Abstract
Huntington disease (HD) is a progressive neurodegenerative disorder caused by expression of polyglutamine-expanded mutant huntingtin protein (mhtt). Most evidence indicates that soluble mhtt species, rather than insoluble aggregates, are the important mediators of HD pathogenesis. However, the differential roles of soluble monomeric and oligomeric mhtt species in HD and the mechanisms of oligomer formation are not yet understood. We have shown previously that copper interacts with and oxidizes the polyglutamine-containing N171 fragment of huntingtin. In this study we report that oxidation-dependent oligomers of huntingtin form spontaneously in cell and mouse HD models. Levels of these species are modulated by copper, hydrogen peroxide, and glutathione. Mutagenesis of all cysteine residues within N171 blocks the formation of these oligomers. In cells, levels of oligomerization-blocked mutant N171 were decreased compared with native N171. We further show that a subset of the oligomerization-blocked form of glutamine-expanded N171 huntingtin is rapidly depleted from the soluble pool compared with “native ” mutant N171. Taken together, our data indicate that huntingtin is subject to specific oxidations that are involved in the formation of stable oligomers and that also delay removal from the soluble pool. These findings show that inhibiting formation of oxidation-dependent huntingtin oligomers, or promoting their dissolution, may have protective effects in HD by decreasing the burden of soluble mutant huntingtin.
Keywords: Copper, Neurodegeneration, Oxidation-Reduction, Polyglutamine Disease, Thiol, Huntingtin, Protein Oligomer
Introduction
Huntington disease (HD)2 is a progressive neurodegenerative disorder caused by a glutamine-encoding CAG expansion in the huntingtin gene (1). Mutant huntingtin protein (mhtt) is cleaved into a number of polyglutamine containing N-terminal fragments that misfold and form soluble monomeric and oligomeric proteins as well as insoluble aggregates. Several publications have shown that soluble N-terminal fragments of mhtt are important toxic factors that drive HD (2–4). Further, in HD and other neurodegenerative diseases, there is evidence that soluble oligomers may be particularly important (2, 5–8).
Soluble oligomers of mhtt may form or be stabilized through a number of mechanisms including noncovalent polyglutamine-dependent interactions (5) and possibly transglutaminase-mediated covalent cross-links (9). In addition, oxidation of huntingtin could also promote oligomerization (10). In general, cysteine oxidation is an important mechanism for forming intermolecular cross-links that may stabilize protein oligomers (11, 12). Site-specific cysteine oxidation may regulate protein function by affecting structure including oligomerization state (13–15). There is growing evidence that a variety of protein residues up and downstream of the mhtt polyglutamine tract impact HD progression through providing sites of post-translational modification that affect huntingtin degradation, cleavage, or cell location (16–19). The N terminus of huntingtin contains several cysteine residues, the most C-terminal of which are within the exon 2-encoded region downstream of the glutamine tract and are present in all but the shortest of N-terminal huntingtin fragments (20). Redox-mediated changes of these residues may modulate huntingtin function and therefore be important modulators of HD.
The cysteine thiol group (-SH) can be oxidized constitutively or in response to oxidative stress even in the reducing cytosolic environment (21, 22). Disulfides (-S-S-) and sulfenic acid (-SOH) can be converted back to thiols by chemical reductants or thiol reductase enzymes. However, sulfinic acid (-SO2H) and sulfonic acid (-SO3H) require enzymatic reduction (15, 23). Susceptibility of cysteine residues to oxidation depends critically on side chain exposure to the surrounding redox environment. The complete structure of N-terminal huntingtin has not been solved. However, intriguingly, the most N-terminal cysteine residue of mammalian huntingtin, Cys115, is predicted to be particularly prone to oxidation due to influences of the adjacent N-terminal histidine, similar to what has been described for Cys111 in human SOD1 (14).
There is considerable evidence that mhtt expression results in increased levels of oxidatively damaged proteins and other macromolecules in HD brain (24–27). Factors that may be implicated include increased levels of pro-oxidants, including copper, iron, and 3-hydroxykynurenine (28–30) and decreased proteasomal function that may contribute to the accumulation of damaged proteins (31). Previously we reported that an N-terminal fragment of huntingtin, called N171, interacts with and chemically reduces copper (II) (28). The aim of this study was to investigate how copper, and the redox environment in general, modulates N-terminal huntingtin structure. Here, we show that oxidation of cysteine residues within N171 huntingtin is required for the formation of one type of oligomeric species. Further, we show slower clearance of oxidizable N171 huntingtin from the soluble cell fraction, demonstrating that modulation of huntingtin oxidation state can alter soluble mhtt levels in HD.
EXPERIMENTAL PROCEDURES
Materials
AB1 antibody was provided by Dr. Marian DiFiglia; MAB5492 and 1C2 (Chemicon). Unless otherwise indicated all other supplies were from Sigma, Fisher, or Invitrogen.
Mice
CAG140 mice were maintained on the B6/CBA background by crossing CAG140 males with F1 females. All protocols were approved by the institutional IACUC.
Bacterial N171 Huntingtin Expression
The procedure was similar to that already described (28). In brief, N171 protein was expressed in BL21 bacteria using 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C (17Q) or 25 °C (40Q). N171 was purified from bacterial extracts using a GST affinity column, and then the GST tag was cleaved using PreScission protease (GE Healthcare). The entire purification procedure was performed in the presence of 1 mm EDTA. Protein was eluted after PreScission cleavage into 20 mm BisTris (pH 6.7) containing 1 mm DTT and 1 mm EDTA then stored at −70 °C. For the metal-binding stoichiometry experiment, samples were loaded onto a DEAE-anion exchange column (GE Healthcare) previously equilibrated in 20 mm BisTris (pH 6.7). The column was washed with 5 ml of 20 mm BisTris (pH 6.7). Protein was then eluted in a continuous gradient of NaCl in 20 mm BisTris. All buffers were Chelex resin-treated and degassed before use. Huntingtin-containing fractions were combined and then processed by buffer exchange into 20 mm Tris (pH 7.2), kept on ice, and used within 1 h. For all other experiments, N171 eluted from the PreScission cleavage step was buffer exchanged directly into 20 mm Tris (pH 7.2) containing 0.15 m NaCl and used immediately.
