Abstract
The HECT-type ubiquitin ligase (E3) Smad ubiquitination regulatory factor 1 (Smurf1) targets various substrates, including Smad1/5, RhoA, Prickle 1, MEKK2, and JunB for degradation and thereby regulates adult bone formation and embryonic development. Here, we identify the endoplasmic reticulum (ER)-localized Wolfram syndrome protein (WFS1) as a specific degradation substrate of Smurf1. Mutations in the WFS1 gene cause Wolfram syndrome, an autosomal recessive disorder characterized by diabetes mellitus and optic atrophy. WFS1 negatively regulates the ER stress response, and WFS1 deficiency in mice increases ER stress and triggers apoptosis. We show that Smurf1 interacts with WFS1 at the ER and promotes the ubiquitination and proteasomal degradation of WFS1. A C-terminal luminal region in WFS1, including residues 667–700, is involved in this degradation. Wild-type WFS1 as well as a subset of WFS1 mutants that include this degron region are susceptible to Smurf1-mediated degradation. By contrast, pathophysiological deletion mutants of WFS1 lacking the degron, such as W648X, Y660X, and Q667X, are resistant to degradation by Smurf1. Depletion of Smurf1 by RNA interference results in increased WFS1 and decreased ATF6α levels. Furthermore, we show that ER stress induces Smurf1 degradation and WFS1 up-regulation. These findings reveal for the first time that Smurf1 targets an ER-localized protein for degradation and that Smurf1 is regulated by ER stress.
Keywords: Endoplasmic Reticulum (ER), Protein Degradation, Protein Stability, Ubiquitin Ligase, Ubiquitination, Smurf1 Ubiquitin Ligase, Wolfram Syndrome
Introduction
Ubiquitin-mediated proteasomal degradation represents the most critical pathway to control the stability and quality of cellular proteins in eukaryotes. Ubiquitin ligases (also called E3) are responsible for substrate recognition and are divided into two major classes: RING finger-type and HECT domain-type (1). Smurf1 belongs to the Nedd4 (neuronal precursor cell-expressed developmentally down-regulated 4) family of HECT-type E3 ligases and plays a critical role in the regulation of embryonic development, cell polarity, and bone homeostasis by targeting the degradation of Smad1/5, TGFβ receptor (TGFβR), RhoA, MEKK2, Prickle 1, and JunB (2–7). TGFβR and RhoA localize to the plasma membrane, Smad1/5, MEKK2, and Prickle 1 localize in the cytoplasm, and JunB is localized in the nucleus. The subcellular localization of Smurf1 in mammalian cells is dynamic under different spatiotemporal contexts. Smurf1 shuttles between the inside and outside of the nucleus (8). Treatment with the CRM1 inhibitor, leptomycin B, results in the retention of Smurf1 in the nucleus. The inhibitory Smad member, Smad7, also helps to export Smurf1 from the nucleus to the cytoplasm and the plasma membrane. Through this mechanism, Smurf1 targets local substrates for degradation and turns off bone morphogenetic protein/TGFβ signaling. The N-terminal C2 domain of Smurf1 also plays a role in its distribution, because the C2 domain possesses the ability to interact with phospholipids in the plasma membrane (8). The recently identified auxiliary factor of Smurf1, CKIP-1 (CK2 interacting protein-1), can enrich Smurf1 at the plasma membrane and promote the degradation of substrates at or close to the plasma membrane, including Smad1/5, MEKK2, and RhoA (9).
As a typical ubiquitin ligase, Smurf1, can mediate self-ubiquitination and then auto-degradation, like most ligases. Unlike its close relative Smurf2, which exhibits intramolecular C2-HECT autoinhibition and thereby is inactive without further activators (10), Smurf1 retains a relatively high level of constitutive ubiquitin ligase activity, although its full activation can be gained by the interaction with CKIP-1 (9). Whether the ligase activity of Smurf1 is regulated by certain physiological or pathological stimuli and stress remains poorly understood.
In this study we found that Smurf1 is significantly and specifically degraded by the proteasome upon ER stress, implicating a possible role for Smurf1 in the ER stress response. Further screening of Smurf1 targets by yeast two-hybrid identified the Wolfram syndrome protein (WFS1) as a specific substrate of Smurf1 at the ER.3
Mutations in the WFS1 gene are the most frequent genetic cause of Wolfram syndrome, which is an autosomal recessive disorder leading to juvenile-onset insulin-dependent diabetes mellitus, optic atrophy, sensorineural deafness, and diabetes (11–13). Wolfram syndrome was first reported in 1938 (14), and the first mutations in the WFS1 gene were identified in Wolfram syndrome patients in 1998 (15). The WFS1 protein contains a cytoplasmic N-terminal domain, a central nine-transmembrane domain, and a luminal C terminus, and the protein is predominantly localized in the ER (16). WFS1 mRNA and protein levels increase upon ER stress, partially through transcriptional activation (17, 18). WFS1 then negatively regulates ER stress signaling by stabilizing the E3 ligase HRD1, recruiting ATF6α (activating transcription factor 6α, a key transcription factor to activate unfolded protein response target genes) to HRD1, and enhancing its ubiquitination and proteasomal degradation (19). WFS1-deficient mice exhibit impaired glucose homeostasis, defective insulin secretion, increased apoptosis of pancreatic islet cells due to enhanced ER stress, and an increase in the unfolded protein response (20–22). Therefore, WFS1 is critical in maintaining ER homeostasis. To date, little is known about how the stability of WFS1 is controlled, especially how it is down-regulated. In this study we provide evidence that the ubiquitin ligase Smurf1 promotes the ubiquitination and degradation of WFS1 and that certain mutations in WFS1 result in resistance against this regulation.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Full-length WFS1 cDNA was a kind gift from Dr. Fumihiko Urano. 6Myc-Smurf1 wild type, 6Myc-Smurf1-C699A, and FLAG-Smurf1 were provided by Dr. Kohei Miyazono. Hemagglutinin (HA)-tagged ubiquitin was a gift from Dr. Yue Xiong. Truncating and missense mutations of WFS1 were created by inserting PCR-amplified fragments into the related vectors. Constructs of other Nedd4 family members were kindly provided by Dr. Wesley I. Sundquist and described previously (23).
