Abstract
The tumor suppressor PTEN (phosphatase and tensin homologue) negatively regulates the PI3K pathway through its lipid phosphatase activity and is one of the most commonly lost tumor suppressors in human cancers. Though the tumor suppressive function involves the lipid phosphatase-dependent and -independent activities of PTEN, the mechanism leading to the phosphatase-independent function of PTEN is understood poorly. Some PTEN mutants have lipid phosphatase activity but fail to suppress cell growth. Here, we use a cancer-associated mutant, G20E, to gain insight into the phosphatase-independent function of PTEN by investigating protein-protein interactions using MS-based stable isotope labeling by amino acids in cell culture (SILAC). A strategy named parallel affinity purification (PAP) and SILAC has been developed to prioritize interactors and to compare the interactions between wild-type and G20E PTEN. Clustering of the prioritized interactors acquired by the PAP-SILAC approach shows three distinct clusters: 1) wild-type-specific interactors, 2) interactors unique to the G20E mutant, and 3) proteins common to wild-type and mutant. These interactors are involved mainly in cell migration and apoptosis pathways. We further demonstrate that the wild-type-specific interactor, NUDTL16L1, is required for the regulatory function of wild-type PTEN in cell migration. These findings contribute to a better understanding of the mechanisms of the phosphatase-dependent and -independent functions of PTEN.
Keywords: Apoptosis, Cell Migration, Mass Spectrometry (MS), Protein-Protein Interactions, Proteomics, Tumor Suppressor
Introduction
PTEN is a tumor suppressor gene that frequently is somatically deleted or mutated in a variety of human cancers, including those of the brain, endometrium, prostate, and lung (1, 2). Germ line mutations of PTEN are the cause of Cowden disease and Bannayan-Riley-Ruvalcaba syndrome, an autosomal dominant hamartoma syndrome with increased risk for the development of tumors in a variety of tissues (1, 2). The PTEN protein consists of N-terminal phosphatase, and C-terminal C2, phosphorylation, and PDZ (PSD-95, DLG1, and ZO-1) binding domains (2). The catalytic domain of PTEN functions to dephosphorylate the 3′ position of the phospholipids PI(3,4,5)P3 (PIP3) and PI(3,4)P2 (3). As such, PTEN lipid phosphatase activity regulates the Akt serine/threonine kinase pathway through modulation of PIP3 levels and hence, regulates cell cycle progression, apoptosis, and migration, cell proliferation and motility, which also are critical for tumor development (4). Thus, deregulation of the PI3K/PTEN/Akt pathway has been found in many malignant cancers.
More than 20% of all known PTEN mutations in tumors are located outside of the catalytic site, and these mutants exhibit normal phosphatase activity (1), suggesting two possibilities. First, mutations in the interacting domain for the regulatory proteins of PTEN may affect its ability for tumor suppression through PTEN phosphatase activity, which is regulated by PTEN interactors (5–10). Indeed, a PTEN mutation within a binding site of PICT1, which controls stability of PTEN, affects the protein level of PTEN and thereby reduces its enzymatic activity in the cells (11). Second, some cancer-associated mutations of PTEN may harbor mutation of a region with tumor suppressive functions distinct from those mediated by the catalytic activity, as these mutants still possess the phosphatase activity (12). Emerging evidence suggests other PTEN functions that are unrelated to PI3K/Akt signaling, indicating PTEN enzymatic activity, will not cover the entire mechanism of its tumor-suppressive ability (13, 14). For example, independent of the phosphatase domain, the C2 domain in the C-terminal region of PTEN can regulate cell migration (15), affect p53 transcriptional activity by competing with MDM2 for direct binding (16, 17), and suppress the transformation of MSP58 (18). Thus, we hypothesize that the inactivation of tumor suppression by somatic mutation of PTEN is caused by lack of, or changes to, key PTEN interaction partners, which regulate the tumor suppressive function of PTEN. Identifying interacting partners of a PTEN mutant retaining phosphatase activity may therefore uncover new PTEN regulatory networks involved in a phosphatase-independent function.
There are several studies that have used comparison of the interacting proteins of the wild-type and cancer-associated mutants to elucidate their specific functions. For example, alteration of protein interactions has been used to explain p53 gain of function (19, 20). The instability of PTEN correlated with its missense mutations and the reduction of HIF1α degradation with the missense mutation of its interacting protein also have been shown to involve protein interactions (11, 21). Protein-protein interactions are likely to be an important mechanism to regulate protein function when the protein has no inherent enzymatic activity. As wild-type and G20E PTEN have no enzymatic activity other than phosphatase activity, the phosphatase-independent function is likely to result from changes in protein-protein interactions, which direct PTEN subcellular localization, modifications, protein stability, and regulate downstream targets of biological processes. Thus, mapping protein interactions between PTEN wild-type and G20E mutant may provide a route to understanding how mutants deregulate tumor suppressor activity although they have phosphatase activity.
Affinity purification coupled with MS is a powerful tool for deciphering protein interaction networks at the proteome level (22–24). The sensitivity and precision of current mass spectrometric technology for protein identification allows the determination of ever larger numbers of proteins in immunoaffinity and pulldown experiments. In addition to bona fide interaction partners, however, these expanding lists may include increased numbers of nonspecific and contaminant proteins. Finding universal methods for removal of nonspecific partners while retaining specific interactions has proven difficult.
In MS-based methods of interactomics, tandem affinity purification (TAP) is a generic two-step affinity purification method that enables the isolation of protein complexes under close to physiological conditions for subsequent analysis by mass spectrometry (25, 26). Proteins from an affinity-purified sample are separated by one-dimensional SDS-PAGE, and after in-gel tryptic digestion, the resulting peptides are analyzed by mass spectrometry. To be successful, the method generally requires stable interactions between bait and prey. Proteins of transient or weaker interactors may be difficult to detect on one-dimensional SDS-PAGE because of masking by high abundance proteins or by loss in the two-step work-up. Thus, a high sensitivity method for the whole interactome with low stringency conditions during purification is needed to capture most or all of the interactors in a given complex.
