Abstract
In kidney, FXYD proteins regulate Na,K-ATPase in a nephron segment-specific way. FXYD2 is the most abundant renal FXYD but is not expressed in most renal cell lines unless induced by hypertonicity. Expression by transfection of FXYD2a or FXYD2b splice variants in NRK-52E cells reduces the apparent Na+ affinity of the Na,K-ATPase and slows the cell proliferation rate. Based on RT-PCR, mRNAs for both splice variants were expressed in wild type NRK-52E cells as low abundance species. DNA sequencing of the PCR products revealed a base alteration from C to T in FXYD2b but not FXYD2a from both untreated and hypertonicity-treated NRK-52E cells. The 172C→T sequence change exposed a cryptic KKXX endoplasmic reticulum retrieval signal via a premature stop codon. The truncation affected trafficking of FXYD2b and its association with Na,K-ATPase and blocked its effect on enzyme kinetics and cell growth. The data may be explained by altered splicing or selective RNA editing of FXYD2b, a supplementary process that would ensure that it was inactive even if transcribed and translated, in these cells that normally express only FXYD2a. 172C→T mutation was also identified after mutagenesis of FXYD2b by error-prone PCR coupled with a selection for cell proliferation. Furthermore, the error-prone PCR alone introduced the mutation with high frequency, implying a structural peculiarity. The data confirm truncation of FXYD2b as a potential mechanism to regulate the amount of FXYD2 at the cell surface to control activity of Na,K-ATPase and cell growth.
Keywords: Membrane Trafficking; Na,K-ATPase; Plasma Membrane; Post-translational Modification; RNA Splicing; Accessory Proteins; Cell Growth; Mutagenesis; Regulation
Introduction
Na,K-ATPase is an enzyme in eukaryotic cells that provides non-equilibrium distribution of Na+ and K+ ions across the plasma membrane. Besides its obligatory α and β subunits, the complex contains a so-called “FXYD” subunit (1), a short single span membrane protein involved in regulation of the kinetic properties of Na,K-ATPase. In mammals, there are seven different FXYD genes that are expressed in a tissue- and cell-specific manner. Remarkably, association of the Na,K-ATPase with each of them leads to specific changes in kinetic parameters of the pump either at the level of K0.5 for the substrates or Vmax (for a review, see Ref. 2).
FXYD2, also called the γ subunit, was the first accessory FXYD protein discovered in connection with the Na,K-ATPase (3). It was identified as a proteolipid in membranes from dog kidney that was labeled with a photoaffinity derivative of the specific inhibitor of the pump, ouabain, along with the α and β subunits (4). Expression of the FXYD2 protein is mostly in kidney (5), although the expressed sequence tag database suggests some other tissues as potential sources for FXYD2 (for instance, pancreas and mammary glands). There are two splice variants of FXYD2, FXYD2a and FXYD2b, which are identical in structure except for the N-terminal segment comprising the first exon (6, 7). Based on results from different heterologous expression systems (Xenopus oocytes or mammalian cell transfectants), FXYD2 reduces the activity of the pump by increasing the K0.5 for Na+ as well as for K+ (8–11). We also demonstrated that both splice variants can be post-translationally modified when expressed in epithelial NRK-52E cells, and those modifications relieved the kinetic effects of splice variants on the activity of Na,K-ATPase (8, 9). Another group implicated FXYD2 in modulation of the Km for ATP when exogenously expressed in HeLa and HEK293 cells (12). However, characterization of renal Na,K-ATPase from FXYD2 knock-out mice (13) confirmed the role of FXYD2 only in the reduction of the affinity for Na+, thus leaving the effects on the apparent affinities for K+ and ATP as cell-specific.
Crystal structures of Na,K-ATPase from pig kidney and shark rectal glands unambiguously revealed the FXYD subunit as a genuine part of the complex (14, 15). Interaction occurs between TM9 of the α subunit and the transmembrane span of the FXYD subunit. In addition, there is a shared interface at the extracellular surface involving all three subunits of the complex, α, β, and FXYD (15). The N-terminal and C-terminal extramembranous segments of FXYD were not resolved. Characterization of FXYD2-FXYD4 chimeras implicated the transmembrane span in modulation of the affinity of the enzyme for Na+ (16), whereas experiments with FXYD4-FXYD5 chimeras implicated it in an increase in Vmax (17). On the other hand, the flanking regions of FXYD2 were implicated in modulation of Km for ATP (18), and the cytoplasmic domain of FXYD1 is the target for the kinase-mediated regulation of Na,K-ATPase (19).
Expression of FXYD2 splice variants in kidney is complex. For instance, FXYD2a is expressed in proximal convoluted tubules, whereas FXYD2b is found in distal convoluted tubules (6, 18). Both splice variants are found in macula densa (6), whereas in inner medulla, FXYD2a is expressed in intercalated cells of the initial portion of collecting duct and in principal cells in the middle and terminal portions of collecting duct (20). Both splice variants are simultaneously expressed in the same cells in medullary thick ascending limb. The functional significance of the localization of the spliced forms is not yet clear.
We showed previously that expression of FXYD2 in NRK-52E cells, either by transfection (FXYD2a or FXYD2b) or by induction with hypertonicity (FXYD2a), correlated with a reduction of Na,K-ATPase activity and the rate of cell growth (8, 21). Selective suppression of FXYD2a with siRNA during exposure to hypertonicity relieved both the inhibition of activity of Na,K-ATPase and the inhibition of cell proliferation (21), thus underlining the physiological significance of the FXYD2 subunit in controlling cell growth via modulation of Na,K-ATPase.
Although expression of the FXYD2b protein can be achieved in NRK-52E cells (proximal convoluted tubule origin) only by transfection (9), here we discovered that mRNA for both FXYD2a and FXYD2b is expressed in low abundance in control NRK-52E cells. Surprisingly, we obtained evidence of selective sequence modification of splice variants: there was a base point substitution, 172C→T, in the RT-PCR product for FXYD2b but not for FXYD2a and as a result generation of a premature stop codon in the FXYD2b protein. The “hot spot” in FXYD2b was further confirmed by random mutagenesis. The truncation exposed a targeting sequence with consequences for FXYD2b distribution and interaction with Na,K-ATPase. Thus, the sequence modification was a loss of function and may be utilized by cells as another level of control of Na,K-ATPase.
EXPERIMENTAL PROCEDURES
Antibodies and Cell Lines
Polyclonal antiserum K3 was used for detection of the α1 and β1 subunits of Na,K-ATPase on blots (22). Monoclonal antibody McK1 was used in immunostaining and Western blot to monitor the α1 subunit (23, 24). 9A7 (a kind gift from Dr. M. McEnery, Case Western Reserve University) is another monoclonal antibody that recognizes the α1 subunit of Na,K-ATPase (25). The RNGB polyclonal antibody raised against the N-terminal peptide of the rat FXYD2b splice variant (6) was used to identify the protein on blots as well as in immunostaining of cells. RCT-G1 antiserum was raised against the 13 amino acids at the shared C terminus of FXYD2a and FXYD2b (8). Calnexin was visualized with polyclonal rabbit anti-calnexin antibodies (Stressgen, Enzo Life Sciences).
The normal rat kidney epithelial cell line (NRK-52E), rat glioblastoma (C6), and rat skeletal muscle myocytes (L6) were purchased from American Type Culture Collection and grown according to the manufacturer's recommendations in Dulbecco's modified Eagle's medium with 10% FBS (300 mOsm). Hypertonic stress of NRK-52E cells was performed by addition of NaCl (200 mOsm) to the medium for 48 h as described previously (21, 26).