Metal-Huntingtin Stoichiometry
N171-17Q huntingtin was purified as described above using the GST tag and DEAE chromatography. Protein was quantified using BSA as standard and then lyophilized prior to analysis by inductively coupled plasma-mass spectrometry. Buffer blanks were analyzed as negative controls.
Cell Culture
COS1 cells were grown in 10% FBS-supplemented DMEM at 37 °C and 5% CO2. Reduced serum medium (Opti-MEM®-1; Invitrogen) and Lipofectamine 2000 (Invitrogen) were used for transfections following standard procedures. Wild-type and polyglutamine-expanded N-terminal huntingtin fragments were expressed using the pcDNA1 vector. Point mutations were generated using the QuikChange® Site-directed Mutagenesis kit (Stratagene). To measure the half-life of soluble huntingtin, cells were transfected, then 18 h later treated with the protein synthesis inhibitor cycloheximide (10 μg/ml) as described (32). To test for autophagy-mediated degradation of soluble huntingtin, we used 10 mm 3-methyladenine for 3 h. In these experiments, total soluble huntingtin was quantified by reducing SDS-PAGE and Western blot analysis.
Fluorescence-lifetime Imaging (FLIM) Analysis
A FLIM assay was used to determine the proximity of GFP- (donor fluorophore) and RFP- (acceptor fluorophore) labeled soluble N-terminal huntingtin species in solution. We studied the wild-type N171 fragment to prevent confounds due to aggregation. N171-17Q huntingtin was subcloned into pDsRedmono and pAcGFP vectors (Clontech) to generate RFP-N171-17Q- and GFP-N171-17Q-expressing constructs, respectively. The GFP-N171-17Q was used as a FLIM negative control and a GFP-RFP fusion as a positive control. The lifetime of a donor fluorophore indicates the proximity between a donor and acceptor fluorophores. If the fluorophores are <10 nm apart, the donor lifetime decreases due to energy transfer to the acceptor fluorophore. COS1 cells were grown on CC2-coated chamber slides (Lab-Tek), transfected with respective constructs, and fixed 48 h later in 2% paraformaldehyde for 10 min. Cells were imaged using a Zeiss LSM510. A Chameleon Ti:sapphire laser was used to excite the donor fluorophore at 840 nm. A Hamamatsu MCP 3809 detector and Becker&Hickl (Berlin, Germany) TCSPC hardware/software were used to obtain donor lifetime information in a pixel-by-pixel basis. Data analysis was performed using SPC Image (Becker&Hickl), in which donor fluorophore lifetimes in the entire cell are determined by fitting the data to one (negative control) or two (experimental conditions) exponential decay curves. In the two-exponential model of lifetime analysis, the longer (no-FRET) lifetime is “fixed” as a t1 value, and the second, shorter lifetime reflecting the presence of FRET is calculated by the system as a t2 value. The t2 lifetime is used for comparisons between different experimental conditions. Thus, “non-FRETing” component (t1) is excluded from the lifetime comparisons. The donor lifetime information was color-coded and displayed in a 128 × 128-pixel matrix as pseudo-color images (33).
Western Blot Analysis
SDS-resistant and reduction-sensitive oligomers were detected by nonreducing SDS-PAGE analysis. At the termination of cell-free experiments, 20 μl of sample was incubated with 10 units of catalase and 1 mm EDTA for 2 min at 25 °C. SDS was added to 1% (w/v) and the sample mixed and incubated for 2 min. Thirty millimolar N-ethylmaleimide (prepared in ethanol) was then added and the sample incubated for 15 min at 25 °C. Samples were mixed with nonreducing sample buffer (Bio-Rad) and analyzed by SDS-PAGE. For cell- and mouse-based experiments, samples were lysed in 20 mm Tris (pH 7.2), 150 mm NaCl, 2 mm EDTA, 1 mm bathocuproine, 1% Triton X-100, 40 mm N-ethylmaleimide, and HaltTM protease inhibitor mixture (Pierce). Lysates were incubated for 10 min at 25 °C then centrifuged for 30 min at 20,000 × g and 4 °C. Protein was quantified using DC protein assay reagents (Bio-Rad) and BSA standards (Sigma). For reducing gels, samples were boiled in 50 mm DTT for 15 min. Protein was transferred to PVDF using standard procedures. HRP-based chemiluminescent detection was with film or a FluorChem® HD2 Imager (Alpha Innotech). Densitometric analysis was undertaken using ImageJ software.
GSH Assay
Glutathione levels in COS1 cells were measured using the monochlorobimane fluorescence assay as described previously (34).
Quantitative PCR
N171 huntingtin mRNA levels were quantified exactly as described previously (34). Primers used were gcaccgaccaaagaaagaac (forward) and tagaaggcacagtcgagg (reverse).
Filter Trap Assay
One μm N171-40Q huntingtin was incubated with various copper concentrations for up to 48 h. Filter trap analysis was performed essentially as described previously (35). Twenty μl of sample (20 pmol) was used for analysis. Trapped huntingtin was detected using a FLAG antibody (M2 from Sigma) and HRP chemiluminescence.
Huntingtin Aggregate Morphology
N171-40Q huntingtin was expressed and purified without a DEAE step. Two micromolar protein was incubated for 48 h in the presence of 20 μm EDTA or 20 μm copper (II) chloride at 37 °C. Protein samples were absorbed onto Formvar-carbon film on 400 mesh copper (Canemco-Marivac; Quebec, QC, Canada). Excess liquid was removed by blotting. Grids were then stained with 0.2 μm of filtered 3% uranyl acetate for 15 min and then washed for 5 min in deionized water. Samples were blotted and air-dried. Images were taken using a JEOL 100CX electron microscope.
Statistical Analysis
Data were analyzed using SAS version 9.1 software by t test or GLM-based ANOVA analysis. p values <0.05 were considered significant.