SILAC2 has been shown to distinguish specific interactions from background using a one-step affinity purification and has been applied to the analysis of complete interactomes (27, 28). In SILAC, the proteome is labeled metabolically by incorporating either a normal or a heavy isotope-substituted amino acid, such as 13C/15N-labeled lysine or arginine. Peptides derived from two otherwise identical samples can be differentiated in a mass spectrometer owing to their mass difference. The ratio of the integrated signal intensities of these peptide “SILAC” pairs accurately reflects the abundance ratio of the corresponding proteins. Thus, the interactors of a target protein can be identified by direct comparison of the peptide ratios between “pulled down” proteins (28). Nonetheless, a one-step affinity purification likely captures both direct interactors and secondary interactors and may result in a long protein list with high SILAC ratios that may not reflect direct interactors. It is therefore important to develop a strategy for prioritizing the interactors from this primary list of SILAC-based interactors to mine critical molecular targets.
Here, we describe specific interactors of PTEN wild-type and a cancer-associated PTEN mutant retaining lipid phosphatase activity by a modified SILAC-based approach, PAP, and analyze the interactors in terms of functional networks involved in oncogenic pathways using bioinformatics. We further show functional relevance of a wild-type PTEN-specific interactor in cell migration.
EXPERIMENTAL PROCEDURES
Cell Culture for SILAC
The U87MG human malignant glioma cell line (obtained from ATCC; HTB14) was used to make stable cell lines that expressed FLAP,2 FLAP-WT PTEN, and -G20E PTEN. Tagging was based on Lap fusions strategy (29). The stable cells were maintained in DMEM with 10% FBS and 1 μg/ml of puromycin. The cells were grown in SILAC DMEM supplemented with 10% dialyzed fetal calf serum. The “light” medium (Lys-0, Arg-0) was prepared from l-arginine (84 μg/ml) and l-lysine (146 μg/ml), whereas the “heavy” medium (Lys-8, Arg-10) containing [13C]l-arginine and [13C]l-lysine (Cambridge Isotope) was prepared at the same concentrations as above. Control cells were grown in the light medium and the other in the heavy medium. Cells were grown for at least six doublings in the labeling medium.
Affinity Purification of PTEN Complexes
The cell lysates were prepared by using Nonidet P-40 lysis buffer (20 mm Tris-HCl, pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40 with protease inhibitors tablet (Roche Applied Science). Protein lysates (50 mg) were incubated with either FLAG beads (Sigma) or GFP-trap-agarose (ChromoTek). Affinity matrices incubated with the same amount of light and heavy lysate were mixed together after precipitation. The endogenous PTEN was precipitated with anti PTEN and mouse IgG beads (Santa Cruz Biotechnology). For tandem affinity purification, the cell lysates were prepared by Nonidet P-40 lysis buffer, and each lysate (50 mg) was incubated with GFP-trap-agarose. The precipitated samples were incubated with tobacco etch virus protease (Invitrogen) at 4 °C overnight. Collected supernatants were incubated with S-tag beads and eluted with SDS loading buffer.
Immunoblotting for Co-precipitation
The precipitated samples of anti-PTEN, anti-FLAG beads, and GFP-trap were immunoblotted and detected by specific antibodies: PTEN A2B1, IQGAP1 H-109 (Santa Cruz Biotechnology), ubiquitin-specific peptidase 7 (USP7) (Bethyl Laboratories), CALU, FLAG M2 (Sigma), and V5 (Invitrogen) with ECL reagent (GE Healthcare). The plasmid expressing V5-tagged NUDT16L1 was constructed into pCI neo vector by PCR (Promega). Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Mass Spectrometry
Eluted protein complexes were separated by one-dimensional SDS-PAGE and digested with trypsin using published procedures (30). Samples were analyzed on an Orbitrap or Orbitrap XL (Thermo Fisher). Survey full scan MS spectra (m/z 300–1400) were acquired with a resolution of r = 60,000 at m/z 400, an AGC target of 1e6, and a maximum injection time of 500 ms The 10 most intense peptide ions in each survey scan with an ion intensity of >2000 counts and a charge state ≥2 were isolated sequentially to a target value of 1e4 and fragmented in the linear ion trap by collisionally induced dissociation using a normalized collision energy of 35%. A dynamic exclusion was applied using a maximum exclusion list of 500 with one repeat count, repeat, and exclusion duration of 30 s.
Identification and Quantification of Peptides and Proteins
Data were searched using Mascot (version 2.2; Matrix Science, London, UK) against a concatenated target/decoy database, prepared appending a sequence-reversed by human International Protein Index (IPI) (version 3.52, 73,928 sequences) and adding common contaminants such as human keratins, porcine trypsin, and proteases to yield a total of 148,380 sequences. Cysteine carbamidomethylation was searched as a fixed modification, and N-acetylation and oxidized methionine were searched as variable modifications. Labeled arginine and lysine were specified as fixed or variable modifications, depending on the prior knowledge about the parent ion. SILAC peptide and protein quantification was performed with MaxQuant (31) using default settings. Maximum false discovery rates were set to 0.01 for both protein and peptide.
Data Analysis of TAP-tagged Sample
All MS/MS samples were analyzed using X!Tandem (version TORNADO (2008.02.01.4)). X!Tandem was set up to search the IPI HUMAN.v3.52 database (73,928 sequences) assuming the digestion enzyme trypsin. A fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 7.0 ppm were used. Iodoacetamide derivative of cysteine was specified as a fixed modification. Pyroglutamic acid from Glu of glutamic acid, pyroglutamic acid from Gln of glutamine, deamidation of asparagine, oxidation of methionine, and acetylation of the N terminus were specified as variable modifications. Scaffold (version Scaffold_2_05_02, Proteome Software, Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications.
siRNA and Cell Migration Assay
Shortly before transfection, 7.5 × 105 cells were seeded into 60-mm dishes in DMEM containing 4500 mg/liter glucose, supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. A final concentration of 25 nm siRNA (all siRNAs purchased from Dharmacon RNAi Technologies) was used in transfection using HiPerFect Transfection Reagent according to the manufacturer's protocol. Cells were incubated at 37 °C under a humidified atmosphere of 5% CO2 for 48 h. After serum starvation for 16 h, 2.5 × 105 cells in DMEM 0.5% FBS were seeded into the upper chamber with medium containing 10% FBS in the lower chamber using 24-well plates and incubated for 8 h under normal cell culture condition. The migrated cells were counted. The assays were performed in nine independent experiments.