RT-PCR Analysis
Total RNA from cells was prepared with the RNeasy system (Qiagen). The cDNA was obtained using 1 μg of total RNA, oligo(dT) as the priming oligonucleotide, and SuperScript II RT (Invitrogen). Alternatively, cDNA was obtained with the SuperScript III Cell Direct cDNA synthesis system (Invitrogen). The PCR was performed in the presence of 3 mm MgCl2 with either Taq polymerase (Fisher) or Platinum Taq polymerase (Invitrogen). The following primers contained nucleotide sequences from the 5′- and 3′-ends of the coding sequence for rat FXYD2b plus EcoRI and BamHI sites for unidirectional cloning, respectively: forward primer, 5′-GCGAATTCCACCATGGACAGGTGGTAC-3′; reverse primer, 5′-CGCGGATCCTCACAGCTCATCTTCATTGAC-3′.
Four independent PCRs were performed. PCR products were separated by electrophoresis in 1.2% agarose gels and purified with a QIAquick gel extraction kit (Qiagen). The PCR products were sequenced at the Massachusetts General Hospital DNA core facility using a fluorescently labeled dideoxynucleotide chain termination method. Sequence information on the entire coding region was obtained.
Construction of cΔ7
Construction of cΔ7 was performed by PCR using the plasmid containing full-length cDNA for rat FXYD2b as a template. The following primers contained nucleotide sequences from FXYD2b (including the new stop codon in the reverse primer) plus EcoRI and BamHI sites for unidirectional cloning: forward, 5′-GCGAATTCCACCATGGACAGGTGGTAC-3′; reverse, 5′-CGCGGATCCCTACCTATGCTTCTTACTGCCCCC-3′.
As designed, the nucleotides for the final 7 amino acids are not present in the expression vector. The PCR was performed with the Fisher Taq polymerase in a RapidCycler instrument (Idaho Technologies). The gel-purified DNA fragment was cloned into a pIRES-neo vector (Clontech), and the mutation was confirmed by DNA sequencing. The plasmid was further transfected into NRK-52E cells, and stable transfectants were selected under pressure of G4182 antibiotic.
Membrane Preparations and Enzyme Purification
Crude membrane preparations were obtained from scraped cells by homogenization and differential centrifugation (6, 26). Partial purification of Na,K-ATPase from cell membranes (1.4 mg/ml) was with SDS extraction (0.56 mg/ml) and sedimentation on 7–30% sucrose gradients. Fractions were collected from the bottom and analyzed on SDS-Tricine gels. Typical specific activity of Na,K-ATPase was 50–150 μmol of Pi/mg of protein/h in preparations from NRK-52E cells (6). Isolation of membranes from rat kidney outer medulla was performed as described elsewhere (27). To obtain NRK cell lysates, a buffer containing 50 mm Tris-Cl, pH 8.0, 5 mm EDTA, 1% Nonidet P-40, and protein phosphatase inhibitor mixture I (Sigma) (1:100, v/v) was used. Cells were scraped and triturated, and insoluble material was removed by centrifugation at 3,000 × g for 10 min (Sorvall SS-34).
Subcellular Fractionation
NRK-52E cells mock-transfected or transfected with full-length FXYD2b or truncated form of FXYD2b were grown to confluence in 75-cm2 flasks in DMEM culture medium supplemented with l-glutamine, penicillin, and 10% fetal bovine serum. Cells were scraped and homogenized in TE buffer (10 mm Tris-HCl, pH 7.3, 1 mm EDTA) containing 250 mm sucrose. Nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min at 4 °C. The supernatant was centrifuged at 19,000 × g, and the pellet was resuspended in TE buffer containing 1.3 m sucrose. It was layered on a cushion consisting of 2 m sucrose and 1.6 m sucrose (both in TE buffer) and covered by another two layers of 1.1 m sucrose and 0.8 m sucrose (in TE buffer). Centrifugation was at 100,000 × g for 2 h (Beckman SW50.1) (adapted from Refs. 18 and 28). Fractions enriched in endoplasmic reticulum, Golgi, and plasma membranes were collected at the interfaces 1.3/1.6, 1.1/1.3, and 0.8/1.1 m sucrose, respectively; diluted with TE buffer; centrifuged at 100,000 × g for 30 min (Beckman Ti 70); and resuspended in TE buffer containing 250 mm sucrose.
Gel Electrophoresis
Membrane fractions were resolved either on SDS-Tricine or on 4–12% Bis-Tris-SDS gels with MES buffer (NuPAGE system, Invitrogen) (8, 26). Proteins were transferred to nitrocellulose and incubated with antibodies, and detection was with chemiluminescence with an LAS 4000 imager. For positive control samples, membrane preparations from kidney were used. Quantification was with ImageQuant TL image analysis software (GE Healthcare).
Enzymatic Assays
Na,K-ATPase activity was measured in crude membranes from either control or transfected cells as a function of Na+ concentration in medium containing 3 mm Tris-ATP, 4 mm MgCl2, and 30 mm histidine, pH 7.4 in the presence of 20 mm K+ (9). All the reactions were performed at 37 °C for 30 min with and without 3 mm ouabain, and ouabain-sensitive Pi release was measured colorimetrically by using the Fiske-Subbarow method. Data were analyzed by nonlinear regression using the Sigma Plot Graph System (Jandel Scientific). Na+ activation curves were fitted according to the Hill model for ligand binding. Alternatively, activity in crude membranes was measured using the so-called “yellow method,” which tolerates more protein and lipid. After quenching, the unreduced phosphomolybdate complex was extracted into an isobutanol organic phase where its absorbance was measured at 380 nm.
Cell Proliferation
NRK-52E cells, either wild type or transfected as described above, were seeded into 96-well flat bottomed plates (5 × 103 cells/well) in quadruplicate and allowed to adhere overnight. Cell proliferation over the following 2–6 days was assayed based on the cleavage of the tetrazolium salt WST-1 (Roche Diagnostics) by mitochondrial dehydrogenases in viable cells. Quantification was with a microtiter plate reader at 450 nm (21, 26).
Immunofluorescence
Immunocytochemistry was performed as described elsewhere in more detail (6). Briefly, cells were fixed with 2% periodate, lysine, paraformaldehyde solution for 30 min at room temperature and washed with PBS followed by 5-min incubation with 1% SDS in PBS, several washes with PBS, and blocking with 5% goat serum in PBS to prevent nonspecific binding of secondary antibodies. McK1 antibody was used for α1 detection, whereas RNGB antibodies were used to probe expression of FXYD2b. Calnexin antibody was used as a positive control for detection of endoplasmic reticulum. The secondary antibodies were either Cy3-conjugated goat anti-mouse IgG (1:300; Accurate Chemical) or fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (1:300; Jackson ImmunoResearch Laboratories). Slides were examined with a Nikon TE300 fluorescence microscope equipped with a Bio-Rad MRC 1024 scanning laser confocal system (version 3.2).
Random Mutagenesis
Random mutagenesis was performed with the GeneMorph PCR Mutagenesis kit (Stratagene) using full-length rat FXYD2b (verified by sequencing) as a template. The following primers included nucleotide sequences for rat FXYD2b, partial 5′ and 3′ sequences from the plasmid, and EcoRI and BamHI restriction sites for unidirectional cloning: RMF, 5′-CTGCGGCCGCGTCGACGGAATTCCACCATG-3′; RMRV, 5′-GCACACTGGCGGCCGTTACTAGTGGATCCTCA-3′.
The expected rate of introduced mutations by Mutazyme was in the range 0–3/200 bp. PCR was performed in a RapidCycler instrument (Idaho Technologies) in glass capillary tubes. The gel-purified DNA cassette was ligated into a pIRES vector as a pool and transfected into NRK-52E cells for selection and subcloning as described earlier. Selection of stable clones was with G418 (8, 9). In control experiments, the random mutagenesis procedure was performed with either Mutazyme or polymerases from different sources (Fisher, Eppendorf, and Applied Biosystems) in parallel and subcloning with Escherichia coli instead of mammalian cell transfection. DNA sequencing was performed at the Massachusetts General Hospital DNA Core facility with the following primers: forward primer: GamB, 5′-GCGAATTCCACCATGGACAGGTGGTAC-3′; reverse primer: pIRESRev, 5′-GCGCAGAAGTCATGCCCGC-3′. Sequence information on the entire coding region of FXYD2b mutants was obtained.