Yeast Two-hybrid Screening
A yeast two-hybrid screen was performed in a human liver cDNA library with the ProQuestTM two-hybrid system (Invitrogen) as described previously (24). Briefly, the WW domains (amino acids 236–340) of human Smurf1 were cloned in-frame with the DNA binding domain (DB) of GAL4 in pDBLeu. MaV203 yeast cells were transformed with Smurf1-WW and the library, which was inserted into the pPC86 vector. Approximately 1 × 106 independent transformants were analyzed. Clones were selected for positive interactions when the candidate DB-Smurf1-WW-AD-Prey pairs activated at least two of three promoters: His, LacZ, and URA3. Positive clones were retested in fresh yeast cells, and the AD-Prey identities were determined with interaction sequence tags by DNA sequencing. The AD-Prey reading frame was verified to avoid the recovery of out-of-frame peptides.
Cell Culture and Transfection
Human embryonic kidney HEK293T cells, human breast cancer MCF7 cells, mouse embryonic fibroblast NIH3T3 cells, and mouse osteoblastic cells MC3T3-E1 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). Human lung adenocarcinoma H1299 cells and rat insulinoma cells INS1 were maintained in RPMI 1640 medium (Hyclone) with 10% FBS. Cells were transfected with Lipofectamine 2000 following the manufacturer's protocol (Invitrogen).
Antibodies and Reagents
The protein synthesis inhibitor cycloheximide, lysosome inhibitor bafilomycin A1, proteasome inhibitors MG132 and lactacystin, ER stress inducers thapsigargin (Tg) and tunicamycin (Tm), and anti-FLAG, anti-NEDL1, and anti-NEDL2 antibodies were purchased from Sigma. Anti-Smurf1, anti-Smurf2, and anti-WFS1 antibodies were purchased from Abcam. The anti-HA antibody was from Roche Applied Science, and the anti-Myc antibody was from MBL. Anti-GAPDH, anti-p53, anti-CHOP, anti-ubiquitin, anti-ATF6α and secondary antibodies were purchased from Santa Cruz Biotechnology.
Immunoprecipitation and Immunoblotting
Forty-eight hours post-transfection cell lysates were prepared in HEPES lysis buffer (20 mm HEPES, pH 7.2, 50 mm NaCl, 0.5% Triton X-100, 1 mm NaF, and 1 mm DTT) supplemented with protease inhibitor mixture (Roche Applied Science). Immunoprecipitations were performed using the indicated primary antibody and protein A/G-agarose beads (Santa Cruz) at 4 °C. Lysates and immunoprecipitates were examined using the indicated primary antibodies followed by detection with the related secondary antibody and the SuperSignal chemiluminescence kit (Pierce).
In Vivo and in Vitro Ubiquitination Assays
For the in vivo ubiquitination assay, cells were treated with lactacystin (30 μm) for 16 h before harvest to avoid the proteasome-mediated degradation. The cell lysate was prepared in HEPES lysis buffer supplemented with protease inhibitors, and proteins were immunoprecipitated with the indicated antibody and detected by immunoblotting with anti-ubiquitin. For the in vitro ubiquitination assay, E1, UbcH5c (E2), HA-Ub (all from Boston Biochem), His-Smurf1 (expressed in bacteria and purified), and FLAG-WFS1 (expressed in HEK293T cells and purified by immunoprecipitation with an anti-FLAG antibody) were incubated at 30 °C for 2 h, and the assay was terminated with sample buffer.
RNA Interference
The Smurf1 siRNA-A (5′-GGGCUCUUCCAGUAUUCUATT-3′), siRNA-B (5′-GCAUCGAAGUGUCCAGAGAAG-3′), and non-targeting siRNAs (5′-UUCUCCGAACGUGUCACGU-3′) were synthesized by Shanghai GenePharm. All siRNAs were transfected into the cells according to the manufacturer's protocol.
Real-time RT-PCR
Total RNA was isolated from the cells using TRIzol (Invitrogen) and reversed-transcribed using 1 μg of total RNA with an oligo-dT primer. The following primers were used for real-time PCR: human GAPDH forward, 5′-GGGAAGGTGAAGGTCGGAGT-3′; GAPDH reverse, 5′-TTGAGGTCAATGAAGGGGTCA-3′; human WFS1 forward, 5′-GTTCCCGACTCAATGCCACA-3′; and WFS1 reverse, 5′-CCGCTGCGTCTCTAACACC-3′.
Fluorescence Analysis
For detection of colocalization by immunofluorescence, cells were fixed with 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 (PBS). Proteins were stained using the indicated antibodies and detected with a TRITC-conjugated or FITC-conjugated secondary antibody. The nuclei were stained with DAPI (Sigma), and images were visualized with a Zeiss LSM 510 Meta inverted confocal microscope.
