Abstract
The NIMA family protein kinases Nek9/Nercc1 and the highly similar Nek6 and Nek7 form a signaling module activated in mitosis, when they are involved in the control of spindle organization and function. Here we report that Nek9, the module upstream kinase, binds to DYNLL/LC8, a highly conserved protein originally described as a component of the dynein complex. LC8 is a dimer that interacts with different proteins and has been suggested to act as a dimerization hub promoting the organization and oligomerization of partially disorganized partners. We find that the interaction of LC8 with Nek9 depends on a (K/R)XTQT motif adjacent to the Nek9 C-terminal coiled coil motif, results in Nek9 multimerization, and increases the rate of Nek9 autoactivation. LC8 binding to Nek9 is regulated by Nek9 activity through the autophosphorylation of Ser944, a residue immediately N-terminal to the (K/R)XTQT motif. Remarkably, LC8 binding interferes with the interaction of Nek9 with its downstream partner Nek6 as well as with Nek6 activation, thus controlling both processes. Our work sheds light into the control of signal transduction through the module formed by Nek9 and Nek6/7 and uncovers a novel manner in which LC8 can regulate partner physiology by interfering with protein complex formation. We suggest that this and other LC8 functions can be specifically regulated by partner phosphorylation.
Keywords: Cellular Regulation, Phosphorylation Enzymes, Protein Kinases, Protein-Protein Interactions, Signal Transduction, LC8, NIMA Family Kinases
Introduction
The Never in Mitosis A (NIMA) family of protein kinases is named after the Aspergillus nidulans NIMA kinase. NIMA is required for entry into mitosis and is involved in the control of chromatin condensation, spindle and nuclear envelope organization, and cytokinesis (1, 2). Mammalian cells contain 11 NIMA family members or Neks that share catalytic domains that are 40% identical to that of NIMA (1, 3). None of the mammalian Neks seem to individually fulfill the broad range of mitotic functions that NIMA performs in Aspergillus, and the different kinases seem to have specialized in performing specific functions related to the control of the microtubule and ciliary machineries (4), or in the case of Nek1, Nek10, and Nek11, the response to DNA damage (5–9).
Nek9, together with the highly similar (∼80% identical) Nek6 and Nek7, form a signaling module that is activated during mitosis and involved in the regulation of the mitotic spindle (2, 10–12). Although Nek6 and Nek7 consist almost exclusively of a catalytic protein kinase domain, Nek9 (also known as Nercc1) is a 120-kDa polypeptide composed of an amino-terminal kinase domain followed by an autoinhibitory domain homologous to RCC1 (the exchange factor for the small G protein Ran) and a C-terminal domain that contains both a Nek6/7 binding region and a coiled coil motif involved in dimerization (13). (Nek6 and Nek7 are 84% identical. Although it is not clear whether both kinases have specific roles in different cell types, they seem to be functionally equivalent in most instances; thus, when adequate, the two kinases will be collectively referred to as Nek6/7.) In vitro, Nek9 is capable of autoactivating through autophosphorylation; in vivo, the kinase is inactive during interphase and is activated at centrosomes and spindle poles during mitosis, when it binds Nek6 and Nek7 and is then able to directly phosphorylate these kinases in the activation loop, in turn inducing their activation (13–15). Nek6/7 binding to Nek9 has in addition been reported to directly increase the activity of Nek6 and Nek7 by disrupting an autoinhibited conformation of these kinases (16). Despite the importance of the interaction between Nek9 and Nek6/7, how it is physiologically regulated has not been described to date.
Interference with Nek6, Nek7, or Nek9 in different systems results in mitotic abnormalities that can be related to a failure to organize a normal spindle (13, 14). Interference with either Nek6 or Nek7 delays cells at metaphase with fragile mitotic spindles (17), and interference with Nek7 has been shown to result in an increased incidence of multipolar spindle phenotypes (18). In mice, lack of Nek7 leads to lethality in late embryogenesis or at early postnatal stages, and Nek7−/− fibroblasts show chromosomal lagging and abnormalities in primary cilia number (19). Nek6-deficient mice die early during embryogenesis.3 Despite the proven importance of Nek9, Nek6, and Nek7 for proper progression through mitosis, a precise description of their physiological roles is presently missing. We have suggested that Nek9 could be controlling spindle organization in part through the action of the downstream Nek6 and Nek7 and their ability to phosphorylate the kinesin Eg5 at a site necessary for normal mitotic progression (15), but possibly more substrates of the signaling module await to be discovered.
LC8 is a cytoplasmic, ubiquitous, essential, and extraordinarily conserved protein (∼90% sequence identity between homologues from mammalians, Drosophila, Caenorhabditis, and Chlamydomonas) that interacts with many different proteins and protein complexes (LC8a and LC8b are highly similar and collectively referred to LC8 herein). In the cytoplasmic dynein complex, two copies of LC8 (also known in this context as dynein light chain, DYNLL4 or DLC) interact with the dynein intermediate chain (DIC), structuring this polypeptide and inducing its dimerization and the organization of the dynein motor complex (20, 21). LC8 seems to be performing a similar function in other protein complexes, dimerizing partially disordered targets, and thus it has been proposed to act as a multifunctional cellular dimerizing hub (22). LC8 function can be better understood from its tridimensional structure (23); two protein monomers associate through their β-sheets, packed against two adjacent α-helixes that lay at the exterior of the dimer. Most of LC8-interacting polypeptides bind to the β-sheet region of the dimer in two opposite hydrophobic grooves in between the α-helixes. This interaction thus relies on LC8 dimerization and is dependent on one of two different protein sequence motifs recognized by LC8 in its target proteins: (K/R)XTQT and G(I/V)QVD (where X is any amino acid). Present knowledge suggests that regulation of LC8 dimerization would be a direct and effective way to regulate its interaction with other proteins, and indeed, signaling inputs such as phosphorylation or changes in the pH or the cellular redox state of the cell have been proposed to control LC8 function in this manner (24–26). Specifically, regarding phosphorylation, it has been shown that modification of LC8 serine 88 promotes dissociation of the dimer, and as a result, LC8 release from some (but not all) binding partners such as DIC (25, 27). One protein kinase able to regulate LC8 in such a manner may be the p21-activated kinase PAK1; this kinase has been described to phosphorylate LC8 at Ser88, resulting in monomerization and the abolition of its interaction with BimL and reduced apoptosis (28). In any case, the physiological relevance of these observations is controversial as structural studies demonstrate that, once bound to PAK1, Ser88 is inaccessible for phosphorylation (29), thus leaving open the identity of the Ser88 kinase.
Although in Saccharomyces cerevisiae (30) and Schizosaccharomyces pombe (31) LC8 null alleles have only minor phenotypes, in Drosophila and zebrafish, loss of LC8 expression is lethal, whereas partial loss of function results in morphogenetic defects and sterility (32–34). Conversely, overexpression of LC8 promotes cancerous properties in mammalians, and LC8 protein levels have been reported to be elevated in more than 90% of human breast tumors (28).
Here we report that Nek9 is an LC8 partner and that both proteins are associated in vivo through a (K/R)XTQT motif present in the extreme C terminus of Nek9. LC8 binding to Nek9 is not necessary for Nek9 oligomerization, although it results in the formation of higher order Nek9 complexes as well as in an increased ratio of Nek9 autoactivation. Remarkably, our work shows that the LC8-Nek9 interaction is regulated by the activation state of Nek9 through autophosphorylation of Ser944, a residue adjacent to the LC8 recognition motif, and that LC8 binding directly interferes with Nek9 binding to its downstream partner Nek6, as well as with Nek6 activation. Thus, the study of the interaction between LC8 and Nek9 uncovers the mechanism that regulates Nek6 (and possibly Nek7) binding to Nek9, shedding light into the control of signal transduction through the module formed by Nek9 and Nek6/7. In addition, it suggests that together with other previously described mechanisms that directly affect LC8 ability to form complexes, changes in partner phosphorylation may be a general mode of regulation of LC8 binding to different polypeptides, specifically controlling LC8 partner selection and recycling in time and space. Finally, the study of the LC8-Nek9 interaction also uncovers a previously unappreciated manner in which LC8 can regulate partner physiology: the negative control of protein complex formation.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Different Nek9 and Nek6 expression plasmids have been described elsewhere (13, 15, 35). cDNAs coding for the human Nek9 Kin (1–346), RCC1 (347–726), and Ct (721–979) were subcloned into pGBKT7 (Clontech). The LC8 cDNA was amplified by PCR and subcloned into pEBG and pET28a Smt3 vectors. Additional Nek9 and LC8 mutants were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All constructs were sequenced after generation.