RESULTS
SILAC-based PAP to Compare Protein Interactions of PTEN-WT and G20E Mutant
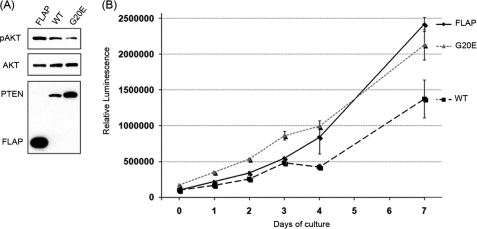
To understand the phosphatase-independent function of PTEN, we sought differences in the protein complexes of PTEN wild-type WT and mutant G20E. The mutation G20E PTEN has been found in patients with endometrial carcinoma and has been reported to show lipid phosphatase activity in vitro (12). We also confirmed this activity through measurement of phospho Akt level by transient transfection (supplemental Fig. S1). The U87MG cell line was used to make these stable cell lines because the cell line does not endogenously express wild-type PTEN allowing expression of the FLAP, FLAP fusion PTEN, or its mutant without the PTEN wild-type background. In these stable cell lines, PTEN WT and G20E mutant carry a FLAG-GFP-S-tag (FLAP) at their N termini, as constructs of G20E PTEN indicate lipid phosphatase activity through the phosphorylated Akt level (Fig. 1A). G20E showed lipid phosphatase activity in the cell but had no effect on the cell growth (Fig. 1B). Thus, we used the G20E mutant to compare the protein binding partners with those of PTEN WT.
FIGURE 1.
PTEN G20E mutant has a lipid phosphatase activity, but lacks growth suppressive ability. A, cell lysates prepared from U87 FLAP, PTEN WT, and G20E stable cells were immunoblotted with antibodies against FLAG and pAKT and AKT. B, each cell line is seeded on 96-well plates, and cell numbers are measured at the time points indicated. Cell growth of individual cell lines was measured by CellTiter-GloTM. Error bar indicates standard deviation of triplicate experiments.
The SILAC approach for capturing specific interactors and our workflow for SILAC-based parallel affinity purification are summarized in Fig. 2. For SILAC, we prepared labeled cells as follows. U87 cells expressing the tagged protein, which is either PTEN WT or G20E, are grown in heavy medium, i.e. containing 13C- and 15N-substituted-arginine and lysine and, for the control, cells expressing free FLAP are grown in light, i.e. natural isotope (12C- and 14N-) media. Protein purifications were performed by tag purification with the lysates from the labeled cells (Fig. 2A). We performed one-step purification using GFP-trap, an Escherichia coli-expressed 16-kDa protein derived from a llama heavy chain antibody that binds with high affinity and specificity to GFP (32). For the wild-type PTEN experiment, immunoprecipitation with anti-PTEN-agarose beads was carried out using heavy-labeled LN229 cell lines and anti-mouse IgG-agarose beads for the control light-labeled cell lysate. For the mutant, G20E immunoprecipitation with anti-FLAG-agarose beads was carried out using U87 G20E cells. Negative light control and experimental heavy samples were mixed and eluted using SDS buffer before mass spectrometric analysis (Fig. 2A). This reduces experimental variability that inevitably results when the samples are processed independently. Samples were mixed after incubation of the control and test samples with affinity matrix (followed by limited washes) to help capture lower affinity interactors (33). The precipitated samples were eluted using SDS-PAGE loading buffer.
FIGURE 2.
PAP-SILAC allows reliable identification and prioritization of PTEN interactors. A, specific interactions can be distinguished from background using SILAC. K0R0, Lys-0, Arg-0; K8R10, Lys-8, Arg-10. B, left: PAP for wild-type was carried out using GFP-trap and anti-PTEN antibody, whereas GFP-trap and anti-FLAG were used for G20E. Only common proteins with higher SILAC ratios (see text for details) from PAP for each experiment were extracted for clustering to identify their specificities. Right, TAP approach was carried out using GFP binder and S-tag beads with tobacco etch virus (TEV) digestion as indicated in the figure. IP, immunoprecipitation.
All eluates were fractionated by one-dimensional SDS-PAGE followed by in-gel tryptic digestion using standard procedures (30). Resulting peptides were further fractionated by nanoliquid chromatography and analyzed by high accuracy mass spectrometry (Orbitrap). SILAC ratios were determined using MaxQuant software (31). Prioritizing the proteins obtained from each SILAC analysis was based on the premise that specific interactors should be present in two samples independently purified with different affinity matrixes (and using different tag binding). In this way, we can minimize false positives that are method-dependent binders.
The experimental outline of data analyses is shown in Fig. 2B (left). We prepared precipitates using GFP-trap from both PTEN WT and G20E. For the interactors of wild-type, we performed immunoprecipitation of SILAC using LN229 cell lysate with anti-PTEN antibody-conjugated beads as the parallel experiment. This allows recognition of PTEN interactors at the endogenous expression level of the bait as well as eliminating cell line-specific interaction for U87 and LN299. For the mutant, we used FLAG pulldown using anti-FLAG antibody coupled to agarose beads as the parallel affinity purification to GFP-trap. Overall, we used two different cell lines and three different pulldowns (GFP-trap, anti-FLAG antibody, and anti-PTEN antibody) to remove method-biased interactors whose interactions with PTEN may be absent in physiological conditions. This approach is termed PAP.
Further filtering of identified proteins from the SILAC-PAP approach was carried out as follows. First, we discarded proteins with SILAC ratio<2 or undetected in at least one pulldown. The filtered list (supplemental table) contained three data sets: SILAC ratio >2 in all pull-downs, SILAC ratio >2 in only two of the wild-type pulldowns, and SILAC ratio >2 in only two of the mutant pull-downs. A heat map was generated by TMEV (TIGR) software for further clustering of the filtered interactors (supplemental table) using the log2 values of their SILAC ratios. Hierarchical clustering of the heat map of PAP data indicates three clusters: wild-type-specific (log2 H/L ratio >1 for both wild-type pulldowns and undetected for both mutant pulldowns), mutant-specific (log2 H/L ratio >1 for both mutant pulldowns and undetected for both wild-type pulldowns), and common (log2 H/L ratio >1 for at least three pulldowns, including WT endogenous) (Fig. 3). These proteins are shown in Table 1 with the associated SILAC ratios. (The heat map shows log2 values of these ratios.) In the common interactor category, USP7 and RNH1 had H/L > 2 in all experiments, whereas the others appeared with H/L > 2 in three experiments and with H/L > 0 in the other pulldowns.
