2 is the main engine of Hsp104 and assigns NBD1 a more regulatory role in mediating the switch between [RT] and [TR] states. This is in agreement with previous findings that NBD2 mutations affect Hsp104 function more severely than corresponding mutations in NBD1 (36, 45). Hsp104 function in vivo requires both AAA+ modules to bind and hydrolyze ATP (46). However, the chemical energy from ATP appears to be used to different ends in both AAA+ modules. In NBD1, it drives a conformational cycle that turns ATP hydrolysis in NBD2 on and off and controls binding and release of polypeptide substrates. In NBD2, it generates the mechanical force necessary to disrupt non-native protein structures. Hence, NBD1 co-ordinates polypeptide binding and power-stroke generation during disaggregation.
Supplementary Material
Acknowledgments
We thank P. Menhorn, J. Buchner, and J. Bardwell for helpful discussions.

The on-line version of this article (available at http://www.jbc.org) contains Figs. S1–S8, Table 1, Experimental Procedures, Results, and additional references.
- NBD
- nucleotide binding domain
- ANS
- 8-anilino-1-naphthalene sulfonic acid
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- FFL
- firefly luciferase
- RCMLa
- reduced carboxymethylated α-lactalbumin
- f-RCMLa
- fluorescently labeled, reduced carboxymethylated α-lactalbumin.
REFERENCES
- 1. Ogura T., Wilkinson A. J. (2001) Genes Cells 6, 575–597 [DOI] [PubMed] [Google Scholar]
- 2. Erzberger J. P., Berger J. M. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 [DOI] [PubMed] [Google Scholar]
- 3. Parsell D. A., Kowal A. S., Singer M. A., Lindquist S. (1994) Nature 372, 475–478 [DOI] [PubMed] [Google Scholar]
- 4. Sanchez Y., Lindquist S. L. (1990) Science 248, 1112–1115 [DOI] [PubMed] [Google Scholar]
- 5. Motohashi K., Watanabe Y., Yohda M., Yoshida M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7184–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., Bukau B. (2004) Cell 119, 653–665 [DOI] [PubMed] [Google Scholar]
- 7. Tessarz P., Mogk A., Bukau B. (2008) Mol. Microbiol. 68, 87–97 [DOI] [PubMed] [Google Scholar]
- 8. Glover J. R., Lindquist S. (1998) Cell 94, 73–82 [DOI] [PubMed] [Google Scholar]
- 9. Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beinker P., Schlee S., Auvula R., Reinstein J. (2005) J. Biol. Chem. 280, 37965–37973 [DOI] [PubMed] [Google Scholar]
- 11. Bösl B., Grimminger V., Walter S. (2006) J. Struct. Biol. 156, 139–148 [DOI] [PubMed] [Google Scholar]
- 12. Schaupp A., Marcinowski M., Grimminger V., Bösl B., Walter S. (2007) J. Mol. Biol. 370, 674–686 [DOI] [PubMed] [Google Scholar]
- 13. Schlieker C., Tews I., Bukau B., Mogk A. (2004) FEBS Lett. 578, 351–356 [DOI] [PubMed] [Google Scholar]
- 14. Weber-Ban E. U., Reid B. G., Miranker A. D., Horwich A. L. (1999) Nature 401, 90–93 [DOI] [PubMed] [Google Scholar]
- 15. Kenniston J. A., Baker T. A., Fernandez J. M., Sauer R. T. (2003) Cell 114, 511–520 [DOI] [PubMed] [Google Scholar]
- 16. Navon A., Goldberg A. L. (2001) Mol. Cell 8, 1339–1349 [DOI] [PubMed] [Google Scholar]
- 17. Benaroudj N., Goldberg A. L. (2000) Nat. Cell Biol. 2, 833–839 [DOI] [PubMed] [Google Scholar]
- 18. Wendler P., Shorter J., Plisson C., Cashikar A. G., Lindquist S., Saibil H. R. (2007) Cell 131, 1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rouiller I., DeLaBarre B., May A. P., Weis W. I., Brunger A. T., Milligan R. A., Wilson-Kubalek E. M. (2002) Nat. Struct. Biol. 9, 950–957 [DOI] [PubMed] [Google Scholar]
- 20. Martin A., Baker T. A., Sauer R. T. (2005) Nature 437, 1115–1120 [DOI] [PubMed] [Google Scholar]
- 21. Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S. (2004) Cell 119, 47–60 [DOI] [PubMed] [Google Scholar]