RESULTS
Base Change in mRNA
The FXYD2 subunit is abundantly expressed in several nephron segments, but NRK-52E and other renal epithelial cells lack its expression as a protein in basal conditions (8, 26, 29). Hypertonic stress (500 mOsm NaCl in culture medium) results in a selective induction of FXYD2a protein without any trace of the FXYD2b protein (21). However, mRNA for both FXYD2a and FXYD2b could be detected by PCR in both control and hypertonicity-treated cells (not shown). Moreover, with RT-PCR, FXYD2b mRNA was also detected in other cell lines, such as C6 (rat glioblastoma) and L6 (rat skeletal muscle myocytes), where the protein was not detected (not shown).
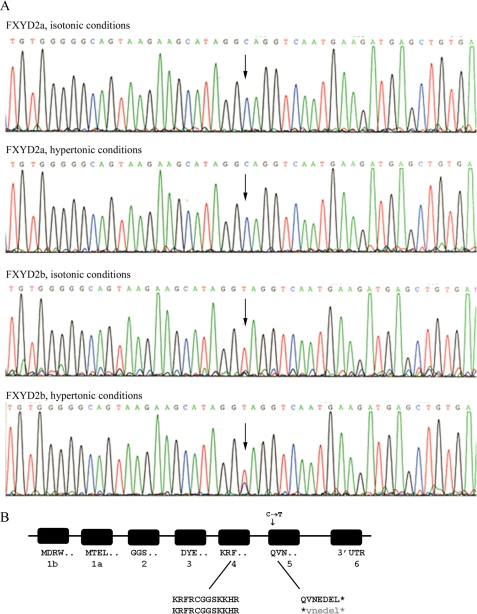
To verify their identity, the PCR products from NRK-52E cells were submitted to sequence analysis. Remarkably, although being the products of alternative splicing of the same gene, FXYD2a and FXYD2b showed a difference in the nucleotide sequence of the shared exon 5: cytidine in FXYD2a was replaced by thymidine in FXYD2b (Fig. 1A). This change in the DNA sequence resulted in a nonsense mutation and appearance of a premature stop codon and, as a result, should produce a truncated form of FXYD2b (Fig. 1B). The premature stop codon prevents translation of the last 7 amino acids. The same transition mutation was identified in FXYD2b from C6 rat glioma cells (not shown). The fact that FXYD2a and FXYD2b have different 5′-UTR sequences supports a potential role for different post-transcriptional processing either via splicing or RNA editing (discussed below).
FIGURE 1.
Sequence differences in mRNA for FXYD2a and FXYD2b in NRK-52E cells. A, chromatograms of cDNA sequence from the PCR products for FXYD2a and FXYD2b from control NRK-52E cells or cells treated with hypertonicity. The single base substitution C to T was observed in the FXYD2b cDNAs, although in hypertonicity-treated cells, both bases were detected, suggesting potential modulation of RNA editing of this site. Arrows indicate the position of the targeted site. The data are representative of three experiments from independent sources of mRNA. B, diagram of the genomic organization of the rat FXYD2 gene showing the location of the alternatively spliced exons and the modified nucleotide that generates a premature stop codon. Asterisks indicate where a stop codon is in the corresponding mRNA. Lower case letters represent sequence that is not translated after the stop codon.
When hypertonicity was used to induce FXYD2 expression, the base substitution was still absent from FXYD2a mRNA and present in FXYD2b mRNA. Heterogeneity seen at the 172C→T site in FXYD2b from the hypertonicity-treated cells (Fig. 1A, bottom panel) suggests that the base alteration was responsive to metabolic changes. Because no FXYD2b protein was observed on Western blots from control or hypertonicity-treated cells in contrast to induction of FXYD2a (21), the data suggest that truncated protein, if made, was below the level of detection in cells that normally express only FXYD2a and that have a low abundance of FXYD2b mRNA transcripts.
In repeated experiments, we detected T or a mixture of C and T in six of six PCR sequence experiments on mRNA isolated from NRK, C6, and L6 cells and in zero of four experiments with rat and mouse renal mRNA. To estimate the efficacy of the base alteration, we performed subcloning of NRK-52E cells and sequenced the PCR products. We detected T instead of C in three of 12 subclones.
Expression of Truncated Form of FXYD2b
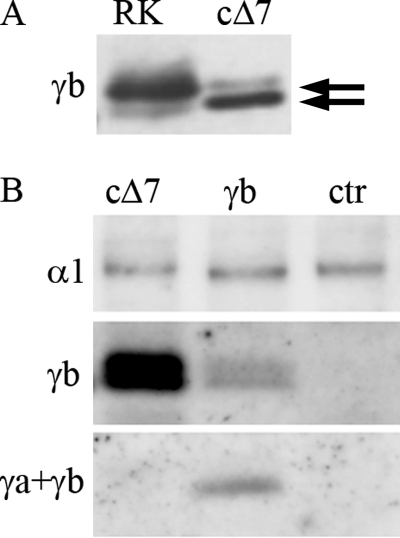
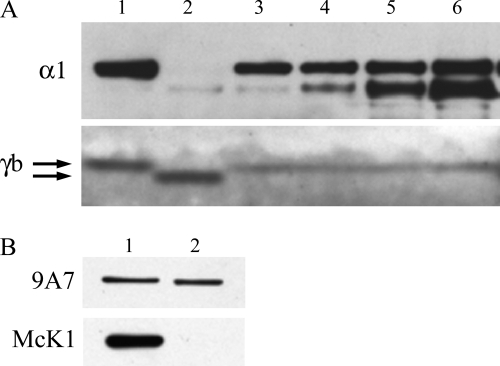
To characterize the truncated form of FXYD2b, we produced it independently by site-directed mutagenesis of C172. Stable transfectants (cΔ7 for C terminus without 7 amino acids) were generated in NRK-52E cells. Analysis of the clones with FXYD2b-specific antibody (N terminus) demonstrated a high level of expression of the truncated protein. As expected, the electrophoretic mobility of the mutant protein was faster than that of the wild type FXYD2b (Fig. 2A). Noticeably, there was also an extra band with a slightly slower electrophoretic mobility that coincided with that of the wild type FXYD2b (Fig. 2A). This suggests a potential processing or post-translational modification of the truncated form of FXYD2b when expressed in mammalian cells. The RCT-G1 antibody, which was raised against the last 13 amino acids in the C terminus of FXYD2, did not recognize the cΔ7 protein (Fig. 2B). Therefore, parallel application of RNGB and RCT-G1 antibodies can be useful as a method to discriminate between the full-length and truncated forms of FXYD2b regardless of the status of their post-translational modification.
FIGURE 2.
Characterization of cΔ7 mutant protein by Western blot. A, rat kidney microsomes (RK) were electrophoresed on a Novex 4–12% MES-SDS gel side by side with crude membranes from cΔ7-transfected cells. Staining with the RNGB antibody (specific for the N terminus of FXYD2b; γb) showed a shift in electrophoretic mobility of the mutant protein and an extra minor band, suggesting post-translational modification of cΔ7 protein. Arrows indicate the position of both bands in the doublet. B, a blot of crude membranes from cΔ7 cells, full-length FXYD2b transfectants (γb), and mock-transfected cells (ctr) was stained with the 9A7 monoclonal antibody (α1), antiserum RNGB (γb), and antiserum RCT-G1, which recognizes the shared C terminus of FXYD2a and FXYD2b (γa+γb). The absence of RCT-G1 staining of cΔ7 indicates that the last 7 amino acids are important for the epitope. The data are representative of three experiments.
To evaluate the effect of truncation on kinetic parameters of the pump, Na,K-ATPase activity was analyzed in crude membranes. No significant changes in the Vmax were seen between mock-transfected cells, NRK-52E cells transfected with the full-length FXYD2b, and cΔ7 cells. However, the apparent affinity for Na+ was significantly higher in the mock-transfected and cΔ7 cells compared with the NRK-52E cells expressing full-length FXYD2b (Table 1). Because the major consequence of association of Na,K-ATPase with the FXYD2 subunit is a reduction in the apparent affinity for Na+ (8, 9, 11, 12), the data imply that 172C→T was a loss-of-function mutation.
