Abstract
In neurons, the proper distribution of mitochondria is essential because of a requirement for high energy and calcium buffering during synaptic neurotransmission. The efficient, regulated transport of mitochondria along axons to synapses is therefore crucial for maintaining function. The trafficking kinesin protein (TRAK)/Milton family of proteins comprises kinesin adaptors that have been implicated in the neuronal trafficking of mitochondria via their association with the mitochondrial protein Miro and kinesin motors. In this study, we used gene silencing by targeted shRNAi and dominant negative approaches in conjunction with live imaging to investigate the contribution of endogenous TRAKs, TRAK1 and TRAK2, to the transport of mitochondria in axons of hippocampal pyramidal neurons. We report that both strategies resulted in impairing mitochondrial mobility in axonal processes. Differences were apparent in terms of the contribution of TRAK1 and TRAK2 to this transport because knockdown of TRAK1 but not TRAK2 impaired mitochondrial mobility, yet both TRAK1 and TRAK2 were shown to rescue transport impaired by TRAK1 gene knock-out. Thus, we demonstrate for the first time the pivotal contribution of the endogenous TRAK family of kinesin adaptors to the regulation of mitochondrial mobility.
Keywords: Kinesin, Mitochondria, Mitochondrial Transport, Neurobiology, Trafficking
Introduction
Mitochondria serve several functions in cells. These functions include the generation of energy in the form of ATP, the buffering of calcium ions, and the regulation of apoptosis. Thus, within cells, mitochondria need to move so they can respond to local needs. In the nervous system, mitochondria and mitochondrial transport are particularly important because of a requirement for high energy and calcium buffering during synaptic neurotransmission. The mitochondrial population in neurons is therefore highly mobile, and the dynamics of their transport are tightly regulated to satisfy these demands. Mitochondria can move in both anterograde and retrograde directions, utilizing motor proteins and the microtubule network (for reviews, see Refs. 1–3). Furthermore, they can be anchored at defined sites; one example is mitochondrial immobilization by a Ca2+-dependent mechanism at synaptic sites (4–7). Recently, there has been significant progress in the understanding of mitochondrial transport processes with the identification of several proteins implicated in their trafficking mechanisms. The best characterized of these include the trafficking kinesin protein (TRAK)2/Milton family of kinesin adaptors; Miro1 and Miro2, atypical Rho GTPases that reside in the mitochondrial outer membrane that are purported receptors for TRAKs; syntabulin, also a kinesin adaptor protein; and syntaphilin, an axonal mitochondrial docking protein (for reviews, see Refs. 3 and 8).
There are two mammalian TRAKs, TRAK1 and TRAK2, that share ∼58% amino acid homology (9, 10). The TRAKs, like their Drosophila orthologue Milton (11), have been shown to function as kinesin adaptors linking kinesin heavy chain (KHC) to mitochondria by their association with Miro1/2. Thus, TRAK1, TRAK2, or Milton each co-immunoprecipitate with KHC from detergent extracts of neuronal tissue (11, 12). The association between the TRAKs and KHC is direct and involves interaction between TRAKs and the KHC non-motor, C-terminal cargo binding domain (13). In heterologous expression, TRAKs and Milton are targeted to mitochondria (11, 12, 14). The co-expression of TRAK1, TRAK2, or Milton with KHC results in the redistribution of mitochondria such that they co-localize with TRAK1, TRAK2, Milton, and kinesin heavy chains at the tips of cellular processes (13, 15, 16). Miro1 and Miro2 co-distribute and co-immunoprecipitate with Milton, TRAK1, or TRAK2 following overexpression in mammalian cells (15, 16, 17). Furthermore, Miro1 co-immunoprecipitates with TRAK2 from brain extracts (18). Overexpression of fluorescently tagged TRAK2 or Miro1 constructs in hippocampal neurons results in an increase in the number of mitochondria transported to the periphery of hippocampal neurons (18). Furthermore, expression of the Miro1 binding domain of TRAK2, uncoupling Miro1 from TRAK2, results in prevention of this induced redistribution of mitochondria into the periphery (18). Finally, in dendrites of hippocampal neurons, overexpression of Miro1 results in a percent increase in the number of moving mitochondria. Conversely, knockdown of Miro1 in hippocampal neurons by targeted shRNAi results in a decrease in the mobile mitochondrial fraction (6). Surprisingly, this was not the case in axons in these same neurons because mitochondrial velocity and mobility were unaffected by overexpression of Miro1 (7). In dorsal root ganglia neurons, knockdown of Miro2 but not Miro1 disrupts axonal mitochondrial transport (19).
All the above studies are supportive of a kinesin-TRAK-Miro mitochondrial trafficking complex. However, apart from the Miro1 shRNAi studies, the findings all result from overexpression of fluorescently tagged TRAK2/Milton, KHC, or Miro constructs. In this study, we investigated the role of endogenous TRAKs in the trafficking of mitochondria in axons of hippocampal pyramidal neurons using both gene silencing by targeted shRNAi and dominant negative approaches. We report that inhibiting the formation or the availability of the TRAK kinesin adaptor resulted in a decrease in mitochondrial mobility. Thus, we show definitively and for the first time that endogenous TRAKs are indeed mediators of axonal mitochondrial transport. Furthermore, we report that the contributions of TRAK1 and TRAK2 to the movement of mitochondria differed, suggesting that possibly the selective binding by TRAK1 and/or TRAK2 to different members of the kinesin transport family or to Miro1 and Miro2 may represent crucial regulatory points in controlling the traffic of mitochondrial cargoes in neurons.
EXPERIMENTAL PROCEDURES
Constructs and Antibodies
pCISTRAK2 (formerly pCISGRIF-1, splice form GRIF-1a; hereafter referred to as TRAK2) was as described previously (12). Full-length rat TRAK1 cDNA (NCBI accession number NM_001134565.1) was amplified from rat brain by Eurogentec Ltd. (Southampton, UK), and the sequence was verified by nucleotide sequencing. It was subcloned into the XhoI and EcoRI sites of pEGFP-C2 to yield pEGFP-ratTRAK1 (pEGFP-rTRAK1). (Note, to simplify nomenclature, plasmids encoding proteins that were tagged with fluorescent proteins at the N terminus were named with the tag name preceding the protein; i.e. pEGFP-TRAK1 encodes TRAK1 with the EGFP tag at the N terminus. Conversely, plasmids encoding proteins that were tagged at the C terminus were named with the tag name following, the protein, e.g. pKIF5C-EYFP.) Three silent base mutations, T1852C, A1855G, and G1858C, were introduced into pEGFP-rTRAK1 using the QuikChangeTM mutagenesis kit (Stratagene, La Jolla, CA) to generate pEGFP-rTRAK1silent. DNA encoding the dominant negative TRAK2(124–283) (hereafter termed TRAK2 DN) was excised from pCMVc-MycTRAK2(124–283) and cloned in-frame in the EcoRI and XhoI sites of the vector pIRES-hrGFP-1a (Agilent Technologies Inc., Basel, Switzerland) to generate pIRES-GFPTRAK2 DN. For the TRAK2 shRNAi studies, two double-stranded oligonucleotide pairs encoding rat TRAK2 bp 785–805 and rat TRAK2 bp 1886–1905 siRNA sequences as in Ref. 20 were purchased. The oligonucleotide sequences were as follows: TRAK2 785–805, 5′-GGAACCATGTCTTATCTGAGC plus the complementary sequence with 5′-GGGAACCTTCTTAAATCTGGC as the scrambled control; and TRAK2-2 1886–1905, 5′-GAACGGAAGCTCCAGATTCT with 5′-TTGCAAGCCAAGGTTCGACA as the scrambled control. The respective oligonucleotides and their complementary counterparts were each subcloned into the BamHI and EcoRI sites of pSIREN-RetroQ-ZsGreen (Clontech) to generate pSIRENGreenTRAK2 (pGreenTRAK2), pSIRENGreenTRAK2-2 (pGrenTRAK2-2), and the scrambled controls pSIRENGreenTRAK2scr (pGreenTRAK2scr) and pSIRENGrenTRAK2-2scr. For TRAK1, the oligonucleotide encoding the siRNA sequence was as in Ref. 21 but adapted for the rat TRAK2 DNA sequence. The oligonucleotide was 5′-CGAAAGAGTTGGCCAGATG corresponding to rat TRAK1 bp 294–312. The scrambled control was 5′-AACCGGTTTGGAACGAGAG. The respective oligonucleotides and their complementary counterparts were each subcloned into the BamHI and EcoRI sites of pSIREN-RetroQ-ZsGreen and pSIREN-RetroQ-ZsRedExpress (Clontech) to generate pSIRENGreenTRAK1 (pGreenTRAK1), pSIRENRedTRAK1 (pRedTRAK1), and the scrambled control pSIRENGreenTRAK1scr (pGreenTRAK1scr). pCMVTag4aTRAK2 (C-terminal FLAG-tagged TRAK2) was as in Ref. 9; pcDNAHisMaxKIF5C (N-terminal His tag), pCMVTRAK2(283–913) (N-terminal c-Myc tag), and pCIShumanTRAK1 (pCIShTRAK1) were as in Ref. 12; pDsRed1-Mito for visualization of mitochondria, pECFP-TRAK2, and pKIF5C-EYFP were as in Ref. 13; and pECFP-hTRAK1 was as in Ref. 16. Rat TRAK2 was cloned in-frame into the EcoRI site of pEGFPC2 to yield pEGFP-rTRAK2. The far-red fluorescent construct pTurboFP635-NMito1 was generated by cloning the DNA mitochondrial targeting domain of cytochrome oxidase 8A into the EcoRI and BamHI sites of pTurboFP635-N (Evrogen, Moscow, Russia). pSynaptophysin-EYFP was a kind gift from Dr. Ann-Marie Craig, Vancouver, Canada. Table 1 contains a descriptive summary of all clones used.