RESULTS
Copper Promotes Oligomerization of N171 Huntingtin Protein through Site-specific Cysteine Oxidation
We investigated the effect of copper on N171 huntingtin structure. N171-17Q was purified from bacteria, subjected to experimental conditions, and then resolved by gel electrophoresis. When resolved by reducing SDS-PAGE, monomeric N171-17Q migrates predominantly at 31 kDa, higher than its actual mass of 19.8 kDa (Fig. 1A). Without copper incubation, a significant amount of N171-17Q migrated at twice the predicted mass, consistent with a dimer when resolved by nonreducing SDS-PAGE. Prior incubation with copper (II) resulted in a significant increase in dimer levels and also higher mass products. These species were sensitive to chemical reduction (Fig. 1A), indicating that thiol oxidation is required for generating SDS-resistant oligomers. Despite loading equal amounts of N171 huntingtin, signal strength is stronger following copper incubation consistent with a copper-induced epitope conformational change. Incubation of N171-17Q huntingtin in the presence of EDTA significantly decreased the amount of oligomerization compared with no EDTA conditions (Fig. 1B, first and second lanes), indicating that an oligomerization promoting metal co-purifies with N171-17Q. Further, we found that as little as 50 nm added copper (II) had a small effect in promoting N171-17Q huntingtin oligomerization and that progressively higher concentrations resulted in a dose-dependent increase (Fig. 1B). Because cysteine thiol groups can be sensitive to a variety of oxidants we next sought to determine the specificity of the effect of copper on oligomerization of N171-17Q huntingtin. Forty nm hydrogen peroxide was able to promote oligomerization of N171-17Q huntingtin. However, metal chelation completely blocked the effect of 2 μm hydrogen peroxide on N171-17Q oligomerization (Fig. 1C), again showing that a metal co-purifies with N171. To address whether the effect of copper on oligomerization is secondary to hydrogen peroxide generation, we tested the effect of copper in the presence of the hydrogen peroxide scavenger catalase. However, there was no inhibitory effect of catalase on N171-17Q oligomerization (data not shown), suggesting that copper (II) may directly oxidize N171-17Q. To elucidate further which metal(s) may co-purify with N171-17Q huntingtin, we evaluated the inhibitory effect of a number of metal chelators on hydrogen peroxide-promoted oligomerization. Bathophenanthroline, an iron (II) and copper (II) chelator, trientine, a copper (II) chelator, and bathocuproine, a copper (I) chelator, all inhibited oligomerization, but the iron (III) chelator deferoxamine did not (Fig. 1D). We then tested whether other redox-active metals promote oligomerization. There was no effect of manganese or iron (Fig. 1E).
FIGURE 1.
Copper promotes oligomerization of cell-free N171 huntingtin. N171-17Q huntingtin was expressed and purified from bacteria (see “Experimental Procedures”). Aliquots containing 2 μm protein were incubated for 1 h at 37 °C under conditions outlined below and then evaluated by Western blot analysis. A, purified N171 huntingtin spontaneously forms reduction-sensitive dimers. Copper incubation (5 μm) promotes formation of additional dimer and higher molecular mass oligomers. B–D, all samples were analyzed by nonreducing SDS-PAGE. B, copper dose-dependently promotes N171 oligomerization. As little as 50 nm added copper (II) promotes N171 oligomerization. C, hydrogen peroxide (2 μm) promotes N171-17Q oligomerization, but the effect is blocked by metal chelation. D, copper-selective chelators inhibit hydrogen peroxide (4 μm)-promoted oligomerization. E, iron and manganese do not promote N171 oligomerization. F, copper co-purifies with N171 huntingtin. N171-17Q huntingtin was expressed in bacteria and then purified using a DEAE column (see “Experimental Procedures”). Protein was quantified using BSA as standards. Metals were quantified by inductively coupled plasma-mass spectrometry. There is an average of 1 copper per 3 N171 huntingtin protein molecules. Manganese is present but in significantly smaller amounts. n = 3; beta-ME, β-mercaptoethanol; BC, bathocuproine.
Our data so far suggested that copper co-purifies with N171 huntingtin and also promotes oligomerization by thiol oxidation. To confirm whether copper co-purifies with bacterial expressed N171-17Q huntingtin, we measured copper, iron, and manganese levels in N171-17Q huntingtin purified from bacteria grown under standard conditions. We used a two-column purification protocol utilizing GST-Sepharose and then DEAE-anion exchange chromatography (see “Experimental Procedures”). Metal levels were measured by inductively coupled plasma-mass spectrometry. Despite a prolonged purification procedure (∼24 h) in the continual presence of 1 mm EDTA we found that there were ∼0.3 copper ions for every N171-17Q protein (Fig. 1F). Iron was not detected. Manganese was present at ∼0.04 ions/N171 protein.
The N171-17Q fragment of huntingtin contains four cysteine residues (Fig. 2A). Because our data indicated that cysteine oxidation is required for oligomerization (Fig. 1), we generated a series of cysteine mutants to characterize the dependence of cysteine residues on oligomerization further. Purified N171 proteins were incubated for 1 h at 37 °C in the presence or absence of added copper (5 μm). We did not identify effects of single cysteine mutations in our cell-free oligomerization assay (data not shown). However, analysis of double cysteine mutants (Fig. 2B) revealed that the C115A/C119A mutant was significantly less prone to dimerization. Double mutants with Cys115 intact were most prone to oligomerization as oligomers were present in the absence of copper treatment. Further, the C137A/C158A mutant dimerized very readily but did not form higher mass oligomers (Fig. 2B), suggesting that these cysteines inhibit dimerization. We next analyzed triple cysteine mutants (Fig. 2C) and found that all four readily form dimers (Fig. 2C). Replacement of all cysteine residues within N171-17Q completely blocked formation of all SDS-resistant oligomers, even after copper treatment (Fig. 2D).
FIGURE 2.
Cysteines 115 and 119 are most important for dimerization of N171 huntingtin. A, sequence of N171-17Q huntingtin is shown. The four cysteine residues (Cys115, Cys119, Cys137, and Cys158) and the FLAG tag are shown in uppercase. A hyphen (-) delineates the boundary of exon 1–2-encoded regions. B, two-way cysteine mutagenesis of N171 demonstrates that cysteines 115 and 119 are important for dimerization. C, three-way cysteine mutagenesis of N171 reveals that single cysteine residues promote rapid dimerization in the presence of copper. D, four-way cysteine mutagenesis blocks oligomerization of N171 even in the presence of copper.