TABLE 1.
Effects of truncation at C terminus of FXYD2b on kinetic parameters of Na,K-ATPase in NRK-52E cells
Apparent affinity for Na+ was determined with crude membrane preparations from the mock-, cΔ7- and FXYD2b (γb)-transfected NRK-52E cells. Na+ activation curves were fitted according to a cooperative model for ligand binding. The table summarizes the data from four to five independent clones. Statistical significance compared with the full-length γb transfectants was identified by the Student's t test. The Na+ affinity data for mock and γb transfectants were essentially identical to published results (9) except that there the Na,K-ATPase was partially purified by SDS treatment, and here crude membrane preparations were used.
| NRK-52E cells, crude membranes | Na+ affinity | Vmax |
|---|---|---|
| mm | μmol Pi/mg/h | |
| Mock transfectants | 5.4 ± 0.1 (n = 5) | 1.2 ± 0.1 (n = 5) |
| cΔ7 transfectants | 5.6 ± 0.8a (n = 4) | 1.6 ± 0.4 (n = 4) |
| γb transfectants | 7.4 ± 0.2 (n = 5) | 1.3 ± 0.3 (n = 5) |
a p < 0.001.
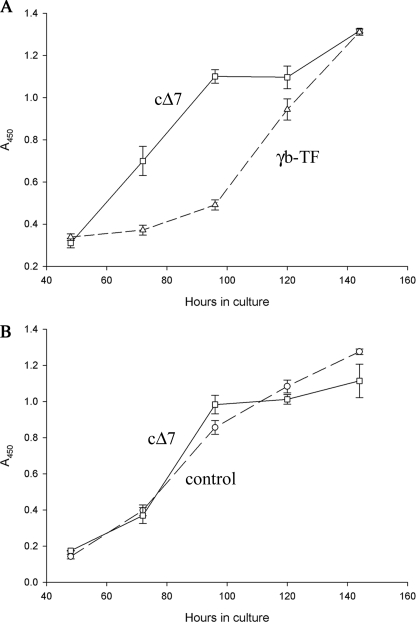
In previous work, we showed that reduction in the affinity for Na+ in cells expressing the FXYD2 subunit correlated with reduction in the rate of cell growth (21). Fig. 3, A and B, demonstrate a comparative analysis of the rate of cell proliferation in wild type NRK-52E cells (control) and cells transfected with either the full-length (γb-TF) or truncated form of FXYD2b (cΔ7). Although expression of the full-length FXYD2b indeed made cells grow at a slower rate similar to what we observed previously (21), expression of the truncated form of FXYD2b did not have any effect on proliferation of NRK-52E cells.
FIGURE 3.
Deletion of 7 amino acids from C terminus relieves inhibitory effect of FXYD2b on cell growth. Full-length FXYD2b- (γb-TF; dashed line in A), cΔ7- (solid line in A and B), and mock-transfected cells (control; dashed line in B) were plated at a density of 5 × 103 cells/well and grown in medium supplemented with 10% FBS for 5 days. Cell proliferation was assayed with WST-1 reagent. Cells transfected with the truncated form of FXYD2b proliferated significantly faster than the full-length FXYD2b transfectants (A) but grew at a rate similar to the control cells (B). The data are representative of multiple experiments (n = 3 for FXYD2b; n = 4 for mock and cΔ7). At the later time points, the cultures approached confluence. Each data point is the mean ± S.E.
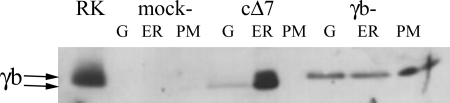
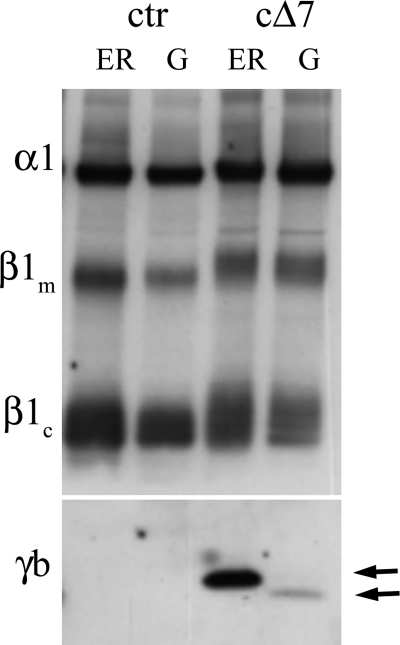
Fig. 4 shows the distribution of protein after subcellular fractionation of mock, FXYD2b, and cΔ7 transfectants. Although full-length FXYD2b was distributed between cellular compartments and had the highest level in plasma membranes, the truncated form was abundant in membranes enriched in endoplasmic reticulum with only traces found in membranes enriched in Golgi apparatus. No truncated FXYD2b was seen in plasma membranes. The data suggest that the truncated protein was retained intracellularly in contrast to the full-length FXYD2b. Retention of cΔ7 did not cause retention of either α1 or β1 subunits (Fig. 5).
FIGURE 4.
Truncated form of FXYD2b is associated with ER membranes. Postnuclear membranes from mock-, cΔ7-, and FXYD2b-transfected (γb) NRK-52E cells were fractionated by differential centrifugation and analyzed by Western blot with the RNGB antibody specific for the N terminus of FXYD2b (γb). Rat kidney microsomes (RK) served as a positive control. Although FXYD2b from cells transfected with normal FXYD2b was distributed in all three fractions, enriched in ER, Golgi (G), and plasma membrane (PM), most of the cΔ7 protein was associated with the ER membranes. The cΔ7 doublet apparently represents a mixture of core and post-translationally modified protein (indicated by arrows). The data are representative of four experiments.
FIGURE 5.
Retention of cΔ7 protein in ER does not affect distribution of α1 and β1 subunits. ER- and Golgi (G)-enriched membranes from control (ctr) and cΔ7-transfected cells were run on a gel. The top two-thirds of the blot was stained with antiserum K3, which recognizes both α1 and β1 subunits, and the bottom one-third was stained with RNGB antibody (γb). Arrows indicate the position of the truncated form of FXYD2b with and without post-translational modification. Although the truncated form of FXYD2b was retained in the ER membranes, there was no retention of α1 or β1 subunits, suggesting separate trafficking of αβ complex to the cell surface. β1m and β1c stand for β1 with mature glycosylation and β1 with core glycosylation, respectively. The data are representative of three experiments. A difference in β1 core and mature form mobilities is apparent in cΔ7 compared with untransfected cells but was also seen with full-length FXYD2b transfectants (not shown). The data may suggest more extensive branching of N-glycans in β1 subunit in FXYD2b-transfected NRK cells. The N-terminal part of FXYD2 that is exposed to the luminal compartment and that associates with the glycosylated domain of the β subunit is not affected by the truncation.