FLAG-Nek9 and FLAG-Nek9[Q948A] polypeptides were expressed in 293T cells and purified by immunoprecipitation with anti-FLAG antibody (Sigma) followed by repeated washes and elution using FLAG peptide (Sigma). For bacterial expression, cDNAs codifying for different Nek9 and LC8 polypeptides were subcloned into pET28a Smt3 and expressed in Escherichia coli Rosetta 2 (DE3) (Novagen) for 16 h at 18 °C after isopropyl-1-thio-β-d-galactopyranoside (Sigma) induction. Poly-His-Smt3 fusion proteins were purified using Ni-NTA beads (Qiagen) following standard protocols. Nek9 and LC8 forms were obtained after digestion with poly-His-Senp2 protease. Poly-His-Senp2 and digested poly-His-Smt3 were eliminated from the sample using Ni-NTA beads.
Yeast Two-hybrid Screening
Yeast two-hybrid screening was performed using the Matchmaker GAL4 system 3 (Clontech Laboratories). Briefly, S. cerevisiae strain AH109 expressing the fusion protein DBD-Nek9 FL (1–979) or Ct (721–979) were used as bait and mated with S. cerevisiae strain Y187 pretransformed with a human testis cDNA library constructed in pACT2 (Clontech Laboratories) and plated on selective medium lacking tryptophan, leucine, histidine, and adenosine. Positive bait-prey interactions were verified using Mel1 expression. DNA was isolated from positive colonies, and the interacting prey was identified by sequencing. Mapping of the LC8 interaction with Nek9 was performed by transforming different pGBKT7 Nek9 forms and pACT2 LC8 in S. cerevisiae strains AH109 and Y187, respectively, and mating these as described before (15). Expression of the different fusion proteins used was confirmed by Western blot in all cases.
Cell Culture and Transfection
HeLa, U2OS, and HEK 293T cells were cultured as described (13). Cells in mitosis were obtained by mitotic shake off of nocodazole-arrested (250 ng/ml, 16 h) cultures. HEK 293T cells were transfected using different expression plasmids with Lipofectamine (Invitrogen) according to the manufacturer's instructions. HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs were transfected using siPORT NeoFX transfection agent (Ambion) according to the manufacturer's instructions. siRNA duplexes (Ambion) were as follows: LC8, 5′-AUGCGGACAUGUCGGAAGA-3′ and 5′-AACAAGGACUGCAGCCUAA-3′ (36); Nek9 UTR, 5′-GCUGCCUUGGGAAUUCAGU-3′ and GCAGCCAAACTTTGATTAA-3′.
Immunoprecipitation and Western Blot Analysis
Immunoprecipitations, GST pulldowns, and Western blotting were performed as described in Ref. 13. Anti-Nek9, anti-Nek9[Thr210-P], anti-Nek6, and anti-Nek6[Ser206-P] polyclonal antibodies have been described in Refs. 13, 14, and 35. Other antibodies used are: anti-LC8 (Abcam), anti-FLAG (Sigma), anti-phospho-cdc2 (Cell Signaling), and anti-GST and normal rabbit IgG (Santa Cruz Biotechnology). Secondary antibodies were from Jackson ImmunoResearch Laboratories and were detected by ECL chemiluminescence (Thermo Scientific).
Gel Filtration
For in vitro gel filtration analysis, Nek9[893–974], Nek9[893–974, Q948A], and LC8 were produced in E. coli Rosseta2 (DE3) in the vector pSmt3. Expression cultures were harvested after 3 h at 30 °C of isopropyl-1-thio-β-d-galactopyranoside induction. Poly-His-SUMO (small ubiquitin-like modifier) fusion proteins were purified using Ni-NTA beads (Qiagen) following standard protocols. After Senp2 cleavage, Nek9 polypeptides and LC8 were purified by gel filtration chromatography using a Superdex 200 column (GE Healthcare). LC8 was subsequently purified from small ubiquitin-like modifier by anion exchange using a ResourceQ column (GE Healthcare). Fractions containing the peaks were collected, and proteins were concentrated in the same column purification buffers. Complex between LC8 and Nek9 Ct (893–974) was made by mixing equimolar concentrations of the two components in a low salt buffer (100 mm NaCl, 20 mm Tris, pH 8, 1 mm β-mercaptoethanol). LC8, Nek9[893–974], complex between LC8 and Nek9[893–974], and complex between LC8 and Nek9[893–974, Q948A] were separated in a gel filtration Superdex 200 column (GE Healthcare) equilibrated with low salt buffer (100 mm NaCl, 20 mm Tris, pH 8, 1 mm BME). Standards for molecular markers were also run under the same buffer conditions.
Kinase Assays
Protein kinase assays were carried out as described previously (13) using 10 or 50 μm ATP with trace amounts of [γ-32P]ATP and myelin basic protein (MBP) as substrates.
Immunofluorescence
Cells were grown on coverslips and fixed and permeabilized as described earlier (15). Primary antibodies used were mouse anti-γ-tubulin (Sigma) and mouse anti-FLAG (Sigma) and were detected with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 555 goat anti-mouse IgG (Invitrogen). DNA was stained with DAPI (Sigma). Images were taken using a Leica TCS SPE confocal system with a DM2500 CSQ upright microscopy and a 63× 1.30 ACS Apo lens and edited using Leica LAS AF software (Leica Microsystems) and Photoshop (Adobe).
RESULTS
Identification and Mapping of the Dynein Light Chain LC8 Interaction with Nek9
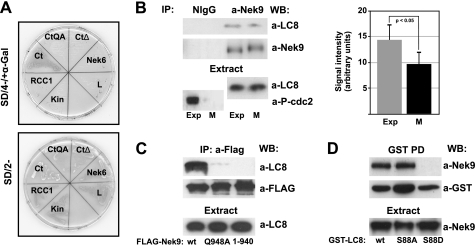
To identify Nek9 regulators and substrates, we performed different yeast two-hybrid experiments. Full-length forms of the two cytoplasmic dynein light chain variants LC8 type 1 (LC8a/DYNLL1/DLC1) and LC8 type 2 (LC8b/DYNLL2/DLC2) were identified as Nek9 interactors from a testis cDNA library using either full-length Nek9 or the C-terminal domain of Nek9 (Nek9[721–979]) as bait. LC8 was originally described as an essential component of the cytoplasmic dynein complex and has subsequently been shown to interact with a diverse array of proteins through one of two specific recognition sequences (22, 37, 38). A search in the Nek9 primary sequence revealed a motif in the C-terminal region of the polypeptide (945KGTQT949) that perfectly conforms to the described LC8 recognition motif KXTQT (where X is any amino acid) (37). We thus hypothesized that LC8 could bind to Nek9 through this sequence. To test this, we assessed the ability of LC8 to interact specifically with different forms of the C-terminal region of Nek9. Fig. 1A shows that in a two-hybrid assay, LC8 binds to Nek9 C-terminal domain (residues 721–979), but not to its kinase (residues 1–346) or RCC1 domains (residues 347–726). Removal of the last 39 residues of Nek9, including the KXTQT motif, results in a polypeptide (Nek9[721–940]) that is not able to interact with LC8. Finally, a point mutation in the putative LC8 binding motif (Nek9[721–979, Q948A]) totally abrogates the LC8-Nek9 interaction. Thus, LC8 interacts with Nek9 through a motif located in the extreme C terminus of the protein kinase that conforms to an LC8 recognition sequence.