Different N-terminal Huntingtin Fragments Form Redox-modulated Oligomers in Cultured Cells
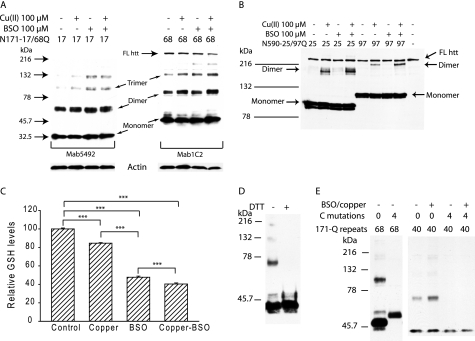
We next sought to determine whether the huntingtin oligomers we had identified exist in cells or form following oxidative stress. We transfected COS1 cells with wild-type and polyglutamine-expanded forms of N171 and N590 huntingtin. Wild-type and mutant N171 protein spontaneously formed soluble oligomers as determined by nonreducing SDS-PAGE analysis (Fig. 3A). Furthermore, the levels of oligomers were increased a small amount if the cells were treated with copper (1 h) and/or the glutathione synthesis inhibitor l-buthionine-sulfoximine (BSO) (24 h) (Fig. 3A). To investigate whether oligomers form with other huntingtin fragment, we assessed the larger N590 (caspase 6) fragment. N590 formed just detectable levels of dimer spontaneously 24 h after transfection. However, copper, but not BSO, significantly enhanced their abundance (Fig. 3B). Copper can interact with and modulate GSH levels (36). We therefore determined how our copper/BSO treatment paradigm modulated GSH. Copper, BSO, and copper/BSO decreased cellular GSH levels by 18, 52, and 60%, respectively (Fig. 3C). The N590 fragment dimerized with copper, but not BSO. Therefore, the effect of copper on dimerization is not secondary to glutathione depletion but could involve a direct interaction with huntingtin. Oligomers of N171-40Q were readily reduced to monomer (Fig. 3D). Further, in agreement with our cell-free experiments (Fig. 2) mutant N171 huntingtin (40 and 68Q) failed to form SDS-resistant oligomers when all cysteines were replaced with alanines, even after treatment with copper (Fig. 3E). These data indicate that N-terminal huntingtin fragments long enough to contain cysteine residues form reducible oligomeric species in cells. To determine whether full-length huntingtin can form the same species, we evaluated the effect of copper on endogenous COS1 huntingtin. Because full-length huntingtin is very large (>300 kDa) we did not try to discriminate between dimers and larger species. We found that transient reducible high molecular mass full-length huntingtin species form after treatment with as little as 10 μm copper (supplemental Fig. S1). Intriguingly, we were unable to generate high molecular mass species of full-length huntingtin when studying COS1 cells transfected with N590 huntingtin (Fig. 3B); the reasons for this are not known. However, taken together our data indicate that reduction-sensitive oligomers of two N-terminal huntingtin fragments and full-length huntingtin can form in cultured cells.
FIGURE 3.
N171 and N590 N-terminal huntingtin fragments form oligomers in cultured cells. A–C, COS1 cells were transfected with various huntingtin-encoding constructs, and the experiments were terminated 48 h later. Treatments were applied 24 h (BSO) and 1 h (copper) prior to termination. A, N171 wild-type and mutant huntingtin spontaneously form oligomers in COS1 cells. Copper and the glutathione synthesis inhibitor (BSO) result in a small increase in oligomer levels. B, copper promotes dimerization of N590 huntingtin in COS1 cells. Full-length huntingtin serves as a loading control. C, copper decreases glutathione levels significantly less than BSO. D, oligomers of N171-40Q huntingtin are lost on reduction with DTT. E, mutagenesis of all four N171-40 and 68Q cysteine residues blocks formation of oligomers even in the presence of copper and BSO. NR, nonreducing gel; r = reducing gel; FL htt, full-length huntingtin.
Because oligomers may be the result of artifacts introduced during cell lysis we used FLIM as an independent method to verify that self-associated N171 protein is present in cells in situ. We chose to study wild-type N171 to exclude the potential for insoluble protein aggregates as a confounding factor. We found that co-transfection of donor and acceptor fluorophore-containing N171-17Q-encoding fragments (see “Experimental Procedures”) resulted in an 83% decrease in donor fluorescence lifetime compared with the GFP-N171-17Q negative control (supplemental Fig. S2). These results, although not confirming oxidation-dependent oligomers, do show that N171-17Q protein is, at least partly, in a self-associated form in this model system consistent with our findings from nonreducing SDS-PAGE gel analysis experiments (Fig. 3).
Oxidation-dependent Species of Mutant Huntingtin Are Present in Mouse HD Brain
We next sought to determine whether reduction-sensitive oligomeric species of huntingtin are present in mouse HD brain. We initially evaluated the N171-82Q transgenic mouse model of HD but were unable to detect monomeric or oligomeric huntingtin by Western blot analysis. We then chose to assess 12-month-old CAG140, a full-length knock-in HD mouse model (37). Our colony of CAG140 mice at this age does not have overt symptoms of HD (28). We undertook Western blot analysis for reduction-sensitive huntingtin species by comparing banding on reducing and nonreducing SDS-PAGE analysis of aliquots from the same mouse. Because banding patterns derived from full-length huntingtin cleavage are complex (20) we used two huntingtin antibodies enabling us to distinguish wild-type from mutant huntingtin N-terminal fragments. In frontal cortex, reducing gels demonstrated mutant huntingtin bands at 192 and 203 kDa that were not present in the nonreducing lanes (Fig. 4A). These bands were very faint in striatum, suggesting much less huntingtin proteolysis in this region (data not shown). Evidence of reduction-sensitive wild-type huntingtin fragments in wild-type or HD mice were not identified using a lower percentage gel (Fig. 4B). Our cell culture data indicate that wild-type and mutant N-terminal huntingtin can form high molecular mass reducible species. However, in genetically accurate CAG140 mice we only found evidence for reduction-sensitive species formed by mutant and not wild-type huntingtin.
FIGURE 4.
Reduction-sensitive species of mutant huntingtin are present in CAG140 knock-in HD mice. Brains of 12-month-old CAG140 and wild-type littermate mice were analyzed for reducible species of wild-type and mutant huntingtin and glutathione levels. Cortices were processed for nonreducing and reducing SDS-PAGE analysis using a 5% gel. Parallel samples were probed with AB1 (detects N-terminal containing wild-type and mutant huntingtin) and 1C2 (detects mutant but not wild-type polyglutamines). A, Western analysis for mutant huntingtin. Two bands are present in AB1 and IC2 blots with sample reduction only (single-headed arrows), indicating mutant huntingtin N-terminal fragments. Calculated molecular masses are 203 and 192 kDa for upper and lower bands. The lower band in 1C2 blots is partly obscured by IgG smear (double-headed arrow). B, Western blot analysis for wild-type huntingtin. Reduction-sensitive species are not detected in cortex of CAG140 or wild-type littermate mice. Two N-terminal fragments of wild-type huntingtin consistent with monomeric species are present with calculated molecular masses of 78 and 153 kDa (single-headed arrows).