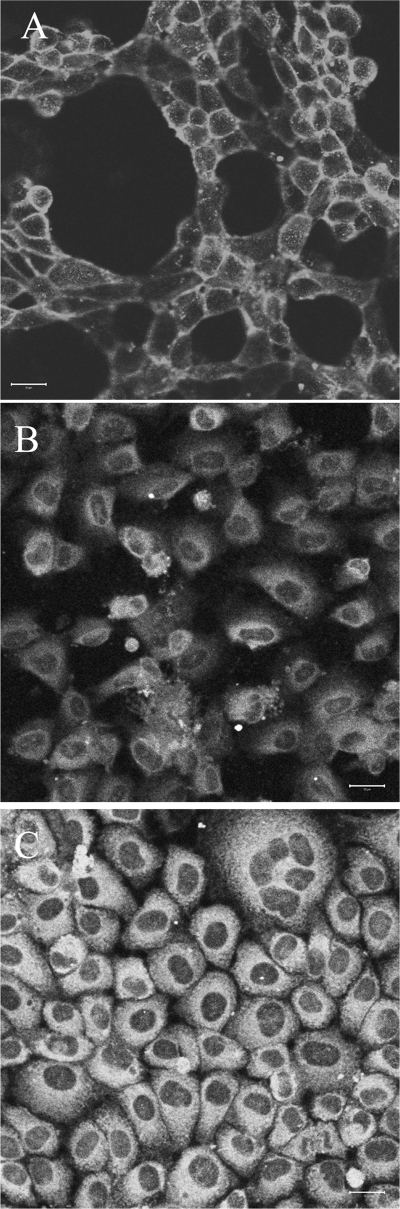
The 4 amino acids at the C terminus of the truncated form, -KKHR, represent a canonical ER retrieval motif, -KKXX, a sorting signal known to interact with COPI coat vesicles involved in retrograde transport of proteins from Golgi back to ER (30). Confocal microscopy (Fig. 6) indeed revealed the transfected truncated form of FXYD2b retained in endoplasmic reticulum: the staining pattern with the RNGB antibodies resembled the pattern obtained with antibodies against calnexin, a permanent resident of endoplasmic reticulum. α1 subunit was found mostly at the plasma membrane, indicating a complete spatial segregation of the truncated form of FXYD2b and the Na,K-ATPase.
FIGURE 6.
Spatial segregation of α1 and truncated FXYD2b in transfected cells. cΔ7 cells were fixed and stained for α1 (A), FXYD2b (B), and calnexin (C). Although α1 was localized at the plasma membrane, truncated FXYD2b was exclusively distributed intracellularly in a pattern similar to calnexin. The data suggest α1 and γ to be in different cellular compartments in cΔ7-transfected cells. Scale bar, 20 μm.
Truncation of FXYD2b in Kidney
As can be seen in Fig. 2, the preparation of rat kidney microsomes used as a positive control was seen as a doublet when stained with the antibody against the N terminus of FXYD2b. Moreover, the mobility of the minor, faster migrating band was similar to that of the major truncated form of FXYD2b. Fig. 7A shows fractionation of kidney microsomes during purification by SDS extraction. Microsomes were pretreated with a low concentration of SDS and separated from detergent-extracted proteins by sedimentation into a 4–30% sucrose density gradient. The fractions collected in the bottom third of the gradient were tested on Western blot with antibodies against α1 and the N terminus of FXYD2b. Intriguingly, in gradients from both medulla (Fig. 7A) and cortex (not shown), a faster migrating form of FXYD2b was detected in the heaviest fractions (either fraction 1 or 2 in different experiments), whereas the full-length FXYD2b was found in the other Na,K-ATPase-containing fractions. We speculate that the heavy fraction contains proteasomes, which have sedimentation coefficients of 26–30. (In this particular experiment, full-length FXYD2b and unmodified α1 appeared in fraction 1, but this was probably due to contamination of the fraction with pelleted undispersed tissue-derived material because fractions were collected from the bottom of the tube. The additional band recognized by McK1 antibody in lighter fractions may represent a proteolytic product of the α subunit but has not been characterized.) Although this short form of FXYD2b may represent a proteolytic product (KKHR constitutes a cluster of basic residues close to the C terminus (1)), truncation of FXYD2b due to modification of the mRNA is also a possible source. Another interesting detail is that the α1 subunit is not actually absent from fraction 2 in Fig. 7A; the short form of FXYD2b was associated with α1 subunit that was post-translationally modified near the N terminus. The α1 antibody used in Fig. 7A, McK1, binds near the N terminus but is blocked either by phosphorylation at the serine in the sequence DKKSKK (residues 15–20 of the mature protein) (24) or blocked by its proteolysis. Fig. 7B shows equal detection of α1 in fractions 1 and 2 with monoclonal antibody 9A7 (which recognizes the protein at a central site (31)) but lack of detection in fraction 2 with monoclonal antibody McK1. Whatever the cause of the modifications of both α1 and FXYD2b, the data suggest that a short form of FXYD2b can be observed in kidney and that this form may be complexed with modified α1, possibly for degradation.
FIGURE 7.
Evidence for physical separation of truncated and full-length FXYD2b in rat kidney. A, rat kidney microsomes from outer medulla were treated with a low concentration of SDS and fractionated on a sucrose density gradient. Fractions were collected from the bottom. The fractions were electrophoresed on an SDS-Tricine gel, and the blot was stained with McK1 (α1) and RNGB (FXYD2b; γb) antibodies. Arrows indicate a difference in electrophoretic mobility for FXYD2b between fraction 2 and the rest of the gradient. Notably, there was no stain for α1 in fraction 2. The extra band seen in other fractions may indicate proteolysis of α1 subunit. B, blots of fractions 1 and 2 from the gradient shown in A were probed with two antibodies against the α1 subunit: McK1, which binds near the N terminus and is sensitive to phosphorylation at Ser-18, and 9A7, which recognizes the protein at a different site. The data suggest that fraction 2 contains α1 subunit that is modified (clipped or phosphorylated) just at the end.
Random Mutagenesis
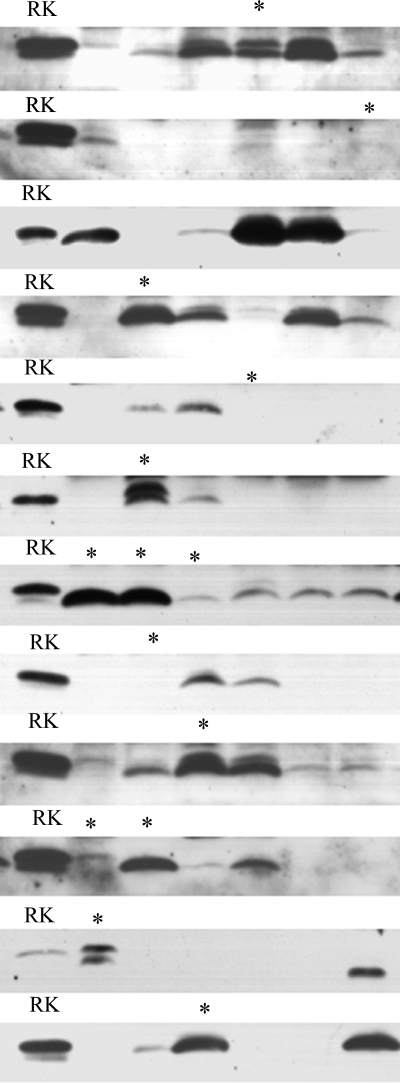
Our previous work showed that expression of FXYD2 is deleterious for cell growth in culture presumably because its ability to reduce Na+ affinity effectively reduces Na,K-ATPase activity (21). Thus, our hypothesis was that screening for the rate of cell growth would allow us to discriminate between deleterious, advantageous, and neutral mutations in cells stably transfected with randomly mutated FXYD2. Because 172C→T is a loss-of-function mutation, we predicted that transfectants expressing it (and other loss-of-function mutations) would develop into visible clones faster than cells expressing wild type FXYD2b. The transfection vector containing FXYD2b was first subjected to mutagenesis with the error-prone DNA polymerase Mutazyme and then stably transfected into NRK-52E cells. Seventy-two clones arising in 11–19 days were analyzed by Western blot. As shown in Fig. 8, based on their appearance in SDS gels, there was a fair representation of clones containing FXYD2b migrating like the truncated form.
FIGURE 8.
Random mutagenesis of FXYD2b revealed unexpectedly high frequency of 172C→T mutation. Clones of NRK-52E cells stably expressing randomly mutated FXYD2b were lysed and electrophoresed on SDS-Tricine gels. The blots were stained with the RNGB antibody. Note the variability of clone FXYD2b mobility in SDS gels. Selected clones (marked with asterisks) were subjected to DNA sequence analysis; all of them contained the 172C→T mutation. RK, rat kidney microsomes used as a positive control for the gel.