FIGURE 1.
Nek9 interacts with LC8. A, LC8 interacts with Nek9 C-terminal tail in the yeast two-hybrid system. Yeast cells were transformed with different plasmids encoding the GAL4 DNA binding domain fused to Nek6 or the Nek9 kinase domain (Kin, Nek9[1–346]), RCC1 domain (RCC1, Nek9[347–726]), C-terminal domain (Ct, Nek9[721–979]), and different C-terminal mutants (CtQA, Nek9[721–979, Q948A]; CtΔ, Nek9[721–940]) and mated to compatible cells transformed with plasmid encoding the GAL4 activation domain fused to LC8. Cells were selected for the presence of the two corresponding plasmids (lower plate, SD/2−), and the existence of an interaction between fusion proteins was assessed by histidine and adenine prototrophy plus α-galactosidase activity (upper plate, SD/4−/+α-Gal). Gal4 DNA binding domain fused with lamin (L) was used as negative control. B, immunoprecipitates (IP) were prepared from exponentially growing (Exp) or nocodazole-arrested mitotic (M) HeLa cell extracts using either normal rabbit IgG (NIgG) or anti-Nek9 antibodies and analyzed by Western blot (WB) with the indicated antibodies. cdc2[Tyr15-P] (P-cdc2; a marker of interphase cells) and LC8 in the corresponding extracts are shown in the lower panels. The amount of LC8 in Nek9 immunoprecipitates was quantified by densitometry of similarly exposed films (mean ± S.E. of five independent experiments; statistical significance was determined using the paired student's t test). C, anti-FLAG immunoprecipitates from HEK293T cells transfected with FLAG-Nek9, FLAG-Nek9[Q948A], or FLAG-Nek9[1–940] were immunoblotted with anti-LC8 and anti-FLAG. LC8 in the corresponding extracts is shown in the lower panel. D, GST pulldowns (GST PD) from HEK293T cells expressing GST-LC8 wild type (wt), GST-LC8 S88A (S88A), or GST-LC8 S88D (S88D) were immunoblotted with the indicated antibodies. Nek9 in the corresponding extracts is shown in the lower panel. Western blots in this and subsequent figures represent significant examples from at least three separate experiments.
Nek9 interaction with LC8 was confirmed in mammalian cells (Fig. 1B). Using extracts from both exponentially growing and mitotic U2OS cells, we found that endogenous LC8 and Nek9 coimmunoprecipitated in both conditions, although consistently, we detected ∼33% less LC8 in mitotic Nek9 immunoprecipitates than in those from exponentially growing cells, suggesting that the observed interaction may be at least partially cell cycle-regulated. Immunoprecipitation of recombinant wild type Nek9, Nek9[Q948A], and Nek9[1–940] from 293T cells confirmed that in mammalian cells, LC8 binding to the kinase was absolutely dependent on the existence of an intact KXTQT motif in the C terminus of Nek9 (Fig. 1C). Additionally, Nek9 was shown to co-precipitate with GST-LC8, but not with GST-LC8[S88D], a GST fusion of a form of LC8 that does not dimerize (25). Substitution of LC8 Ser88 to alanine, predicted to only have a minor effect on LC8 dimerization, did not interfere with LC8 binding to Nek9 (Fig. 1D).
LC8 Is Not Necessary for Nek9 Dimerization but Affects Nek9 Oligomeric State
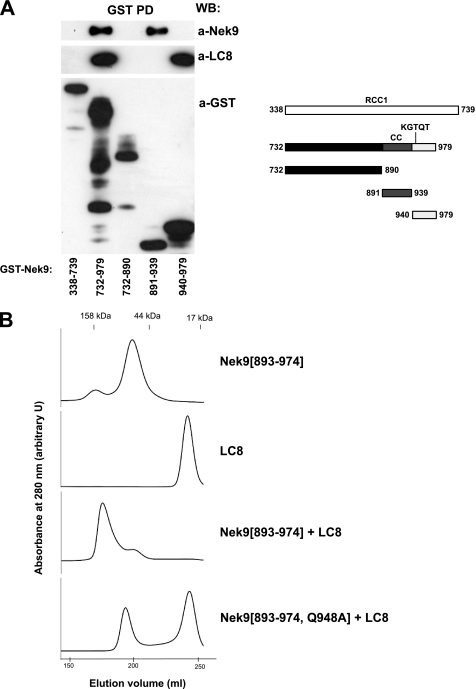
In the context of the dynein motor complex, LC8 interacts with DIC, inducing its structural organization as well as dimerization. LC8 has been proposed to have a similar function when interacting with other proteins, thus organizing and aligning different interacting partners and promoting their dimerization through domains such as helical or coiled coil regions (22). Nek9 does not coimmunoprecipitate with DIC (data not shown), making it unlikely that LC8 connected this kinase to the dynein complex. Thus, we sought to determine whether LC8, independently of its function as a dynein light chain, could be influencing Nek9 quaternary structure. Our previous experiments have shown that Nek9 oligomerizes through its C-terminal coiled coil motif (residues 891–940), immediately upstream of the LC8 binding site, and that this motif is both necessary and sufficient for oligomerization (13). These data are confirmed by Fig. 2A, showing that LC8 binding to Nek9 is not necessary for oligomerization. Thus, GST-Nek9[891–939], corresponding exclusively to Nek9 coiled coil motif and not able to bind LC8, interacted with full-length Nek9 as efficiently as the complete Nek9 C-terminal domain (Nek9[732–979], containing both the coiled coil and the LC8 binding motif). Conversely, GST-Nek9[940–979], corresponding to the extreme C-terminal region of Nek9, was not able to bind full-length Nek9, regardless of the fact that it strongly interacted with LC8. We conclude that LC8 is not required for Nek9 oligomerization.
FIGURE 2.
LC8 effect on Nek9 oligomerization. A, GST pulldowns (GST PD) from HEK293T cells transfected with the indicated GST-Nek9 fragments were immunoblotted with anti-Nek9, anti-LC8, or anti-Nek9 antibodies. WB, Western blot. A graphic of the Nek9 fragments used is shown (right). B, the indicated Nek9 fragments and LC8 were expressed in bacteria and purified. The elution profile after gel filtration measured at 280 nm for Nek9[893–974], LC8, the combined Nek9[893–974] + LC8, or the combined Nek9[893–974, Q948A] + LC8 is shown. arbitrary U, arbitrary units.
We next sought to determine whether LC8 could be involved in the formation of Nek9 higher-order oligomers (for example, inducing the association of different coiled coil-induced dimers). For this, we expressed both LC8 and Nek9[893–974], a C-terminal fragment of Nek9 containing both the coiled coil and the LC8 binding motifs, in bacteria and purified them to homogeneity. Using gel filtration, we estimated the molecular weight of the complexes formed by LC8, Nek9[893–974], and a mix of both polypeptides (Fig. 2B). LC8 alone showed a molecular mass of ∼20 kDa, corresponding to a dimer and consistent with published data (39). Nek9[893–974], with a predicted molecular mass of ∼10 kDa, oligomerized with an apparent molecular mass of ∼50 kDa. This could be due to the formation of tetramers, or alternatively, an overestimation of the molecular weight of dimers due to the probable elongated shape of the polypeptide. Interestingly, the addition of an equimolar amount of LC8 to Nek9[893–974] very efficiently induced the appearance of higher-order complexes that included both polypeptides with an estimated molecular mass of ∼150 kDa. This effect was not observed when Nek9[893–974, Q948A] was mixed with LC8. We thus conclude that Nek9 coiled coil is sufficient for dimerization or possibly tetramerization but that LC8 binding to Nek9 KXTQT motif induces the appearance of higher-order oligomers (tetramers or octamers) of the protein kinase by interacting with two Nek9 units and inducing their multimerization.