FIGURE 3.
PAP-SILAC allows categorizing of PTEN wild-type and G20E interactors. A heat map under hierarchical clustering using Cosine correlation (TMEV software, TIGR) was generated for above common PAP protein list.
TABLE 1.
Prioritized PTEN interactors in PAP approach
ND, not determined.
| Gene name | WT PAP-SILAC |
G20E PAP-SILAC |
TAP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Endo | GFP | GFP | FLAG | No. of peptides | |||||||
| Ratio H/L | No. of peptides | Ratio H/L | No. of peptides | Ratio H/L | No. of peptides | Ratio H/L | No. of peptides | Flap | WT | G20E | |
| WT-specific | |||||||||||
| MYO1D | 4.4 | 2 | 33.7 | 2 | ND | ND | |||||
| GTF3C1 | 5.4 | 6 | 8.5 | 2 | ND | ND | |||||
| PGAM5 | 9.2 | 13 | 2.8 | 1 | ND | ND | |||||
| NUDT16L1 | 3.0 | 5 | 2.6 | 8 | ND | ND | |||||
| G20E-specific | |||||||||||
| AIM1 | ND | ND | 27.4 | 48 | 4.3 | 34 | 0 | 1 | 2 | ||
| GLB1 | ND | ND | 8.8 | 5 | 2.2 | 9 | |||||
| CALU | ND | ND | 11.0 | 3 | 3.6 | 3 | |||||
| RCN1 | ND | ND | 7.5 | 6 | 3.4 | 1 | |||||
| IMMT | ND | ND | 2.3 | 15 | 5.5 | 13 | |||||
| Common | |||||||||||
| LDHA | 2.7 | 8 | 1.4 | 1 | 5.2 | 10 | 4.1 | 5 | |||
| ARF4 | 2.1 | 4 | 1.3 | 4 | 2.3 | 5 | 2.1 | 2 | |||
| YWHAQ | 2.8 | 3 | 1.0 | 6 | 2.5 | 7 | 2.1 | 5 | |||
| IQGAP1 | 3.4 | 18 | 1.2 | 50 | 2.8 | 60 | 2.8 | 5 | 0 | 1 | 5 |
| CAV1 | 6.8 | 3 | 1.5 | 4 | 4.6 | 4 | 16.4 | 9 | |||
| GNB2L1 | 3.1 | 10 | 1.3 | 2 | 2.8 | 3 | 3.6 | 2 | |||
| ANXA2 | 2.4 | 19 | 1.0 | 17 | 2.3 | 18 | 2.9 | 13 | |||
| PTEN | 11.1 | 11 | 24.2 | 32 | 39.1 | 22 | 22.8 | 24 | 0 | 87 | 98 |
| USP7 | 3.2 | 8 | 7.2 | 7 | 7.3 | 12 | 5.4 | 9 | 0 | 14 | 12 |
| TUFM | 3.4 | 8 | 2.9 | 7 | 1.9 | 11 | 3.8 | 5 | |||
| RNH1 | 3.8 | 6 | 2.2 | 11 | 2.9 | 17 | 3.5 | 4 | 3 | 10 | 4 |
| CFL1 | 3.4 | 6 | 1.7 | 14 | 2.0 | 9 | 3.1 | 6 | 2 | 6 | 6 |
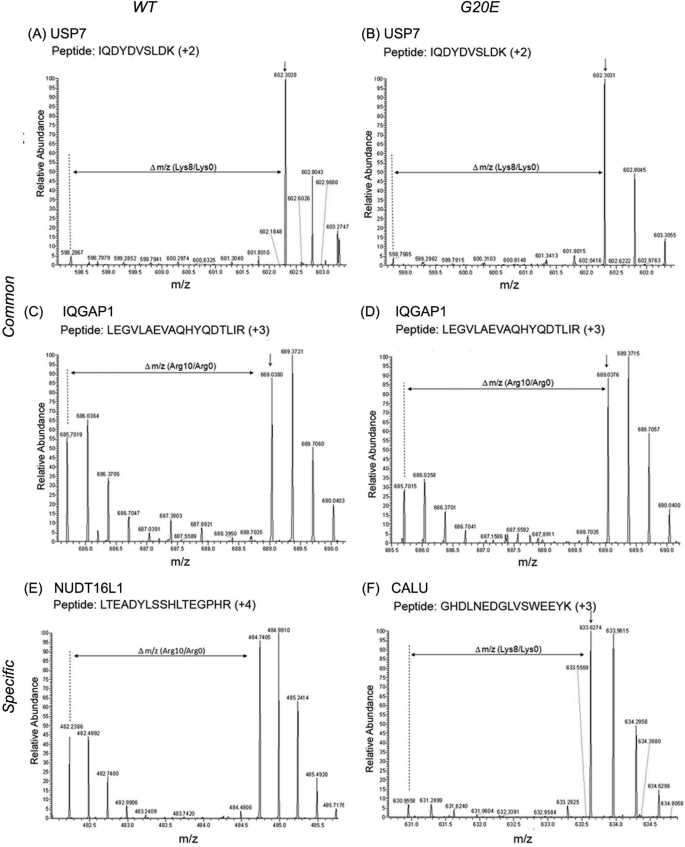
PAP analyses indicated four wild-type-specific interacting proteins (myosin 1D (Myo1D), general transcription factor 3C1 (GTF3C1), phosphoglycerate mutase family member 5 (PGAM5), and Nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1/Syndesmos (NUDT16L1/SDOC)) and five G20E mutant-specific interactors (AIM1 (absent in melanoma 1), GLB1 (galactosidase, β1), calumenin (CALU), reticulocalbin 1 (RCN1), and mitofilin (IMMT). Eleven common interactors include two known interacting proteins: USP7 and caveolin 1 (Cav1), and nine new interacting proteins: ribonuclease/angiogenin inhibitor 1 (RNH1), cofilin 1 (CFL1), Tu translation elongation factor (TUFM), annexin A2 (ANXA2), guanine nucleotide binding protein β polypeptide 2-like 1/receptor for activated C kinases (GNB2L1/RACK1), YWHAQ/14-3-3 θ, ADP-ribosylation factor 4 (ARF4), and lactate dehydrogenase A. Mass spectra of typical SILAC peptide pairs of representative interactors from each category are depicted in Fig. 4. These candidates are interacting partners for PTEN WT and PTEN G20E.
