Abstract
In yeast, the two main FO proton-translocating subunits of the ATP synthase (subunits 6/a and 9/c) are encoded by mitochondrial DNA (mtDNA). Unfortunately, mutations that inactivate the FO typically result in loss of mtDNA under the form of ρ−/ρ0 cells. Thus, we have designed a novel genetic strategy to circumvent this problem. It exploits previous findings that a null mutation in the nuclear ATP16 gene encoding ATP synthase subunit δ results in massive and lethal FO-mediated protons leaks across the inner mitochondrial membrane. Mutations that inactivate the FO can thus, in these conditions, be selected positively as cell viability rescuing events. A first set of seven mutants was analyzed and all showed, as expected, very severe FO deficiencies. Two mutants carried nuclear mutations in known genes (AEP1, AEP2) required for subunit c expression. The five other mutations were located in mtDNA. Of these, three affect synthesis or stability of subunit a transcripts and the two last consisted in a single amino acid replacement in subunit c. One of the subunit c mutations is particularly interesting. It consists in an alanine to valine change at position 60 of subunit c adjacent to the essential glutamate of subunit c (at position 59) that interacts with the essential arginine 186 of subunit a. The properties of this mutant suggest that the contact zone between subunit a and the ten subunits c-ring structure only involves critical transient interactions confined to the region where protons are exchanged between the subunit a and the c-ring.
Keywords: F1Fo ATPase, Membrane Biogenesis, Mitochondrial DNA, Protein-protein Interactions, Yeast Genetics
Introduction
The energy needs of living organisms are essentially met through the action of an F1FO-type ATP synthase, a complex found in the plasma membrane of bacteria, in the inner membrane of mitochondria, and in the thylakoid membrane of chloroplasts. The ATP synthase catalyzes the synthesis of ATP from ADP and orthophosphate using a chemiosmotic energy, most commonly that of a proton gradient, across its host membrane (1–3). The ATP synthase consists of two distinct domains, a globular extramembrane catalytic unit called F1, and a membrane-embedded proton-translocating domain known as FO. The current model for the ATP synthase energy coupling is the binding change mechanism, according to which affinity changes for substrates and products at the catalytic sites are coupled to proton transport through the rotation of a subcomplex of the enzyme (1).
In mitochondria, the F1 is an assembly of five different subunits, with a α3β3γδϵ stoichiometry, that contains three catalytic sites located in the β-subunits (4–6). The synthesis of ATP by the β subunits depends upon rotation of the F1 subcomplex (called the central stalk) formed by subunits γ, δ, and ϵ. Evidence for this rotation has been obtained by optical microscopy (7). The main components involved in proton translocation are a ring of c subunits (ten in yeast where this subunit is also referred to as Atp9p, (8)) and a single subunit a (also referred to as Atp6p in yeast) (9, 10). Direct contacts between the c-ring and subunits γ and δ enable the c-ring and the central stalk to rotate together as a fixed ensemble (8, 11). The F1 is also physically connected to FO via its external surface by the so-called peripheral stalk that acts as a stator to counter the tendency of the α3β3 subcomplex to follow the rotation of the subunit γ during catalysis. In yeast mitochondria, the peripheral stalk consists of single copies of subunits OSCP (Oligomycin Sensitivity Conferring Protein), 4, d, h, f, 8, and i (2, 12, 13). In mitochondria, the ATP synthase exists as a dimer (14), a structure mediated by subunits e and g that is important for cristae formation (15, 16). The mitochondrial ATP synthase also contains peptides (If1p, Stf1p, and Stf2p in yeast) that have been implicated in the regulation of the enzyme by modulating its hydrolytic activity (17). In most eukaryotes, the mitochondrial ATP synthase has a dual genetic origin, nuclear and mitochondrial. In Saccharomyces cerevisiae, the mitochondrial genome encodes three subunits of the FO (subunits a, c, and 8). Studies in yeast have revealed that the assembly pathway of the ATP synthase is particularly complex with a number of protein factors having specific actions in the expression of the mitochondrial DNA (mtDNA)4-encoded subunits and in the establishment of proper subunit interactions (reviewed in Refs. 2, 13, 18).
As a genetically tractable eukaryote with a good fermenting capacity, yeast is a good system to study the ATP synthase. Both nuclear and mitochondrial genes can be manipulated in this organism (19), making it possible to introduce virtually any change in ATP synthase structure and in proteins involved in the assembly of this enzyme. However, defects in ATP synthase often result, in yeast, in loss of mtDNA under the form of ρ−/ρ0 cells (20). Only mutants deficient in α-F1 and β-F1 proteins do not produce ρ−/ρ0 cells because of the inability of these mutants to survive the loss of the respiratory proteins encoded by the mitochondrial genome (21). The most prominent impact on mtDNA stability, with 100% ρ−/ρ0 cells, has been observed in mutants lacking the γ-F1 and δ-F1 proteins (22, 23). Using a regulatable subunit δ gene (ATP16), we have shown that a lack in this subunit results in the formation of partial F1FO assemblies freely transporting protons across the mitochondrial inner membrane (24). Beyond a 50% deficit in subunit δ, the cells are unable to maintain any electrical potential (ΔΨ) across the mitochondrial inner membrane (25). As a consequence, the cells need then to inactivate the FO to survive, which explains the 100% conversion into ρ−/ρ0 cells of strains lacking the subunit δ gene. Mutations in the FO have usually also a strong impact on the mtDNA, but the reason for this is still unknown. There is no reported case of mutations of the FO that largely uncouple the mitochondrion like mutations in the central stalk. A recent study has revealed that in bacteria specific subunit a mutations may generate massive protons leaks through the FO (26). In yeast, such mutations would render maintenance of a complete (ρ+) mtDNA not compatible with cell viability, like in strains deficient in subunit δ, which may explain the failure to isolate such mutants.