Cysteine Oxidation-blocked N171-40Q Huntingtin Shows Increased Clearance from the Soluble Pool
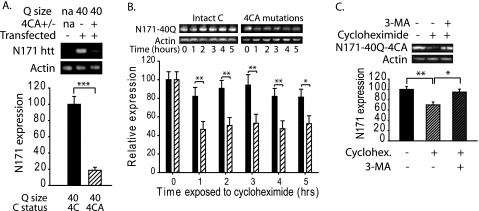
In our comparison of native and cysteine-mutated N171 protein we noted that the cysteine mutant form was present at a lower level in transfected cells (Fig. 3F). As modulation of soluble mutant huntingtin levels is one important approach to modify the course of HD we investigated the mechanism by which this occurs. Using reducing SDS-PAGE and Western blot analysis we verified significantly lower N171-40Q protein levels with the variant lacking cysteine residues (Fig. 5A). There was no difference in transcript expression level as determined by quantitative PCR (not shown). To assess whether cysteine mutagenesis altered soluble N171-40Q half-life we used the translational inhibitor cycloheximide to investigate soluble protein clearance, essentially as has been used previously for other proteins (32). Results show that the cysteine nonoxidizable form of N171-40Q huntingtin (N171-40Q-4CA) is cleared more rapidly from the soluble fraction than native N171-40Q protein. Approximately 50% of N171-40Q-4CA is cleared within 1 h of cycloheximide treatment, whereas decreases in soluble levels of native N171-40Q were significantly less (Fig. 5B). We further show that N171-40Q-4CA huntingtin is cleared by macroautophagy (Fig. 5C), consistent with delayed clearance of N171-40Q oligomers being due to decreased macroautophagic capacity for this substrate. Because this experiment does not fully rule out the possibility that soluble N171-40Q-4CA huntingtin may be rapidly cycled into an insoluble form rather than degraded, we compared the cell-free aggregation properties of native N171-40Q and N171-40Q-4CA. We used the previously described filter trap assay to measure insoluble aggregates (>0.2 μm) (35). We found time- and copper concentration-dependent effects on the level of trapped huntingtin (Fig. 6A). Incubation of N171-40Q huntingtin in the presence of EDTA resulted in slow formation of oligomeric species; copper promoted the rate of this process. We did not detect clear differences in levels of entrapped N171-40Q-4CA and N171-40Q. However, we found that cysteine mutagenesis modulated the ultrastructural morphology of self-associated mutant huntingtin in a copper-dependent and cysteine-dependent manner (Fig. 6B). N171-40Q formed spherical and fibrillar oligomers after a 48-h incubation in EDTA or copper, respectively. N171-40Q-4CA formed amorphous and spherical oligomers after a 48-h incubation in EDTA or copper, respectively (Fig. 6B).
FIGURE 5.
Cysteine-oxidation blocked N171 mutant huntingtin has decreased soluble protein half-life in COS1 cells. A, monomeric N171-40Q with all four cysteine residues mutated is present at lower levels. Western blot analysis was 24 h after transfection by reducing SDS-PAGE. n = 4. B, protein synthesis inhibition using cycloheximide demonstrates that the soluble protein half-life is significantly lower for the oxidation-blocked form of N171-40Q huntingtin. n = 5 at each time point. C, cysteine oxidation-blocked soluble huntingtin (N171-40Q-4CA) is degraded by the macroautophagy pathway. 3-Methyladenine (3-MA)-mediated inhibition of macroautophagy inhibits N171-40Q-4CA huntingtin degradation. n = 6. *, p < 0.05; **, p <0.01; ***, p < 0.001.
FIGURE 6.
Cysteine residues within N171-40Q huntingtin modulate the structure but not the amount of aggregation. A, copper promotes trapping of N171-40Q huntingtin species in a dose- and time-dependent manner. Aliquots of 1 μm N171-40Q or cysteine-mutated N171-40Q protein were incubated with various concentrations of copper (II) chloride at 37 °C. Aggregated material was quantified using a filter trap assay. Incubation for 24–48 h without copper is sufficient to promote filter trapping. There is no difference in the response to copper or time of incubation for N171-40Q versus N171-40Q-4CA. Each set of four graph bars represents, left to right: 0, 0.25, 2, and 10 μm copper, respectively. Bars indicate means ± S.E. (error bars) of three independent experiments each in triplicate. B, N171-40Q huntingtin, after 48-h incubation at 37 °C in the presence of EDTA or copper, forms spherical and fibrillar species, respectively. N171-40Q-4CA huntingtin forms amorphous species after incubation with EDTA and spherical species after incubation with copper. N171-40Q-4CA = cysteine to alanine mutated form of N171-40Q. Scale bars, 500 nm; inset, 250 nm wide. n = 3.
DISCUSSION
Polyglutamine expansion within the N terminus of huntingtin protein is the proximal cause of HD (1). Full-length huntingtin protein gives rise to numerous polyglutamine-containing N-terminal huntingtin fragments that are the main drivers of disease expression (20). These fragments exist as soluble monomers and oligomers as well as insoluble protein aggregates. Our study provides experimental evidence for an oligomeric N-terminal huntingtin species that is stabilized by thiol oxidation-mediated disulfide formation. Further, we show that this species is cleared more slowly from the soluble fraction than non-thiol-oxidizable protein and accumulates to higher levels. Thiol oxidation in vivo could promote the formation and inhibit the clearance of toxic mutant huntingtin oligomers, and thus its inhibition or reversal could be neuroprotective.