FIGURE 4.
Intensities of SILAC peptide pairs in MS/MS spectra show specificity of PTEN interactors. MS/MS peptide spectra of a known wild-type PTEN interactor USP7 (A and B), IQGAP1, a previously unknown interactor to PTEN (C and D), WT-specific interactor NUDT16L1 (E), and G20E-specific interactor CALU (F).
TAP to Capture Stable Interactors
The TAP method involves fusion of the TAP tag to the target protein and introduction of the construct into the host cell. The fusion protein and associated components are recovered from cell extracts by two sequential purification steps. Potential drawbacks of this approach are low overall yield and loss of weakly bound proteins through the two-step purification. Despite these limitations, it effectively removes background and provides identification of stable interactors (26). As both stable and less stable interactors have been captured by PAP approach, we used TAP to validate the more stable interactions revealed by the PAP data.
In the TAP experiment, the GFP portion of the TAP tag was used to bind GFP-trap in the first purification, and complexes were released by tobacco etch virus protease cleavage (Fig. 2B, left). In the second step, the S-tag was bound to S-tag affinity beads and then eluted with SDS buffer. FLAP only-expressing cells were used as the negative control. Samples were analyzed by in-gel digestion and mass spectrometry, and inferred proteins were compared with the PAP data. Interestingly, TAP data overlap with the highest ranked proteins of the common cluster (USP7, IQGAP1, RNH1, and CFL1), suggesting these are possible stable interactors. In the mutant specific interactors, AIM1 appears to be enriched in the mutant pulldown compared with the wild-type. Proteins in the PAP data WT-specific category were not observed in the TAP experiment. These interactors might be weaker binding or transient with a short lifetime in the complex.
Validation of Interactions
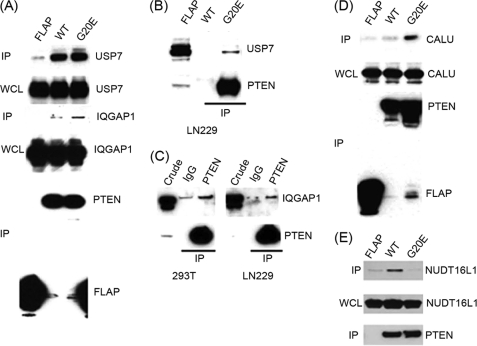
To further verify the interactions and their specificity, we carried out co-immunoprecipitation assays for selected interactors obtained from the PAP approach. The selection was based on possible physiological role of the interactions and availability of commercial antibodies. We confirmed the interactions of USP7 and IQGAP1with both wild-type and the mutant in U87 stable cell lines (Fig. 5A). IQGAP1 and USP7 also were precipitated with the endogenous PTEN in LN229 and/or 293T cell lines, indicating the interactions of USP7 and IQGAP1 with wild-type in physiological condition (Fig. 5, B and C). The SILAC ratios in PAP and number of peptides in TAP of USP7 are comparable between the wild-type and the mutant (Fig. 4, A and B). We observed that the SILAC ratios of IQGAP1 with mutant are higher than those of the wild-type (Table 1 and Fig. 4, C and D), indicating that the mutant may have a higher propensity to interact with IQGAP1. The intensities of immunoblot analysis for IQGAP1 in mutant and wild-type were consistent with the SILAC ratio intensities of IQGAP1 in PAP, indicating the reliability of PAP data (Fig. 5A). Moreover, we confirmed the specificity of CALU as mutant-specific (Fig. 5D). To analyze wild-type-specific interaction of NUDT16L1, we prepared lysates of 293T cells to which the expression vector of V5-tagged NUDT16L1 was transfected with expression plasmids of FLAP, FLAP PTEN wild-type, and G20E. NUDT16L1 preferentially interacted with wild-type PTEN (Fig. 5E). Taken together, the PAP approach successfully enriches and prioritizes target proteins of PTEN WT and G20E mutant.
FIGURE 5.
The interaction between PTEN and its interactors in PAP is confirmed by co-precipitation. Cell extracts from U87 FLAP, FLAP PTEN WT, and FLAP PTEN G20E cells were incubated with a FLAG beads, and precipitates were analyzed with the indicated antibodies: immunoblot with anti-USP7 and anti-IQGAP1 (A) and anti-CALU (D). Lysates from HEK293T and LN 229 cells were incubated with anti-PTEN and mouse IgG-conjugated beads. The co-precipitates were detected with anti-USP7 (B) and anti-IQGAP1 (C) antibodies. GFP-trap beads were incubated with lysates of HEK293T transfected the expression plasmids of FLAP, FLAP PTEN WT, and FLAP PTEN G20E with V5-tagged NUDT16L1. The precipitates were detected with V5 and FLAG antibodies (E). IP, immunoprecipitation; WCL, whole cell lysate.
Pathway Analysis of Interactors
Prioritized interactors of PTEN WT and the cancer-associated G20E mutant have been subjected to pathway analysis to find functional relevance of those interacting partners. This analysis indicated that the PTEN interactors could be subdivided into two groups in terms of their functions: migration and apoptosis (Fig. 6).
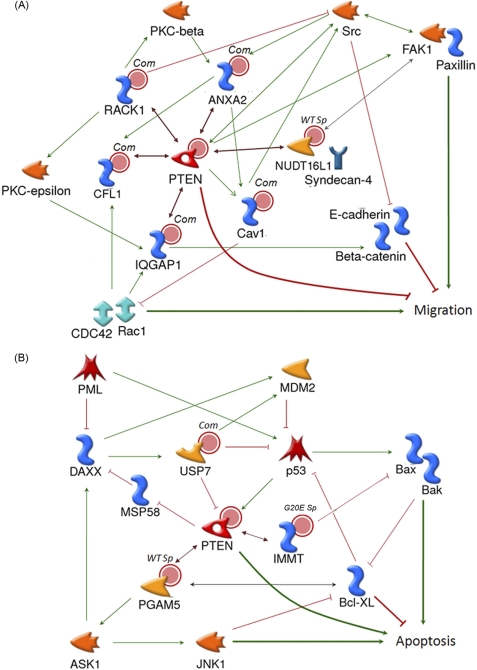
FIGURE 6.