LC8 Increases the Rate of Nek9 Activation by Autophosphorylation
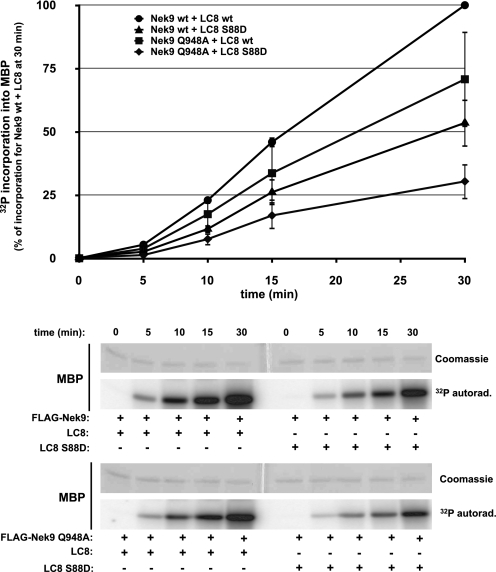
We have previously shown that the ability of Nek9 to activate through autophosphorylation depends on oligomerization (13). We therefore sought to determine whether LC8 binding (and thus Nek9 multimerization) could affect the Nek9 autoactivation rate. For this, we performed Nek9 activation assays using either Nek9 wild type or Nek9[Q948A] incubated with an excess of LC8 wild type or LC8[S88D]. Fig. 3 shows that although autoactivation occurred in all cases, wild type Nek9 incubated with LC8 displayed a higher activation rate than both wild type Nek9 incubated with LC8[S88D] (that does not bind Nek9) and Nek9[Q948A] (not able to bind LC8) incubated with either form of LC8. Thus, we conclude that although not absolutely required for Nek9 autoactivation, LC8 binding to Nek9 increases the efficiency of this process.
FIGURE 3.
LC8 binding to Nek9 increases Nek9 autoactivation rate. FLAG-Nek9 and FLAG-Nek9[Q948A] were immunoprecipitated from HEK293T cells, washed, eluted with FLAG peptide, and incubated for the indicated times at 25 °C in phosphorylation buffer with either LC8 or LC8[S88D] (1 μg/40 μl) plus 50 μm ATP (supplemented with trace amounts of [γ-32P]ATP) and MBP (1 μg/40 μl). The reaction was stopped by the addition of SDS sample buffer followed by SDS-PAGE and Coomassie Blue staining. Lower panels, MBP Coomassie Blue staining and 32P incorporation into MBP are shown. Upper panel, 32P incorporation into MBP was quantified by PhosphorImager. Data are mean ± S.E. of four independent experiments. 32P incorporation into MBP at 30 min for Nek9 wt + LC8 wt is set as 100%. ●, FLAG-Nek9 wild type (wt) + LC8 wt; ▴, FLAG-Nek9 wt + LC8[S88D]; ■, FLAG-Nek9[Q948A] + LC8 wt; ♦, FLAG-Nek9[Q948A] + LC8 [S88D]. 32P autoradiography, 32P autoradiography.
LC8 Binding to Nek9 Is Regulated by Nek9 Autophosphorylation at Ser944
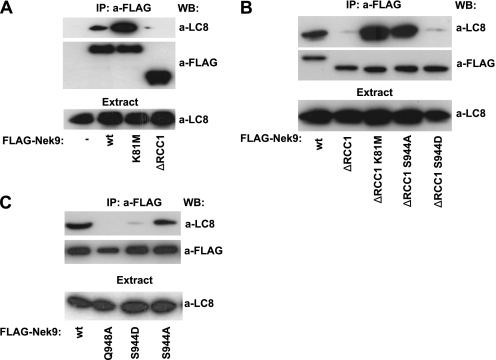
During our characterization of the LC8-Nek9 binding, we observed that the activation state of Nek9 had a direct effect on this interaction. To investigate this, we expressed wild type, kinase-inactive, and constitutively active forms of Nek9 in 293T cells and assessed their ability to bind LC8 by immunoprecipitation. Fig. 4A shows that Nek9[K81M], devoid of protein kinase activity, coimmunoprecipitated with a significantly bigger amount of LC8 than Nek9 wild type. Conversely, LC8 interaction with Nek9[Δ346–732], a constitutively active form of Nek9 (13), was greatly reduced. A possible explanation of these data would be that Nek9 (or its downstream kinases Nek6/7) could phosphorylate LC8, disrupting its ability to dimerize and thus to bind Nek9. We thus attempted to phosphorylate bacterially expressed LC8 in vitro using Nek9 or Nek6 purified from mammalian cells. Although in the conditions used we observed strong phosphorylation of model substrates such as MBP, we did not observe in any case phosphate incorporation into LC8 (data not shown). Additionally, LC8 was not phosphorylated in vivo during mitosis (as determined by 32P labeling of cells, data not shown), when Nek9, Nek6, and Nek7 are active (13, 17, 35). Therefore, we conclude that LC8 is not a Nek9 or Nek6/7 in vitro or in vivo substrate.
FIGURE 4.
LC8 binding to Nek9 is modulated by Nek9[Ser944] autophosphorylation. A, HEK293T cells were transfected with empty FLAG vector (−), FLAG-Nek9 (wt), FLAG-Nek9[K81M] (K81M), or FLAG-Nek9[Δ346–732] (ΔRCC1), and anti-FLAG immunoprecipitates (IP) were immunoblotted with the indicated antibodies. WB, Western blot. B, HEK293T cells were transfected with the indicated FLAG-Nek9 forms, and FLAG immunoprecipitates were immunoblotted with the indicated antibodies. C, HEK293T cells were transfected with FLAG-Nek9 (wt) or the indicated FLAG-Nek9 mutants, and FLAG immunoprecipitates were immunoblotted with the indicated antibodies. In all cases, LC8 in the corresponding extracts is shown in the lower panel.
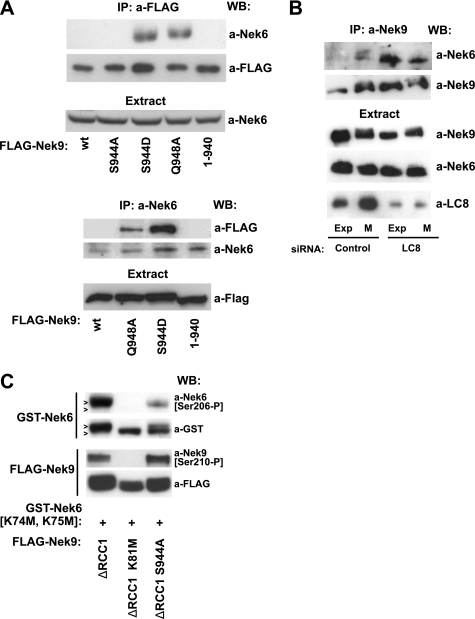
Nek9 activation is paralleled by autophosphorylation of several residues (13, 14). One of these residues, Ser944, is located immediately N-terminal to the LC8 binding motif (as determined by mass spectrometry analysis of in vitro activated Nek9,5 and of endogenous Nek9 (40)). We thus sought to determine whether Nek9 Ser944 autophosphorylation could directly interfere with the interaction of LC8 with Nek9. For this, we compared the amount of LC8 that coimmunoprecipitated with different forms of Nek9 wild type and Nek9[Δ346–732], including the constitutively active Nek9[Δ346–732], a kinase-deficient form, Nek9[Δ346–732, K81M], an active form not modifiable by phosphorylation at Ser944, Nek9[Δ346–732, S944A], and a form with a phosphomimetic residue at position 944, Nek9[Δ346–732, S944D] (Fig. 4B). As observed before, Nek9[Δ346–732] coimmunoprecipitation with LC8 was severely impaired when compared with Nek9 wild type. In contrast, Nek9[Δ346–732, K81M] coimmunoprecipitated with a significant amount of LC8, thus confirming that Nek9 activity interferes with LC8 binding. Nek9[Δ346–732, S944A], although reaching levels of activation equal to those of Nek9[Δ346–732] (as determined by direct activity measurements and the use of a phosphospecific antibody that exclusively recognizes active Nek9 (14), see supplemental Fig. 1 and Fig. 5C), interacted with a markedly bigger amount of LC8 than its counterpart. Finally, the phosphomimetic Nek9[Δ346–732, S944D] behaved similarly to Nek9[Δ346–732]. These results thus support that phosphorylation of Ser944 (or its replacement by a negatively charged residue) interferes with the interaction of LC8 with Nek9. To further confirm this, in a complementary experiment, we produced a phosphomimetic mutant of full-length Nek9, Nek9[S944D], and compared its ability to bind LC8 with that of Nek9 wild type, Nek9[Q948A], and Nek9[S944A]. Fig. 4C shows that although Nek9[S944A] interacted with LC8 with a similar efficiency to that of Nek9 wild type (as expected as they are both inactive and thus with unphosphorylated Ser944), Nek9[S944D], by mimicking Nek9 autophosphorylation at Ser944, almost completely lost its ability to interact with LC8, resembling active Nek9[Δ346–732] (or Nek9[Q948A]). Altogether, our results indicate that LC8 binding to Nek9 is negatively regulated by phosphorylation at Ser944 and that Nek9 activation disrupts the interaction of LC8 with the kinase through the autophosphorylation of this residue.