RESULTS
ER Stress Induces Smurf1 Degradation
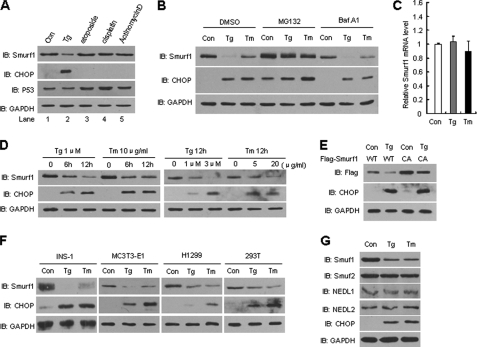
During the analysis of Smurf1 steady-state levels upon various stresses, we found that treatment of cells with Tg, an ER Ca2+ pump inhibitor (18), resulted in the down-regulation of endogenous Smurf1 protein levels (Fig. 1A, second lane). Tg treatment induced ER stress, as indicated by the dramatic increase in CHOP expression, a well defined ER stress response marker. By contrast, DNA damage stress induced by etoposide (third lane) or cisplatin (fourth lane) and ribosomal stress induced by actinomycin D (fifth lane) had no significant effects on the expression levels of Smurf1 or CHOP, although they induced the accumulation of the tumor suppressor p53 as expected. To verify that the down-regulation of Smurf1 was in response to ER stress, another ER stress inducer Tm, an N-glycosylation inhibitor (18), was used to treat the cells. Similarly, we observed the down-regulation of Smurf1 accompanied by the up-regulation of CHOP in Tm-treated cells (Fig. 1B, third lane). The ER stress-triggered Smurf1 down-regulation was blocked by treatment with the proteasome inhibitor MG132 but not by the lysosome inhibitor bafilomycin A1 (Fig. 1B), indicating that Smurf1 protein levels are regulated by proteasome-mediated protein degradation. Indeed, the mRNA level of Smurf1 was not affected by these stress inducers (Fig. 1C). Tg or Tm treatment induced Smurf1 degradation in a dose- and time-dependent manner (Fig. 1D). To further substantiate the role of Smurf1 E3 ligase activity in degradation, wild-type FLAG-Smurf1 (WT) or a ligase-inactive C699A mutant were expressed and stimulated with Tg. As shown in Fig. 1E, ER stress only slightly inhibited the degradation of the C699A Smurft1 mutant, implicating that ER stress triggers the degradation of Smurf1 largely independent of its ligase activity. Importantly, the ER stress-induced Smurf1 degradation was detected in INS-1, MC3T3-E1, H1299, and HEK293T cells (Fig. 1F). This regulation of Smurf1 is also highly specific, because other members of the Nedd4 family, including Smurf2, NEDL1, and NEDL2, were not down-regulated by ER stress (Fig. 1G). Taken together, these results indicate that ER stress induces the proteasomal degradation of Smurf1 independently of cell type.
FIGURE 1.
ER stress induces specific degradation of Smurf1. A, ER stress down-regulates Smurf1. NIH3T3 cells were treated for 12 h with each of the indicated drugs at the following concentrations: thapsigargin (1 μm), etoposide (40 μm), cisplatin (100 μm), and actinomycin D (0.4 g/ml). Whole cell extracts were analyzed by immunoblotting with anti-Smurf1, CHOP, p53, and GAPDH antibodies. The expression of CHOP was used as an indication of the UPR. B, ER stress induces proteasome-mediated degradation of Smurf1. NIH3T3 cells cultured in 12-well plates were treated for 12 h with the ER stress inducers thapsigargin (1 μm) or tunicamycin (10 μg/ml) with or without 40 μm MG132 (proteasome inhibitor) or 10 μg/ml Baf A1 (lysosomal inhibitor). Immunoblots were probed using Smurf1, CHOP, and GAPDH antibodies. C, ER stress does not influence Smurf1 mRNA levels. Total RNA was extracted from NIH3T3 cells treated with 1 μm Tg or 10 μg/ml Tm for 12 h. Smurf1 and GAPDH mRNA levels were determined by quantitative real-time PCR. D, regulation of Smurf1 protein levels by ER stress in a dose- and time-dependent manner is shown. NIH3T3 cells were treated with Tg or Tm at the indicated concentrations and times. Whole cell extracts were prepared, and the expression of endogenous Smurf1, CHOP, and GAPDH was analyzed by Western blot. E, both wild-type and C699A mutant Smurf1 protein levels were affected by ER stress. After 24 h of transfection with FLAG-tagged WT Smurf1 or C699A (CA, ligase-inactive mutant), cells were treated with 1 μm Tg for 12 h. Smurf1 protein expression was analyzed. F, ER stress induces degradation of Smurf1 independent of cell type. INS-1, MC3T3-E1, H1299, and 293T cells were treated with 1 μm Tg or 10 μg/ml Tm for 12 h followed by immunoblotting of cell extracts with Smurf1, CHOP, and GAPDH antibodies. G, NIH3T3 cells were treated for 12 h with ER stress inducers Tg (1 μm) and Tm (10 μg/ml). Protein extracts were immunoblotted with Smurf1, Smurf2, NEDL1, NEDL2, CHOP, and GAPDH antibodies.
Smurf1 Interacts with WFS1
To investigate the possible role of Smurf1 in the ER stress response, we sought to identify Smurf1 targets. A yeast two-hybrid screen was performed in a human liver library using the WW domains of Smurf1 as bait because the WW domains are responsible for substrate recognition (25). One of the candidate interactors of Smurf1 was WFS1, and the prey clone encoded the N-terminal 260 residues of WFS1 (Fig. 2A). The N-terminal part of WFS1 represents the cytoplasmic segment. At present, no known protein domains have been identified within the WFS1 protein. Additionally, WFS1 does not possess a PPXY motif, which is the typical recognition motif for Nedd4 family ligases (25). We confirmed the interaction between WFS1 and Smurf1 in mammalian cells. As shown in Fig. 2B, endogenous WFS1 co-immunoprecipitated with endogenous Smurf1, but not control IgG, from NIH3T3 cells. A further co-immunoprecipitation assay revealed that WFS1 associated with the ligase-inactive C699A mutant and wild-type Smurf1 (Fig. 2C), suggesting that the E3 ligase activity is not required for the interaction.