TABLE 1.
Summary of mammalian expression clones generated
FP, fluorescent protein.
| Clone | Properties |
|---|---|
| pCIShTRAK1 | Human TRAK1 |
| pECFP-hTRAK1 | Human TRAK1 with an N-terminal ECFP tag |
| pEGFP-rTRAK1 | Rat TRAK1 with an N-terminal EGFP tag |
| pECFP-rTRAK1silent | Rat TRAK1 with an N-terminal ECFP tag and with 3 silent base mutations |
| pCISTRAK2 | Rat TRAK2 |
| pEGFP-rTRAK2 | Rat TRAK2 with an N-terminal EGFP tag |
| pECFP-rTRAK2 | Rat TRAK2 with an N-terminal ECFP tag |
| pCMVTag4aTRAK2 | Rat TRAK2 with a C-terminal FLAG tag |
| pCMVTRAK2(283–913) | TRAK2(283–913) with an N-terminal c-Myc tag |
| pIRES-GFPTRAK2 DN | TRAK2(124–283) and the reporter GFP |
| pKIF5C-EYFP | Human KIF5C with a C-terminal tag |
| pcDNAHisMaxKIF5C | Human KIF5C with an N-terminal His tag |
| pGreenTRAK1 | TRAK1 shRNA and a green FP reporter protein |
| pGreenTRAK1scr | TRAK1 control, scrambled shRNA and green FP |
| pRedTRAK1 | TRAK1 shRNA and red FP reporter protein |
| pGreenTRAK2 | TRAK2 shRNA and green FP reporter protein |
| pGreenTRAK2-2 | TRAK2-2 shRNA and green FP reporter protein |
| pGreenTRAK2scr | TRAK2 control, scrambled shRNA and green FP reporter protein |
| pDsRed1-Mito | Red mitochondrial targeting vector |
| pTurboFP635-NMito1 | Far-red mitochondrial targeting vector |
| pSynaptophysin-EYFP | Synaptophysin with a C-terminal EYFP tag, a kind gift from Dr. Ann-Marie Craig, Vancouver, Canada |
Affinity-purified sheep anti-TRAK2(874–889) (formerly anti-GRIF-1(874–889)) antibodies were generated as in Ref. 12; anti-TRAK2(8–633) (formerly anti-GRIF-1(8–633)) antibodies generated as in Ref. 9, and in-house rabbit anti-FLAG antibodies were generated as in Ref. 13. Anti-His G and anti-rabbit Alexa Fluor 680 antibodies were from Invitrogen; anti-GFP antibodies were from Abcam Ltd. (Cambridge, UK); anti-β-actin antibodies were from Sigma-Aldrich; and rabbit, mouse, and sheep horseradish peroxidase-linked secondary antibodies were from Amersham Biosciences.
Culturing and Transfection of Hippocampal Neurons
Cultures of rat hippocampal neurons were prepared at a density of ∼30,000 cells/cm2 on poly-d-lysine- (1 μg/ml) and laminin (2 μg/ml)-coated coverslips from hippocampi that were dissected from P0 rat brain (22). Cultures were grown for 3–4 days in complete Neurobasal medium, which was Neurobasal medium (Invitrogen) containing a 1 in 50 dilution of B27 (Invitrogen), 0.5 mm GlutaMax (Invitrogen), and 0.4% (w/v) glucose. Transfection of neurons was by a calcium phosphate method adapted from Ref. 23 always using DNA clones that had been purified using EndoFree purification kits for the maxipreparation of plasmid DNA (Qiagen, Crawley, Sussex, UK). The transfection mixture was prepared by addition of the appropriate plasmid DNAs to a solution containing 0.25 m CaCl2. This was added dropwise to an equal volume of 2× HEPES-buffered saline, pH 7.14. The culture medium was removed and stored at 37 °C in 5% CO2. Fresh complete Neurobasal medium was added to the neurons followed by dropwise addition of the transfection mixture. Neurons were incubated at 37 °C in 5% CO2 for 30 min. The cell culture medium was aspirated, and neurons were washed quickly once with fresh Neurobasal medium. Fresh complete Neurobasal medium that had been preincubated at 37 °C in 10% CO2 was added, and the neurons were incubated at 37 °C and 5% CO2 for 20 min. The cell culture medium was replaced with the original stored medium, and neurons were maintained at 37 °C in the presence of 5% CO2 until analysis by live imaging confocal microscopy. Transfections were carried out for the shRNAi knockdown studies at 3 days in vitro (DIV) and for the DN knockdown studies at 4 DIV with imaging for both at 6 DIV. For all transfections, 1 μg of DNA was added per dish, i.e. per ∼30,000 cells. The ratio of DNAs for transfection mixtures for TRAK2 DN studies was 25 pIRES-GFPTRAK2 DN or pIRES-GFP to 1 pDsRed1-Mito. For shRNAi knockdown studies, the ratio was 25 pGreenTRAK1, pGreenTRAK2, pGreenTRAK1scr, or pGreenTRAK2scr to 1 pDsRed1-Mito. For the shRNAi rescue experiments, the ratio was 12.5 pRedTRAK1 to 12.5 pEGFP-rTRAK1silent or 12.5 pEGFP-TRAK2 to 1 pTurboFP635-N. For the synaptophysin studies, the DNA ratio was 25 pEYFP-synaptophysin to 1 pDsRed1-Mito. Transfection efficiencies were in the range of 0.1–1%, which corresponded to between four and 10 transfected neurons per dish.
Live Cell Imaging and Analysis
Confocal imaging was carried out using a Zeiss LSM 510 or a Zeiss LSM 710 oil immersion 40× objective with sequential acquisition setting. Neurons were maintained for imaging at 37 °C in 5% CO2. Transfected cells were identified under UV light with the fluorescein isothiocyanate (FITC) filter to enable visualization of the green fluorescence of the respective reporter followed by visualization under UV light with the rhodamine filter to check for the presence of DsRed1-Mito. For the identified transfected neuron, live imaging was carried out using λ = 561-nm excitation at 2% of the intensity of the diode-pumped solid-state laser to minimize bleaching and damage to neurons. Images of 512 × 512-pixel resolution with the pinhole at a setting of 3 μm were taken at 6-s intervals for 50 frames in a single focal plane. The short interval was used to minimize laser-induced neuronal damage. The axons of selected transfected neurons were identified morphologically by their uniform diameter, length, and lack of branching. Only those processes that satisfied these criteria were imaged. The polarity was determined by tracing the axon back to the cell body. The section of the axon selected for imaging was ∼70–100 μm in length, was at least 100 μm from the cell body, and contained at least eight mitochondria. Imaging data were analyzed using VolocityTM software (PerkinElmer Life Sciences). Individual mitochondria were analyzed using the Volocity point tool to track objects manually to yield velocity, displacement (i.e. distance moved), and bearing (direction). Oscillating mitochondria are defined as those that are displaced >2 μm from one site. Fusion events were determined by visualization of two mitochondria merging with a subsequent change in both velocity and direction for each. Fission events were determined by visualization of one mitochondrion becoming two. Videos of live imaging were created by import of the original confocal image file into Volocity and then exported as an AVI movie or as a kymograph to depict mitochondrial movement.
Mammalian Cell Transfections
For immunoprecipitation assays, human embryonic kidney (HEK) 293 cells were transfected with the constructs shown by the calcium phosphate method using 10 μg of DNA/250-ml culture flask. For double transfections, a 1:1 ratio for a total of 10 μg of DNA was used, and for triple transfections, a 1:1:1 ratio was used again with 10 μg of DNA/250-ml culture flask. Cells were harvested 24–48 h post-transfection, and either cell homogenates were analyzed by immunoblotting, or transfected cell homogenates were 1% (v/v) Triton X-100 detergent-solubilized, and extracts were collected following centrifugation for 40 min at 4 °C at 100,000 × g (9).
For confocal microscopy studies, COS-7 cells were plated onto poly-d-lysine (0.1 mg/ml)-coated coverslips and transfected using the calcium phosphate method and plasmid DNA ratios as above. Transfections using pDsRed1-Mito to visualize mitochondria in triple transfections used a ratio of 1 pDsRed1-Mito to 12.5 pECFP-TRAK2 to 12.5 pKIF5C-EYFP or 1 pDsRed1-Mito to 12.5 pECFP-TRAK2 DN to 12.5 pKIF5C-EYFP for a total of 10 μg of DNA. Transfections using pDsRed1-Mito to visualize mitochondria in quadruple transfections used a ratio of 1 pDsRed1-Mito to 8.3 pECFP-TRAK2 to 8.3 pKIF5C-EYFP to 8.3 pIRES-GFPTRAK2 DN; 1 pDsRed1-Mito to 8.3 pKIF5C-EYFP to 8.3 pEGFP-rTRAK1 to 8.3 pEGFP-rTRAK1silent, 8.3 pGreenTRAK1 or pGreenTRAK1scr; or 1 pDsRed1-Mito to 8.3 pKIF5C-EYFP to 8.3 pEGFP-TRAK2 to 8.3 pGreenTRAK2 or pGreenTRAK2scr for a total of 10 μg of DNA.