We describe in this study a novel genetic strategy for the study of yeast FO. The screen is designed for the isolation of mutations that inactivate the FO, like point mutations in the proton channel subunits or lesions in proteins needed to assemble the FO. The mutations are selected for their ability to rescue the viability of cells lacking the subunit δ. The screen is thus positive and targeted on the FO. To avoid rescue of δ-deficient cells by the ρ−/ρ0mutation, which is much more frequent than specific mutations of the FO (∼10−2 versus <10−5), we used a host strain in which a non-respiratory genetic marker, ARG8m (27), has been integrated into an intergenic region of the mitochondrial genome (25). This is a mitochondrial version of the nuclear gene ARG8 that encodes a mitochondrial protein involved in arginine biosynthesis (27). Consequently, without external arginine and in a Δarg8 nuclear background, ρ−/ρ0 cells can no longer grow and only FO inactive mutants with a complete (ρ+) mtDNA are selected as arginine prototrophs. We report the analysis of a first series of seven mutants isolated with this system.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
The S. cerevisiae strains used and their genotypes are listed in Table 1. The following rich media were used for the growth of yeast: 1% (w/v) yeast extract, 1% (w/v) peptone, 40 mg/l adenine, supplemented with 2% (w/v) glucose, 2% (w/v) galactose, or 2% (w/v) glycerol, respectively. The glycerol medium was buffered at pH 6.2 with 50 mm potassium phosphate. We also used minimal medium W0 (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 2% (w/v) glucose) and complete synthetic medium CSM medium (0.17% (w/v) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (w/v) ammonium sulfate, 2% (w/v) glucose, and 0.8% (w/v) of a mixture of amino acids and bases from BIO-101). Solid media contained 2% (w/v) agar.
TABLE 1.
Genotypes of yeast strains
| Strain | Nuclear genotype | mtDNA | Source |
|---|---|---|---|
| BY4741 | Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ρ+ | Euroscarf |
| KL14–4a/60 | Mata his1 trp2 | ρ0 | Gif collection |
| DFS160a | Mata ade2–101 ura3–52 leu2Δ arg8::URA3 kar1–1 | ρ0 | This study |
| MR6/b-3 | Matα his3–11,15 ade2–1 ura3–1 leu2–3,112 trp1–1 CAN1 arg8::HIS3 | ρ0 | This study |
| SDC17–4c | Mata met6 lys2 his3 ura3 leu2 arg8::hisG atp4::URA3 | ρ+Arg8m | This study |
| SDC17–31b | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 | ρ+Arg8m | (25) |
| FG3 | Mata ade2–101 ura3–52 leu2Δ arg8::URA3 kar1–1 | ρ+Arg8m | This study |
| FG4 | Matα his3–11,15 ade2–1 ura3–1 leu2–3,112 trp1–1 CAN1 arg8::HIS3 | ρ+Arg8m | This study |
| SDC25/1 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 aep1 FOi mutant | ρ+Arg8m | This study |
| SDC25/5 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 aep2 FOi mutant | ρ+Arg8m | This study |
| FG18 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::ura3::LYS2 + pATP4-HIS3 aep1 FOi mutant | ρ+Arg8m | This study |
| FG23 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::ura3::LYS2 + pATP4-HIS3 aep2 FOi mutant | ρ+Arg8m | This study |
| FG22 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::ura3::LYS2 + pATP4-HIS3 + pATP16-URA3 (pSDC13) aep1 FOi mutant | ρ+Arg8m | This study |
| FG24 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::ura3::LYS2 + pATP4-HIS3 + pATP16-URA3 (pSDC13) aep2 FOi mutant | ρ+Arg8m | This study |
| SDC25/2 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 | ρ+Arg8m atp9-A60V | This study |
| FG12 | Mata ade2–101 ura3–52 leu2Δ arg8::URA3 kar1–1 | ρ+Arg8m atp9-A60V | This study |
| FG13 | Matα his3–11,15 ade2–1 ura3–1 leu2–3,112 trp1–1 CAN1 arg8::HIS3 | ρ+Arg8m atp9-A60V | This study |
| SDC25/4 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 | ρ+Arg8m Δ3′UTR of ATP6 | This study |
| SDC25/6 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 | ρ+Arg8m Δ3′UTR of ATP6 | This study |
| SDC25/14 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 | ρ+Arg8m atp9-M67K | This study |
| FG1 | Mata ade2–101 ura3–52 leu2Δ arg8::URA3 kar1–1 | ρ+Arg8m atp9-M67K | This study |
| FG2 | Matα his3–11,15 ade2–1 ura3–1 leu2–3,112 trp1–1 CAN1 arg8::HIS3 | ρ+Arg8m atp9-M67K | This study |
| SDC25/26 | Matα met6 lys2 his3 ura3 arg8::hisG atp16::KanMX atp4::URA3 + pATP4-HIS3 | ρ+Arg8m Δ cox1-atp8-atp6-ens2 | This study |
| SDC30/2a | Mata ade2–101 ura3–52 leu2Δ arg8::URA3 kar1–1 | ρ- ATP6 | (50) |
| FG8 | Mata ade2–101 ura3–52 leu2Δ Arg8::URA3 kar1–1 | ρ- ATP8 | This study |
| FG9 | Mata ade2–101 ura3–52 leu2Δ Arg8::URA3 kar1–1 | ρ- ATP9 | This study |
Cytoduction of the atp9-A60V and atp9-M67K Mutations in a Wild-type Nuclear Context
We studied the properties of the atp9-A60V and atp9-M67K mutations in a wild-type nuclear context. To do this, we transferred the mitochondrial genomes of SDC25/2 and SDC25/14 by cytoduction, first into DFS160a and then into MR6/b-3. The final cytoductants were named FG12/13 and FG1/2, respectively (Table 1). As a wild-type control for FG12/13 and FG1/2, we used strain FG3/4 similarly obtained by two steps cytoduction of the parental strain SDC17–31b from which SDC25/2 and SDC25/14 were isolated.
Restoration of ATP16 Expression in SDC25/1 and SDC25/5 Mutants
To reintroduce the ATP16 gene into mutants SDC25/1 and SDC25/5, we first used plasmid M2660 carrying a DNA cassette including LYS2 to replace the URA3 marker used to delete ATP4 (28). SDC25/1 and SDC25/2 were transformed with plasmid M2660 cut with HindIII and transformed cells were selected on WO supplemented with methionine and uracil, and named, respectively, FG18 and FG23. The plasmid pSDC13 containing ATP16 under its native promoter (24) was then introduced in FG18 and FG23 to give FG22 and FG24, respectively (Table 1).