FIGURE 2.
Smurf1 interacts with WFS1. A, the “bait” and “prey” regions of Smurf1 and WFS1, respectively, used in yeast two-hybrid screening are shown. B, co-immunoprecipitation (IP) of endogenous Smurf1 and WFS1 is shown. NIH3T3 cell lysates were prepared and subjected to immunoprecipitation with Smurf1 antibody or IgG and analyzed by immunoblotting using Smurf1 or WFS1 antibodies. C, E3 ligase activity is not required for the interaction between Smurf1 and WFS1. WT or C699A (CA) Smurf1 was co-expressed with WFS1 in HEK293T cells. Smurf1 was immunoprecipitated, and WFS1 protein was detected by immunoblotting. D, co-localization of Smurf1 and WFS1 in the ER of H1299 cells is shown. Myc-Smurf1 and FLAG-WFS1 were transfected into H1299 cells, and indirect immunofluorescence analysis was performed. The cells were visualized by confocal microscopy, and nuclei were stained with DAPI. E, shown is mapping the Smurf1 binding region on WFS1. Cell lysates from HEK293T cells transfected with FLAG-tagged Smurf1 and Myc-tagged WFS1 deletion mutants were immunoprecipitated with anti-FLAG followed by immunoblotting with anti-FLAG or anti-Myc. F, shown is a schematic representation of WFS1-Smurf1 interaction in co-immunoprecipitation assay. TM, transmembrane. G, FLAG-tagged WFS1-N or WFS1-ΔC truncate was transfected into H1299 cells, and indirect immunofluorescence analysis was performed. The ER was stained with anti-PDI antibody. H, GST pulldown assays of Smurf1 with GST or GST-WFS1 truncates as indicated are indicated. PDI, protein disulfide isomerase.
To be functionally linked, proteins must colocalize at least in part. Indirect immunofluorescence assays revealed that WFS1 and Smurf1 colocalized predominantly at the ER (Fig. 2D). To map the Smurf1-interacting region in WFS1, a series of WFS1 deletion mutants were generated. Both the N-terminal cytoplasmic part (Fig. 2A) and the C-terminal ER luminal part of WFS1 contained the Smurf1-interacting information (Fig. 2, E and F). However, if the N-terminal part was expressed alone in cells, it could not co-immunoprecipitate with Smurf1 (Fig. 2E, second lane). Careful examination of WFS1-N localization revealed that it was predominantly localized to the nucleus (Fig. 2G), explaining why it did not bind to Smurf1 in co-immunoprecipitation assays. Additionally, a GST pulldown assay showed that both the WFS1-N and the WFS1-C interacted with Smurf1 in vitro (Fig. 2H). These results suggest that within the full-length WFS1 protein, both the N- and the C-terminal parts contain the Smurf1-interacting module or motif.
Smurf1 Targets WFS1 for Ubiquitination and Proteasomal Degradation
The fact that the WW domains of Smurf1 are usually responsible for substrate recognition prompted us to investigate whether WFS1 is a substrate of Smurf1. Overexpressed wild-type Smurf1, but not the ligase-inactive C699A mutant, drastically decreased the protein levels of WFS1 (Fig. 3A). The dose-dependent down-regulation of WFS1 by Smurf1 was blocked by treatment with lactacystin, an inhibitor of the proteasome (Fig. 3B). Depletion of endogenous Smurf1 in MCF7 and H1299 cells resulted in a significant increase in WFS1 protein levels (Figs. 3, C and E) but had no significant effects on the mRNA levels of WFS1 (Fig. 3, D and F). Importantly, the regulation of WFS1 by Smurf1 was specific. Among all nine members of the Nedd4 family ligases, only Smurf1 specifically down-regulated WFS1 protein levels (Fig. 3G).
FIGURE 3.
Smurf1 negatively regulates WFS1 protein levels. A, Smurf11 expression decreases the level of WFS1 protein. HEK293T cells were transfected with a constant amount of WFS1 together with increasing amounts of Smurf1 WT or C699A. After 48 h, cells were lysed, and steady-state protein levels were determined by immunoblotting. B, Smurf1 down-regulates WFS1 in a proteasome-dependent manner. Increasing amounts of FLAG-Smurf1 plasmids were co-transfected with Myc-WFS1 into HEK293T cells. Cells were treated with lactacystin (30 μm) or DMSO for 16 h, and the cell lysates were analyzed by immunoblotting. C and E, knock-down of Smurf1 by siRNA transfection both in MCF7 (C) and H1299 (E) cells increased the expression of WFS1 protein. D and F, knock-down of Smurf1 did not affect the expression level of WFS1 mRNA in MCF7 (D) or H1299 (F) cells, as determined by RT-PCR. G, Smurf1 decreases WFS1 protein specifically. The indicated Nedd4 family of E3 ligases were co-transfected with WFS1 into HEK293T cells, and cell lysates were analyzed.