Immunoblotting and Immunoprecipitations
Both immunoblotting and immunoprecipitations were carried out exactly as previously described in Refs. 12, 13, and 16.
RESULTS
Validation of TRAK2 DN construct: Demonstration That TRAK2 DN Prevents Co-immunoprecipitation of Full-length TRAK1 and TRAK2 with Prototypic Kinesin-1, KIF5C
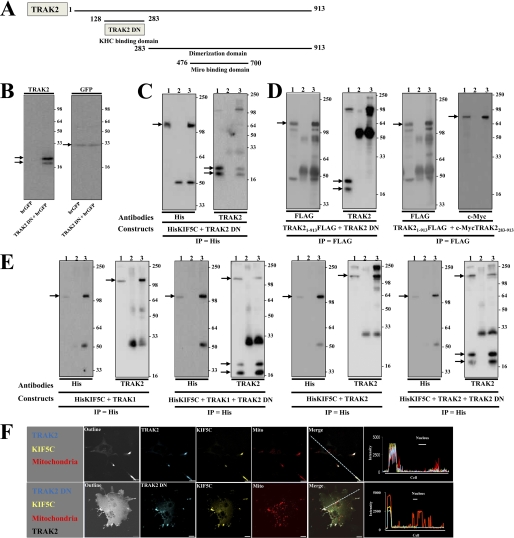
We previously showed by both yeast two-hybrid interaction assays and co-immunoprecipitations from transfected cells that the kinesin binding domain of TRAK2 maps to TRAK2(128–283) (12). The Miro binding domain of TRAK2 maps to TRAK2(476–700) (18). TRAK2(128–283) should behave as a dominant negative by inhibiting the binding of TRAK2 to kinesin and thereby preventing the formation of the kinesin-TRAK-Miro mitochondrial trafficking complex. Thus, the construct pIRES-GFPTRAK2(128–283), i.e. pIRES-GFPTRAK2 dominant negative (pIRES-GFPTRAK2 DN), was generated. The vector was selected because it is dicistronic. It will express the cloned insert, i.e. TRAK2 DN, under the control of the cytomegalovirus promoter and a reporter protein, humanized recombinant green fluorescent protein (hrGFP), by virtue of the encephalomyocarditis virus internal ribosomal entry site. hrGFP has weaker expression compared with TRAK2 DN, but it enables identification of transfected cells. Fig. 1 shows the characterization and experiments designed to validate the use of TRAK2 DN in heterologous expression.
FIGURE 1.
Validation of TRAK2 DN: TRAK2 DN inhibits co-immunoprecipitation of full-length TRAK1 and TRAK2 with KIF5C and does not yield mitochondrial redistribution in KIF5C/TRAK2 DN-transfected cells. A, a schematic diagram of key domains of TRAK2. B, HEK 293 cells were transfected with either pIRES-GFP or pIRES-GFPTRAK2 DN, and cell homogenates were prepared and analyzed by immunoblotting using anti-TRAK2 or anti-GFP antibodies as shown. C, HEK 293 cells were co-transfected with pcDNAHisMaxKIF5C + pIRESTRAK2 DN, cell homogenates were prepared and solubilized, immunoprecipitations were carried out with anti-His or a non-immune control antibody, and pellets were analyzed by immunoblotting using antibodies as shown. D, HEK 293 cells were co-transfected with either pCMVTag4aTRAK2 + pIRESTRAK2 DN or pCMVTag4aTRAK2 + pCMVTag4aTRAK2(283–913), cell homogenates were prepared and solubilized, immunoprecipitations were carried out with anti-FLAG or a non-immune control antibody, and immune pellets were analyzed by immunoblotting using antibodies as shown. E, HEK 293 cells were co-transfected with pcDNAHisMaxKIF5C + pCIShTRAK1 + pIRES-GFP, pcDNAHisMaxKIF5C + pCIShTRAK1+ pIRES-GFPTRAK2 DN, pcDNAHisMaxKIF5C + pCISTRAK2 + pIRES-GFP, or pcDNAHisMaxKIF5C + pCISTRAK2+ pIRES-GFPTRAK2 DN; cell homogenates were prepared and solubilized; immunoprecipitations were carried out with an anti-His or a non-immune control antibody; and immune pellets were analyzed by immunoblotting using antibodies as shown. For C–E, all immunoblots have the same layout where lane 1 is detergent-solubilized HEK 293 cell homogenates, lane 2 is non-immune pellet, and lane 3 is immune pellet. Note that for double transfections 6% of immune pellets was analyzed for detection of immunoprecipitating (IP) antibody protein, whereas 94% of immune pellets was analyzed for detection of co-associating proteins. For triple transfections, 6% of immune pellets was analyzed for detection of immunoprecipitating antibody protein, whereas 47% of immune pellets was analyzed for detection of co-associating proteins. Immunoblots are representative of at least n = 3 separate immunoprecipitations from three independent transfections. → denotes KIF5C, TRAK1, TRAK2, TRAK2 DN, or hrGFP as appropriate. The positions of molecular mass standards (kDa) are on the right. F, COS-7 cells were transfected with pKIF5C-EYFP + pECFP-TRAK2 + pDsRed1-Mito or pKIF5C-EYFP + pECFP-TRAK2 + pIRESTRAK2 DN + pDsRed1-Mito. Cells were fixed 24–40 h post-transfection, stained with anti-TRAK2(8–633) antibodies and anti-rabbit Alexa Fluor 680 for detection of TRAK2 DN, and imaged by confocal microscopy. Outline refers to images with saturated fluorescence intensity to show complete cell outlines. The fluorophore is indicated on each image: yellow, KIF5C; blue, TRAK2 or TRAK2 DN; red, mitochondria. Merge shows respective merged images for each panel. Images are a single confocal section of a selected cell. Scale bars are 20 μm. Images are representative of at least n = 20 cells from at least n = 3 independent transfections. Note that the TRAK2/KIF5C/mitochondrial images are taken from Ref. 13 and are shown for comparison.
TRAK2 DN Co-immunoprecipitates with Full-length KIF5C
Fig. 1B shows that transfection of HEK 293 cells with pIRES-GFPTRAK2 DN results in expression of TRAK2 DN and GFP. Two TRAK2-immunoreactive bands with Mr values of 24,000 ± 1000 and 22,000 ± 1000 (n = 10) were always detected for TRAK2 DN. The higher molecular weight band was always the strongest, and the molecular size compared well with the predicted weight for TRAK2 DN, i.e. Mr = 20,000. TRAK2 DN co-immunoprecipitated with KIF5C similarly to that described previously for the co-immunoprecipitation of c-Myc-tagged TRAK2 and KIF5C (Fig. 1C and Ref. 12). (Note also that the band at ∼110 kDa in the immunoblot probed with anti-TRAK2(8–633) antibodies is His tagged-kinesin; anti-TRAK2 antibodies were raised against a His-tagged TRAK2(8–633) fusion protein and have previously been observed to recognize His-tagged proteins (12).)
TRAK2 DN Does Not Homodimerize with Full-length TRAK2
TRAK2 is known to form at least homodimers (14, 24). If TRAK2 DN was able to dimerize with full-length TRAK2, this may negate its use as a dominant negative because full-length TRAK2 may retain the ability to bind Miro1/2. It was necessary therefore to investigate this caveat. Full-length FLAG tagged-TRAK2 was co-expressed with either TRAK2 DN or c-Myc-tagged TRAK2(283–913), expressed proteins were immunoprecipitated by anti-FLAG antibodies, and the immune pellets were screened for reactivity with anti-TRAK2(8–633) or anti-c-Myc antibodies. Immunoreactivity in the precipitates was observed for c-Myc-TRAK2(283–913) but not for TRAK2 DN despite robust activity being detected in the input (Fig. 1D). Thus, TRAK2 DN does not dimerize with full-length TRAK2.
TRAK2 DN Inhibits Association of TRAK1 and TRAK2 with Full-length KIF5C
TRAK1 and TRAK2 are members of the same gene family and share an overall 48% amino acid identity and 58% amino acid sequence similarity. TRAK2 DN has 58 and 78% amino acid similarity and identity, respectively, with the aligned TRAK1 sequence; thus, TRAK2 DN may inhibit the binding of both TRAK2 and TRAK1 to kinesin. To investigate this possibility, HEK 293 cells were co-transfected with the following clones: pcDNAHisMaxKIF5C encoding the prototypic kinesin-1 KHC KIF5C, pIRES-hrGFP-1a (control), or pIRES-GFPTRAK2 DN plus either pCISTRAK2 or pCIShTRAK1. Immunoprecipitations were carried out with anti-His antibodies (KIF5C), and immune pellets were analyzed for KIF5C, TRAK1, and TRAK2 immunoreactivities (Fig. 1E). In the presence of TRAK2 DN, TRAK2 and TRAK1 immunoreactivities in immune pellets were significantly decreased, demonstrating that indeed TRAK2 DN inhibits the binding of both TRAK1 and TRAK2 to KIF5C.