Construction of the Synthetic ρ− Strain FG8 and FG9
The wild-type ATP8 locus, from nucleotide position −271 upstream from the ATP8 initiator codon to nucleotide position +153 downstream from the ATP8 stop codon, was amplified by PCR- using mtDNA from strain SDC17–31b as a template and primers ATP8.3 (5′-CGGGATCCCGCTCCGCAAAGCCGGATTAATG-3′) for the sense strand and ATP8.120 (5′-ATATAAATATATAGTCCGTAAGGA-3′) for the antisense strand. The PCR product was blunt-end ligated into the EcoRV site of pMOS-Blue (pMOS Blue Blunt Ended Cloning Kit from GE Healthcare) to give plasmid pMOS-ATP8. The insert was removed from pMOS-ATP8 by digestion with BamHI-XbaI and ligated into the vector pJM2; this vector contains COX2 as a genetic marker for mitochondrial transformation (19)) creating plasmid pJM2-ATP8. This plasmid was introduced by biolistic transformation into DFS160a mitochondria with the nuclear selectable LEU2 plasmid pFL46 by microprojectile bombardment using a biolistic PDS-1000/He particle delivery system (Bio-Rad) as described previously (19). Mitochondrial transformants were identified among the Leu+ nuclear transformants by their ability to produce respiring clones when mated with the non-respiring NB40–3C strain bearing a deletion in the mitochondrial COX2 gene (19). One such mitochondrial transformant was isolated and named FG8.
The wild-type ATP9 locus, from nucleotide position −228 upstream from the ATP9 initiator codon to nucleotide position +98 downstream from the ATP9 stop codon, was amplified by PCR using mtDNA from strain SDC17–31b (25) as a template and primers ATP9.1 (5′-AATAAGATATATAAATAAGTCCC-3′) for the sense strand and ATP9.67 (5′-GAATGTTATTAATTTAATCAAATGAG-3′) for the antisense strand. The PCR product was blunt-end ligated into the EcoRV site of pMOS-Blue (pMOS Blue Blunt Ended Cloning kit from GE Healthcare) to give plasmid pMOS-ATP9. The insert was removed from pMOS-ATP9 by digestion with BamHI-XbaI and ligated in the vector pJM2 to create plasmid pJM2-ATP9. This plasmid was introduced into mitochondria by biolistic transformation as described above. One mitochondrial transformant was isolated and named FG9.
In Vivo Labeling of Mitochondrial Translation Products
These experiments were performed as previously described (29). Briefly, strains were grown to early exponential phase (107 cells/ml) in 10 ml of galactose-rich media. The cells were harvested by centrifugation and washed twice with a low sulfate medium (W0-AS, W0 without ammonium sulfate (30)). Cells were resuspended in 10 ml of W0-AS containing 1% galactose, supplemented with the corresponding auxotrophic markers but without methionine and cysteine and incubated for 2 h. These cultures were centrifuged, and the cells were resuspended in 0.5 ml of W0-AS containing 1% galactose and 1 mm of a freshly prepared solution of cycloheximide and incubated at 20 °C for 5 min; then 50 μCi of [35S]methionine-[35S]cysteine mix (1000 Ci/mmol, Perkin Elmer) was added. The reaction was terminated after 20 min by the addition of 75 μl of 1.85 m NaOH, 1 m β-mercaptoethanol, and 0.01 m phenylmethylsulfonyl fluoride. An equal volume of 50% trichloroacetic acid was added, and the mixture was centrifuged in a microcentrifuge for 5 min at 14,000 rpm. The pellet consisting of precipitated proteins was washed once with water and resuspended in 50 μl of sample buffer (2% SDS, 10% glycerol, 2.5% β-mercaptoethanol, 0.06 m Tris-HCl, pH 6.8, and 0.002% bromphenol blue). The proteins were then separated on a 16.5% polyacrylamide gel (31).
Miscellaneous Procedures
Isolated mitochondria were prepared by the enzymatic method (32). Protein amounts were determined by the procedure of Lowry (33) in the presence of 5% SDS. SDS-PAGE was as described by Laemmli and Schägger (31, 34). BN-PAGE experiments were carried out as described previously (35). Western blot analyses were performed as described (36). Polyclonal antibodies raised against yeast ATP synthase were used at a dilution of 1:50,000 for subunits β; 1:10,000 for subunit Atp6p and 1:5,000 for subunit Atp9p. Monoclonal antibodies against yeast porin (from Molecular Probes) were used at a dilution of 1:5000. Nitrocellulose membranes were incubated with peroxidase-labeled antibodies at a 1:10,000 dilution and revealed with the ECL reagent (GE Healthcare).
RESULTS
Procedure for Random Isolation of Mutants with an Inactive FO
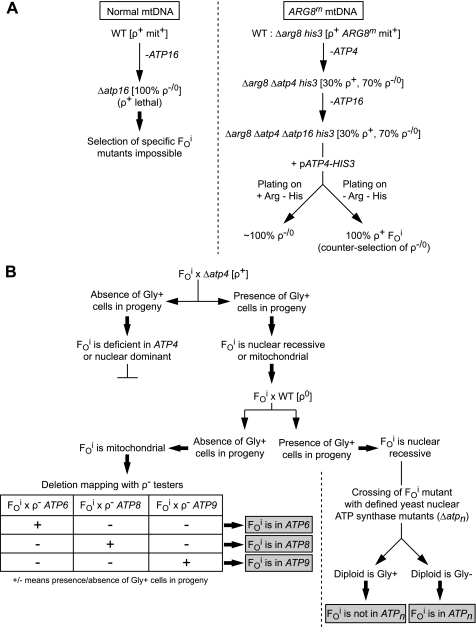
Retention of functional mtDNA in Δδ cells is not viable due to massive FO-mediated proton leaks across the inner mitochondrial membrane (25). As a result, 100% of Δδ cells are found as ρ−/ρ0 petites (22). Specific mutations of the FO (FOi) can thus not be selected for directly from Δδ cells. We therefore designed a strategy based on the use of a null mutation of Atp4p (Δatp4), a subunit of the peripheral stalk of the ATP synthase. A lack of Atp4p prevents the incorporation of Atp6p into the FO (37). In these conditions, the absence of subunit δ can no longer uncouple the mitochondrial membrane and subunit δ-deficient cells with a complete mtDNA are viable (25). Transformation of Δδ + Δatp4 cells with a plasmid-borne wild-type ATP4 gene will restore the assembly of a functional FO and the reconstituted Δδ cells will die. However, if the Δδ + Δatp4 cells acquired a mutation that alone inactivates the FO prior to their transformation with ATP4, the reconstituted Δδ cells are expected to be viable. To eliminate the ρ−/ρ0 mutation from the screen, we used cells with modified mtDNA in which ARG8m, a non-respiratory genetic marker (27), has been integrated into an intergenic region (25). This is a mitochondrial version of the nuclear gene ARG8 encoding a mitochondrial protein involved in arginine biosynthesis (27). As ρ−/ρ0 cells always lack at least one of the mitochondrial genes encoding components of the organelle protein synthesis system (tRNAs, rRNAs, and Var1p), they are unable to express ARG8m and hence unable to grow in the absence of external arginine (in a nuclear Δarg8 context). Only those FOi mutants that contain a complete mitochondrial genome (ρ+), can therefore be selected for directly by their ability to grow in the absence of arginine.