FIGURE 5.
LC8 binding to Nek9 regulates Nek6 interaction with Nek9 and activation. A, upper panels, the indicated FLAG-Nek9 forms were immunoprecipitated from transfected HEK293T cells and immunoblotted with anti-FLAG or anti-Nek6 antibodies. Nek6 in the corresponding extracts is shown in the lower panel. Lower panels, reciprocal Nek6 immunoprecipitation (IP) using anti-Nek6 antibody; the corresponding FLAG fusion proteins in the extracts are shown. WB, Western blot. B, HeLa cells were transfected with control or LC8 siRNA, and immunoprecipitates were prepared from exponentially growing (Exp) or nocodazole-arrested mitotic (M) extracts, using anti-Nek9 antibody, and immunoblotted with the indicated antibodies. The corresponding amount of Nek9, Nek6, and LC8 in the extracts is shown. C, HEK293T cells were cotransfected with GST Nek6[K74M, K75M], and the indicated forms of FLAG-Nek9[Δ346–732] (ΔRCC1), and total and active forms of the recombinant kinases were detected by Western blot using a-GST and a-Nek6[Ser206-P] for Nek6 and a-FLAG and a-Nek9[Thr210-P] for Nek9. Note that phosphorylated forms of GST-Nek6[K74M, K75M] show a higher apparent molecular weight (arrowheads).
The Interaction of LC8 with Nek9 Negatively Controls the Binding of Nek6
The NIMA family protein kinases Nek6 and the highly similar Nek7 interact with the C-terminal tail of Nek9, where they are activated by direct phosphorylation by Nek9 (13, 35), and we have previously shown that Nek6 binding to Nek9 is highly regulated and occurs exclusively during mitosis, when a small fraction of Nek9 is activated (15). We reasoned that LC8 binding to the C-terminal region of Nek9 could modulate the interaction of Nek6/7 with Nek9. To test that hypothesis, we expressed different forms of Nek9 in 293T cells and observed their ability to interact with endogenous Nek6 by immunoprecipitation of either kinase (Fig. 5A). As described previously, in exponentially growing cells, wild type Nek9 did not coimmunoprecipitate with Nek6. Nek9[S944A], a mutant without any discernible effect on LC8 binding (Fig. 4C), behaved similarly. In contrast to this, two different Nek9 mutants with impaired LC8 binding, Nek9[S944D] and Nek9 [Q948A], readily coimmunoprecipitated with Nek6. Interestingly, removal of the extreme C terminus of Nek9 (Nek9[1–940]), a modification that eliminates the LC8 binding motif but also the last 29 residues of Nek9, totally abrogated Nek6 binding to Nek9, thus indicating that the C-terminal residues of Nek9 (Nek9[941–979]) are necessary not only for interaction with LC8, but also for Nek6 binding to Nek9.
To further support a negative regulatory role of LC8 on Nek6 binding to Nek9, we next used a mix of two LC8 siRNAs to down-regulate LC8 and observed the effect of this down-regulation on the amount of Nek6 bound to Nek9 (Fig. 5B). As expected, in cells transfected with control siRNA oligonucleotides, a small amount of Nek6 interacted with Nek9 exclusively during mitosis, and thus Nek9 immunoprecipitates from interphase cells did not contain any significant amounts of Nek6. In contrast, Nek9 immunoprecipitates from interphase cells transfected with an LC8 siRNA (and thus depleted of this polypeptide) contained Nek6 levels that were similar to these of Nek9 immunoprecipitates from mitotic cells and equal to or bigger than those observed from control mitotic cells. Thus, LC8 depletion deregulates Nek6 binding to Nek9.
Expression of active forms of Nek9 induces Nek6 phosphorylation in the activation loop, and as a result, Nek6 activation, and this can be monitored by the use of an antibody that specifically recognizes Nek6 phosphorylated at Ser206 (35). To determine whether LC8 could interfere not only with Nek6 binding to Nek9 but also with Nek6 activation by Nek9 in vivo, we next tested the ability of different forms of Nek9 to phosphorylate Nek6[Ser206]. For this, we cotransfected kinase-deficient GST-Nek6[K74M, K75M], devoid of the ability to autophosphorylate (35), with different forms of Nek9. Fig. 5C shows that active Nek9[Δ346–732] (but not the kinase-deficient Nek9[Δ346–732, K81M]) readily induced GST-Nek6[K74M, K75M] phosphorylation at Ser206. Nek9[Δ346–732, S944A], in contrast, showed a greatly impaired capacity to phosphorylate Nek6 despite the fact that it is as active as Nek9[Δ346–732], as shown by similar levels of activation loop phosphorylation at Nek9[Thr210] (and by their ability to phosphorylate a model substrate such as MBP in vitro, see supplemental Fig. 1).