TRAK2 DN Impairs Redistribution of Mitochondria in TRAK2/KIF5C-transfected Cells
Finally, we previously showed (similarly to Ref. 15) that co-expression of KIF5C and TRAK1 or TRAK2 resulted in a redistribution of mitochondria such that they co-localized at the tips of processes with KIF5C and TRAK1/TRAK2 (13, 16). Here we compared the redistribution pattern of mitochondria between COS-7 cells transfected with KIF5C and TRAK2 clones and COS-7 cells transfected with KIF5C, TRAK2, and TRAK2 DN. The confocal images in Fig. 1F show clearly that although TRAK2 DN and KIF5C are co-distributed their co-localization with mitochondria is less evident. Many mitochondria remain within the cell cytoplasm (Fig. 1F). Thus, TRAK2 DN was shown to act as a dominant negative for both TRAK1- and TRAK2-mediated mitochondrial transport in model heterologous expression systems by virtue of its ability to inhibit kinesin/TRAK association.
TRAK2 DN Reduces Mitochondrial Transport in Axons of Hippocampal Neurons
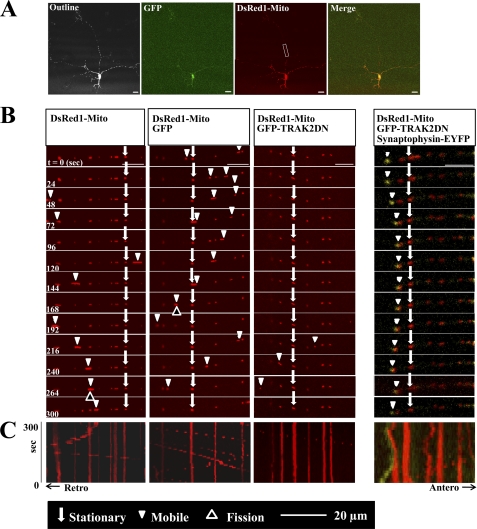
To investigate the role of endogenous TRAK1 and TRAK2 in mitochondrial transport in neurons, hippocampal pyramidal cells were transfected in parallel at 4 DIV with either pDsRed1-Mito alone or pDsRed1-Mito + pIRES-hrGFP-1a or pDsRed1-Mito + pIRES-GFPTRAK2 DN. At 6 DIV, mitochondrial mobility in axons was studied by live imaging confocal microscopy. Representative images and kymographs are shown in Fig. 2. Table 2 summarizes the effects of TRAK2 DN on the parameters of mitochondrial dynamics. Accompanying representative videos are available as supplemental Videos 1–3.
FIGURE 2.
TRAK2 DN decreases mitochondrial mobility in axons of hippocampal pyramidal neurons: demonstration by live imaging. Hippocampal neurons prepared from P0 rat brain were transfected at 4 DIV with either pDsRed1-Mito, pDsRed1-Mito + pIRES-GFP, pDsRed1-Mito + pIRES-GFPTRAK2 DN, or pDsRed1-Mito + pIRES-GFPTRAK2 DN and pEYFP-synaptophysin and imaged at 6 DIV all as described under “Experimental Procedures.” A is a representative example of a transfected neurone where Outline refers to an image with saturated fluorescence intensity to show the complete cell outline, GFP shows the green fluorescence enabling identification of the transfected neuron, DsRed1-Mito shows the distribution of mitochondria, and Merge shows a merge of GFP + DsRed1-Mito fluorescence. B is a representative series of time lapse images for each of the above transfection conditions taking a cropped area typical of that shown in A. C, kymographs of the time lapse images. The parameters of mitochondrial dynamics are summarized in Table 2.
TABLE 2.
Summary of effect of TRAK DN on mitochondrial dynamics in axons of hippocampal pyramidal neurons
Hippocampal neurons were transfected at 4 DIV with either pDsRed1-Mito, pDsRed1-Mito + pIRES-hrGFP-1a, or pDsRed1-Mito + pIRES-GFPTRAK2 DN. At 6 DIV, co-transfected cells were identified, mitochondrial dynamics were imaged by confocal microscopy, and the results were analyzed using Volocity software all as described under “Experimental Procedures.” Oscillating mitochondria are defined as those that are displaced >2 μm from one site. Values are the means ± S.E. from n = 33 images from n = 23 neurons for pDsRed1-Mito (295 mitochondria imaged), n = 18 images from n = 14 neurons for pDsRed1-Mito + pIRES-hrGFP-1a transfections (267 mitochondria imaged), and n = 33 images from n = 21 neurons for pDsRed1-Mito + pIRES-GFPTRAK2 DN (406 mitochondria imaged) for n = 4 independent transfection experiments. Statistical significances were obtained using the Students t test.
| Clones transfected | Mitochondrial density | Velocity | Fusion events/10 mitochondria | Fission events/10 mitochondria | Oscillating mitochondria | Stationary mitochondria | Mobile mitochondria | Anterograde mitochondria | Retrograde mitochondria |
|---|---|---|---|---|---|---|---|---|---|
| μm−1 | μm/s | % | % | % | % | % | |||
| pDsRed1-Mito | 0.18 ± 0.03 | 0.46 ± 0.17 | 1.1 | 1.0 | 1 ± 1 | 68 ± 4 | 32 ± 4 | 17 ± 3 | 14 ± 3 |
| pDsRed1-Mito pIRES-GFP | 0.16 ± 0.03 | 0.66 ± 0.25 | 0.9 | 0.9 | 2 ± 2 | 60 ± 10 | 40 ± 10 | 19 ± 2 | 19 ± 4 |
| pDsRed1-Mito pIRES-FPTRAK2 DN | 0.18 ± 0.03 | 0.48 ± 0.18 | 0.6a | 0.5a | 1 ± 1 | 89 ± 4b | 11 ± 4b | 4 ± 2c | 7 ± 2a |
a p < 0.05.
b p < 0.001.
c p < 0.0005.
For all the mitochondrial parameters measured, no differences were found between neurons transfected with pDsRed1-Mito alone or pDsRed1-Mito + the control vector pIRES-hrGFP-1a. The percentage of mobile mitochondria (∼30%) agreed with values determined for axons of hippocampal neurons (e.g. Refs. 25–27) and for axons of cortical neurons (5). Similarly, the mean mitochondrial velocities were in agreement with published works, i.e. 0.46 ± 0.17 μm/s (summarized in Ref. 3). Furthermore, the axonal mitochondria were pleomorphic, showing traits previously described; i.e. they were a mixture of different lengths ranging from approximately <1 to 7 μm with a uniform diameter of ∼0.5 μm and more rounded, compact structures with an approximate diameter of 0.2 μm. Mitochondria also underwent fusion and fission.
TRAK2 DN significantly decreased the percentage of mobile mitochondria compared with both controls (Table 2). A 71 ± 7% decrease in the percentage of mobile mitochondria in pIRES-hrGFP-1aTRAK2 DN- compared with pIRES-hrGFP-1a-transfected controls was found. Interestingly, significant effects on the percentage of mobile mitochondria moving in both anterograde and retrograde directions were evident. Percent decreases were 85 ± 6% for anterograde and 59 ± 10% for retrograde transport. No significant differences were induced by TRAK2 DN over the 5-min time frame of study in mitochondrial density in the imaged section of axon, in the mean velocities, and in the percentage of oscillating mitochondria (Table 2). Further analysis found no significant difference between the mean velocities for anterograde versus retrograde transport. The values were 0.44 ± 0.2 μm/s for anterograde velocity and 0.53 ± 0.22 μm/s for retrograde velocity. There were no obvious changes in mitochondrial morphology. Interestingly, the number of fusion and fission events was decreased, although the significance was less than that found for the TRAK2 DN-induced mitochondrial mobility (Table 2).
Synaptophysin-EYFP mobility was also imaged in neurons co-transfected with this construct, pIRESGFP-TRAK2 DN, and pDsRed1-Mito to determine whether the transport of other organelles was affected by TRAK2 DN. A representative example of a transfected neuron is shown in Fig. 2. As above, little movement of mitochondria was evident due to TRAK2 DN, but fluorescence associated with synaptophysin was observed to move albeit at a reduced velocity compared with mitochondria.
TRAK1 and TRAK2 Knockdown Studies
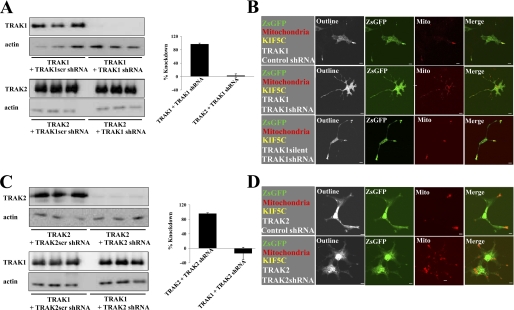
Because we showed that the TRAK2 DN inhibited the association of both TRAK1 and TRAK2 to kinesin, it was of interest to investigate the contribution of each to axonal mitochondrial transport. Thus, a gene knockdown approach was used utilizing TRAK1- and TRAK2-gene targeted shRNAs. Each shRNAi (and the respective scrambled control) was validated by immunoblotting and confocal microscopy imaging following expression in HEK 293 cells and COS-7 cells, respectively (Fig. 3). By quantitative immunoblotting, TRAK1 shRNAi yielded a 97 ± 3% (n = 3) knockdown of exogenously expressed TRAK1 with respect to the TRAK1scr shRNAi control (Fig. 3). There was no significant knockdown of exogenously expressed TRAK2; the value for TRAK2 expression was 115 ± 17% (n = 3) for TRAK1 shRNAi compared with 100% for TRAK1scr shRNAi. Similarly, TRAK2 shRNAi yielded a 96 ± 2% (n = 3) knockdown of exogenously expressed TRAK2 with respect to the TRAK2scr shRNAi control. There was no significant knockdown of exogenously expressed TRAK1; the value for TRAK1 expression was 86 ± 17% (n = 3) for TRAK2 shRNAi compared with 100% for TRAK2scr shRNAi (Fig. 3). TRAK2-2 shRNAi yielded a knockdown of exogenous TRAK2 similar to that found for TRAK2 shRNAi, i.e. ∼ 95% (data not shown).