In practice, the FOi ρ+ mutants were selected as follows (Fig. 1A). Freshly grown Δδ + Δatp4 cells (strain SDC17–31b (25)) were transformed with a HIS3 plasmid containing ATP4 under control of its native promoter. The transformed cells were plated on a medium lacking both histidine and arginine (CSM-RH), i.e. conditions selective for both the plasmid (histidine synthesis) and the ρ+ state (arginine synthesis). As to estimate the total number of transformed cells (ρ+ and ρ−/ρ0 petites) that were plated on CSM-RH, an aliquot of transformed cells was plated on a medium lacking only histidine (CSM-H). On this medium, those transformed cells that were petites, i.e. by far the most frequent (70%), are able to grow. Out of 108 transformed cells plated on CSM-RH, about 30 viable clones were obtained, indicating that, excluding the ρ−/ρ0 petites, mutations rescuing cells lacking subunit δ are rare (10−6 to 10−7).
FIGURE 1.
Procedure for the isolation and mapping of yeast mutants with an inactive FO (FOi). Panel A describes the procedure for the isolation of FOi mutants using a host strain where the nuclear ARG8 gene has been replaced by a mitochondrial version of that gene (ARG8m) integrated into an intergenic region of the mitochondrial genome (mtDNA); see text for details. Panel B describes the procedure for mapping the mutations in nuclear or mitochondrial DNA, see text for details. Gly+/− refers to yeast cells able/unable to grow on glycerol; WT, wild-type; ρ−/0 refers to large (>50%) deletions (ρ−) or complete loss (ρ0) of the mtDNA; ρ+, complete mtDNA; ATPn is a known nuclear gene required for ATP synthase expression.
Genetic Origin, Nuclear, or Mitochondrial, of the FOi Mutants
Plasmid-borne genes are more exposed to mutation or rearrangement than chromosomal genes. We therefore primarily tested whether the FOi isolates resulted from the inactivation of the plasmid-borne ATP4 gene. FOi isolates were crossed with a Δatp4 strain containing a wild-type mitochondrial genome (SDC17–4c, see Fig. 1B): when the FOi mutation is located in the plasmid-borne ATP4 gene, the progeny will be entirely deficient for respiratory growth whereas other mutations, either nuclear or mitochondrial, will result in respiratory competent progeny. Out of 28 FOi isolates, only 7 produced respiratory competent cells when crossed with SDC17–4c. Note that the absence of complementation by SDC17–4c may alternatively be the consequence of an FOi mutation being nuclear dominant affecting a gene other than ATP4. However, although this type of rescuing event could theoretically occur, it must be very rare as the screen is designed to isolate loss-of-function mutations, which are expected to be mostly recessive if located in nuclear DNA. We therefore decided not to analyze further the FOi isolates that were not rescued by the Δatp4 strain.
The seven FOi isolates that proved to be competent for ATP4 expression (ATP4+) were further analyzed by crossing with a wild-type strain lacking mtDNA (KL14–4a/60). This cross should restore respiratory competence in any nuclear recessive FOi mutant but fail to rescue any mutation in the mtDNA. This genetic test indicated that two of the seven FOi ATP4+ isolates (SDC25/1 and SDC25/5) were nuclear recessive mutants and that the five others (SDC25/2, SDC25/4, SDC25/6, SDC25/14, and SDC25/26) were mitochondrial mutants.
Nuclear FOi Mutants
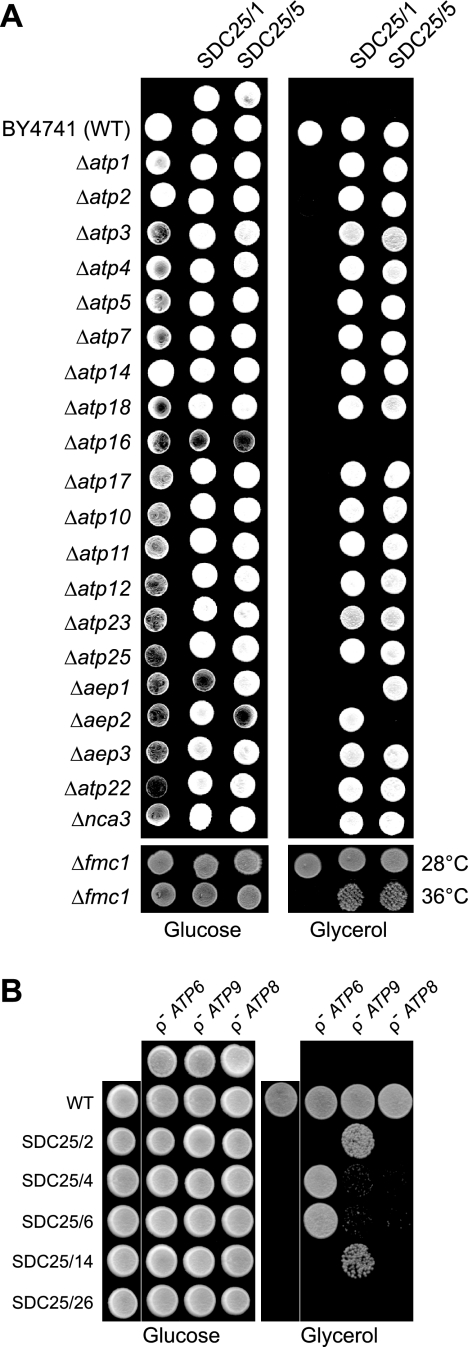
To determine whether SDC25/1 and SDC25/5 carried mutations in a nuclear gene already known to be necessary for the expression of a functional FO, we tested these mutants for rescue in crosses with defined nuclear ATP synthase mutants. The tester strains were deficient in a gene encoding either a structural subunit (ATP4, ATP5, ATP7, ATP14, ATP18, ATP17) or an accessory factor (ATP10, ATP23, AEP1, AEP2, AEP3, ATP22, ATP25, NCA3) of the FO. We also tested mutants lacking α-F1 (ATP1) or β-F1 (ATP2) or assembly factors of these subunits (ATP11, ATP12, FMC1), since α-F1 and/or β-F1 deficient mutants exhibit a secondary defect in FO with notably a failure to accumulate subunit a (38–40). When a FOi isolate carries a mutation in one of these tester genes, the corresponding homozygous diploid strain will be unable to grow on glycerol, whereas all the other crosses will produce respiratory competent progenies. The only testers that failed to complement SDC25/1 and SDC25/5 were those deficient in AEP1 or AEP2, respectively (Fig. 2A). As expected these mutants were not complemented also by crossing with a Δatp16 strain since they lack the ATP16 gene encoding subunit δ. AEP1 and AEP2 encode mitochondrial proteins required for the synthesis of subunit c (41–46). Upon reintroduction of AEP1 into SDC25/1 and of AEP2 into SDC25/5, complementation was restored in crosses with Δaep1 and Δaep2 testers (not shown), confirming that AEP1 and AEP2 were actually inactivated in SDC25/1 and SDC25/5 respectively. DNA sequencing revealed a single nucleotide deletion in AEP1, -A at position 547 of the coding sequence, which leads to a truncated protein of 201 residues (the wild-type Aep1p protein being 518 residues long). The AEP2 gene in SDC25/5 carries a single nucleotide substitution (A781T) introducing a premature stop codon, which results in a truncated protein of 260 residues (the wild-type Aep2p protein being 580 residues long). Consistent with these genetic data, pulse labeling of mitochondrial translation products revealed that SDC25/1 and SDC25/5 failed to synthesize subunit c, while all the other mitochondrially encoded proteins were properly synthesized (not shown).