Intermolecular Disulfides Are Necessary for Redox-sensitive Huntingtin Oligomer Formation
We demonstrated that SDS-resistant and reduction-sensitive oligomers of N171 huntingtin form in cell-free conditions and within COS1 cells. This finding suggested to us that an understanding of cell-free oxidation-mediated oligomerization of N171 is relevant to an understanding of huntingtin protein in vivo. We therefore undertook a number of cell-free experiments to characterize the oligomerization process. N171 with just one of each of the four cysteines intact retained the ability to form dimers (Fig. 2C), clearly showing that these oligomers are stabilized by intermolecular disulfides. Further mutagenesis experiments demonstrated that whereas each of the four cysteine residues within N171 huntingtin can mediate oligomerization they do not behave equally. Cysteines 115 and 119 were most important because their combined removal almost completely blocked oligomerization (Fig. 2B) whereas together cysteines 137 and 158 inhibited oligomerization (Fig. 2B). We found that even under our most oxidizing conditions there were always significant amounts of monomeric N171 huntingtin remaining (Fig. 1B). Based on these findings we propose a model whereby internal disulfides between Cys115/Cys119 and Cys137/Cys158 block SDS-resistant oligomerization that is mediated by intermolecular disulfides primarily involving cysteines 115 and 119.
Redox-sensitive N-terminal Huntingtin Oligomeric Species Are Present in Cells
Huntingtin protein is expressed throughout the brain and is located primarily in neurons; cytosolic, nuclear, and membrane-associated locations have been described (38, 39). Our FLIM analysis shows primarily cytosolic location of N171 huntingtin in COS1 cells (supplemental Fig. S2). Despite a strongly reducing cytoplasmic environment, there are numerous cytosolic proteins that form disulfides, either stably (for example, SOD1) (22) or during oxidative stress (21). We found that cell-free N171 huntingtin is sensitive to oxidation by very low concentrations of copper and hydrogen peroxide. In COS1 cells, we found the same oligomeric species of N171 huntingtin for both wild-type and polyglutamine-expanded fragments (Fig. 3). We also found that oligomers of N171-40Q form in the HN10 hippocampal neuron cell line, demonstrating that findings are relevant to neuronal cells (data not shown). Further, the SDS-resistant oligomers are not specific for the N171 huntingtin fragment. These experiments demonstrate that similar oligomeric species are formed by wild-type and mutant N590 huntingtin (Fig. 3B) and full-length huntingtin (supplemental Fig. S1). These findings indicate that huntingtin is very susceptible to oxidation-mediated oligomerization and that the oxidation-sensitive residues are present within the N terminus where the polyglutamine tract is located.
We used knock-in HD mice (CAG140 line) to determine whether reduction-sensitive huntingtin species are present in vivo. Our results strongly suggest that only mutant N-terminal fragments of huntingtin form reduction-sensitive species in mouse brain. Although the oligomeric species of mutant huntingtin were not identified directly, monomeric species generated by sample reduction were present. This finding is most consistent with the presence of a very high molecular mass oligomeric mutant huntingtin species that does not enter the gel or is not transferred to membrane. Disulfide association with insoluble aggregates is possible but unlikely because the high centrifugal force used after brain lysis to prepare supernatant removes nearly all aggregated species. In contrast to our findings in CAG140 mice (Fig. 4), in cell-free conditions and using COS1 cells we identified oligomers of wild-type huntingtin (Figs. 1 and 3). This apparent discrepancy can be explained when the concentration of huntingtin protein in these different systems is taken into account. Legleiter et al. (5) demonstrated that oligomers of the exon 1 huntingtin fragment form in a concentration- and glutamine length-dependent manner. They also reported formation of nonfibrillar oligomers of wild-type exon 1 huntingtin under cell-free conditions. Huntingtin concentration in our cell-free conditions and with plasmid-mediated expression is significantly higher compared with endogenous expression in mouse brain. Therefore, high levels of expression of wild-type huntingtin may promote transient polyglutamine-dependent interactions allowing for oxidative cross-linking under appropriate redox conditions. In the genetically accurate CAG140 mouse model of HD, huntingtin expression is driven by the huntingtin promoter. Here, much lower wild-type huntingtin concentration and polyglutamine size will result in insufficient polyglutamine-dependent interactions for oxidative cross-linking to occur.
Promotion of Huntingtin Oligomerization by Copper
Our experiments demonstrate that copper (II) is capable of promoting oligomerization of cell-free and cell-associated huntingtin whereas other redox active metals do not. Copper reactivity necessitates tight regulation of its metabolism to prevent unwanted redox chemistry. For this reason, copper in cells is present bound to other molecules and is not thought to exist free in solution (40). We have previously reported elevated brain copper in R6/2 HD mice in late-stage disease, but not in 12-month CAG140 mice (28). Given the very low concentrations of copper required to promote huntingtin oligomers in cell-free conditions, it is possible that currently undetected defects of copper distribution or levels may be sufficient to promote oligomerization in CAG140 brain (Fig. 4). More studies are needed to understand better how these oligomers form in cells and why this is polyglutamine length-dependent effect in CAG140 mouse HD.
We provide data supporting our previous finding that N171 huntingtin has copper binding properties (28) (Fig. 1F). Currently, it is not clear whether N171 huntingtin is a copper metalloprotein or whether it has low affinity copper sites involved in transient interactions. Interestingly, the CXXXC motif present in N171 huntingtin (Fig. 2A) is also part of a low affinity copper site in the Sco family of copper chaperone proteins (41). Copper may therefore have a role in promoting or regulating huntingtin oligomerization through direct interaction at this site. This is consistent with our finding that combined mutation of Cys115 and Cys119 had a strong inhibitory effect on copper-induced oligomerization (Fig. 2B). Our data additionally provide insight into the mechanism(s) involved in copper-induced oligomerization. Cell-free experiments utilized extensively degassed buffers, implying that copper may oxidize N171 thiols directly in a reaction that does not require oxygen. We also found that hydrogen peroxide promoted oligomerization of N171 huntingtin in a partly metal-dependent manner (Fig. 1, C and D), indicating that oxygen-dependent chemistries may also be involved. Together our findings suggest that copper promotes oligomerization of N-terminal huntingtin by binding to a copper site within the protein; when weak noncovalently cross-linked oligomeric species form, an unknown factor may initiate oxidation of Cys115 and/or Cys119, which then forms intermolecular disulfides. Further studies will be needed to confirm this proposed mechanism.