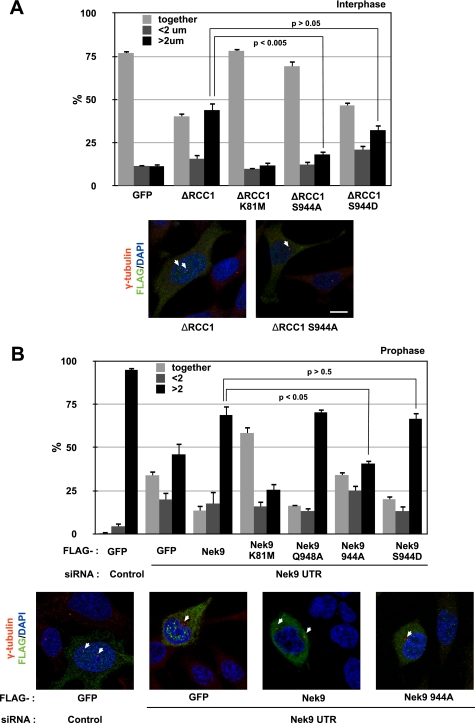
We conclude that LC8 binding, in addition to modulating Nek9 oligomerization and autophosphorylation, controls the interaction of Nek6 with Nek9. In response to Nek9 activation and autophosphorylation, LC8 binding to this kinase is disrupted, thus allowing Nek9 to interact with Nek6, phosphorylate Nek6 in the activation loop, and activate it. Ultimately, LC8 controls signal transduction through the Nek9/Nek6 mitotic module by ensuring that Nek6 is activated in a coordinated manner with Nek9. To test the physiological importance of this control, we sought to determine whether known Nek9/Nek6 functions depend on the ability of Nek9 to disengage LC8 and thus to be able to activate Nek6. We have recently found that the Nek9/Nek6 module is necessary for normal early mitotic centrosome separation and that constitutively active forms of Nek9 are able to induce centrosome separation even in interphase in a Nek6-dependent manner.6 We thus compared the ability to separate centrosomes of different recombinant Nek9 forms (Fig. 6A). We observed that active Nek9[Δ346–732] induced centrosome separation in a significant number of cells, with 44% of cells showing centrosomes that were separated >2 μm (versus 11% in control cells). In contrast, kinase-deficient Nek9[Δ346–732, K81M] resulted in only 12% of cells showing that phenotype. Nek9[Δ346–732, S944A], although as active as Nek9[Δ346–732], showed an impaired ability to separate centrosomes (resulting in 18% of cells with centrosomes separated >2 μm). Finally, Nek9[Δ346–732, S944D], not phosphorylatable at Ser944 but with a phosphomimetic residue in this position, induced centrosome separation in 32% of the cells, a value not significantly different from that of Nek9[Δ346–732]. Complementary results were obtained by determining the ability of different Nek9 forms to rescue endogenous Nek9 depletion by RNAi during early centrosome separation (Fig. 6B). Transfection of Nek9 3′-UTR siRNA resulted in a significant number of prophase cells with unseparated centrosomes (34% versus <1% in control cells), and expression of wild type Nek9 was able to partially rescue this effect, resulting in only 14% of cells with unseparated centrosomes. In contrast, expression of a control protein or the kinase-deficient Nek9[K81M] did not rescue the RNAi phenotype, with Nek9[K81M] acting as a dominant negative and resulting in 58% of prophase cells with centrosomes separated <2 μm. Nek9[Q948A] and Nek9[S944D], with an impaired binding to LC8 and interacting constitutively with Nek6, behaved similarly to Nek9 wild type. In contrast, Nek9[S944A], which is predicted to bind LC8 constitutively and thus not to be able to bind Nek6, was not capable of rescuing the observed phenotype, resulting in a percentage of cells with unseparated centrosomes that is not significantly different from that of cells expressing GFP (34% in both cases).
FIGURE 6.
LC8 binding to Nek9 regulates its physiological activity. A, exponentially growing HeLa cells transfected either with FLAG-GFP or with different FLAG-Nek9 [Δ346–732] (ΔRCC1) forms were fixed and stained with antibodies against FLAG and γ-tubulin to detect centrosomes and DAPI to visualize DNA. The percentages of FLAG-positive cells showing two unseparated centrosomes (together), two centrosomes separated less than 2 μm (<2 μm), and fully separated centrosomes (>2 μm) are shown in the upper panel (mean ± S.E. of three independent experiments; ∼50 cells were counted in each experiment, and statistical significance was determined using the standard Student's t test). Representative examples of the observed phenotypes are shown below (bar, 10 μm). B, exponentially growing HeLa cells were transfected with either control or Nek9 3′-UTR siRNAs, and 24 h later, they were transfected with expression plasmids for the indicated FLAG-tagged proteins. After an additional 24 h, cells were fixed and processed as in A. FLAG-GFP was used as a control protein. Prophase cells (showing condensed chromosomes and intact nuclei as assessed by the shape of the DNA and a γ-tubulin exclusion) were categorized according to centrosome separation (mean ± S.E. of three independent experiments; ∼50 cells were counted in each experiment, and statistical significance was determined using the standard Student's t test). Representative examples of the observed phenotypes are shown below (bar, 10 μm).
In conclusion, phosphorylation at Nek9[Ser944] and thus disengagement from LC8 is necessary for Nek9 to perform at least some of its physiological functions. We thus put forward LC8 as a controller of signal transduction through the Nek9/Nek6 module and suggest that LC8, independently from its function as a dimerization hub, may be capable of performing similar regulatory roles in the context of other protein complexes.
DISCUSSION
The importance of the NIMA family protein kinases Nek9, Nek6, and Nek7 for mitotic spindle formation and normal progression through mitosis has been underscored in several experimental systems (13, 14, 17–19, 41). Interference with the function of these kinases can lead to aberrant mitosis and aneuploidy (13, 17, 19), and at least for Nek6, a connection between their deregulation and cell transformation has been established (42, 43). Nek9 and Nek6/7 are activated in a coordinated manner, thus forming a signaling module. The module upstream kinase, Nek9, is activated in early mitosis, when it binds Nek6/7 through its non-catalytic C-terminal region (13, 14). Once active, Nek9 is able to directly phosphorylate Nek6 and Nek7 in the activation loop (respectively at Nek6[Ser206] and Nek7[Ser195]), directly activating both kinases (35). In addition, the binding of Nek6/7 to Nek9 has been described to release Nek6/7 from an autoinhibited conformation and thus to directly contribute to their activation (16).
Despite all the reported data, a complete picture of Nek6/7 activation by Nek9 still lacks an account of the mechanism that regulates the interaction of these kinases during the cell cycle. Here we propose that this interaction is regulated by the small protein LC8 in conjunction with Nek9 autophosphorylation. LC8 (also known as DYNLL and DLC) was first described as a light chain subunit of the dynein complex and has since been shown to interact with multiple proteins, helping to organize and oligomerize them and thus effectively acting as a dimerization hub (22). We have identified the two mammalian LC8 variants, LC8a/DYNLL1/DLC1 and LC8b/DYNLL2/DLC2, in a screen for Nek9 interactors. LC8a and LC8b are highly similar, differing in only 6 out of 89 amino acids near the N terminus of the sequence, with only three of the substitutions being non-homologous (44), and are referred collectively as to LC8 herein. Our results show that LC8 interacts with the non-catalytic C-terminal part of Nek9 and that this interaction is dependent on the existence of a sequence (945KGTQT949) that conforms to the previously described LC8 recognition motif (KR)XTQT (37, 38). Interestingly, in the original study describing (K/R)XTQT as an LC8 recognition motif, Lo et al. (37) unknowingly identified a C-terminal fragment of Nek9 (named C82, as full-length Nek9 was yet to be identified and described) as an LC8-interacting protein, mapping the binding in vitro to the sequence 940VGMHSKGTQTAKEE953 and thus agreeing with our findings. Our results show that Nek9 binds to dimeric LC8 as the observed binding is disrupted by an LC8 mutation that interferes with LC8 dimerization by mimicking phosphorylation, LC8[S88D]. Phosphorylation of LC8 Ser88 has been proposed to differentially interfere with the binding of LC8 to different partners, selecting more tightly bound polypeptides and releasing weaker interactors such as DIC (25, 27). We have not detected phosphorylation of LC8 by [32P]orthophosphate in vivo labeling of unstimulated exponentially growing or mitotic cells, and thus assume that in the conditions used in this study, most LC8 would be present as a dimer and thus available for binding to partners such as Nek9.