FIGURE 2.
Test crosses for the mapping of the FOi mutants. Panel A, the nuclear FOi mutants SDC25/1 and SDC25/5 were crossed with a series of yeast strains each carrying a null mutation in one of known nuclear genes (indicated on the left) required for ATP synthase expression. The various strains were crossed drop on drop on rich glucose medium (YPGA) for 2 days at 28 °C, and then replica plated on glycerol medium (N3). The N3 plate was incubated at 28 or 36 °C, as indicated, for 4 days and photographed. As controls the non-mated strains were spotted on the YPGA plate. Panel B, the mitochondrial FOi mutants SDC25/2, SDC25/4, SDC25/6, SDC25/14 and SDC25/26 were crossed with synthetic ρ− testers carrying only the ATP6 (ρ− ATP6, strain SDC30/2a), the ATP8 (ρ− ATP8, strain FG8) or the ATP9 (ρ− ATP9, strain FG9) gene in their mitochondria. The various strains were crossed drop on drop on YPGA for 2 days at 28 °C, with as controls the non-mated strains. A N3 replicate of the YPGA plate was incubated for 4 days at 28 °C and then photographed.
Mitochondrial FOi Mutants
The five FOi mutations that were localized to mtDNA were expected to be located in one of the three mitochondrial ATP synthase genes encoding subunit a (ATP6 gene), subunit c (ATP9 gene), and subunit 8 (ATP8 gene). We therefore tried to map these mutations by crossing them with three synthetic ρ− strains each carrying only one of these genes in their mitochondria (ρ−ATP9/strain FG9, ρ−ATP6/strain SDC30/2a, and ρ−ATP8/strain FG8). The rationale for these crosses is as follows (see Figs. 1B and 2B): when the FOi mutation is, for example, in the ATP9 gene, it can be corrected by mtDNA recombination in crosses with FG9 but not in crosses with FG8 or SDC30/2a. The presence/absence of wild-type mtDNA recombinants can be determined by testing for the presence/absence of glycerol-positive (Gly+) clones in the cross progenies (Fig. 2B). On this basis, two of the five FOi mutations of mitochondrial origin (SDC25/2 and SDC25/14) were mapped to the ATP9 locus (see below for a detailed description of these mutants). Two others (SDC25/4 and SDC25/6) mapped to the ATP6 locus and consisted in small deletions within the 3′-UTR of ATP6 leading to a strong decrease in the synthesis of subunits a and 8 (supplemental Fig. S1). The last mutant (SDC25/26) was not rescued in any of the three crosses (Fig. 2B), and this mutant later proved to be lacking both the ATP8 and ATP6 genes due to a deletion in the transcription unit containing these two genes (supplemental Fig. S1).
The Subunit c Mutants SDC25/2 and SDC25/14
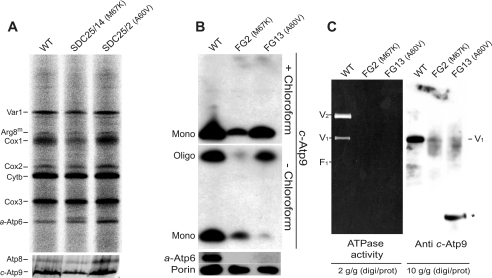
The ATP9 genes in SDC25/2 and SDC25/14 were sequenced: both carried a single nucleotide substitution leading to an amino acid substitution in subunit c, A60V (in SDC25/2) and M67K (in SDC25/14). The two mutated residues are located in the second transmembrane helix of subunit c (Fig. 3, A and B) and both are rather well evolutionary conserved (Fig. 3B). These subunit c mutations had a moderate if any impact on subunit c synthesis (Fig. 4A). We tested their consequences on the ATP synthase after transferring the mtDNA from SDC25/2 and SDC25/14 into the wild-type nuclear genetic background of the MR6 strain (the original isolates lack the ATP16 gene). The resulting cytoductants were named FG13 and FG2, respectively. The mtDNA of the SDC17–31b strain, from which SDC25/2 and SDC25/14 were isolated, was similarly cytoducted into MR6 to provide a wild-type control (FG4) for FG2 and FG13. In view of previous findings showing that mutations of the ATP synthase often destabilize the mitochondrial genome in yeast (20), we first determined the influence of the two subunit c mutations on mtDNA stability by scoring the percentage of ρ−/ρ° cells in glucose cultures of the different strains. The FG2 and FG13 cultures contained 60 and 20% of ρ−/ρ° cells, respectively, versus less than 5% for the corresponding wild-type control (FG4). The steady-state accumulation of subunit c in these strains was then assessed by Western blotting of mitochondrial protein extracts treated with chloroform (conditions that completely dissociate the c-ring; Fig. 4B). In FG13, subunit c accumulation was 70% of wild-type levels and in FG2 it was only 40%. The decreases in subunit c content can be in part attributed to the increased propensities of FG13 and FG2 to produce ρ−/ρ0 cells (20 and 60%, respectively, see above). When the mitochondrial protein extracts were not treated with chloroform, a substantial proportion of subunit c both in FG13 and in the wild-type was detected as assembled oligomers, whereas in FG2 only trace amounts of the c-ring were detected (Fig. 4B). This indicated that the M67K mutation in subunit c severely compromised the assembly/stability of the c-ring while the A60V mutation had no significant impact on the ability of subunit c to form stable oligomers.
FIGURE 3.