Huntingtin Oxidation Delays Clearance from the Soluble Pool
There is considerable evidence that soluble mhtt species are important in disease pathogenesis (2–4). The toxic effects of a given soluble mhtt species will depend on the combined effects of its toxic potency and cellular levels. We therefore addressed whether the cysteine-oxidizable versus nonoxidizable forms of soluble (monomeric and oligomeric) N171 protein were present at different levels in cells. In transfection experiments we found significantly decreased steady-state soluble levels of the N171-40Q protein lacking cysteine residues as determined by reducing Western blot analysis (Fig. 5A). To investigate the effect of N171 oxidation on clearance of soluble protein we used the protein synthesis inhibitor cycloheximide. We found that soluble cysteine nonoxidizable N171-40Q (N171-40Q-4CA) was cleared significantly more rapidly than native N171-40Q (Fig. 5B). We cannot fully rule out the possibility that cysteine to alanine mutagenesis alters N171 structure to promote clearance by an oxidation-independent mechanism. Future studies will address in more detail how these mutations block the oligomers and promote huntingtin clearance. However, the simplest explanation for our findings is that blocking cysteine oxidation within N171 promotes clearance of soluble protein. Because only ∼50% of N171-40Q-4CA was rapidly cleared, these data further suggest that this protein may exist in two pools within COS1 cells. Our cell-free aggregation experiments found no evidence to support the possibility that N171-40Q-4CA aggregation may occur more rapidly than N171-40Q (Fig. 6A), supporting our interpretation that more rapid clearance of N171-40Q-4CA from the soluble pool involves a degradative pathway rather than rapid partitioning into the insoluble pool. We did find significant differences in the formation of previously described ultrastructural oligomeric morphologies (5) of N171-40Q versus N171-40Q-4CA. These data show although the level of copper-promoted self-association of N171-40Q is independent of cysteine status there are cysteine-dependent effects on morphology.
CONCLUSION
Despite a number of studies showing increased markers of oxidative stress in mouse and human HD (24, 27, 34) there has been uncertainty as to whether oxidized macromolecules contribute directly to HD progression or are only downstream markers of other more proximal disease pathways. These results demonstrate that macromolecular oxidation in HD also affects huntingtin protein. This suggests that altered redox biology may be an important upstream modulator of HD by altering huntingtin structure directly. This study has demonstrated one relevant pathway by which huntingtin oxidation may modulate HD. Our proposed role of oxidation-dependent mutant huntingtin oligomers in HD pathogenesis is summarized in Fig. 7. Findings suggest that interventions that inhibit huntingtin oxidation or promote conversion of oxidized oligomers to monomer may be protective. Finally, recent studies have provided evidence that oxidized forms of α-synuclein and wild-type SOD1 have important roles in Parkinson disease and sporadic amyotrophic lateral sclerosis, respectively (42, 43). This suggests that oxidized forms of key disease-associated proteins may be important upstream mediators in a number of neurodegenerative diseases.
FIGURE 7.
Proposed role of oxidized-mutant huntingtin in HD pathogenesis. Full-length mutant huntingtin is cleaved into N-terminal fragments most of which contain cysteine 115 (20). Age-related or mhtt-induced elevations of oxidative stress promote oxidation-dependent oligomerization of some N-terminal mhtt fragments. Oxidation-dependent mhtt oligomers are cleared slowly compared with monomeric mhtt and accumulate in cells promoting disease progression. The very short exon 1 mhtt fragment lacks cysteine residues and cannot form thiol-dependent oligomers. Huntingtin colors: red, expanded polyglutamine tract; blue, polyproline tract; green, oxidation-dependent intermolecular cross-links. Blue arrows demonstrate proposed disease pathways. Red arrows represent potential therapeutic targets: 1, thioltransferase-mediated conversion to monomer; 2, inhibition of huntingtin oxidation.
Supplementary Material
Acknowledgment
We thank Professor Hermann Schätzl for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants NS045806 (to S. H.), 5P20RR015640-10 (to University of Wyoming Neuroscience COBRE), and 1R21NS072372 (to J. F.). This work was also supported by Huntington's Disease Society of America awards (to S. H. (Coalition for the Cure) and J. F.) and also by the Cure Huntington's Disease initiative (to S. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- HD
- Huntington disease
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- BSO
- l-buthionine-sulfoximine
- FLIM
- fluorescence-lifetime imaging
- mhtt
- mutant huntingtin protein
- RFP
- red fluorescent protein.
REFERENCES
- 1. Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Sánchez I., Mahlke C., Yuan J. (2003) Nature 421, 373–379 [DOI] [PubMed] [Google Scholar]
- 3. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 4. Slow E. J., Graham R. K., Osmand A. P., Devon R. S., Lu G., Deng Y., Pearson J., Vaid K., Bissada N., Wetzel R., Leavitt B. R., Hayden M. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11402–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Legleiter J., Mitchell E., Lotz G. P., Sapp E., Ng C., DiFiglia M., Thompson L. M., Muchowski P. J. (2010) J. Biol. Chem. 285, 14777–14790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hillmer A. S., Putcha P., Levin J., Högen T., Hyman B. T., Kretzschmar H., McLean P. J., Giese A. (2010) Biochem. Biophys. Res. Commun. 391, 461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerber R., Tahiri-Alaoui A., Hore P. J., James W. (2007) J. Biol. Chem. 282, 6300–6307 [DOI] [PubMed] [Google Scholar]