FIGURE 3.
Validation of TRAK1- and TRAK2-targeted shRNAis in heterologous expression. A and B demonstrate the specificity of the TRAK1 shRNAi probe, and C and D demonstrate the specificity of the TRAK2 shRNAi probe. In A and C, HEK 293 cells were co-transfected with TRAK1 or TRAK2 clones and either pGreenTRAK1 or pGreenTRAK1scr (A) or pGreenTRAK2 or pGreenTRAK2scr (C). Cell homogenates were collected 24 h post-transfection, and aliquots were analyzed in triplicate by immunoblotting using either anti-β-actin or anti-TRAK1(8–633) antibodies that recognize both TRAK1 and TRAK2 as indicated. The histograms show the percentage of knockdown of the exogenous TRAK. Values were normalized using actin expression. Values are presented as the percentage of knockdown with respect to shRNAi scrambled controls and are the means ± S.E. for n = 3 independent transfection experiments. The percentage of knockdown of TRAK1 by TRAK1 shRNAi was 97 ± 3%, and for knockdown of TRAK2 by TRAK2 shRNAi, the value was 96 ± 2%. TRAK1 shRNAi had no significant effect on TRAK2 expression and vice versa for TRAK2 shRNAi. In B and D, COS-7 cells were co-transfected with pDsRed1-Mito + pKIF5C-EYFP + pEGFP-rTRAK1 and either pGreenTRAK1 or pGreenTRAK1scr (B) or pGreenTRAK2 or pGreenTRAK2scr (D) as indicated in the left-hand squares. In B also, COS-7 cells were co-transfected with pDsRed1-Mito + pKIF5C-EYFP + pEGFP-rTRAK1silent (bottom panel) + pTRAK1. Cells were imaged by confocal microscopy 24 h post-transfection. Outline refers to images with saturated fluorescence intensity to show complete cell outlines. The fluorophore is indicated on each image: green, ZsGFP; red, mitochondria. Merge shows respective merged images for each panel. Images are a single confocal section of a selected cell. Scale bars are 20 μm. Images are representative of at least n = 20 cells from at least n = 3 independent transfections.
The shRNAi probes were also characterized with respect to their ability to inhibit kinesin/TRAK-mediated redistribution of mitochondria in COS-7 cells co-transfected with pDsRed1-Mito + pKIF5C-EYFP + pEGFP-rTRAK1 and either pGreenTRAK1/pGreenTRAK2 or the respective scrambled controls, i.e. pGreenTRAK1scr/pGreenTRAK2scr. Thus, cells expressing the various test and control shRNAis were identified by green fluorescence, mitochondria were identified by red fluorescence, and the presence of both KIF5C-EYFP and TRAK1/2 was identified by the characteristic pattern of green and red fluorescence at the tips of processes, the latter due to the KIF5C/TRAK1(and 2) redistribution of mitochondria. Representative results are shown in Fig. 3. For both TRAK1 and TRAK2, the respective scrambled shRNAis had no effect on KIF5C/TRAK-mediated redistribution of mitochondria; i.e. mitochondria were enriched at the tips of processes, reminiscent of earlier findings (13, 16). No such redistribution of mitochondria was observed for TRAK1 and TRAK2 test shRNAis. Here, mitochondria remained within the cell cytoplasm (Fig. 3) despite the presence of KIF5C-EYFP as deduced by its characteristic distribution profile.
The expression of EGFP-rTRAK1silent (used in rescue experiments) was also characterized with respect to sensitivity to TRAK1 shRNAi. In the confocal microscopy imaging studies, co-expression of EGFP-rTRAK1silent with DsRed1-Mito, KIF5C-EYFP, and TRAK1 shRNAi resulted in the redistribution of mitochondria to the tips of processes (Fig. 3B).
Knockdown of TRAK1 Reduces Mitochondrial Transport in Axons of Hippocampal Neurons
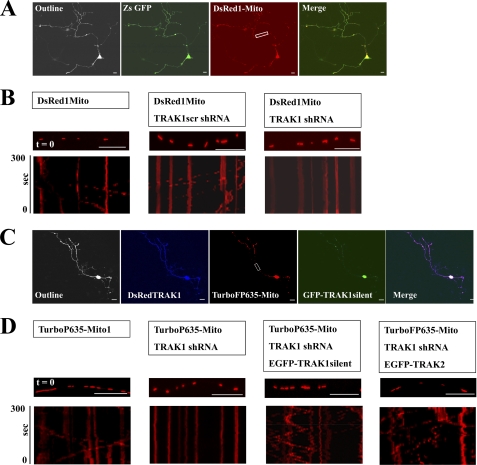
For the targeted gene knockdown of TRAK1, initially hippocampal neurons were transfected in parallel at 3 DIV with pDsRed1-Mito alone, pDsRed1-Mito + pGreenTRAK1scr, or pDsRed1-Mito + pGreenTRAK1. At 6 DIV, the mobility of mitochondria in axons of identified co-transfected neurons was studied by live imaging using confocal microscopy. A representative transfected neuron and representative kymographs are shown in Fig. 4, A and B. Table 3 summarizes the effects of TRAK1 knockdown by shRNAi on the parameters of mitochondrial dynamics. Accompanying representative videos are available as supplemental Videos 4–6.
FIGURE 4.
Knockdown of TRAK1 by targeted TRAK1 shRNAi reduces mitochondrial transport in axons of hippocampal neurons. Hippocampal neurons prepared from P0 rat brain were transfected at 3 DIV with either pDsRed1-Mito alone, pDsRed1-Mito + the scrambled shRNAi, pGreenTRAK1scr, or pDsRed1-Mito + pGreenTRAK1 or for the rescue experiments with pTurboFP635-NMito, pTurboFP635-NMito + pRedTRAK1, pTurboFP635-NMito + pRedTRAK1 + pEGFP-rTRAK1silent, or pTurboFP635-NMito + pRedTRAK1 + EGFPTRAK2 and imaged at 6 DIV all as described under “Experimental Procedures.” A and C are representative examples of transfected neurons where Outline refers to an image with saturated fluorescence intensity to show the complete cell outline. In A, ZsGFP shows the green fluorescence enabling identification of transfected neurons, DsRed1-Mito shows the distribution of mitochondria, and Merge shows the merge of ZsGFP + DsRed1-Mito fluorescence. In C, DsRedTRAK1 shows the red fluorescence (colored here blue) for the TRAK1 shRNAi vector, TurboP635-Mito shows the distribution of mitochondria, GFP-TRAK1silent shows the fluorescence due to EGFP-rTRAK1silent, and Merge shows a merge of all three. B and D show a section of an axon at time t = 0 for each condition as labeled with the respective kymographs below. Scale bars are 20 μm. The parameters of mitochondrial dynamics are summarized in Table 2.
TABLE 3.
Summary of effects of TRAK1 gene knockdown on mitochondrial dynamics in axons of hippocampal pyramidal neurons
Hippocampal neurons were transfected in parallel at 3 DIV with either pDsRed1-Mito, pDsRed1-Mito + pGreenTRAK1scr, pDsRed1-Mito + pGreenTRAK1, pTurboFP635-NMito1, pTurboFP635-NMito1 + pRedTRAK1, pTurboFP635-NMito1 + pRedTRAK1 + pEGFP-rTRAK1silent, or pTurboFP635-NMito1 + pRedTRAK1 + pEGFP-TRAK2. At 6 DIV, co-transfected neurons were identified, mitochondrial dynamics were imaged by confocal microscopy, and the results were analyzed using Volocity software all as described under “Experimental Procedures.” Values are the means ± S.E. from n = 17 images from n = 8 neurons for pDsRed1-Mito (180 mitochondria imaged), n = 29 images from n = 14 neurons for pDsRed1-Mito + pGreenTRAK1s transfections (299 mitochondria imaged), n = 42 images from n = 23 neurons for pDsRed1-Mito + pGreenTRAK1 (406 mitochondria imaged), n = 28 images from n = 18 neurons for pTurboFP635-NMito1 (310 mitochondria imaged), n = 33 images from n = 16 neurons for pTurboFP635-NMito1 + pRedTRAK1 (335 mitochondria imaged), n = 35 images from n = 19 neurons for pTurboFP635-NMito1 + pRedTRAK1 + pEGFP-ratTRAK1 (323 mitochondria imaged), and n = 29 images from n = 17 neurons for pTurboFP635-NMito1 + pRedTRAK + pEGFP-TRAK2 (333 mitochondria imaged) for at least n = 3 independent transfection experiments except where noted. Statistical significances were obtained using the Student's t test.