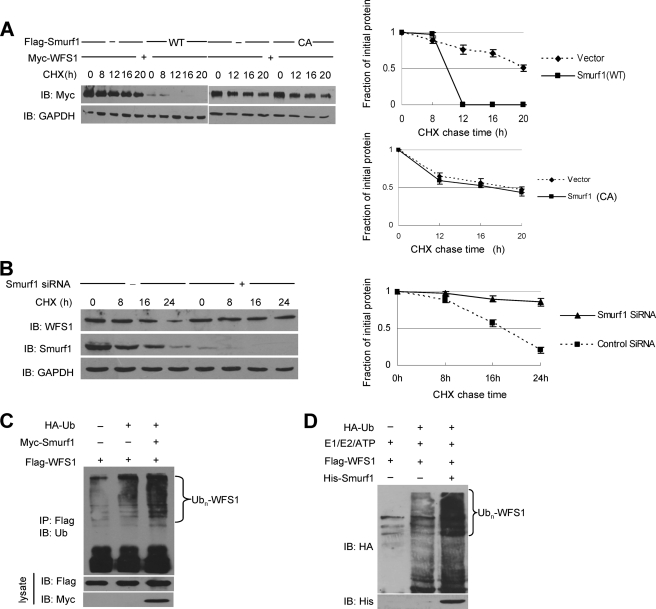
To confirm whether Smurf1 affected WFS1 protein levels by reducing its stability, we measured steady-state levels of WFS1 in the presence or absence of Smurf1 using the cycloheximide (CHX) chase assay. The half-life of exogenously expressed WFS1 was greatly reduced by expression of wild-type Smurf1 but not by expression of the ligase-inactive Smurf1 mutant (Fig. 4A). Importantly, marked stabilization of endogenous WFS1 protein was observed with siRNA against Smurf1, compared with a scrambled siRNA (Fig. 4B), suggesting that the stability of endogenous WFS1 protein was controlled by Smurf1 ligase.
FIGURE 4.
Smurf1 promotes the ubiquitination and proteasomal degradation of WFS1. A, the effect of Smurf1 on the half-life of WFS1 is shown. Smurf1 WT or C699A (CA) was transfected together with a WFS1 expression vector, and cells were treated with CHX at 10 μg/ml for the indicated times. The half-life of WFS1 was measured by Western blot. B, CHX-chase experiments of WFS1 in MCF7 cells transfected with scrambled siRNA or Smurf1 siRNA are shown. Cells were treated with CHX at 10 μg/ml for the indicated times. The half-life of WFS1 was measured by a Western blot. C, Smurf1 promotes WFS1 ubiquitination in mammalian cells. HEK293T cells were transfected with HA-tagged ubiquitin (HA-Ub), FLAG-tagged WFS1, control vector, or Myc-tagged Smurf1. Sixteen hours before cell harvest, the cells were treated with the potent proteasome inhibitor lactacystin (30 μm) to avoid the proteasome-mediated degradation. Cell lysates were then prepared and immunoprecipitated with an anti-FLAG antibody. The immunoprecipitates (IP) were analyzed by immunoblotting with the anti-ubiquitin antibody to indicate the polyubiquitinated WFS1. The lysates were also analyzed by immunoblotting with anti-FLAG and anti-Myc antibodies. D, Smurf1 enhances WFS1 ubiquitination in vitro. E1, UbcH5c (E2), HA-Ub (all from Boston Biochem), His-Smurf1 (expressed in bacteria and purified), and FLAG-WFS1 (expressed in HEK293T cells and purified by immunoprecipitation with an anti-FLAG antibody) were incubated at 30 °C for 2 h in ubiquitination buffer. Ubiquitinated WFS1 was visualized by immunoblotting with an anti-HA antibody.
We next asked whether Smurf1 functions as an E3 ligase to promote the ubiquitination of WFS1. Overexpressed Smurf1 enhanced the polyubiquitination of WFS1 in cultured mammalian cells (Fig. 4C). In addition, bacteria-expressed and purified Smurf1 protein promoted the polyubiquitination of WFS1 directly in vitro (Fig. 4D). Taken together, these data indicate that Smurf1 acts as a biologically relevant E3 ligase for WFS1 to promote the ubiquitination and degradation of WFS1.
Differential Stability of Various WFS1 Mutants
We next investigated which region of WFS1 is critical for Smurf1-mediated degradation. To maintain the ER localization of WFS1 truncates, we utilized ΔN and ΔC forms of WFS1 to analyze their protein levels in the presence of increasing doses of Smurf1. As shown in Fig. 5A, ΔN was down-regulated by Smurf1, whereas ΔC was resistant to this degradation, suggesting that the C-terminal part, which extends to the ER lumen, was critical for Smurf1-mediated degradation.
FIGURE 5.
Differential stability of wild-type WFS1 and various mutants. A, the C-terminal part of WFS1 is important for degradation by Smurf1. HEK293T cells were transfected with Smurf1 and deletion mutants of WFS1, and the cell lysates were analyzed by immunoblot analysis. B, shown is a half-life analysis of wild-type and mutant WFS1. Wild-type and the indicated WFS1 mutants were transfected into HEK293T cells, and the cells were treated with a 40 μg/ml cycloheximide chase for the indicated times. The half-lives of wild-type and WFS1 mutants were determined by Western blot. Data are the mean ± S.D. (n = 3). Quantitative analysis was performed by measuring integrated optical density using the program Gel-Pro analyzer. The exposure time of the Western blot analysis of certain mutants was adjusted to show a comparable expression level at time point zero (lane 1 of each group).
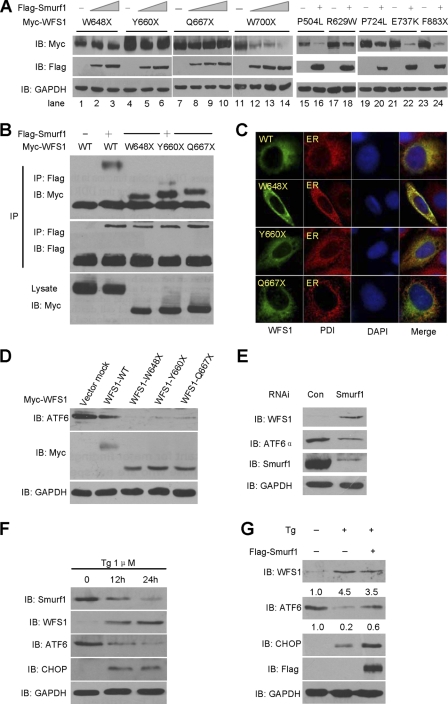
We further analyzed a series of WFS1 mutants that are mutated in the C-terminal part. Seven point mutation mutants were randomly selected. Among them, five represent non-sense mutations leading to deletion mutants lacking different lengths of the extreme C terminus, and they include: W648X, Y660X, Q667X, W700X, F883X (Fig. 5B). Two other mutants, R629W and P724L, represent the missense mutations, and these mutations cause decreased protein stability (26). These mutants were ectopically expressed in HEK293T cells because clinical samples were unavailable, and the half-lives of the proteins were analyzed by CHX chase assay. Consistent with a previous report (26), we found that the half-lives of R629W, P724L, W700X, and F883X were similar to or shorter than that of wild-type WFS1 (Fig. 5B, lower Western blots and degradation curve). Strikingly, the half-lives of mutants W648X, Y660X, and Q667X were longer than WFS1-WT (Fig. 1B, upper Western blots and degradation curve). These results implicated that the C-terminal region consisting of amino acids 667–700 might be important for the stability of the WFS1 protein.