The new interactors of PTEN are involved in the migration and apoptosis signaling pathways. Network maps of cell migration (A) and apoptosis (B) for PTEN interactors from SILAC-based PAP-SILAC were created by using information gathered from published literature and MetaCoreTM (version 6.0). The double-headed arrow indicates interaction of proteins. Facilitation is a green arrow (single-headed arrow), and inhibition is a red line. The known partner proteins of PTEN are implicated in the control of migration and apoptosis. Com, common interactors; WTsp, wild type specific interactors; G20Esp, G20E mutant interactors; PML, promyelocytic leukemia protein.
Proteins of IQGAP1, RACK1, ANXA2, CFL1, and NUDT16L1 (Syndesmos) are involved in a cell migration pathway (Fig. 6A). IQGAP1 (a common interactor) has roles in migration and control of cell polarity through its interaction with numerous proteins, such as E-cadherin, β-catenin, Rac1, and Cdc42. It modulates the actin cytoskeleton by activating Rac1 and Cdc42, and cell-cell adhesion though E-cadherin and β-catenin. RACK1 regulates directional cell migration through regulation of Src activity and paxillin dynamics at focal complexes (34, 35). ANXA2 is a Src substrate, a lipid-, calcium-, and actin-binding protein (36). The ANXA2-induced scattering is mediated via the actin-severing protein CFL1 (37). NUDT16L1 is a PTEN WT-specific interactor. It interacts with the cytoplasmic domain of syndecan-4 in focal contacts and works as an adapter protein, which may link syndecan-4 and paxillin (38). Thus, NUDT16L1 is a candidate protein to test its functional involvement in wild-type PTEN-dependent cell migration.
We identified that USP7, PGAM5, and IMMT are in the apoptosis pathway (Fig. 6B). USP7 is a known interactor to wild-type, and our study shows that it also interacts with G20E. USP7 regulates the nuclear localization of PTEN (39) and also stabilizes p53 and Foxo4 by an interaction and deubiquitination activity. The monoubiquitination of PTEN is required for its nuclear localization and induces apoptosis; thus, USP7 is essential in the regulation of apoptosis (39). The nuclear localization of PTEN also is required during p53-dependent apoptosis by oxidative stress (40). PAGM5, a WT-specific interactor, is a serine/threonine-specific phosphatase and a positive regulator of apoptosis. It activates apoptosis signal-regulating kinase 1 (ASK1) through the interaction during oxidative stress. PTEN might control ASK1 activity by regulating PGAM5 activity, as the JNK pathway, a downstream target of ASK1, is activated in PTEN knockdown cells (41). IMMT, which is a mutant-specific interactor, also negatively regulates apoptosis (42). Its interaction with the mutant could positively affect on the IMMT-derived inhibition of apoptosis. Of note, it has been shown that USP7 plays a critical role in apoptosis through the control of PTEN subcellular localization (39). Hence, the presence of USP7 in our data provides robust evidence for functional involvement of the interactors found in our PAP-SILAC approach.
Functional Validation of NUDT16L1 Interaction in Cell Migration
We chose to focus on the migration pathway to show the biological relevance of interactors revealed by the PAP-SILAC approach. Interestingly, we observed that G20E mutant cells promote cell migration (Fig. 7B), although G20E retains its intact C2 domain that inhibits migration in a lipid phosphatase-independent manner (15, 43). Thus, it was interesting to test the specific interactor either WT or G20E mutant.
FIGURE 7.
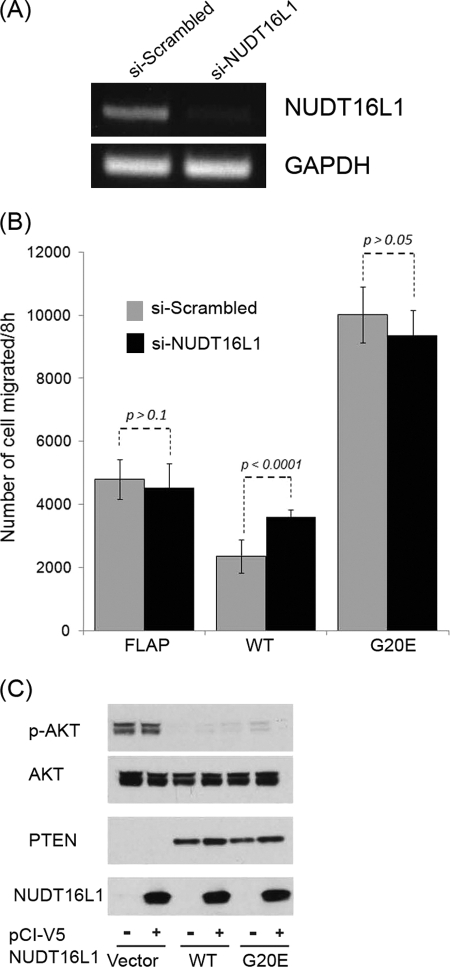
NUDT16L1 is required for PTEN-dependent regulation of cell migration. A, RT-PCR for NUDT16L1 expression was carried out with NUDT16L1 (si-NUDT16L1) and nontarget scrambled siRNA (si-Scrambled). B, Transwell migration assays were performed for U87 FLAP, WT and G20E stable cell lines. Numbers of migrated cells were measured under transfection of NUDT16L1 siRNA (si-NUDT16L1) in comparison with scrambled siRNA (si-Scrambled). Error bar indicates standard error from nine independent experiments. C, phosphorylated Akt levels were compared by immunoblotting. In the presence and absence of V5 NUDT16L1 expression plasmid indicated as + and − at the bottom, respectively, the blot with phospho-Akt (Ser473) antibody, which indicated as P-Akt, was performed with plasmids of vector control, PTEN WT, and G20E using U87 MG cells.