| Clones transfected | Mitochondrial density | Velocity | Fusion events/10 mitochondria | Fission events/10 mitochondria | Oscillating mitochondria | Stationary mitochondria | Mobile mitochondria | Anterograde mitochondria | Retrograde mitochondria |
|---|---|---|---|---|---|---|---|---|---|
| μm−1 | μm/s | % | % | % | % | % | |||
| pDsRed1-Mito | 0.21 ± 0.03 | 0.40 ± 0.06 | 0.7 | 0.7 | 2 ± 1 | 64 ± 2 | 36 ± 2 | 17 ± 2 | 16 ± 3 |
| pDsRed1-Mito pGreenTRAK1scr | 0.16 ± 0.02 | 0.45 ± 0.05 | 0.7 | 0.8 | 1 ± 1 | 64 ± 3 | 36 ± 3 | 16 ± 1 | 18 ± 3 |
| pDsRed1-Mito pGreenTRAK1 | 0.14 ± 0.02 | 0.38 ± 0.07 | 0.4a | 0.3b | 2 ± 1 | 84 ± 3c | 16 ± 3d | 7 ± 2d | 7 ± 1d |
| pTurboFP635-NMito1 | 0.18 ± 0.01 | 0.33 ± 0.05 | 0.6 | 0.7 | 0 ± 0 | 68 ± 3 | 32 ± 3 | 16 ± 2 | 16 ± 3 |
| pTurboFP635-NMito1 pRedTRAK1 | 0.13 ± 0.02 | 0.34 ± 0.05 | 0.3b | 0.3b | 2 ± 1 | 84 ± 4c | 14 ± 4c | 7 ± 2d | 7 ± 2d |
| pTurboFP635-NMito1 pRedTRAK1 pEGFP-rTRAK1silent | 0.15 ± 0.02 | 0.42 ± 0.04 | 0.6 | 0.7 | 4 ± 3 | 49 ± 7d | 51 ± 7d | 26 ± 4c | 21 ± 5a |
| pTurboFP635-NMito1e pRedTRAK1 pEGFP-TRAK2 | 0.13 ± 0.01 | 0.46 ± 0.04 | 0.6 | 0.7 | 3 | 46 | 54 | 23 | 28 |
a p < 0.05.
b p < 0.01.
c p < 0.0005.
d p < 0.005.
e n = 2 transfections were carried out.
All the parameters for the control transfections, i.e. pDsRed1-Mito alone or pDsRed1-Mito + the scrambled shRNAi, pGreenTRAK1scr, were comparable with those found for the controls of the TRAK2 DN study. Knockdown of TRAK1 resulted in a significant percent decrease in mobile mitochondria. A 56 ± 6% decrease in the percentage of mobile mitochondria was observed. Interestingly and similar to the findings for the TRAK2 DN study, significant decreases in the percentage of mobile mitochondria moving in both anterograde (56 ± 4%) and retrograde (64 ± 7%) directions were found. Furthermore, the number of fusion and fission events was also decreased, although again, the significance was less than that found for the effect of knockdown of TRAK1 on mitochondrial mobility (Table 3). No differences in mitochondrial density, mean velocities, and the percentage of oscillating mitochondria were induced by TRAK1 gene knockdown. There were no obvious changes in mitochondrial morphology.
Experiments designed to rescue the decrease in TRAK1 gene knockdown mitochondrial mobility were also carried out. To do this, it was necessary to be able to visualize neurons in which three fluorescent constructs were expressed. Thus, the new mitochondrial targeting vector pTurboFP635-NMito1 was generated. This enabled visualization of mitochondria in the far-red fluorescence by setting the emission filter detection bands between λ = 650 and 700 nm. The TRAK1 shRNAi sequence was subcloned into the vector pSIRENRed for visualization of red fluorescence, setting the emission filter detection bands between λ = 550 and 590 nm. The pEGFP-rTRAK1silent construct was visualized by setting the emission filter detection bands between λ = 500 and 550 nm. Fig. 4C shows a representative transfected neuron. EGFP fluorescence (i.e. rTRAK1) was enriched in the cell body and at the tips of some processes, probably the growth cones. It was also evident at lower concentrations as puncta throughout most processes. The live imaging results are summarized in Fig. 4B, Table 3, and supplemental Videos 7–10. First, the parameters for mitochondrial dynamics were similar for the control pTurboFP635-NMito1 compared with those for pDsRed1-Mito. Second, transfection with pRedTRAK1 resulted in an overall 57 ± 9% decrease of mobile mitochondria with decreases found for both anterograde (64 ± 9%) and retrograde (59 ± 14%) transport. The number of fusion and fission events was also significantly decreased. Co-expression of pEGFP-rTRAK1 resulted in the rescue of all induced changes (Table 3). Thus, there was an increase of 337 ± 123% in the number of mobile mitochondria in EGFP-rTRAK1+ TRAK1 shRNAi silent-transfected neurons compared with TRAK1 shRNAi-transfected neurons. The effects were more pronounced on anterograde (i.e. 440 ± 210%) versus retrograde (i.e. 290 ± 50%) transport, and now, there was no significant difference between fusion and fission events in test versus controls. The percentage of mobile mitochondria in the rescue experiments was higher than under basal, control conditions (Table 3). The percent increase in mobile mitochondria comparing pTurboFP635-NMito1 + pRedTRAK1 + pEGFP-rTRAK1silent- with pTurboFP635-NMito1-transfected neurons was 63 ± 8%.
Rescue experiments were also attempted using EGFP-TRAK2. Findings similar to those obtained for TRAK1 were obtained; i.e. TRAK2 was able to reverse the TRAK1 shRNAi-induced arrest of mitochondria. The percentage of mobile mitochondria in the rescue experiments was higher than under basal, control conditions with increases in both anterograde and retrograde transport. The decrease in the number of fusion and fission events induced by TRAK1 shRNAi was now reversed such that no differences were evident between controls and EGFP-TRAK2 rescue transfections. Furthermore, no change in mean mitochondrial velocity or mitochondrial density and morphology over the 5-min period of live imaging was observed (Table 3).
Effects of Knockdown of TRAK2 on Mitochondrial Dynamics in Axons of Hippocampal Neurons
Experiments to determine the effects of TRAK2 gene knockdown on mitochondrial dynamics in axons of hippocampal neurons were also carried out. The findings are summarized in Table 4 and supplemental Fig. 1 and Videos 11–13. There were no significant effects of either the TRAK2 or TRAK2-2 shRNAs on any of the parameters of mitochondrial dynamics despite the comparable activity between TRAK1 and TRAK2 shRNAis in the validation experiments.
TABLE 4.
Summary of effects of TRAK2 gene knockdown on mitochondrial dynamics in axons of hippocampal pyramidal neurons
Hippocampal neurons were transfected in parallel at 3 DIV with either pDsRed1-Mito, pDsRed1-Mito + pGreenTRAK2scr, pDsRed1-Mito + pGreenTRAK2, or pDsRed1-Mito + pGreenTRAK2-2. At 6 DIV, co-transfected cells were identified, mitochondrial dynamics were imaged by confocal microscopy, and the results were analyzed using Volocity software all as described under “Experimental Procedures.” Oscillating mitochondria are defined as those that are displaced >2 μm from one site. Values are the means ± S.E. from n = 35 images from n = 18 neurons for pDsRed1-Mito (343 mitochondria imaged), n = 29 images from n = 15 neurons for pDsRed1-Mito + pGreenTRAK2scr transfections (251 mitochondria imaged), n = 39 images from n = 23 neurons for pDsRed1-Mito + pGreenTRAK2 (325 mitochondria imaged), and n = 27 images from n = 15 neurons for pDsRed1-Mito + pGreenTRAK2-2 (306 mitochondria imaged) for at least n = 3 independent transfection experiments.
| Clones transfected | Mitochondrial density | Velocity | Fusion events/10 mitochondria | Fission events/10 mitochondria | Oscillating mitochondria | Stationary mitochondria | Mobile mitochondria | Anterograde mitochondria | Retrograde mitochondria |
|---|---|---|---|---|---|---|---|---|---|
| μm−1 | μm/s | % | % | % | % | % | |||
| pDsRed1-Mito | 0.17 ± 0.03 | 0.41 ± 0.1 | 0.8 | 0.8 | 2 ± 1 | 69 ± 5 | 31 ± 5 | 14 ± 3 | 15 ± 3 |
| pDsRed1-Mito pGreenTRAK2scr | 0.14 ± 0.03 | 0.43 ± 0.11 | 0.7 | 0.7 | 1 ± 1 | 65 ± 5 | 35 ± 4 | 17 ± 2 | 17 ± 2 |
| pDsRed1-Mito pGreenTRAK2 | 0.15 ± 0.03 | 0.42 ± 0.07 | 0.7 | 0.7 | 1 ± 1 | 67 ± 3 | 33 ± 3 | 16 ± 1 | 16 ± 2 |
| pDsRed1-Mito pGreenTRAK2-2 | 0.18 ± 0.03 | 0.34 ± 0.05 | 0.6 | 0.8 | 1 ± 1 | 68 ± 3 | 32 ± 3 | 16 ± 2 | 15 ± 2 |
Analysis of Velocity in TRAK-impaired Mitochondrial Transport in Axons of Hippocampal Neurons
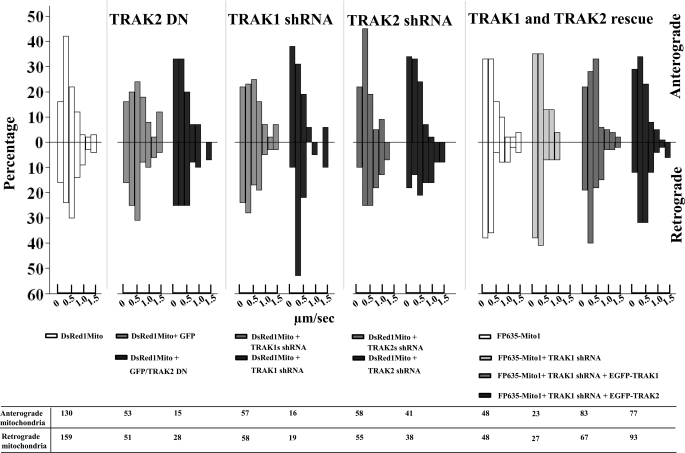
The velocities of all moving mitochondria were compared in axons of hippocampal neurons that had been transfected with TRAK2 DN, TRAK1 shRNAi, TRAK2 shRNAi, and shRNAi + EGFP-TRAK1 or EGFP-TRAK2 (for rescue experiments) together with appropriate controls (data from Tables 2, 3, and 4). The results are summarized in Fig. 5. The main features are that, in control transfections, the majority of mitochondria move in both anterograde and retrograde directions within the range of 0–0.8 μm/s. In TRAK2 DN and TRAK1 shRNAi experiments where there was a decrease in mitochondrial mobility, there was a shift toward increased numbers of mitochondria moving with the lower velocities, i.e. 0–0.4 μm/s. The maximum velocity recorded was 2.6 μm/s in the anterograde direction and 2.4 μm/s in the retrograde direction. It was notable that of mitochondria within the >1.2 μm/s range the majority were in the retrograde direction, i.e. 21 of 29. Mitochondria with velocity >1.2 μm/s were seen in both anterograde and retrograde directions in neurons in which the mobility of mitochondria was impaired by a TRAK-dependent mechanism. It was observed that these fast moving mitochondria in the TRAK-impaired conditions were rounded with a small, 0.2-μm diameter.