Differential Susceptibility of WFS1 Mutants to Degradation by Smurf1
Trp-648, Tyr-660, and Gln-667 residues are all located within the C-terminal ER luminal region. We then asked whether the higher stability of the corresponding mutants is due to resistance against degradation by Smurf1. Indeed, when co-expressed with an increasing amount of Smurf1, the protein levels of W648X, Y660X, and Q667X were maintained at a relatively constant level and clearly exhibited a resistant effect against Smurf1 (Fig. 6A, lanes 1–10). By contrast, W700X (lanes 11–14), R629W (lanes 17–18), P724L (lanes 19–20), and F883X (lanes 23–24) were significantly decreased by Smurf1 similar to wild-type WFS1. In addition, two other missense mutants that possess an intact C-terminal part, including P504L (lanes 15 and 16) and E737K (lanes 21 and 22), were also down-regulated by Smurf1. These results suggest that the region Gln-667 through Trp-700 of WFS1 might be important for Smurf1-mediated degradation.
FIGURE 6.
Degradation of different WFS1 mutants by Smurf1. A, Smurf1 regulates WFS1 mutants. The indicated mutants were co-expressed with FLAG-tagged Smurf1, and the protein expression of WFS1 mutants and Smurf1 was determined by anti-Myc and anti-FLAG antibodies. B, co-immunoprecipitation (IP) of Smurf1 and WFS1 truncating mutants. The indicated wild-type WFS1 or mutants were co-transfected with FLAG-Smurf1 or CMV-FLAG. Cell lysates were immunoprecipitated with an anti-FLAG antibody followed by immunoblotting with anti-FLAG and anti-Myc antibodies. C, co-localization of Smurf1 and WFS1 truncating mutants in the ER is shown. MCF7 cells transiently transfected with Myc-WFS1 (WT), Myc-WFS1 (W648X), Myc-WFS1 (Y660X), and Myc-WFS1 (Q667X) were subjected to indirect immunofluorescence. Fixed and permeabilized cells were stained with antibodies against Myc (green) and PDI (endoplasmic reticulum marker, red); nuclei were stained with DAPI. PDI, protein disulfide isomerase. D, overexpression of WFS1 truncating mutants decreases ATF6α protein levels. The indicated wild-type WFS1 or mutants were transfected into MCF7 cells, and ATF6α protein levels were analyzed by Western blot. E, knock-down of Smurf1 leads to WFS1 up-regulation and ATF6α down-regulation. MCF7 cells were transfected with siRNAs against Smurf1. Cell lysates were analyzed by immunoblotting with anti-Smurf1, anti-WFS1, anti-ATF6α, and anti-GAPDH antibodies. F, HepG2 cells were treated with Tg for the indicated times. Cell lysates were prepared and immunoblotted with Smurf1, WFS1, ATF6α, CHOP, and GAPDH antibodies. G, overexpression Smurf1 partially reversed the Tg effects. HepG2 cells were transfected with FLAG-tagged Smurf1 or vector control. The cells were then treated with Tg for 24h. The protein levels of WFS1, ATF6α, CHOP, Smurf1, and GAPDH were analyzed by immunoblotting with the indicated antibodies. Quantification of the WFS1 and ATF6α amounts is shown.
We also performed a co-immunoprecipitation assay and subcellular localization analysis to test whether the resistance of the mutants W648X, Y660X, and Q667X against Smurf1-mediated degradation was caused by abrogated interaction and/or mislocalization in cells. The results showed that these truncates retained the ability to interact with Smurf1 (Fig. 6B), and all are normally localized at the ER (Fig. 6C), ruling out these possibilities.
WFS1 was recently demonstrated to promote the degradation of ATF6α and negatively control ER stress (19). Notably, ectopic expression of W648X, Y660X, and Q667X exhibited higher basal levels and suppressed the levels of ATF6α protein to a lower extent than wild-type WFS1 (Fig. 6D), implicating that the higher ability of these mutants to regulate ATF6α might correlate with their resistance against Smurf1.
Finally, we examined the possible role of endogenous Smurf1 in the regulation of ATF6α. Depletion of Smurf1 by RNAi resulted in an increase in WFS1 and a decrease in ATF6α (Fig. 6E). Additionally, in the HepG2 cells treated with Tg, Smurf1 was gradually degraded accompanied by an significant increase of WFS1 level and a decrease of ATF6α level (Fig. 6F). As a marker, the expression of CHOP was induced upon Tg treatment. These results suggested that ER stress induced the degradation of Smurf1, and then as a substrate of Smurf1, WFS1 was released from degradation. Consistent with previous report that WFS1 recruits HRD1 ligase to promote the degradation of ATF6α (19), the degradation of ATF6α was further enhanced.