Topological location and evolutionary conservation of the Ala-60 and Met-67 residues of yeast subunit c. Panel A, three-dimensional structure of yeast subunit c monomer according to (48); the view is perpendicular to the membrane plane. The positions of the c-E59, c-A60, and c-M67 residues are indicated by arrowheads. H1 and H2 helices of subunit c are colored in bright and dark gray, respectively. The gray bars mark the membrane borders. IMS, intermembrane space. Panel B, alignments of subunits c from various species around helix H2: S. cerevisiae (NP_009319.1), Candida glabrata (NP_818784.1), Schizosaccharomyces pombe (NP_039507.1), Neurospora crassa (0808299A), Arabidopsis thaliana (NP_651852.1), Nicotiana tabacum (NC_006581.1), Mus musculus (NP_778180.1), Xenopus laevis (NP_001080083.1), Drosophila melanogaster (NP_651852.1), and Homo sapiens (NP_001002031.1). The Ala-60 and Met-67 residues of yeast subunit c are replaced by Val and Lys in mutants SDC25/2 and SDC25/14, respectively.
FIGURE 4.
Influence of the c-A60V and c-M67K mutations on mitochondrial protein synthesis and ATP synthase assembly. Panel A, pulse labeling of mtDNA-encoded proteins. Freshly grown cells of the mutants SDC25/14 (c-M67K) and SDC25/2 (c-A60V), and corresponding parental strain SDC17–31b, were incubated for 20 min in the presence of a mixture of [35S]methionine + [35S]cysteine and cycloheximide to inhibit cytosolic protein synthesis. Total cellular protein extracts were prepared from the radiolabeled cells and run in SDS-PAGE gels containing either 12% acrylamide-4 m urea (for a good separation of Cox3p and subunit a) or 17% acrylamide (for good resolution of Atp8 and subunit c). Each lane was loaded with 30000 disintegrations/min, which in this experiment corresponded to about the same numbers of cells for each analyzed strain. The gel was dried and analyzed with a PhosphorImager. The mitochondrial translation products corresponding to the various radioactive bands are indicated on the left. Panels B and C, effects of the c-M67K and c-A60V mutations on ATP synthase assembly. Mitochondria were extracted from the mutants FG2 (c-M67K) and FG13 (c-A60V) and the corresponding wild-type strain (FG4) grown in a synthetic (CSM) galactose medium lacking arginine. Panel B, 15 μg of mitochondrial proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with antibodies against the indicated proteins. Prior to loading on the gel, the samples were preincubated in presence or absence of chloroform as indicated. In absence of chloroform, some subunit c proteins migrate as c-ring oligomers whereas in the presence of chloroform all c-rings dissociate into monomeric subunit c. Panel C, the mitochondria were treated with 2 g or 10 g of digitonin (digi) per g of protein, as indicated, and then centrifuged to remove insoluble materials. The mitochondrial complexes were separated by BN-PAGE and probed in-gel for ATPase activity (left) or transferred to polyvinylidene difluoride membranes and probed with antibodies against subunit c (right). V1 and V2 correspond to monomeric and dimeric ATP synthase, respectively.
Although the subunit a was synthesized at normal rates (Fig. 4A), it fails to accumulate both in FG2 and in FG13 (Fig. 4B). This suggests that both subunit c mutations prevent the incorporation of subunit a into the ATP synthase such that unassembled subunit a is subsequently eliminated from the cells. Consistent with this, BN-PAGE analyses of mitochondrial extracts revealed the absence in FG2 and FG13 of fully assembled F1FO complexes (Fig. 4C). Western blotting of the BN-PAGE gels revealed the presence in FG13 of a subunit c-immunoreactive complex that had the size of the c-ring (Fig. 4C). This further confirmed that the c-A60V mutation does not prevent the assembly of the c-ring but may impair the interaction of the c-ring with subunit a.
DISCUSSION
We report a genetic screen designed to randomly isolate mutations that impair the FO component of the ATP synthase. A first set of seven mutants was analyzed and all proved as intended to be severely deficient in the FO. Two mutants fail to synthesize subunit c due to alterations in AEP1 or AEP2, two nuclear genes encoding mitochondrial proteins required for expression of subunit c (41–46). The five other mutations are in the mtDNA. Of these, three impair the synthesis of both subunits a and 8 due to restricted deletions within the transcriptional unit encoding these two proteins; the last two involve single point mutations leading to an amino acid substitution in subunit c, Ala-60 by Val and Met-67 by Lys.
The c-M67K mutation severely compromises the assembly/stability of the c-ring, which is not very surprising because it introduces a charged residue in the membrane (in the second transmembrane segment of subunit c, see Fig. 3A). The mutated methionine is particularly well conserved, in 95% of known subunits c from various origins, and in those few species where it is not conserved only non-polar residues (leucine or valine) are found at the corresponding position (Fig. 3B). Not surprisingly, despite a substantial synthesis, the subunit a fails to accumulate in the c-M67K mutant, which is in line with previous work showing that the subunit a is rapidly degraded in yeast cells when it cannot properly associate with its partner subunits (2, 18).
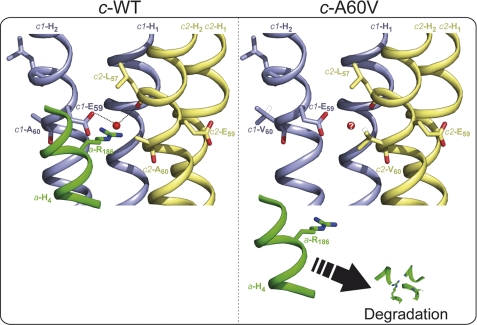
The second subunit c mutation, c-A60V, is particularly interesting. Unlike the c-M67K mutation, it has no visible impact on the assembly/stability of the c-ring. However, it has dramatic consequences on subunit a that despite a normal synthesis is no longer detectable at steady state. It can be inferred that the c-A60V mutation must impair the interaction of the c-ring with subunit a, such that unassembled subunit a is subsequently eliminated from the cells. The mutated c-A60 residue is located besides the c-E59 residue that is essential for the proton translocating activity of the FO (Fig. 3B). This glutamate interacts with an arginine residue of subunit a (a-R186), which is hypothesized to lower the pKa of the glutamate thereby enabling it to release a proton (47). According to a recently solved structure of the yeast c-ring (48), the c-A60 residue is at the external surface of the c-ring and its substitution by valine is predicted not to create significant clashes with neighboring residues of subunit c, which is consistent with the ability of the mutated subunit c to form stable oligomers.