FIGURE 5.
Velocity profiles for mobile mitochondria in axons of hippocampal neurons: effect of TRAK1-induced arrest and TRAK1 and TRAK2 rescue of movement. The velocities of all mobile mitochondria in axons of hippocampal pyramidal neurons from experiments described in Tables 1–3 were determined as described under “Experimental Procedures.” Velocities were sorted into the following ranges: 0–0.2, 0.2–0.4, 0.4–0.6, 0.6–0.8, 0.8–1, 1.0–1.2, and >1.2 μm/s. The figure shows the percentage for each velocity range in both anterograde and retrograde directions for each condition together with the number of mobile mitochondria analyzed.
DISCUSSION
In this study, we have shown using both targeted gene knockdown and dominant negative studies that disruption of the kinesin-TRAK-Miro mitochondrial trafficking complex resulted in a reduction of basal mitochondrial mobility in the axons of hippocampal neurons. These findings prove a role for TRAK1 as an endogenous mediator of mitochondrial trafficking in axons of hippocampal neurons.
Furthermore, we report the ability of TRAK1silent and TRAK2 to rescue TRAK1 shRNAi-induced arrest of mitochondrial mobility at 6 DIV. Our findings support a central role for TRAK1 and TRAK2 in mitochondrial trafficking mechanisms but suggest that the contributions of each to the mediation and regulation of this transport may be distinct.
The most marked effect of shRNAi gene knockdown and dominant negative studies was the percent decrease in the mobile fraction of mitochondria. If both TRAK1 and TRAK2 contribute to the transport of mitochondria, it would be expected that TRAK2 DN would yield an increased percentage of immobile mitochondria compared with each TRAK shRNA because it was shown to block the binding of both TRAK1 and TRAK2 to kinesin (Fig. 1). But, although the percent decrease in mobility for TRAK2 DN was higher than that found for TRAK1 shRNAi, suggesting a possible involvement of TRAK2, the values were not significant. Furthermore, transfection of neurons with two different TRAK2 shRNAis had no effect on mitochondrial mobility despite the fact that both had an efficacy comparable with TRAK1 shRNAi in heterologous expression. Failure to attain a complete arrest of mitochondrial movement may be explained by the efficacy of the TRAK1 shRNAi and TRAK2 DN reagents. Alternatively, there may be TRAK-dependent and TRAK-independent modes of mitochondrial transport. Macaskill et al. (6) showed by in vitro pulldown assays that Miro can associate directly with KIF5A, KIF5B, and KIF5C, implying that not all mitochondrial transport occurs via kinesin adaptor-mediated mechanisms. Alternatively, different modes of transport may be distinguished by the use and availability of different motors and different adaptors. For example, the syntaxin-binding protein, syntabulin, is implicated in mitochondrial transport. In hippocampal neurons in culture, syntabulin co-localizes with mitochondria, it co-migrates with mitochondria along processes, and siRNA knockdown of syntabulin results in a significant reduction in the mitochondrial density in processes (26). Syntabulin utilizes the KIF5B motor (28), whereas TRAKs (both 1 and 2) co-immunoprecipitate from brain extracts predominantly with KIF5A (12).
When the decrease in mitochondrial mobility was analyzed further, it was apparent that TRAK2 DN- and TRAK1 shRNAi-induced arrest of mitochondria mobility resulted in significant decreases in both anterograde and retrograde transport (Tables 2 and 3). This was unexpected because TRAKs bind directly to kinesin (13), and it is generally accepted that kinesins mediate anterograde transport. Association of TRAKs with dynein has been investigated by immunoprecipitations from brain extracts; however, no dynein was detected in anti-TRAK1/2 immune pellets (12). In Drosophila, however, it was found that Miro (and by implication TRAKs/Milton) was required for both anterograde and retrograde transport of axonal mitochondria (29). In investigating mediators of mitochondrial transport, some published reports do not discriminate between the anterograde and retrograde modes of transport, focusing only on the fact that there is an overall reduction in mobility (e.g. Ref. 26 for syntabulin siRNA-mediated arrest and Ref. 6 for Miro1 shRNAi-mediated arrest). But Misko et al. (19) recently reported that knock-out of the outer mitochondrial protein mitofusin, a dynamin family GTPase that is commonly mutated in Charcot-Marie-Tooth disease, resulted in an increase in the time paused for both anterograde and retrograde moving mitochondria in axons of dorsal root ganglia neurons. It has been proposed that there may be an efficient regulatory scheme whereby the direction of mobility of intracellular organelles is determined by the number of motors bound to the cargo, i.e. the stochastic tug-of-war model (for a review, see Ref. 30). By this model, when TRAKs are down-regulated or their binding to KHC is inhibited, then the number of kinesin motors associated with mitochondria will be decreased, resulting in the predominance of dynein-mediated, retrograde transport. However, the empirical observation here was that the arrest of mitochondrial mobility resulted in a decrease in retrograde transport. It may be that there is an intrinsic mechanism perhaps involving cross-talk between dynein and kinesin to maintain homeostasis of mitochondrial movement under basal conditions. It was notable that concomitant with the decrease in both anterograde and retrograde mobility there was a tendency toward an increase in the number of mitochondria moving with slow velocities (Fig. 5). This may be a result of a decrease in the number of kinesin motors attached indirectly to mitochondria.
A second unexpected finding was that the decreased mobility of mitochondria induced by TRAK2 DN and TRAK1 shRNAi was accompanied by decreases in the frequency of mitochondrial fusion and fission events (Tables 2 and 3). These significant changes were reversed in the TRAK1silent and TRAK2 rescue experiments (Table 3). TRAK1 and TRAK2 co-immunoprecipitate with mitofusins 1 and 2 following overexpression of each in HEK 293 cells (19), suggesting an in vivo association between TRAKs and mitofusins, although this may not necessarily be direct. Mitofusins are implicated in facilitating fusion of mitochondria in addition to playing a role in axonal transport, suggesting possible interplay between the two mechanisms. Furthermore, overexpression of Miro enhanced the fusion state of mitochondria under basal Ca2+ concentrations, again reinforcing the concept of cross-talk between mitochondrial transport and fusion/fission mechanisms (31). Evidence suggests that Miro is not directly involved in fusion or fission but that it may modulate these processes (29, 31). Because of the known association between Miro and TRAKs, this implicates a role for TRAKs in this modulation.
Transfection of hippocampal neurons with TRAK1shRNAi but not TRAK2 shRNAi resulted in a significant impairment of mitochondrial mobility. The conservation in amino acid sequence between TRAK1 and TRAK2 and the similarity in their known functional properties (i.e. their direct binding to the cargo domains of KHC; their ability to co-distribute with mitochondria, resulting in the collapse of the mitochondrial network when overexpressed in cells; the redistribution of mitochondria to the tips of processes when either TRAK1 or TRAK2 are co-expressed with KHC; and the co-immunoprecipitation of Miro1 or Miro2 with either TRAK1 or TRAK2 following overexpression in mammalian cells) suggest that both proteins have similar functions (9, 12, 13, 15, 17). Yet despite the finding that TRAK1 and TRAK2 shRNAs had similar efficacies in heterologous expression, TRAK2 shRNAis had no effect on mitochondrial mobility. There are several possible explanations for this apparent inconsistency. The most obvious is that TRAK2 is not expressed in axons of hippocampal pyramidal neurons at this early stage in development, i.e. 6 DIV from P0 animals. However, we have previously described the distribution of TRAK2 using specific anti-TRAK2 antibodies in neuronal cultures prepared from hippocampi of E18 rats at 14 DIV (equivalent to 11 DIV for P0 cultures as used here; Ref. 18). Anti-TRAK2 immunoreactivity was present throughout all neuronal processes, and ∼50% was co-distributed with mitochondria (18). The developmental profiles of TRAKs 1 and 2 were determined by analyzing their time-dependent expression in P0 hippocampal neurons in culture by immunoblotting using anti-TRAK2(8–633) (which recognizes TRAK1 and TRAK2) and anti-TRAK2(874–889) (which recognizes TRAK2 only) antibodies (supplemental Fig. 2). Immunoreactive signals were observed at all DIV for both antibodies, but the signal was more robust for anti-TRAK2(8–633), suggesting that although both TRAK1 and TRAK2 are expressed TRAK1 is present at higher levels. Other possible explanations for the observed differences between the effects of TRAK1 versus TRAK2 shRNAs on mitochondrial mobility may include their distinct subcellular distributions such as dendritic versus axonal localization within a single neuronal cell type, different turnover times of the two proteins, and the availability and specificity of kinesin motor proteins.