LC8 was originally attributed an adaptor function for dynein-interacting proteins that could in this manner be transported by the motor complex as cargo. Structural studies strongly argue against this as different partners (including DIC) occupy the same LC8 binding region, thus impeding the simultaneous assembly of a dynein-cargo complex (for a review, see Ref. 22). Indeed we have not detected any interaction between Nek9 and DIC, the subunit of dynein that directly interacts with LC8, and we thus rule out that LC8 is attaching Nek9 to dynein. LC8 dimeric structure, with dual symmetric binding sites, has been proposed to be the basis of its function, that is, to promote partner dimerization through its ability to align proteins in the proximity of each other (22). The presence of coiled coil or other dimerization domains together with disordered and flexible regions in the partners would facilitate LC8 role as a dimerization engine. Nek9 is a homooligomer and has a putative C-terminal coiled coil motif (residues 891–940) (13) that is immediately N-terminal to a region, residues 933–979, that contains the LC8 binding motif and that is predicted to be disordered (45). It is thus possible that LC8 could facilitate Nek9 dimerization or affect its oligomeric state. Our present and past (13) results indicate that Nek9 coiled coil is both necessary and sufficient for Nek9 oligomerization and that this is independent of LC8. Nevertheless, in vitro experiments using purified proteins and size-exclusion gel chromatography suggest that LC8 is indeed able to affect the quaternary structure of Nek9. Our results are consistent with a role for LC8 in Nek9 tetramerization, and endogenous Nek9 (with a predicted molecular mass of ∼120 kDa) has an apparent molecular mass of ∼600 kDa, also compatible with a tetramer (13). We thus propose that Nek9 coiled coil is sufficient for dimerization and that LC8 binding to Nek9 KXTQT motif, besides possibly ordering the extreme C terminus of the protein, induces the dimerization of Nek9 dimers and thus its tetramerization. We hope that our ongoing efforts to resolve the tridimensional structure of different Nek9 polypeptides, including Nek9[893–974]-LC8 complexes, will determine the validity of this hypothesis.
We have previously shown that the oligomerization state of Nek9 is directly related to its autoactivation mechanism; Nek9 is able to activate in vitro through autophosphorylation, and this activation completely depends on oligomerization through Nek9 coiled coil (13). We now show that LC8, although not necessary for Nek9 activation, significantly increases the rate of autoactivation of the kinase in vitro. Our current data indicate that Nek9 is activated in vivo by phosphorylation of a combination of mitotic kinases and that autophosphorylation of the kinase could have a role locally amplifying this activation.6 LC8 may have a role during this amplification step, facilitating Nek9 trans-autophosphorylation in the context of a higher-order oligomer.
In addition to its effects on Nek9 quaternary structure and activation, LC8 binding to the kinase has an additional, novel, and probably physiologically more important role in the control of signal transduction through the Nek9/Nek6/7 module. This conclusion comes from the observation that LC8 binding to Nek9 depends on the activation state of the kinase, with kinase-defective forms of Nek9 interacting with increased amounts of LC8 and constitutively active Nek9 forms almost totally devoid of LC8 interaction. This cannot be attributed to LC8 phosphorylation by Nek9 or the downstream Nek6 as LC8 is not a substrate of these kinases. However, once active, Nek9 autophosphorylates at several sites (13, 14), and this modification could interfere with LC8 binding to Nek9. We have confirmed this and identified Ser944, an autophosphorylation site located immediately N-terminal to the LC8 recognition motif, as the residue that controls LC8 binding to Nek9. Ser944 autophosphorylation during Nek9 activation results in disruption of LC8 interaction. Ser944 location ideally suits it for this role as this residue is directly adjacent to the LC8 recognition motif. A structural model (see supplemental Fig. 2, based on structural data of LC8-peptide complex, PDB ID: 3E2B (46)) shows that the interaction between LC8 and Nek9 is made by main chain contacts, forming a new antiparallel β-strand with LC8, and more specifically, by the side chains buried in the interface with LC8. Our oligomerization model proposes two Nek9 chains interacting with one LC8 dimer. Adjacent or within the LC8 recognition motif, only two residues of Nek9 are in contact with the second LC8 molecule from the dimer, Ser944 and Gln948 (see supplemental Fig. 2). Although Gln948 contacts to LC8 are essential and seem to contribute to dimer interaction, phosphorylated Ser944 would possibly have a negative effect on the stability of the LC8 dimer bound to Nek9 as the negative charges of two phosphorylated Ser944 residues from both Nek9 chains might interfere with the LC8 dimer and promote its dissociation from Nek9.
The tight regulation of LC8 binding to the C terminus of Nek9 suggests that this interaction has an important physiological role. Our results lead us to propose this role to be the control of the interaction between Nek9 and its downstream effector Nek6 (and possibly Nek7). Endogenous Nek6/7 binding to Nek9 C-terminal domain is highly regulated, occurring only during mitosis, when a fraction of Nek9 is activated (15) and when we have observed that the kinase interacts with a reduced amount of LC8. Accordingly, our data show that mutations that interfere with LC8 binding to Nek9 (by mimicking Nek9 phosphorylation at Ser944 or directly disrupting the LC8 recognition motif) directly result in ectopic Nek6 binding to Nek9 in exponentially growing cells. A causal relationship between LC8 binding and Nek6 interaction with Nek9 can be further proven by down-regulating the cellular pool of LC8 by RNAi, which results in the deregulation of the Nek6-Nek9 binding and the observation of Nek9/Nek6 coimmunoprecipitates both in interphase and in mitosis.
Our data show that Nek6 binding to Nek9 is necessary for Nek6 phosphorylation by Nek9, activation, and function. Nek9[Δ346–732, S944A], an active form of Nek9 that is constitutively bound to LC8 and is thus not able to bind Nek6, has a severely impaired ability to phosphorylate Nek6 in the activation loop, a modification that we have previously shown that parallels kinase activation (35). Additionally, this and other LC8-bound S944A Nek9 mutants (but not the corresponding S944D phosphomimetic mutants that do not bind LC8) are impaired in eliciting cellular effects that depend on Nek6/7 activation, such as centrosome separation.6 Thus, regulated binding of LC8 to Nek9 controls signal transduction through the Nek9/Nek6/7 signaling module, not only because it affects Nek9 oligomeric structure favoring its activation, but also and perhaps more importantly, because it is responsible for the control of Nek6 (and presumably Nek7) binding to Nek9 and thus of Nek6/7 activation (by phosphorylation but also probably through the induction of a conformational change (16)).
While describing the mechanism that regulates the binding of Nek6 to Nek9, our work has uncovered both a novel function and a novel regulatory mechanism for LC8. A negative effect of LC8 on protein complex formation has to our knowledge never been described and could be a general function of this protein. Similarly, the modification by phosphorylation of an LC8 partner near the LC8 recognition site could be a common form of control of LC8 interaction with different partners. A Swiss-Prot database search using the (ST)(KR)XTQT pattern shows that there are at least 49 human proteins that could be regulated in this manner, including confirmed LC8 partners such as DIC1 and DIC2. The number of candidates would significantly increase if the incorporation of phosphate to other residues in or near the motif could also interfere with LC8 binding. In fact, it has been shown that phosphorylation of the Bcl2-related BimL inside the LC8 recognition motif (at Thr56 in the sequence 52KSTQT56) releases the protein from LC8 (47), suggesting that this may be the case. Conceivably, this mechanism could regulate in both space and time the interaction of different proteins with LC8 in response to both autophosphorylation and phosphorylation by different kinases, as well as their dephosphorylation by phosphatases. The binding of LC8 to partners could thus be regulated globally by changes in LC8 dimerization (induced by direct phosphorylation of LC8[Ser88]) or by changes in the pH or redox state of the cell, or in a more subtle and partner-specific manner, by partner phosphorylation. This hypothesis awaits a complete characterization of the LC8 interactome, the identification of possible sites of phosphorylation located around the different LC8 recognition sequences, and the determination of the effects of this modification on the interaction of LC8 with its partners.