Next, we tested whether overexpression of Smurf1 could abrogate the effect of ER stress inducer. Upon Tg treatment, the level of endogenous WFS1 protein was up-regulated to 4.5-fold of that in the control untreated cells (Fig. 6G, second lane versus the first lane). Smurf1 overexpression resulted in attenuation to 3.5-fold (third lane). Consistent with these alterations, the level of ATF6α was down-regulated to 0.2-fold that of control upon Tg treatment and then reversed to 0.6-fold when Smurf1 was overexpressed. The expression of ER stress marker CHOP was increased significantly upon Tg treatment and further increased when Smurf1 was overexpressed (Fig. 6G), which is consistent with the further up-regulation of ATF6α and the fact that CHOP is a transcriptional target gene of ATF6α (27, 28). Thus, overexpression of Smurf1 can partially reverse the effect of Tg treatment.
In summary, the results above suggested a feedback loop between Smurf1, WFS1, and ATF6α. Smurf1 functions as a ligase to promote WFS1 degradation, whereas WFS1 recruits HRD1 ligase to promote ATF6α degradation (19). ER stress induced Smurf1 degradation and WFS1 up-regulation. Up-regulated WFS1 further down-regulates the expression level of ATF6α. This feedback loop might contribute to turn off the ER stress response to avoid effects due to overloading.
DISCUSSION
The ER plays a central role in the productive folding of secretory proteins and the degradation of misfolded proteins (27). Perturbations in ER function cause ER stress and activate a series of unfolded protein response (UPR) signaling events, including ATF6α transactivation and CHOP up-regulation (27, 28). WFS1 deficiency in mice leads to enhanced ER stress signaling and triggers apoptosis of pancreatic islet cells (20–22), indicating that WFS1 is required for maintaining ER homeostasis. Therefore, controlling the levels of WFS1 should be crucial for ER homeostasis. Upon ER stress, WFS1 expression is up-regulated, at least partially through transcriptional activation (17, 18). The synthesized WFS1 proteins interact with Hrd1 ubiquitin ligase, a RING finger-type E3 and a critical regulator in the ER, to form a functional complex to target ATF6α for proteasomal degradation (19). By this mechanism, WFS1 contributes to preventing the overload response. On the other hand, how WFS1 is degraded remains largely unclear. Here, we identify the degradation mechanism of WFS1. The HECT domain-type E3 ligase Smurf1 interacts with WFS1 and targets WFS1 for proteasomal degradation. Interestingly, the ER lumen-targeting C terminus of WFS1 is important for Smurf1-mediated degradation. This result implies that WFS1 might be degraded by the newly synthesized Smurf1 proteins, which are distributed in the ER lumen. Thus, the steady-state level of WFS1 should be maintained at a balanced level and controlled by simultaneous synthesis and degradation in normal, healthy, unstressed cells. Consistent with this hypothesis, depletion of endogenous Smurf1 resulted in a significant up-regulation of WFS1 in different cells (Fig. 3).
Upon ER stress, the WFS1 protein dissociates from Hrd1 and releases ATF6α from degradation (19). ATF6α then transactivates the expression of downstream target genes and induces the UPR to eliminate misfolded proteins or attenuate protein translation (27, 28). At a later stage, WFS1 expression is up-regulated, and it re-inhibits the activity of ATF6α. The negative feedback loop of WFS1 on the ER stress response is similar to the Mdm2-p53 response to DNA damage. In unstressed cells, Mdm2 functions as an E3 ligase to promote p53 degradation. In response to DNA damage, Mdm2 dissociates from p53, and p53 accumulates and promotes the transactivation of downstream genes. At a later stage, Mdm2, a target of p53, is up-regulated and re-binds to p53 to maintain the low activity of p53 (29, 30). We propose that ER stress-triggered Smurf1 degradation might also contribute to the up-regulation of WFS1. Smurf1 degrades proteins in a time- and dose-dependent manner (Fig. 1), which means that at a later stage of ER stress a dramatic decrease in Smurf1 would result in a more significant increase of WFS1. The latter might contribute to re-inhibiting ATF6α activity.
We also showed that a subset of WFS1 mutants, including W648X, Y660X, and Q667X, exhibit resistance against Smurf1-mediated degradation (Fig. 6) and possess longer half-lives (Fig. 5). These mutants lack a proposed Smurf1 degradation region (degron, amino acids 667–700) and maintain ATF6α at lower levels (Fig. 6), at least under ectopic expression. Further investigations should be performed to compare the ability of these mutants and wild-type WFS1 to regulate UPR under ER stress.
Smurf1 has been demonstrated to play a critical role in the regulation of embryonic development, cell polarity, and bone homeostasis by targeting the degradation of substrates, including Smad1/5, RhoA, MEKK2, and Prickle 1 (2–7). Different from the known substrates, WFS1 is embedded in the endoplasmic reticular membrane. Smurf1 was degraded by the proteasome upon ER stress largely independent of its ligase activity (Fig. 1). Our study also provides new insight into the role of Smurf1 in ER homeostasis maintenance.
The role of Smurf1 in the control of WFS1 stability raises many questions. For example, what is the E3 ligase involved in Smurf1 degradation that is specifically activated upon ER stress stimulation? What is the physiological and pathological role of Smurf1 in mediating the degradation of WFS1 in Wolfram syndrome patients? Whether the down-regulation of Smurf1 plays a critical role in the UPR response and WFS1-Hrd1 activity control is worthy of further investigation. In conclusion, we have provided evidence that Smurf1 regulates WFS1 protein levels through ubiquitination and proteasomal degradation. Through this function, Smurf1 is involved in the ER stress response.
Acknowledgments
We thank Drs. Fumihiko Urano, Kohei Miyazono, Wesley I. Sundquist, and Yue Xiong for providing materials.