We hypothesize that the c-A60V mutation disrupts the interaction between a-R186 and c-E59 and that the loss of this interaction is responsible for the defect in subunit a. Indeed, the c-A60 and c-E59 residues of two adjacent subunits c are in close proximity (Fig. 5). It is thus possible that the c-A60V mutation is responsible for a steric hindrance preventing entrance of a-R186 into the proton binding site. Alternatively the mutation may destabilize the c-E59 residue. Indeed, there is strong evidence that its stabilization requires a water molecule in the yeast ATP synthase (48), like in the H+ ATP synthase of Bacillus pseudofirmus OF4 (49). Subunit c residues surrounding the glutamate are involved in the binding of the water molecule (49). Thus if, as believed (48), a water molecule is present also in the H+ binding site of the yeast c-ring, its correct positioning or even its access to this site might be compromised by the c-A60V mutation. As a consequence the c-E59 residue may not reach the position that allows it to properly interact with a-R186.
FIGURE 5.
The loss of the interaction between the essential c-E59 and a-R186 residues is hypothesized to be responsible for the degradation of subunit a in the yeast c-A60V mutant. The figure on the left shows a ribbon diagram built with PyMOL of two adjacent c subunits (c1 in blue and c2 in yellow) around the proton binding site in wild-type yeast ATP synthase, according to the recently solved structure of the yeast c-ring/F1 subcomplex (48). The two transmembrane segments of subunit c are labeled H1 and H2. The proton translocating activity of the ATP synthase depends on the interaction of the glutamate residue 59 of subunit c (E59) with the arginine residue 186 of subunit a (a-R186). The latter residue belongs to the fourth transmembrane segment of subunit a (a-H4, represented in green color). It is believed (48) that proper interaction of c-E59 and a-R186 depends on a water molecule (represented by a red sphere), like in the H+ and Na+ ATP synthases of B. pseudofirmus OF4 (49) and I. tartaricus (54), respectively. In one of the FO inactive mutants isolated in this study, the alanine 60 of yeast subunit c is replaced by valine (right panel). The valine substituting c-A60 is shown in two rotameric conformations (solid and dashed line). The c-A60V mutation does not compromise the ability of subunit c to form stable oligomers but leads to elimination from the cells of the subunit a, presumably because of disruption of the interaction between c-E59 and a-R186 (see text for details). The question mark indicates that the water molecule is perhaps prevented to enter the proton binding site.
It might be that the ionic interaction between c-E59 and a-R186 is a key determinant to maintain subunit a associated with the c-ring, even though this interaction is constantly disrupted and reformed during rotation of the c-ring. If permanently interrupted, subunit a may become loosely attached to the complex thereby increasing its susceptibility to proteolytic degradation. Another possibility is that subunit a cannot fold properly if a-R186 cannot interact with c-E59. This is an interesting hypothesis according to which the folding of subunit a would be confined in front of the c-ring, possibly as a means to minimize non-productive interactions of subunit a during its assembly.
The strong impact of the c-A60V mutation on subunit a is somewhat surprising. Indeed, much more radical mutations at the interface between the c-ring and subunit a, like the a-L183R mutation located one helix turn from a-R186 (50), were found to have no effect on the assembly/stability of the ATP synthase. Although the a-L183R mutation severely compromises the functioning of the ATP synthase proton channel, the assembly/stability of subunit a is unaffected (50). Altogether these findings suggest that the contact zone between subunit a and the c-ring only involves critical transient interactions confined to the region of the FO where protons are exchanged between the two subunits.
The results show that our genetic screen is perfectly appropriate for the selection of mutations specifically affecting the FO component of the ATP synthase. Both nuclear and mitochondrial mutants were obtained, and the seven analyzed isolates were all different and each showed a very severe F0 deficiency. We have now started a more extensive hunt for FO inactive mutants, with two main goals: (i) the identification of amino-acids of subunits a and c that are critical for a proper operation of the proton channel of the ATP synthase; (ii) a better comprehension of the assembly of FO. Regarding the second goal, although several factors involved in the assembly of the FO are already known (18), it is likely that others remain to be discovered. This is evident when one considers the extraordinary complexity of the assembly pathway of complex IV with at least 30 different assembly factors (51) whereas only a dozen of proteins are known to be implicated in the biogenesis of the ATP synthase, an enzyme whose structure is certainly not lesser sophisticate than that of complex IV (2, 18). The strong instability of mtDNA in mutants with a defective FO (with up to 90–95% ρ−/ρ0 cells, see (52) for an example) may have hampered the identification of factors required for FO expression. Our genetic screen allows one to circumvent that problem because ρ−/ρ0 cells are strictly eliminated.
Our screen may also be exploited to better define the cis-elements that control expression of the mitochondrial ATPase genes. In this respect, recent work from Tzagoloff's laboratory has revealed a complex interplay between assembly and expression of these genes aiming to balance the production of different assembly intermediates, possibly to avoid FO-mediated protons leaks across the inner mitochondrial membrane (39, 53). Our screen may lead to the isolation of mutants defective in these mechanisms, like those showing a diminished synthesis of subunits a and 8 (SDC25/4 and SDC25/6) due to restricted deletions in the 3′-UTR of the transcripts encoding these proteins.
Supplementary Material
Acknowledgments
We thank J. Velours for the generous gift of antibodies against yeast subunit a, and M-F. Giraud and A. Dautant for helpful discussions.
This work was supported by grants from the Conseil Régional d'Aquitaine and the Agence Nationale de la Recherche (ANR).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- mtDNA
- mitochondrial DNA.