NUDT16L1 is the only specific interactor for wild-type PTEN in the migration pathway (Fig. 6A). To analyze its effect in cell migration, we carried out a Transwell migration assay using U87 FLAP, WT, and G20E with either NUTD16L1-specific siRNA or with scrambled siRNA. RT-PCR confirmed that the knocking down of NUDT16L1 using siRNA was highly efficient compared with its expression in the cells transfected with scrambled siRNA (Fig. 7A). U87 WT cells have suppressed migration in comparison to the U87 FLAP cells, whereas the ability of U87 G20E cell migration is higher than U87 FLAP cells (Fig. 7B). We compared the effect of si-NUDT16L1 on cell migration in the U87 FLAP, WT, and G20E cells. The absence of NUDT16L1 in U87 WT cells increased migration by ∼1.5-fold compared with the cells transfected with scrambled siRNA control, whereas the knockdown of NUDT16 in U87 FLAP and G20E showed weak reduction (<10%) of the migration. To further confirm the effect of this interaction in cell migration, we knocked down NUDT16L1 in LN229 cells that have a PTEN wild-type background. The migration was increased ∼1.8-fold in the siRNA transfected cells of NUDT16L1 compared with the control with the scrambled siRNA (supplemental Fig. S2). This result is consistent to the results of U87 cells. Thus, our observation supports the interaction of PTEN WT and NUDT16L1 being involved in the regulation of cell migration.
Moreover, to test whether NUDT16L1 affects PTEN lipid phosphatase activity, we performed a PTEN lipid phosphatase assay by monitoring phosphorylated Akt with and without NUDT16L1 overexpression. The results showed that the levels of phosphorylated Akt were equal in both conditions (Fig. 7C). These observations indicate that the regulation of migration through the PTEN-NUDT16L1 complex is independent on PTEN lipid phosphatase activity.
DISCUSSION
The functional role of multisubunit protein assemblies as part of complex dynamic protein interaction networks is well recognized and is an area of active research. The topology of such networks and the association and dissociation of protein complexes are of broad biological importance. However, determining the composition of dynamic protein complexes is a challenge. Prioritization of putative interaction partners identified by any large scale study is important as a first step in confirming their physiological relevance. In this study, we identified the interacting proteins of PTEN from wild-type and mutant by a parallel affinity purification with quantitative mass spectrometry proving it to be a useful strategy for reducing false positives, capturing true interactors, and prioritizing them.
Quantitative mass spectrometry-based proteomic methods such as SILAC remove much of the “noise” of nonspecific interactors found in a standard immunoprecipitation experiments and allow a first round of in silico analysis to provide information on biological function. We further improved the SILAC screening method and made it more informative by using a parallel combination of different affinity purification tags, matrices, and cell lines and further enhancing downstream data analysis. We were able to remove some method-biased false positives, when we used two types of affinity matrices (antibody and GFP-trap) to capture different but related interactor sets. In this way, PRMT5 is found in the FLAG mutant fraction but not in the GFP-trap mutant. Although it has been reported that PRMT5 preferentially interacts with FLAG antibody (44), PRMT5 showed specific interaction with FLAP PTEN with FLAG-immunoprecipitated samples (high SILAC ratio and strong signal in Western blot) but was very weak in the sample from GFP-trap (supplemental Fig. S3). These observations suggest that some interactors may be incorrectly attributed as true interactors if only a single affinity experiment is used to capture interactors. PAP with SILAC accounts only for common high ratios; thus, in comparison with a single SILAC affinity purification, we are able to remove many interactors as false positives.
Prioritized interactors acquired from PAP-SILAC are useful entry points for bioinformatic analysis of PTEN function. Such analysis of the new PTEN interactors showed that they are mainly involved in migration and apoptosis processes (Fig. 6). We therefore further analyzed cell migration to show the functional relevance of interactors revealed from our approach. PTEN has been reported to have a role in cell migration in a lipid phosphatase-dependent as well as -independent manner (15, 43). We demonstrated that knockdown of NUDT16L1, a new and PTEN WT-specific interactor, attenuates the suppression of migration by PTEN WT, and NUDT16L1 fails to affect the lipid phosphatase activity of PTEN. These observations suggested that the lack of affinity of NUDT16L1 to cancer-associated mutant G20E may increase migration of the cells with the mutant, supporting our initial hypothesis that the mutant G20E is unable to interact with a wild-type specific PTEN interactor, which impinges directly or indirectly on cellular pathways. Thus, we infer that G20E mutant exerts phosphatase-independent functions on the cellular machinery through changing the complement of interacting proteins.
In conclusion, the PAP-SILAC is a powerful method to infer the unknown functions of a protein through interactome analyses. We believe that the SILAC mass spectrometry approach, particularly with the improvement herein reported, is now at a stage where secondary validation may be reserved for those candidates that will reveal the details of networks and their deregulated hubs in human cancers.
Supplementary Material
Acknowledgments
We thank Dr. Koji Yamanaka for FLAP vector and Suat Peng Neo for technical assistance. We are grateful to Dr. Ayumi Yamada for providing some reagents and Dr. Chan Shing-Leng for cell sorting.
This work was supported by the grant of National Medical Research Council (NMRC), Singapore and Agency for Science, Technology, and Research (A*STAR), Singapore.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and a table.
- SILAC
- stable isotope labeling by amino acids in cell culture
- PAP
- parallel affinity purification
- TAP
- tandem affinity purification
- PI(3,4,5)P
- phosphatidylinositol 3,4,5-trisphosphate
- Flap
- Flag combined Localization and Affinity Purification.