This work was supported by National Basic Research Programs 2011CB910602, 2007CB914601, and 2010CB912202, National Natural Science Foundation Projects (30830029), and National Key Technologies R&D Program for New Drugs (2009ZX09503-002, 2009ZX09301-002).
- ER
- endoplasmic reticulum
- Tg
- thapsigargin
- Tm
- tunicamycin
- TRITC
- tetramethylrhodamine isothiocyanate
- CHX
- cycloheximide
- UPR
- unfolded protein response
- IB
- immunoblot
- CHOP
- C/EBP homologous protein.
REFERENCES
- 1. Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 2. Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 3. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. (2001) J. Biol. Chem. 276, 12477–12480 [DOI] [PubMed] [Google Scholar]
- 4. Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., Wrana J. L. (2003) Science 302, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 5. Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. (2005) Cell 121, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Narimatsu M., Bose R., Pye M., Zhang L., Miller B., Ching P., Sakuma R., Luga V., Roncari L., Attisano L., Wrana J. L. (2009) Cell 137, 295–307 [DOI] [PubMed] [Google Scholar]
- 7. Zhao L., Huang J., Guo R., Wang Y., Chen D., Xing L. P. (2010) J. Bone Miner. Res. 25, 1246–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tajima Y., Goto K., Yoshida M., Shinomiya K., Sekimoto T., Yoneda Y., Miyazono K., Imamura T. (2003) J. Biol. Chem. 278, 10716–10721 [DOI] [PubMed] [Google Scholar]
- 9. Lu K., Yin X., Weng T., Xi S., Li L., Xing G., Cheng X., Yang X., Zhang L., He F. (2008) Nat. Cell Biol. 10, 994–1002 [DOI] [PubMed] [Google Scholar]
- 10. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., Forman-Kay J. D. (2007) Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 11. Barrett T. G., Bundey S. E., Macleod A. F. (1995) Lancet 346, 1458–1463 [DOI] [PubMed] [Google Scholar]
- 12. Barrett T. G., Bundey S. E. (1997) J. Med. Genet. 34, 838–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sandhu M. S., Weedon M. N., Fawcett K. A., Wasson J., Debenham S. L., Daly A., Lango H., Frayling T. M., Neumann R. J., Sherva R., Blech I., Pharoah P. D., Palmer C. N., Kimber C., Tavendale R., Morris A. D., McCarthy M. I., Walker M., Hitman G., Glaser B., Permutt M. A., Hattersley A. T., Wareham N. J., Barroso I. (2007) Nat. Genet. 39, 951–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolfram D. J., Wagener H. P. (1938) Mayo Clin. Proc. 13, 715–718 [Google Scholar]
- 15. Inoue H., Tanizawa Y., Wasson J., Behn P., Kalidas K., Bernal-Mizrachi E., Mueckler M., Marshall H., Donis-Keller H., Crock P., Rogers D., Mikuni M., Kumashiro H., Higashi K., Sobue G., Oka Y., Permutt M. A. (1998) Nat. Genet. 20, 143–148 [DOI] [PubMed] [Google Scholar]
- 16. Takeda K., Inoue H., Tanizawa Y., Matsuzaki Y., Oba J., Watanabe Y., Shinoda K., Oka Y. (2001) Hum. Mol. Genet. 10, 477–484 [DOI] [PubMed] [Google Scholar]
- 17. Ueda K., Kawano J., Takeda K., Yujiri T., Tanabe K., Anno T., Akiyama M., Nozaki J., Yoshinaga T., Koizumi A., Shinoda K., Oka Y., Tanizawa Y. (2005) Eur. J. Endocrinol. 153, 167–176 [DOI] [PubMed] [Google Scholar]
- 18. Yamaguchi S., Ishihara H., Tamura A., Yamada T., Takahashi R., Takei D., Katagiri H., Oka Y. (2004) Biochem. Biophys. Res. Commun 325, 250–256 [DOI] [PubMed] [Google Scholar]
- 19. Fonseca S. G., Ishigaki S., Oslowski C. M., Lu S., Lipson K. L., Ghosh R., Hayashi E., Ishihara H., Oka Y., Permutt M. A., Urano F. (2010) J. Clin. Invest. 120, 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishihara H., Takeda S., Tamura A., Takahashi R., Yamaguchi S., Takei D., Yamada T., Inoue H., Soga H., Katagiri H., Tanizawa Y., Oka Y. (2004) Hum. Mol. Genet. 13, 1159–1170 [DOI] [PubMed] [Google Scholar]
- 21. Riggs A. C., Bernal-Mizrachi E., Ohsugi M., Wasson J., Fatrai S., Welling C., Murray J., Schmidt R. E., Herrera P. L., Permutt M. A. (2005) Diabetologia 48, 2313–2321 [DOI] [PubMed] [Google Scholar]
- 22. Yamada T., Ishihara H., Tamura A., Takahashi R., Yamaguchi S., Takei D., Tokita A., Satake C., Tashiro F., Katagiri H., Aburatani H., Miyazaki J., Oka Y. (2006) Hum. Mol. Genet. 15, 1600–1609 [DOI] [PubMed] [Google Scholar]
- 23. Nie J., Xie P., Liu L., Xing G., Chang Z., Yin Y., Tian C., He F., Zhang L. (2010) J. Biol. Chem. 285, 22818–22830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li S., Lu K., Wang J., An L., Yang G., Chen H., Cui Y., Yin X., Xie P., Xing G., He F., Zhang L. (2010) Mol. Cell Biochem. 338, 11–17 [DOI] [PubMed] [Google Scholar]
- 25. Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 26. Hofmann S., Bauer M. F. (2006) FEBS Lett. 580, 4000–4004 [DOI] [PubMed] [Google Scholar]
- 27. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 28. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 30. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]