A further consideration is that TRAKs may serve different functions. TRAK1 and TRAK2 have both been implicated in regulating endosome to lysosome trafficking by virtue of their association in HeLa and pheochromocytoma 12 cells with hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs; Refs. 20 and 21). Also, overexpression of Hrs recruits TRAK2 to Hrs-positive endosomal compartments (18). Thus, TRAK1 and TRAK2 may participate in microtubule-based transport of early endosomes by acting as an adaptor linking Hrs-containing endosomes to kinesin (20, 21). TRAKs have also been linked to the forward trafficking of inhibitory GABAA neurotransmitter receptors (9, 32) and Kir2.1 inwardly rectifying potassium channels (33). TRAKs may therefore be promiscuous in terms of the cargo they transport. Cargoes may be determined by availability and the needs of the cell. It is of note that although at any one time all mitochondria contain Miro under resting conditions (i.e. in the culture conditions in which the neurons were grown) only ∼50% are co-distributed with TRAK2 immunoreactivity (18). This value is comparable with the ∼35% of the mitochondrial population that are mobile. Increasing the level of Miro enhances association of TRAKs with mitochondria as does regulating Miro GTPase activity (18). It is not clear whether these manipulations are sufficient to engage kinesin and to enhance mobility via the formation of the kinesin-TRAK-Miro trafficking complex. Additional studies are necessary to elucidate the signaling mechanisms that initiate the formation and indeed the dissociation of the kinesin-TRAK-Miro trafficking complex. Furthermore, it will be important to determine the role of TRAK2 compared with TRAK1 in regulating mitochondrial transport. It may be that both TRAK1 and TRAK2 mediate transport but that there is a preference for TRAK1, so although TRAK1 is present, this is dominant. But should TRAK1 levels be compromised as in the shRNAi studies, TRAK2 has the capacity to compensate for the lack of TRAK1 availability. Finally, it is of importance to determine whether neuronal function is compromised in neurons in which mitochondrial mobility is impaired. This may be central to understanding the pathogenesis of neurodegenerative diseases in which mitochondrial mislocalization has been observed.
Supplementary Material
Acknowledgments
We thank Dr. Karine Pozo for help and advice on the culturing and transfection of hippocampal pyramidal neurons. We also thank Dr. Sarah Cousins for generating the clones pTurboFP635-NMito1, pEGFP-ratTRAK1silent, and pIRES-GFPTRAK2 DN; Tom Randall for help with Fig. 5; and Dr. Carolyn Moores, Birkbeck College, London, UK for insightful discussions. We gratefully acknowledge the financial contribution from The Wellcome Trust for the purchase of the Zeiss 710 confocal microscope.
This work was supported by the Biotechnology and Biological Sciences Research Council (United Kingdom).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Videos 1–13.
- TRAK
- trafficking kinesin protein
- DIV
- days in vitro
- DN
- dominant negative
- EGFP
- enhanced green fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- hrGFP
- humanized recombinant green fluorescent protein
- hrs
- hepatocyte growth factor-regulated tyrosine kinase substrate
- KHC
- kinesin heavy chain
- scr
- scrambled
- rTRAK
- rat TRAK.
REFERENCES
- 1. Boldogh I. R., Pon L. A. (2007) Trends Cell Biol. 17, 502–510 [DOI] [PubMed] [Google Scholar]
- 2. Detmer S. A., Chan D. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 870–879 [DOI] [PubMed] [Google Scholar]
- 3. MacAskill A. F., Kittler J. T. (2010) Trends Cell Biol. 20, 102–112 [DOI] [PubMed] [Google Scholar]
- 4. Yi M., Weaver D., Hajnóczky G. (2004) J. Cell Biol. 167, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang D. T., Honick A. S., Reynolds I. J. (2006) J. Neurosci. 26, 7035–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macaskill A. F., Rinholm J. E., Twelvetrees A. E., Arancibia-Carcamo I. L., Muir J., Fransson A., Aspenstrom P., Attwell D., Kittler J. T. (2009) Neuron 61, 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang X., Schwarz T. L. (2009) Cell 136, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stephenson F. A., Brickley K. (2010) in Protein Folding for the Synapse (Wittenbach A., O'Connor V. eds) pp. 105–119, Springer, New York [Google Scholar]
- 9. Beck M., Brickley K., Wilkinson H. L., Sharma S., Smith M., Chazot P. L., Pollard S., Stephenson F. A. (2002) J. Biol. Chem. 277, 30079–30090 [DOI] [PubMed] [Google Scholar]
- 10. Iyer S. P., Akimoto Y., Hart G. W. (2003) J. Biol. Chem. 278, 5399–5409 [DOI] [PubMed] [Google Scholar]
- 11. Stowers R. S., Megeath L. J., Górska-Andrzejak J., Meinertzhagen I. A., Schwarz T. L. (2002) Neuron 36, 1063–1077 [DOI] [PubMed] [Google Scholar]
- 12. Brickley K., Smith M. J., Beck M., Stephenson F. A. (2005) J. Biol. Chem. 280, 14723–14732 [DOI] [PubMed] [Google Scholar]
- 13. Smith M. J., Pozo K., Brickley K., Stephenson F. A. (2006) J. Biol. Chem. 281, 27216–27228 [DOI] [PubMed] [Google Scholar]
- 14. Koutsopoulos O. S., Laine D., Osellame L., Chudakov D. M., Parton R. G., Frazier A. E., Ryan M. T. (2010) Biochim. Biophys. Acta 1803, 564–574 [DOI] [PubMed] [Google Scholar]
- 15. Glater E. E., Megeath L. J., Stowers R. S., Schwarz T. L. (2006) J. Cell Biol. 173, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brickley K., Pozo K., Stephenson F. A. (2011) Biochim. Biophys. Acta 1813, 269–281 [DOI] [PubMed] [Google Scholar]
- 17. Fransson S., Ruusala A., Aspenström P. (2006) Biochem. Biophys. Res. Commun. 344, 500–510 [DOI] [PubMed] [Google Scholar]
- 18. MacAskill A. F., Brickley K., Stephenson F. A., Kittler J. T. (2009) Mol. Cell. Neurosci. 40, 301–312 [DOI] [PubMed] [Google Scholar]
- 19. Misko A., Jiang S., Wegorzewska I., Milbrandt J., Baloh R. H. (2010) J. Neurosci. 30, 4232–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kirk E., Chin L. S., Li L. (2006) J. Cell Sci. 119, 4689–4701 [DOI] [PubMed] [Google Scholar]
- 21. Webber E., Li L., Chin L. S. (2008) J. Mol. Biol. 382, 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goslin K., Asmussen H., Banker G. (1998) in Culturing Nerve Cells (Banker G., Goslin K. eds) 2nd Ed., pp. 339–370, MIT Press, Cambridge, MA [Google Scholar]
- 23. Jiang M., Chen G. (2006) Nat. Protoc. 1, 695–700 [DOI] [PubMed] [Google Scholar]
- 24. Ojla G., Beck M., Brickley K., Stephenson F. A. (2003) Br. Neurosci. Abstr. 17, P26.12 [Google Scholar]
- 25. Ligon L. A., Steward O. (2000) J. Comp. Neurol. 427, 340–350 [DOI] [PubMed] [Google Scholar]
- 26. Cai Q., Gerwin C., Sheng Z. H. (2005) J. Cell Biol. 170, 959–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang J. S., Tian J. H., Pan P. Y., Zald P., Li C., Deng C., Sheng Z. H. (2008) Cell 132, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai Q., Pan P. Y., Sheng Z. H. (2007) J. Neurosci. 27, 7284–7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Russo G. J., Louie K., Wellington A., Macleod G. T., Hu F., Panchumarthi S., Zinsmaier K. E. (2009) J. Neurosci. 29, 5443–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W., 3rd, Tokito M., Holzbaur E. L. (2010) Curr. Biol. 20, 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saotome M., Safiulina D., Szabadkai G., Das S., Fransson A., Aspenstrom P., Rizzuto R., Hajnóczky G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20728–20733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gilbert S. L., Zhang L., Forster M. L., Anderson J. R., Iwase T., Soliven B., Donahue L. R., Sweet H. O., Bronson R. T., Davisson M. T., Wollmann R. L., Lahn B. T. (2006) Nat. Genet. 38, 245–250 [DOI] [PubMed] [Google Scholar]
- 33. Grishin A., Li H., Levitan E. S., Zaks-Makhina E. (2006) J. Biol. Chem. 281, 30104–30111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.