REFERENCES
- 1. Eng C. (2003) Hum. Mutat. 22, 183–198 [DOI] [PubMed] [Google Scholar]
- 2. Maehama T., Taylor G. S., Dixon J. E. (2001) Annu. Rev. Biochem. 70, 247–279 [DOI] [PubMed] [Google Scholar]
- 3. Maehama T., Dixon J. E. (1998) J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 4. Furnari F. B., Lin H., Huang H. S., Cavenee W. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12479–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao J., Schulte J., Knight A., Leslie N. R., Zagozdzon A., Bronson R., Manevich Y., Beeson C., Neumann C. A. (2009) EMBO J. 28, 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fine B., Hodakoski C., Koujak S., Su T., Saal L. H., Maurer M., Hopkins B., Keniry M., Sulis M. L., Mense S., Hibshoosh H., Parsons R. (2009) Science 325, 1261–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z., Dong X., Dong X., Wang Z., Liu W., Deng N., Ding Y., Tang L., Hla T., Zeng R., Li L., Wu D. (2005) Nat. Cell Biol. 7, 399–404 [DOI] [PubMed] [Google Scholar]
- 8. Okahara F., Itoh K., Nakagawara A., Murakami M., Kanaho Y., Maehama T. (2006) Mol. Biol. Cell 17, 4888–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okumura K., Mendoza M., Bachoo R. M., DePinho R. A., Cavenee W. K., Furnari F. B. (2006) J. Biol. Chem. 281, 26562–26568 [DOI] [PubMed] [Google Scholar]
- 10. Yim E. K., Peng G., Dai H., Hu R., Li K., Lu Y., Mills G. B., Meric-Bernstam F., Hennessy B. T., Craven R. J., Lin S. Y. (2009) Cancer Cell 15, 304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Okahara F., Ikawa H., Kanaho Y., Maehama T. (2004) J. Biol. Chem. 279, 45300–45303 [DOI] [PubMed] [Google Scholar]
- 12. Han S. Y., Kato H., Kato S., Suzuki T., Shibata H., Ishii S., Shiiba K., Matsuno S., Kanamaru R., Ishioka C. (2000) Cancer Res. 60, 3147–3151 [PubMed] [Google Scholar]
- 13. Planchon S. M., Waite K. A., Eng C. (2008) J. Cell Sci. 121, 249–253 [DOI] [PubMed] [Google Scholar]
- 14. Salmena L., Carracedo A., Pandolfi P. P. (2008) Cell 133, 403–414 [DOI] [PubMed] [Google Scholar]
- 15. Maier D., Jones G., Li X., Schönthal A. H., Gratzl O., Van Meir E. G., Merlo A. (1999) Cancer Res. 59, 5479–5482 [PubMed] [Google Scholar]
- 16. Li A. G., Piluso L. G., Cai X., Wei G., Sellers W. R., Liu X. (2006) Mol. Cell 23, 575–587 [DOI] [PubMed] [Google Scholar]
- 17. Freeman D. J., Li A. G., Wei G., Li H. H., Kertesz N., Lesche R., Whale A. D., Martinez-Diaz H., Rozengurt N., Cardiff R. D., Liu X., Wu H. (2003) Cancer Cell 3, 117–130 [DOI] [PubMed] [Google Scholar]
- 18. Okumura K., Zhao M., Depinho R. A., Furnari F. B., Cavenee W. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2703–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Agostino S., Strano S., Emiliozzi V., Zerbini V., Mottolese M., Sacchi A., Blandino G., Piaggio G. (2006) Cancer Cell 10, 191–202 [DOI] [PubMed] [Google Scholar]
- 20. Buchhop S., Gibson M. K., Wang X. W., Wagner P., Stürzbecher H. W., Harris C. C. (1997) Nucleic Acids Res. 25, 3868–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ang S. O., Chen H., Hirota K., Gordeuk V. R., Jelinek J., Guan Y., Liu E., Sergueeva A. I., Miasnikova G. Y., Mole D., Maxwell P. H., Stockton D. W., Semenza G. L., Prchal J. T. (2002) Nat. Genet. 32, 614–621 [DOI] [PubMed] [Google Scholar]
- 22. Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A. M., Schirle M., Schlegl J., Schwab M., Stein M. A., Bauer A., Casari G., Drewes G., Gavin A. C., Jackson D. B., Joberty G., Neubauer G., Rick J., Kuster B., Superti-Furga G. (2004) Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 23. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 24. Major M. B., Camp N. D., Berndt J. D., Yi X., Goldenberg S. J., Hubbert C., Biechele T. L., Gingras A. C., Zheng N., Maccoss M. J., Angers S., Moon R. T. (2007) Science 316, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 25. Cox D. M., Du M., Guo X., Siu K. W., McDermott J. C. (2002) BioTechniques 33, 267–268, 270 [DOI] [PubMed] [Google Scholar]
- 26. Gunzl A., Schimanski B. (2009) Curr. Protoc. Protein Sci. Chapter 19, Unit 19 19 [DOI] [PubMed] [Google Scholar]
- 27. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 28. Selbach M., Mann M. (2006) Nat. Methods 3, 981–983 [DOI] [PubMed] [Google Scholar]
- 29. Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., 3rd, Oegema K., Desai A. (2004) Genes Dev. 18, 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 31. Cox J., Mann M. (2008) Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 32. Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M. C., Leonhardt H. (2008) Mol. Cell Proteomics 7, 282–289 [DOI] [PubMed] [Google Scholar]
- 33. Wang X., Huang L. (2008) Mol. Cell Proteomics 7, 46–57 [DOI] [PubMed] [Google Scholar]
- 34. Chen S., Lin F., Shin M. E., Wang F., Shen L., Hamm H. E. (2008) Mol. Biol. Cell 19, 3909–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Doan A. T., Huttenlocher A. (2007) Exp. Cell Res. 313, 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayes M. J., Moss S. E. (2009) J. Biol. Chem. 284, 10202–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Graauw M., Tijdens I., Smeets M. B., Hensbergen P. J., Deelder A. M., van de Water B. (2008) Mol. Cell. Biol. 28, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baciu P. C., Saoncella S., Lee S. H., Denhez F., Leuthardt D., Goetinck P. F. (2000) J. Cell Sci. 113, 315–324 [DOI] [PubMed] [Google Scholar]
- 39. Song M. S., Salmena L., Carracedo A., Egia A., Lo-Coco F., Teruya-Feldstein J., Pandolfi P. P. (2008) Nature 455, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang C. J., Mulholland D. J., Valamehr B., Mosessian S., Sellers W. R., Wu H. (2008) Mol. Cell. Biol. 28, 3281–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vivanco I., Palaskas N., Tran C., Finn S. P., Getz G., Kennedy N. J., Jiao J., Rose J., Xie W., Loda M., Golub T., Mellinghoff I. K., Davis R. J., Wu H., Sawyers C. L. (2007) Cancer Cell 11, 555–569 [DOI] [PubMed] [Google Scholar]
- 42. John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M., Rangell L., Bennett M. J., Zha J. (2005) Mol. Biol. Cell 16, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. (2004) Science 303, 1179–1181 [DOI] [PubMed] [Google Scholar]
- 44. Nishioka K., Reinberg D. (2003) Methods 31, 49–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.