Fourteen clones, marked with asterisks in Fig. 8, were selected for sequence analysis, including apparently truncated clones, clones with doublets, and clones with little or no detected protein. The sequence of the plasmid insert was obtained with the GamB and pIRESRev primers (“Experimental Procedures”). All 14 clones showed a single base transition mutation, 172C→T. Notably, the premature stop codon was identified even in clones where no protein was detected in SDS gels. The latter suggests that there are differences in expression efficiency among stable transfectants, which is generally consistent with random insertion of the vector into active and inactive DNA regions.
Such a high representation of clones with the premature stop codon was surprising. We expected to see other deleterious mutations. It was also surprising in view of the fact that aminoglycoside antibiotics and G418 in particular are known to effectively suppress nonsense mutations in mammalian cells by promoting translational read-through (32). It is possible that the combination of nucleotides adjacent to the translation termination sequence here (ATAGG) is not favored for G148 to override the nonsense mutation.
To test whether C172 is a hot spot for mutation by this error-prone PCR enzyme, two control experiments were performed. First, the mutagenesis step was repeated with vector containing full-length, verified FXYD2b but using Taq polymerase (Fisher, Eppendorf, or Applied Biosystems enzymes were used in independent experiments), and the subcloning steps were performed in E. coli instead of mammalian cells. Thirteen of 13 resulting subclones had no change at C172. Second, the mutagenesis was repeated with Mutazyme, but the pooled PCR product was sequenced directly. We also took into account the fact that the amount of substrate DNA during the reaction affects the frequency of introduced mutations because a smaller amount of input DNA goes through more rounds of PCR. Serial dilutions (1:10) of template from 1 to 10,000 were used. Interestingly, the nonsense 172C→T mutation was identified in every pool, implying that the FXYD2b DNA sequence has a high susceptibility to mutation by Mutazyme in this particular spot. The formulation of Mutazyme that was used does have a bias that favors a C-to-T transition mutation, but the high conversion of a specific site implies something special about the structure of the DNA.
Conservation of FXYD2 Splice Forms and Retention Motif
If modification of FXYD2b mRNA to generate an ER retention signal is functionally significant, we would predict that it would be evolutionarily conserved. Supplemental Fig. 1 illustrates the sequences of the alternative N-terminal exons and the predicted retention signals in all three subclasses of mammals. It is notable first that there is high sequence variation among the FXYD2a exons, including in predicted length, whereas the FXYD2b exons are highly conserved. The retention signal spans a splice site; one base of the arginine codon and the following glutamine codon that can be mutated to a stop codon are in the acceptor exon. Again, the level of conservation is extremely high. All species preserve dibasic or multibasic motifs. Only one species, the domestic pig, has a sequence that would not produce a truncated form by RNA editing or shifted splicing.
DISCUSSION
FXYD2 Gene Product with Modified RNA Sequence
In this study, we identified a novel form of the FXYD2b regulatory subunit that is retained intracellularly when expressed. This is because it lacks the amino acids normally encoded by the final exon and exposes an ER retrieval motif. The fact that mRNA for this form was found in several established cell lines of different origin (NRK-52E, C6, and L6) generalizes the phenomenon. However, the substitution in the FXYD2b mRNA was not confirmed by BLAST search of human (87 hits), mouse (100 hits), or rat (13 hits) expressed sequence tags from which we infer that it is not a major component of FXYD2 in normal tissue. Functionally, expression of the novel form of FXYD2b abrogated both the inhibitory effect of FXYD2 on activity of Na,K-ATPase and its ability to slow cell growth. Hypothetically, the modification of the transcript could play a protective role, ensuring that the protein is inactive even if made in cells where it may be deleterious. It may be more likely that ER-retained FXYD2b has an undiscovered biological role. Because only FXYD2b but not FXYD2a was found in truncated form, the data suggest a selective post-transcriptional control of the FXYD splice forms and a potential indirect mechanism contributing to their differential regulation of Na,K-ATPase. Two possible mechanisms for producing the modified transcript are discussed below, either of which would result in indistinguishable RNA sequences.
Diversity of FXYDs
Alternative Splicing
Each member of the family of FXYD proteins differentially regulates the kinetic properties of the pump (2). The diversity of functional effects of FXYDs is raised by alternative splicing. Although alternative splicing of FXYD1 occurs in the 5′-UTR region (1), splice forms of FXYD3 (33) and FXYD5 (17) introduce changes within the coding sequence with consequences for regulation of Na,K-ATPase. Splice variants of FXYD2 are well established and have been quite thoroughly investigated (1, 7, 18, 34, 35).
Although the majority of gene splicing follows the GT/AG paradigm, it is known that splicing can occur at other nucleotides (36). Engagement of both canonical and non-canonical splice sites was observed for other members of the FXYD family. Two splice forms of FXYD3 were described in connection with differentiation of Caco-2 cells (33). The prominent short form was the correctly (GT/AG) spliced FXYD3, and this form was characteristic of differentiated cells. The minor long form represented an unusual alternative splicing (GT/CT). It contained an additional 78 nucleotides from an intron and was mostly identified in undifferentiated cells. Interestingly, the insertion of additional amino acids immediately after the transmembrane span almost completely abrogated the effects of FXYD3 on the kinetic properties of Na,K-ATPase, not unlike the effects seen here with truncation of FXYD2b. At least one splice variant has been identified for FXYD5 with a non-canonical GT/CC splice site (17). This variant eliminated the original stop codon and produced a longer transcript encoding 10 additional C-terminal residues. Functional effects of this splice variant have not yet been tested.
One potential mechanism for substituting T for C172 in FXYD2b may be aberrant splicing of FXYD2b by simultaneously shifting two nucleotides downstream from both the acceptor and donor splice sites (Fig. 9). This would keep the transcript unchanged except for the single base substitution encoding the nonsense mutation. Although shown here for the rat, similar shifts in splicing with generation of a premature stop codon would be feasible for mouse and human FXYD2b as well. Unlike the variant splice forms reported for FXYD3 and FXYD5, this one would be hard to prove because the change is subtle.
FIGURE 9.
Hypothetical scheme of potential aberrant splicing of FXYD2b resulting in change of mRNA sequence. Splice sites are marked with slashes. Sequences in italics indicate the intron. The sequences with translation indicated are the products of splicing.
An apparently significant observation is that the premature stop codon was observed only in the FXYD2b but not in FXYD2a mRNAs. However, alternative splicing is transcriptionally coupled. There are a number of reports suggesting interaction (direct or indirect) between transcription factors and splicing factors (RNA-associated proteins involved in formation of the spliceosome complex) (for a review, see Ref. 37). The structure of the promoter may influence recruitment of splicing factors to the nascent pre-mRNA, thus modulating the efficiency of splicing. Because FXYD2a and FXYD2b have different promoters and 5′-UTRs (34), it is conceivable that they may also possess a different splicing pattern at a site distal to the first exon. However, full-length FXYD2b is clearly made in some cells in vivo, so if modified splicing is the cause of the truncation, its manifestation is limited to particular circumstances.
RNA Editing
As an alternative to aberrant splicing, the single base substitution observed in the FXYD2b mRNA may be a result of RNA editing, a process that changes the structure of mRNA through enzymatic modification of nucleotides. This may entail insertions of nucleotides not encoded in template DNA as well as modification of C to uridine (U) or adenine (A) to inosine (I) with specific Zn2+-dependent deaminases. RNA editing may have multiple consequences, such as influence on translation, alternative splicing, RNA stability, and transport as well as a change in amino acid sequence. Although the effects look similar to mutation, the quantitative range for RNA editing is much wider, and there is good evidence that the process is regulated.
Recent genome-wide screening revealed several hundreds of human RNA A-to-I editing sites from seven tissues of a single individual, suggesting an extensive representation of this type of RNA modification in humans (38). A-to-G RNA editing was demonstrated for the α subunit of Na,K-ATPase from squid neurons: RNA editing of three different codons significantly modulated the turnover of the pump and Na+ release (39). In contrast, C-to-U editing has been more widely studied in plants and prokaryotes (for a review, see Ref. 40). There are four well characterized examples of C-to-U editing in vertebrates. Editing of apoB mRNA by Apobec-1 (apoB-editing cytidine deaminase), site-specific deamination of NF1 (neurofibromatosis 1) mRNA, and editing of NAT1 factor in hepatocytes all have a notable feature in common with FXYD2b: in all three cases, replacement of cytidine with uridine resulted in creation of premature stop codons and formation of truncated proteins. In the case of glycine receptor GlyRα3, C-to-U editing changed a proline to leucine and increased receptor affinity (41).