- 22. Beuron F., Dreveny I., Yuan X., Pye V. E., McKeown C., Briggs L. C., Cliff M. J., Kaneko Y., Wallis R., Isaacson R. L., Ladbury J. E., Matthews S. J., Kondo H., Zhang X., Freemont P. S. (2006) EMBO J. 25, 1967–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S., Sowa M. E., Watanabe Y. H., Sigler P. B., Chiu W., Yoshida M., Tsai F. T. (2003) Cell 115, 229–240 [DOI] [PubMed] [Google Scholar]
- 24. Werbeck N. D., Schlee S., Reinstein J. (2008) J. Mol. Biol. 378, 178–190 [DOI] [PubMed] [Google Scholar]
- 25. Hoskins J. R., Doyle S. M., Wickner S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22233–22238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Watanabe Y. H., Motohashi K., Yoshida M. (2002) J. Biol. Chem. 277, 5804–5809 [DOI] [PubMed] [Google Scholar]
- 27. Bösl B., Grimminger V., Walter S. (2005) J. Biol. Chem. 280, 38170–38176 [DOI] [PubMed] [Google Scholar]
- 28. Bendz H., Ruhland S. C., Pandya M. J., Hainzl O., Riegelsberger S., Braüchle C., Mayer M. P., Buchner J., Issels R. D., Noessner E. (2007) J. Biol. Chem. 282, 31688–31702 [DOI] [PubMed] [Google Scholar]
- 29. Grimminger V., Richter K., Imhof A., Buchner J., Walter S. (2004) J. Biol. Chem. 279, 7378–7383 [DOI] [PubMed] [Google Scholar]
- 30. Beinker P., Schlee S., Groemping Y., Seidel R., Reinstein J. (2002) J. Biol. Chem. 277, 47160–47166 [DOI] [PubMed] [Google Scholar]
- 31. Doyle S. M., Shorter J., Zolkiewski M., Hoskins J. R., Lindquist S., Wickner S. (2007) Nat. Struct. Mol. Biol. 14, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S., Sielaff B., Lee J., Tsai F. T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8135–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sielaff B., Tsai F. T. (2010) J. Mol. Biol. 402, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haslberger T., Bukau B., Mogk A. (2010) Biochem. Cell Biol. 88, 63–75 [DOI] [PubMed] [Google Scholar]
- 35. Barends T. R., Werbeck N. D., Reinstein J. (2010) Curr. Opin Struct. Biol. 20, 46–53 [DOI] [PubMed] [Google Scholar]
- 36. Hattendorf D. A., Lindquist S. L. (2002) EMBO J. 21, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schirmer E. C., Ware D. M., Queitsch C., Kowal A. S., Lindquist S. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Augustin S., Gerdes F., Lee S., Tsai F. T., Langer T., Tatsuta T. (2009) Mol. Cell 35, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wendler P., Shorter J., Snead D., Plisson C., Clare D. K., Lindquist S., Saibil H. R. (2009) Mol. Cell 34, 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Osorio H., Carvalho E., del Valle M., Günther Sillero M. A., Moradas-Ferreira P., Sillero A. (2003) Eur. J. Biochem. 270, 1578–1589 [DOI] [PubMed] [Google Scholar]
- 41. Schirmer E. C., Queitsch C., Kowal A. S., Parsell D. A., Lindquist S. (1998) J. Biol. Chem. 273, 15546–15552 [DOI] [PubMed] [Google Scholar]
- 42. Glynn S. E., Martin A., Nager A. R., Baker T. A., Sauer R. T. (2009) Cell 139, 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cha S. S., An Y. J., Lee C. R., Lee H. S., Kim Y. G., Kim S. J., Kwon K. K., De Donatis G. M., Lee J. H., Maurizi M. R., Kang S. G. (2010) EMBO J. 29, 3520–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hersch G. L., Burton R. E., Bolon D. N., Baker T. A., Sauer R. T. (2005) Cell 121, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 45. Lum R., Tkach J. M., Vierling E., Glover J. R. (2004) Journal of Biological Chemistry 279, 29139–29146 [DOI] [PubMed] [Google Scholar]
- 46. Parsell D. A., Sanchez Y., Stitzel J. D., Lindquist S. (1991) Nature 353, 270–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.