REFERENCES
- 1. Boyer P. D. (1997) Annu. Rev. Biochem. 66, 717–749 [DOI] [PubMed] [Google Scholar]
- 2. Ackerman S. H., Tzagoloff A. (2005) Prog. Nucleic Acids Res. Mol. Biol. 80, 95–133 [DOI] [PubMed] [Google Scholar]
- 3. Senior A. E., Nadanaciva S., Weber J. (2002) Biochim. Biophys. Acta 1553, 188–211 [DOI] [PubMed] [Google Scholar]
- 4. Bianchet M. A., Hullihen J., Pedersen P. L., Amzel L. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11065–11070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 6. Kabaleeswaran V., Puri N., Walker J. E., Leslie A. G., Mueller D. M. (2006) EMBO J. 25, 5433–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 8. Stock D., Leslie A. G., Walker J. E. (1999) Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 9. Stock D., Gibbons C., Arechaga I., Leslie A. G., Walker J. E. (2000) Curr. Opin. Struct. Biol. 10, 672–679 [DOI] [PubMed] [Google Scholar]
- 10. Fillingame R. H., Dmitriev O. Y. (2002) Biochim. Biophys. Acta 1565, 232–245 [DOI] [PubMed] [Google Scholar]
- 11. Tsunoda S. P., Aggeler R., Yoshida M., Capaldi R. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 898–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Velours J., Arselin G. (2000) J. Bioenerg. Biomembr. 32, 383–390 [DOI] [PubMed] [Google Scholar]
- 13. Devenish R. J., Prescott M., Rodgers A. J. (2008) Int. Rev. Cell Mol. Biol. 267, 1–58 [DOI] [PubMed] [Google Scholar]
- 14. Arnold I., Pfeiffer K., Neupert W., Stuart R. A., Schägger H. (1998) EMBO J. 17, 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Allen R. D. (1995) Protoplasma 189, 1–8 [Google Scholar]
- 16. Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brèthes D., di Rago J. P., Velours J. (2002) EMBO J. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong S., Pedersen P. L. (2002) Arch. Biochem. Biophys. 405, 38–43 [DOI] [PubMed] [Google Scholar]
- 18. Rak M., Zeng X., Brière J. J., Tzagoloff A. (2009) Biochim. Biophys. Acta 1793, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonnefoy N., Fox T. D. (2001) Methods Cell Biol. 65, 381–396 [DOI] [PubMed] [Google Scholar]
- 20. Contamine V., Picard M. (2000) Microbiol. Mol. Biol. Rev. 64, 281–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lefebvre-Legendre L., Balguerie A., Duvezin-Caubet S., Giraud M. F., Slonimski P. P., Di Rago J. P. (2003) Mol. Microbiol. 47, 1329–1339 [DOI] [PubMed] [Google Scholar]
- 22. Giraud M. F., Velours J. (1997) Eur. J. Biochem. 245, 813–818 [DOI] [PubMed] [Google Scholar]
- 23. Lai-Zhang J., Xiao Y., Mueller D. M. (1999) EMBO J. 18, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duvezin-Caubet S., Caron M., Giraud M. F., Velours J., di Rago J. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13235–13240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duvezin-Caubet S., Rak M., Lefebvre-Legendre L., Tetaud E., Bonnefoy N., di Rago J. P. (2006) J. Biol. Chem. 281, 16305–16313 [DOI] [PubMed] [Google Scholar]
- 26. Mitome N., Ono S., Sato H., Suzuki T., Sone N., Yoshida M. (2010) Biochem. J. 430, 171–177 [DOI] [PubMed] [Google Scholar]
- 27. Steele D. F., Butler C. A., Fox T. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Voth W. P., Jiang Y. W., Stillman D. J. (2003) Yeast 20, 985–993 [DOI] [PubMed] [Google Scholar]
- 29. Zeng X., Neupert W., Tzagoloff A. (2007) Mol. Biol. Cell 18, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Claisse M. L., Pajot P. F. (1974) Eur. J. Biochem. 49, 49–59 [DOI] [PubMed] [Google Scholar]
- 31. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Guérin B., Labbe P., Somlo M. (1979) Methods Enzymol. 55, 149–159 [DOI] [PubMed] [Google Scholar]
- 33. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 34. Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 35. Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 36. Arselin G., Vaillier J., Graves P. V., Velours J. (1996) J. Biol. Chem. 271, 20284–20290 [DOI] [PubMed] [Google Scholar]
- 37. Paul M. F., Velours J., Arselin de Chateaubodeau G., Aigle M., Guerin B. (1989) Eur. J. Biochem. 185, 163–171 [DOI] [PubMed] [Google Scholar]
- 38. Lefebvre-Legendre L., Vaillier J., Benabdelhak H., Velours J., Slonimski P. P., di Rago J. P. (2001) J. Biol. Chem. 276, 6789–6796 [DOI] [PubMed] [Google Scholar]
- 39. Rak M., Tzagoloff A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18509–18514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paul M. F. (1992) Disruption et Mutagenèse du Gène ATP4, Codant pour une Sous-unité du Secteur Fo de I'ATP Synthase Mitochondriale de Saccharomyces cerevisiae. Ph.D. thesis, Université de Bordeaux II, Bordeaux, France [Google Scholar]
- 41. Finnegan P. M., Ellis T. P., Nagley P., Lukins H. B. (1995) FEBS Lett. 368, 505–508 [DOI] [PubMed] [Google Scholar]
- 42. Finnegan P. M., Payne M. J., Keramidaris E., Lukins H. B. (1991) Curr. Genet 20, 53–61 [DOI] [PubMed] [Google Scholar]
- 43. Ackerman S. H., Gatti D. L., Gellefors P., Douglas M. G., Tzagoloff A. (1991) FEBS Lett. 278, 234–238 [DOI] [PubMed] [Google Scholar]
- 44. Payne M. J., Schweizer E., Lukins H. B. (1991) Curr. Genet 19, 343–351 [DOI] [PubMed] [Google Scholar]
- 45. Payne M. J., Finnegan P. M., Smooker P. M., Lukins H. B. (1993) Curr. Genet 24, 126–135 [DOI] [PubMed] [Google Scholar]
- 46. Ziaja K., Michaelis G., Lisowsky T. (1993) J. Mol. Biol. 229, 909–916 [DOI] [PubMed] [Google Scholar]
- 47. Fillingame R. H., Angevine C. M., Dmitriev O. Y. (2003) FEBS Lett. 555, 29–34 [DOI] [PubMed] [Google Scholar]
- 48. Dautant A., Velours J., Giraud M. F. (2010) J. Biol. Chem. 285, 29502–29510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Preiss L., Yildiz O., Hicks D. B., Krulwich T. A., Meier T. (2010) PLoS Biol. 8, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rak M., Tetaud E., Duvezin-Caubet S., Ezkurdia N., Bietenhader M., Rytka J., di Rago J. P. (2007) J. Biol. Chem. 282, 34039–34047 [DOI] [PubMed] [Google Scholar]
- 51. Fontanesi F., Soto I. C., Horn D., Barrientos A. (2006) Am. J. Physiol. Cell Physiol. 291, C1129–C1147 [DOI] [PubMed] [Google Scholar]
- 52. Ellis T. P., Helfenbein K. G., Tzagoloff A., Dieckmann C. L. (2004) J. Biol. Chem. 279, 15728–15733 [DOI] [PubMed] [Google Scholar]
- 53. Rak M., Gokova S., Tzagoloff A. (2011) EMBO J. 30, 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meier T., Krah A., Bond P. J., Pogoryelov D., Diederichs K., Faraldo-Gómez J. D. (2009) J. Mol. Biol. 391, 498–507 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.