RNA editing is highly dependent on the secondary structure of mRNA (for a review, see Ref. 40). Cytidine deaminase is thought to act on imperfectly base-paired double-stranded mRNA. Computer modeling and mutagenesis of the apoB mRNA predict a highly conserved local stem loop secondary structure for the deaminase substrate. Secondary structure may be different after splicing as a consequence of the use of alternative exons. Although this is only a hypothesis requiring further investigation, the different 5′-UTR or exon 1 sequences of FXYD2a and FXYD2b may make them targets for selective control by RNA editing. RNA secondary structure alone should not account for the control of editing, however, because editing is presumably regulated via proteins. Thus, it is not surprising that edited forms do not predominate in mRNA from normal kidney samples.
Mislocalization of Truncated FXYD2b
Several lines of evidence indicate that the truncated form of FXYD2b was accumulated intracellularly. Based on results of subcellular fractionation from full-length FXYD2b and cΔ7 transfectants, there was a clear indication of different distribution between cellular compartments. No cΔ7 was seen in the plasma membrane fraction, whereas the full-length FXYD2b was identified in all three membrane-containing fractions from ER, Golgi, and plasma membranes. Therefore, the deletion of 7 amino acids from the C terminus of FXYD2b clearly affected the trafficking of the protein. Conversely, no obvious changes in the α1 subunit trafficking was observed between the cΔ7- and mock-transfected cells, suggesting no retention of α1 and β1 subunits by the truncated form of FXYD2b in intracellular compartments. Thus, the data confirmed the previous observations that association of the αβ complexes with the FXYD protein (here, FXYD2b) occurs at the plasma membrane but not earlier in ER membranes (42). Immunocytochemistry supported the results obtained by cellular fractionation. The entire staining for FXYD2b was in a pattern similar to calnexin, a resident of the ER. In these experiments, we also did not observe an accumulation of the α1 subunit anywhere but at the plasma membrane.
A hypothesis could be made that truncated FXYD2b does not pass through the quality control point in Golgi due to incorrect folding. However, in other experiments, deletion of 4 or 10 amino acids from the C terminus of FXYD2 apparently did not cause misfolding: the mutants were expressed in HeLa cells and showed effects on Na+ affinity similar to full-length FXYD2 (18). Protein traffic is governed by the molecular recognition of signals often encoded in protein structure. Generation of the premature stop codon in FXYD2b unmasked the short C-terminal -KKXX motif, which plays a crucial role for the localization of type I membrane proteins in the endoplasmic reticulum of eukaryotic cells. If they escape, those proteins are retrieved to the ER from Golgi via retrograde transport of COPI vesicles: the KKXX sequence directly binds to coatomer protein I via the dilysine motif (43). Therefore, retention of cΔ7 inside the cells and loss of association with the α1 subunit are not likely to have been dictated by the C-terminal deletion per se but rather exposure of the specific ER retrieval motif. Although it is true that dibasic retrieval motifs do not have to be within a few residues of the C terminus, it is likely that those are well exposed in the structure. In the case of intact FXYD2, we infer that the dibasic motif is normally not fully accessible to COPI.
Post-translational Modification
Post-translational modification is a common feature in the FXYD family, although the nature and functional consequences of modification vary. Phospholemman (FXYD1) is a proven target for regulation by multiple kinases (19, 44). Dysadherin (FXYD5) is a heavily O-glycosylated protein. The extent of its glycosylation may vary based on cell type and be different in normal and malignant tissues (26, 45–47). FXYD7 is uniquely expressed in brain and is post-translationally modified at Thr-3, Thr-5, and Thr-9 presumably by O-glycosylation that affects the stability of the FXYD7 protein and its routing to the plasma membrane (48). Both splice variants of FXYD2 can be post-translationally modified when stably expressed in NRK-52E cells, and the presence or absence of post-translational modification(s) (the nature of which is still unclear) correlated with modulation of the kinetic properties of Na,K-ATPase (8, 9). Post-translational modification was found in wild type but not G41R (aberrantly trafficked) mutant FXYD2b in HeLa cells (18). All together, the data suggest that cell/tissue-specific post-translational modification of FXYD proteins is a major feature of their regulation.
ER-retained cΔ7 possessed a post-translational modification: the protein migrated as a doublet in SDS gels where the electrophoretic mobility of the minor upper band corresponded to the electrophoretic mobility of full-length FXYD2b. Although the extent of modification (i.e. the distribution between the bands in the cΔ7 doublet) varied in different preparations, the general tendency was that the band with the slower electrophoretic mobility was more abundant in the ER-enriched fractions (Figs. 4 and 5). In contrast to the O-glycosylation of FXYD7, this modification did not allow FXYD2 to escape the retrograde transport from Golgi to ER. Previous analysis of dilysine-tagged marker proteins in mammalian cells and yeast showed that most of those proteins were modified in post-ER compartments (for a review, see Ref. 49) namely in cis- or even medial Golgi. Palmitoylation and O-linked glycosylation (addition of GalNAc molecules) were reported as the common modifications (43). Interestingly, endogenous ER proteins lack such modifications, and they may therefore be considered as special tags for escaped proteins. Based on electrophoretic mobility of the bands in the cΔ7 doublet, the molecular mass of post-translational modification could be about 800 Da and might accommodate up to four GalNAc molecules. Further investigation is required to follow the biosynthesis of cΔ7 and elucidate the nature of its post-translational modification.
Sequence Change Significance
In conclusion, our experimental evidence suggests that modification of the FXYD2b mRNA may represent an additional level of control for expression of FXYD2b in the right place and at the right time, thus increasing the variety of the multiple mechanisms for regulation of Na,K-ATPase. The dibasic motif is apparently not normally accessible to COPI but can be exposed by truncation. It then effectively keeps truncated FXYD2b in the ER while permitting α1β1 to traffic to the plasma membrane. The motif is highly conserved among mammals. It has recently been shown that the α1 subunit also has a dibasic motif (in this case close to the N terminus) (50). This motif only interacts with COPI when α1 fails to complex with β1 and appears to route free α1 to the proteasome for degradation.
We have observed FXYD2b mRNA with the premature stop codon, protein that comigrates with cΔ7, and cytoplasmic FXYD2b staining in mouse pancreas.3 Consequently, it appears that this is a physiologically relevant molecular modification with an in vivo role that may be more prominent in normal pancreas than in normal kidney. The specific mechanisms that control whether the full-length or truncated forms are produced remain to be investigated.
Supplementary Material
Acknowledgment
We thank Natalya K. Asinovski for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL036271, NS045083, and NS050696 and an American Recovery and Reinvestment Act supplement to Grant NS050696. This work was also supported by a grant from the Harvard Medical School 50th Anniversary Scholars in Medicine Margaret H. Walter and Daughters Fellowship in memory of Carl W. Walter, M.D., '32.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
E. Arystarkhova and K. J. Sweadner, unpublished observations.
- G418
- Geneticin
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ER
- endoplasmic reticulum
- BLAST
- Basic Local Alignment Search Tool.