- 9. Zainelli G. M., Ross C. A., Troncoso J. C., Muma N. A. (2003) J. Neuropathol. Exp. Neurol. 62, 14–24 [DOI] [PubMed] [Google Scholar]
- 10. Stadtman E. R., Levine R. L. (2006) in Redox Proteomics: From Protein Modifications to Cellular Dysfunction and Diseases (Dalle-Donne I., Scaloni A., Butterfield D. A. eds) p. 10, Wiley-Interscience, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 11. Sahara N., Maeda S., Murayama M., Suzuki T., Dohmae N., Yen S. H., Takashima A. (2007) Eur. J. Neurosci. 25, 3020–3029 [DOI] [PubMed] [Google Scholar]
- 12. Nadanaka S., Okada T., Yoshida H., Mori K. (2007) Mol. Cell. Biol. 27, 1027–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hashemy S. I., Johansson C., Berndt C., Lillig C. H., Holmgren A. (2007) J. Biol. Chem. 282, 14428–14436 [DOI] [PubMed] [Google Scholar]
- 14. Fujiwara N., Nakano M., Kato S., Yoshihara D., Ookawara T., Eguchi H., Taniguchi N., Suzuki K. (2007) J. Biol. Chem. 282, 35933–35944 [DOI] [PubMed] [Google Scholar]
- 15. Woo H. A., Jeong W., Chang T. S., Park K. J., Park S. J., Yang J. S., Rhee S. G. (2005) J. Biol. Chem. 280, 3125–3128 [DOI] [PubMed] [Google Scholar]
- 16. Jeong H., Then F., Melia T. J., Jr., Mazzulli J. R., Cui L., Savas J. N., Voisine C., Paganetti P., Tanese N., Hart A. C., Yamamoto A., Krainc D. (2009) Cell 137, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. (2006) Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 18. Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., Hayden M. R. (2006) Nat. Neurosci. 9, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson L. M., Aiken C. T., Kaltenbach L. S., Agrawal N., Illes K., Khoshnan A., Martinez-Vincente M., Arrasate M., O'Rourke J. G., Khashwji H., Lukacsovich T., Zhu Y. Z., Lau A. L., Massey A., Hayden M. R., Zeitlin S. O., Finkbeiner S., Green K. N., LaFerla F. M., Bates G., Huang L., Patterson P. H., Lo D. C., Cuervo A. M., Marsh J. L., Steffan J. S. (2009) J. Cell Biol. 187, 1083–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L. M., Trottier Y., Richards W. G., Osmand A., Paganetti P., Bates G. P. (2010) J. Biol. Chem. 285, 8808–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cumming R. C., Andon N. L., Haynes P. A., Park M., Fischer W. H., Schubert D. (2004) J. Biol. Chem. 279, 21749–21758 [DOI] [PubMed] [Google Scholar]
- 22. Furukawa Y., Torres A. S., O'Halloran T. V. (2004) EMBO J. 23, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. (2002) J. Biol. Chem. 277, 38029–38036 [DOI] [PubMed] [Google Scholar]
- 24. Browne S. E., Bowling A. C., MacGarvey U., Baik M. J., Berger S. C., Muqit M. M., Bird E. D., Beal M. F. (1997) Ann. Neurol. 41, 646–653 [DOI] [PubMed] [Google Scholar]
- 25. Browne S. E., Ferrante R. J., Beal M. F. (1999) Brain Pathol. 9, 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bogdanov M. B., Andreassen O. A., Dedeoglu A., Ferrante R. J., Beal M. F. (2001) J. Neurochem. 79, 1246–1249 [DOI] [PubMed] [Google Scholar]
- 27. Sorolla M. A., Rodríguez-Colman M. J., Tamarit J., Ortega Z., Lucas J. J., Ferrer I., Ros J., Cabiscol E. (2010) Free Radic. Biol. Med. 49, 612–621 [DOI] [PubMed] [Google Scholar]
- 28. Fox J. H., Kama J. A., Lieberman G., Chopra R., Dorsey K., Chopra V., Volitakis I., Cherny R. A., Bush A. I., Hersch S. (2007) PLoS ONE 2, e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dexter D. T., Carayon A., Javoy-Agid F., Agid Y., Wells F. R., Daniel S. E., Lees A. J., Jenner P., Marsden C. D. (1991) Brain 114, 1953–1975 [DOI] [PubMed] [Google Scholar]
- 30. Sathyasaikumar K. V., Stachowski E. K., Amori L., Guidetti P., Muchowski P. J., Schwarcz R. (2010) J. Neurochem. 113, 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Wang C. E., Orr A., Tydlacka S., Li S. H., Li X. J. (2008) J. Cell Biol. 180, 1177–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Azzu V., Mookerjee S. A., Brand M. D. (2010) Biochem. J. 426, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berezovska O., Lleo A., Herl L. D., Frosch M. P., Stern E. A., Bacskai B. J., Hyman B. T. (2005) J. Neurosci. 25, 3009–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fox J. H., Barber D. S., Singh B., Zucker B., Swindell M. K., Norflus F., Buzescu R., Chopra R., Ferrante R. J., Kazantsev A., Hersch S. M. (2004) J. Neurochem. 91, 413–422 [DOI] [PubMed] [Google Scholar]
- 35. Wanker E. E., Scherzinger E., Heiser V., Sittler A., Eickhoff H., Lehrach H. (1999) Methods Enzymol. 309, 375–386 [DOI] [PubMed] [Google Scholar]
- 36. Freedman J. H., Ciriolo M. R., Peisach J. (1989) J. Biol. Chem. 264, 5598–5605 [PubMed] [Google Scholar]
- 37. Menalled L. B., Sison J. D., Dragatsis I., Zeitlin S., Chesselet M. F. (2003) J. Comp. Neurol. 465, 11–26 [DOI] [PubMed] [Google Scholar]
- 38. De Rooij K. E., Dorsman J. C., Smoor M. A., Den Dunnen J. T., Van Ommen G. J. (1996) Hum. Mol. Genet. 5, 1093–1099 [DOI] [PubMed] [Google Scholar]
- 39. Kegel K. B., Sapp E., Yoder J., Cuiffo B., Sobin L., Kim Y. J., Qin Z. H., Hayden M. R., Aronin N., Scott D. L., Isenberg G., Goldmann W. H., DiFiglia M. (2005) J. Biol. Chem. 280, 36464–36473 [DOI] [PubMed] [Google Scholar]
- 40. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., O'Halloran T. V. (1999) Science 284, 805–808 [DOI] [PubMed] [Google Scholar]
- 41. Horng Y. C., Leary S. C., Cobine P. A., Young F. B., George G. N., Shoubridge E. A., Winge D. R. (2005) J. Biol. Chem. 280, 34113–34122 [DOI] [PubMed] [Google Scholar]
- 42. Bosco D. A., Morfini G., Karabacak N. M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B. A., Lemay N., McKenna-Yasek D., Frosch M. P., Agar J. N., Julien J. P., Brady S. T., Brown R. H., Jr. (2010) Nat. Neurosci. 13, 1396–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsika E., Moysidou M., Guo J., Cushman M., Gannon P., Sandaltzopoulos R., Giasson B. I., Krainc D., Ischiropoulos H., Mazzulli J. R. (2010) J. Neurosci. 30, 3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.