Supplementary Material
Acknowledgments
We thank M. Nicolás for in vivo 32P labeling of cells. We thank C. Vila and other members of the extended Cell Signaling group as well as the staff of the IRB Barcelona Advanced Digital Microscopy Core Facility for help. We also thank I. Vernos (Centre de Regulació Genòmica, Barcelona, Spain) and J. Lüders (IRB Barcelona) for comments and C. Lima (Sloan-Kettering Institute, New York, NY) for the pSmt3 vector.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
M. T. Bertran and J. Roig, unpublished results.
J. Avruch and J. Roig, unpublished results.
M. T. Bertran, S. Sdelci, L. Regué, J. Avruch, C. Caelles, and J. Roig, submitted.
- DYNLL
- dynein light chain
- DLC
- dynein light chain
- DIC
- dynein intermediate chain
- MBP
- myelin basic protein
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. O'Connell M. J., Krien M. J., Hunter T. (2003) Trends Cell Biol. 13, 221–228 [DOI] [PubMed] [Google Scholar]
- 2. O'regan L., Blot J., Fry A. M. (2007) Cell Div. 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roig J., Avruch J. (2006) in SIGMA-RBI Handbook of Receptor Classification and Signal Transduction (Watling K. J. ed) 5th Ed., pp. 286–290, Sigma-RBI, Natickm, MA [Google Scholar]
- 4. Quarmby L. M., Mahjoub M. R. (2005) J. Cell Sci. 118, 5161–5169 [DOI] [PubMed] [Google Scholar]
- 5. Noguchi K., Fukazawa H., Murakami Y., Uehara Y. (2002) J. Biol. Chem. 277, 39655–39665 [DOI] [PubMed] [Google Scholar]
- 6. Chen Y., Chen P. L., Chen C. F., Jiang X., Riley D. J. (2008) Cell Cycle 7, 3194–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Melixetian M., Klein D. K., Sørensen C. S., Helin K. (2009) Nat. Cell Biol. 11, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 8. Moniz L. S., Stambolic V. (2011) Mol. Cell. Biol. 31, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pelegrini A. L., Moura D. J., Brenner B. L., Ledur P. F., Maques G. P., Henriques J. A., Saffi J., Lenz G. (2010) Mutagenesis 25, 447–454 [DOI] [PubMed] [Google Scholar]
- 10. Roig J. (2010) UCSD-Nature Molecule Pages, 10.1038/mp.a003029.01 [DOI] [Google Scholar]
- 11. Roig J. (2010) UCSD-Nature Molecule Pages, 10.1038/mp.a003428.01 [DOI] [Google Scholar]
- 12. Roig J. (2010) UCSD-Nature Molecule Pages, 10.1038/mp.a003631.01 [DOI] [Google Scholar]
- 13. Roig J., Mikhailov A., Belham C., Avruch J. (2002) Genes Dev. 16, 1640–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roig J., Groen A., Caldwell J., Avruch J. (2005) Mol. Biol. Cell 16, 4827–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rapley J., Nicolàs M., Groen A., Regué L., Bertran M. T., Caelles C., Avruch J., Roig J. (2008) J. Cell Sci. 121, 3912–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richards M. W., O'Regan L., Mas-Droux C., Blot J. M., Cheung J., Hoelder S., Fry A. M., Bayliss R. (2009) Mol Cell 36, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Regan L., Fry A. M. (2009) Mol. Cell. Biol. 29, 3975–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yissachar N., Salem H., Tennenbaum T., Motro B. (2006) FEBS Lett. 580, 6489–6495 [DOI] [PubMed] [Google Scholar]
- 19. Salem H., Rachmin I., Yissachar N., Cohen S., Amiel A., Haffner R., Lavi L., Motro B. (2010) Oncogene 29, 4046–4057 [DOI] [PubMed] [Google Scholar]
- 20. Makokha M., Hare M., Li M., Hays T., Barbar E. (2002) Biochemistry 41, 4302–4311 [DOI] [PubMed] [Google Scholar]
- 21. Nyarko A., Hare M., Hays T. S., Barbar E. (2004) Biochemistry 43, 15595–15603 [DOI] [PubMed] [Google Scholar]
- 22. Barbar E. (2008) Biochemistry 47, 503–508 [DOI] [PubMed] [Google Scholar]
- 23. Liang J., Jaffrey S. R., Guo W., Snyder S. H., Clardy J. (1999) Nat. Struct. Biol. 6, 735–740 [DOI] [PubMed] [Google Scholar]
- 24. Mohan P. M., Barve M., Chatterjee A., Hosur R. V. (2006) Protein Sci. 15, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song Y., Benison G., Nyarko A., Hays T. S., Barbar E. (2007) J. Biol. Chem. 282, 17272–17279 [DOI] [PubMed] [Google Scholar]
- 26. Jung Y., Kim H., Min S. H., Rhee S. G., Jeong W. (2008) J. Biol. Chem. 283, 23863–23871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song C., Wen W., Rayala S. K., Chen M., Ma J., Zhang M., Kumar R. (2008) J. Biol. Chem. 283, 4004–4013 [DOI] [PubMed] [Google Scholar]
- 28. Vadlamudi R. K., Bagheri-Yarmand R., Yang Z., Balasenthil S., Nguyen D., Sahin A. A., den Hollander P., Kumar R. (2004) Cancer Cell 5, 575–585 [DOI] [PubMed] [Google Scholar]
- 29. Lightcap C. M., Sun S., Lear J. D., Rodeck U., Polenova T., Williams J. C. (2008) J. Biol. Chem. 283, 27314–27324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dick T., Surana U., Chia W. (1996) Mol. Gen. Genet. 251, 38–43 [DOI] [PubMed] [Google Scholar]
- 31. Miki F., Okazaki K., Shimanuki M., Yamamoto A., Hiraoka Y., Niwa O. (2002) Mol. Biol. Cell 13, 930–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dick T., Ray K., Salz H. K., Chia W. (1996) Mol. Cell. Biol. 16, 1966–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Phillis R., Statton D., Caruccio P., Murphey R. K. (1996) Development 122, 2955–2963 [DOI] [PubMed] [Google Scholar]
- 34. Lightcap C. M., Kari G., Arias-Romero L. E., Chernoff J., Rodeck U., Williams J. C. (2009) PLoS One 4, e6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belham C., Roig J., Caldwell J. A., Aoyama Y., Kemp B. E., Comb M., Avruch J. (2003) J. Biol. Chem. 278, 34897–34909 [DOI] [PubMed] [Google Scholar]
- 36. Traer C. J., Rutherford A. C., Palmer K. J., Wassmer T., Oakley J., Attar N., Carlton J. G., Kremerskothen J., Stephens D. J., Cullen P. J. (2007) Nat. Cell Biol. 9, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 37. Lo K. W., Naisbitt S., Fan J. S., Sheng M., Zhang M. (2001) J. Biol. Chem. 276, 14059–14066 [DOI] [PubMed] [Google Scholar]
- 38. Rodríguez-Crespo I., Yélamos B., Roncal F., Albar J. P., Ortiz de Montellano P. R., Gavilanes F. (2001) FEBS Lett. 503, 135–141 [DOI] [PubMed] [Google Scholar]
- 39. Benashski S. E., Harrison A., Patel-King R. S., King S. M. (1997) J. Biol. Chem. 272, 20929–20935 [DOI] [PubMed] [Google Scholar]
- 40. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 41. Yin M. J., Shao L., Voehringer D., Smeal T., Jallal B. (2003) J. Biol. Chem. 278, 52454–52460 [DOI] [PubMed] [Google Scholar]
- 42. Nassirpour R., Shao L., Flanagan P., Abrams T., Jallal B., Smeal T., Yin M. J. (2010) Mol. Cancer Res. 8, 717–728 [DOI] [PubMed] [Google Scholar]
- 43. Jeon Y. J., Lee K. Y., Cho Y. Y., Pugliese A., Kim H. G., Jeong C. H., Bode A. M., Dong Z. (2010) J. Biol. Chem. 285, 28126–28133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilson M. J., Salata M. W., Susalka S. J., Pfister K. K. (2001) Cell Motil. Cytoskeleton 49, 229–240 [DOI] [PubMed] [Google Scholar]
- 45. Yang Z. R., Thomson R., McNeil P., Esnouf R. M. (2005) Bioinformatics 21, 3369–3376 [DOI] [PubMed] [Google Scholar]
- 46. Benison G., Karplus P. A., Barbar E. (2007) J. Mol. Biol. 371, 457–468 [DOI] [PubMed] [Google Scholar]
- 47. Lei K., Davis R. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2432–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.