REFERENCES
- 1. Sweadner K. J., Rael E. (2000) Genomics 68, 41–56 [DOI] [PubMed] [Google Scholar]
- 2. Geering K. (2006) Am. J. Physiol. Renal Physiol. 290, F241–F250 [DOI] [PubMed] [Google Scholar]
- 3. Collins J. H., Leszyk J. (1987) Biochemistry 26, 8665–8668 [DOI] [PubMed] [Google Scholar]
- 4. Forbush B., 3rd, Kaplan J. H., Hoffman J. F. (1978) Biochemistry 17, 3667–3676 [DOI] [PubMed] [Google Scholar]
- 5. Mercer R. W., Biemesderfer D., Bliss D. P., Jr., Collins J. H., Forbush B., 3rd (1993) J. Cell Biol. 121, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arystarkhova E., Wetzel R. K., Sweadner K. J. (2002) Am. J. Physiol. Renal Physiol. 282, F393–F407 [DOI] [PubMed] [Google Scholar]
- 7. Kuster B., Shainskaya A., Pu H. X., Goldshleger R., Blostein R., Mann M., Karlish S. J. (2000) J. Biol. Chem. 275, 18441–18446 [DOI] [PubMed] [Google Scholar]
- 8. Arystarkhova E., Wetzel R. K., Asinovski N. K., Sweadner K. J. (1999) J. Biol. Chem. 274, 33183–33185 [DOI] [PubMed] [Google Scholar]
- 9. Arystarkhova E., Donnet C., Asinovski N. K., Sweadner K. J. (2002) J. Biol. Chem. 277, 10162–10172 [DOI] [PubMed] [Google Scholar]
- 10. Zouzoulas A., Dunham P. B., Blostein R. (2005) J. Membr. Biol. 204, 49–56 [DOI] [PubMed] [Google Scholar]
- 11. Béguin P., Wang X., Firsov D., Puoti A., Claeys D., Horisberger J. D., Geering K. (1997) EMBO J. 16, 4250–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pu H. X., Cluzeaud F., Goldshleger R., Karlish S. J., Farman N., Blostein R. (2001) J. Biol. Chem. 276, 20370–20378 [DOI] [PubMed] [Google Scholar]
- 13. Jones D. H., Li T. Y., Arystarkhova E., Barr K. J., Wetzel R. K., Peng J., Markham K., Sweadner K. J., Fong G. H., Kidder G. M. (2005) J. Biol. Chem. 280, 19003–19011 [DOI] [PubMed] [Google Scholar]
- 14. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 15. Shinoda T., Ogawa H., Cornelius F., Toyoshima C. (2009) Nature 459, 446–450 [DOI] [PubMed] [Google Scholar]
- 16. Lindzen M., Aizman R., Lifshitz Y., Lubarski I., Karlish S. J., Garty H. (2003) J. Biol. Chem. 278, 18738–18743 [DOI] [PubMed] [Google Scholar]
- 17. Lubarski I., Karlish S. J., Garty H. (2007) Am. J. Physiol. Renal. Physiol. 293, F1818–F1826 [DOI] [PubMed] [Google Scholar]
- 18. Pu H. X., Scanzano R., Blostein R. (2002) J. Biol. Chem. 277, 20270–20276 [DOI] [PubMed] [Google Scholar]
- 19. Fuller W., Howie J., McLatchie L. M., Weber R. J., Hastie C. J., Burness K., Pavlovic D., Shattock M. J. (2009) Am. J. Physiol. Cell Physiol. 296, C1346–C1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pihakaski-Maunsbach K., Vorum H., Honoré B., Tokonabe S., Frøkiaer J., Garty H., Karlish S. J., Maunsbach A. B. (2006) Am. J. Physiol. Renal Physiol. 291, F1033–F1044 [DOI] [PubMed] [Google Scholar]
- 21. Wetzel R. K., Pascoa J. L., Arystarkhova E. (2004) J. Biol. Chem. 279, 41750–41757 [DOI] [PubMed] [Google Scholar]
- 22. Sweadner K. J., Gilkeson R. C. (1985) J. Biol. Chem. 260, 9016–9022 [PubMed] [Google Scholar]
- 23. Felsenfeld D. P., Sweadner K. J. (1988) J. Biol. Chem. 263, 10932–10942 [PubMed] [Google Scholar]
- 24. Feschenko M. S., Sweadner K. J. (1997) J. Biol. Chem. 272, 17726–17733 [DOI] [PubMed] [Google Scholar]
- 25. Choi Y., Dubel S. J., Pacioaiou M. L., Omori A., Ito T., Copeland T. D., Takahashi M., McEnery M. W. (1997) Arch. Biochem. Biophys. 344, 165–175 [DOI] [PubMed] [Google Scholar]
- 26. Arystarkhova E., Donnet C., Muñoz-Matta A., Specht S. C., Sweadner K. J. (2007) Am. J. Physiol. Cell Physiol. 292, C1179–C1191 [DOI] [PubMed] [Google Scholar]
- 27. Jorgensen P. L. (1974) Methods Enzymol. 32, 277–290 [PubMed] [Google Scholar]
- 28. Laughery M. D., Todd M. L., Kaplan J. H. (2003) J. Biol. Chem. 278, 34794–34803 [DOI] [PubMed] [Google Scholar]
- 29. Capasso J. M., Rivard C., Berl T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13414–13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackson M. R., Nilsson T., Peterson P. A. (1990) EMBO J. 9, 3153–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donnet C., Arystarkhova E., Sweadner K. J. (2001) J. Biol. Chem. 276, 7357–7365 [DOI] [PubMed] [Google Scholar]
- 32. Murphy G. J., Mostoslavsky G., Kotton D. N., Mulligan R. C. (2006) Nat. Med. 12, 1093–1099 [DOI] [PubMed] [Google Scholar]
- 33. Bibert S., Roy S., Schaer D., Felley-Bosco E., Geering K. (2006) J. Biol. Chem. 281, 39142–39151 [DOI] [PubMed] [Google Scholar]
- 34. Sweadner K. J., Wetzel R. K., Arystarkhova E. (2000) Biochem. Biophys. Res. Commun. 279, 196–201 [DOI] [PubMed] [Google Scholar]
- 35. Arystarkhova E., Wetzel R. K. (2003) Ann. N.Y. Acad. Sci. 986, 416–419 [DOI] [PubMed] [Google Scholar]
- 36. Thanaraj T. A., Clark F. (2001) Nucleic Acids Res. 29, 2581–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen M., Manley J. L. (2009) Nat. Rev. Mol. Cell Biol. 10, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J. B., Levanon E. Y., Yoon J. K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G. M. (2009) Science 324, 1210–1213 [DOI] [PubMed] [Google Scholar]
- 39. Colina C., Palavicini J. P., Srikumar D., Holmgren M., Rosenthal J. J. (2010) PLoS Biol. 8, e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blanc V., Davidson N. O. (2003) J. Biol. Chem. 278, 1395–1398 [DOI] [PubMed] [Google Scholar]
- 41. Meier J. C., Henneberger C., Melnick I., Racca C., Harvey R. J., Heinemann U., Schmieden V., Grantyn R. (2005) Nat. Neurosci. 8, 736–744 [DOI] [PubMed] [Google Scholar]
- 42. Pihakaski-Maunsbach K., Nonaka S., Maunsbach A. B. (2008) Acta Histochem. Cytochem. 41, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson M. R., Nilsson T., Peterson P. A. (1993) J. Cell Biol. 121, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bibert S., Roy S., Schaer D., Horisberger J. D., Geering K. (2008) J. Biol. Chem. 283, 476–486 [DOI] [PubMed] [Google Scholar]
- 45. Lubarski I., Pihakaski-Maunsbach K., Karlish S. J., Maunsbach A. B., Garty H. (2005) J. Biol. Chem. 280, 37717–37724 [DOI] [PubMed] [Google Scholar]
- 46. Hirohashi S., Kanai Y. (2003) Cancer Sci. 94, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ino Y., Gotoh M., Sakamoto M., Tsukagoshi K., Hirohashi S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crambert G., Li C., Swee L. K., Geering K. (2004) J. Biol. Chem. 279, 30888–30895 [DOI] [PubMed] [Google Scholar]
- 49. Teasdale R. D., Jackson M. R. (1996) Annu. Rev. Cell Dev. Biol. 12, 27–54 [DOI] [PubMed] [Google Scholar]
- 50. Morton M. J., Farr G. A., Hull M., Capendeguy O., Horisberger J. D., Caplan M. J. (2010) J. Biol. Chem. 285, 33737–33746 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.