Abstract
Prader-Willi syndrome (PWS) is a multisystem disorder caused by genetic loss of function of a cluster of imprinted, paternally expressed genes. Neonatal failure to thrive in PWS is followed by childhood-onset hyperphagia and obesity among other endocrine and behavioral abnormalities. PWS is typically assumed to be caused by an unknown hypothalamic-pituitary dysfunction, but the underlying pathogenesis remains unknown. A transgenic deletion mouse model (TgPWS) has severe failure to thrive, with very low levels of plasma insulin and glucagon in fetal and neonatal life prior to and following onset of progressive hypoglycemia. In this study, we tested the hypothesis that primary deficits in pancreatic islet development or function may play a fundamental role in the TgPWS neonatal phenotype. Major pancreatic islet hormones (insulin, glucagon) were decreased in TgPWS mice, consistent with plasma levels. Immunohistochemical analysis of the pancreas demonstrated disrupted morphology of TgPWS islets, with reduced α- and β-cell mass arising from an increase in apoptosis. Furthermore, in vivo and in vitro studies show that the rate of insulin secretion is significantly impaired in TgPWS β-cells. In TgPWS pancreas, mRNA levels for genes encoding all pancreatic hormones, other secretory factors, and the ISL1 transcription factor are upregulated by either a compensatory response to plasma hormone deficiencies or a primary effect of a deleted gene. Our findings identify a cluster of imprinted genes required for the development, survival, coordinate regulation of genes encoding hormones, and secretory function of pancreatic endocrine cells, which may underlie the neonatal phenotype of the TgPWS mouse model.
Keywords: β-cell, α-cell, hypoglycemia, genomic imprinting
prader-willi syndrome (PWS) is a complex developmental and multisystem disorder characterized by endocrine, neurological, and behavioral/psychiatric abnormalities (10). The syndrome progresses through distinctive clinical stages, beginning with a neonatal period characterized by hypotonia associated with poor feeding and failure to thrive followed by a childhood/adulthood period characterized by hyperphagia and severe obesity, short stature, hypogonadism, characteristic facial appearance, and an obsessive-compulsive behavior (9, 10, 32, 56). Endocrine features of PWS include low growth hormone (GH) response in stimulation tests (9, 50), deficient gonadotropin secretion (8, 9, 25), low insulin-like growth factor (IGF)-I (9, 50), and low IGF-binding protein-3 (23). Plasma levels of insulin are lower than expected in children and adults with PWS relative to the degree of obesity and are generally assumed to be secondary to a deficient GH-IGF axis (23, 25, 27, 39, 49, 54, 61). In addition, levels of circulating pancreatic polypeptide (PP) in response to meals are reduced in PWS (63, 74, 75). In contrast, fasting levels of plasma ghrelin are grossly elevated in patients with PWS (15, 18) possibly because of relative hypoinsulinemia and preserved insulin sensitivity (27). Most of the hormonal, metabolic, and behavioral deficiencies in PWS are believed to be caused by an unknown hypothalamic-pituitary dysfunction (9, 10, 44), but the underlying pathophysiological basis of the clinical features in PWS is unknown.
The most frequent genetic mechanism that underlies PWS in about two-thirds of cases is de novo deletion of a 5- to 7-Mb region in chromosome 15q11–q13 (10, 48). The deletion results in loss of function of a cluster of at least 12 imprinted, paternally expressed genes that encode six proteins, including an RNA-splicing factor (SmN) and two MAGE family proteins (NECDIN and MAGEL2), as well as non-protein-coding RNAs, including five classes of box C/D small nucleolar RNAs (snoRNAs) predicted to regulate the level, splicing, or modification of other RNAs (37, 38, 45, 48). Two other genetic mechanisms, maternal uniparental disomy or an imprinting defect generating a maternal imprint on the paternally inherited chromosome, also lead to loss of function of the entire imprinted gene cluster and result in the classical PWS phenotype (10, 48). Several smaller deletions of snoRNA genes occur in PWS-like subjects and have been suggested to pinpoint the SNORD116 snoRNAs as key genes (17, 53); however, at least four considerations suggest that the molecular etiology of PWS may be a contiguous gene disorder. First, deletions limited to the orthologous Snord116 snoRNA genes in mouse have only a mild phenotype (20, 55). Second, more proximal deletions in human, including NDN and MAGEL2, generate a subset of clinical features of PWS (35). Third, the majority of PWS imprinted genes are coordinately regulated and predominantly or exclusively expressed in neurons (Refs. 12, 41, 48, and 70 and Stefan M and Nicholls RD, unpublished data). Fourth, in the smaller PWS-like deletions (17, 53), the effect on gene expression in neurons is not known. Thus, the contribution of the individual loci to the clinical features of the syndrome is not known, but based on their proposed functions (37, 38, 45, 48), PWS region imprinted genes are likely to regulate genetic and molecular pathways that are dysregulated in PWS.
In a transgenic deletion mouse model of PWS (TgPWS) that recapitulates the most common genetic basis (an ∼5- to 7-Mb deletion) and the neonatal stage of the human disease, we found extremely low levels of plasma insulin and glucagon in both fetal and postnatal life (58), suggesting a defect affecting islet cell development and/or function. We demonstrate here that two distinct mechanisms, increased apoptosis associated with reduced α- and β-cell mass and deficient insulin secretion, contribute to the severe reductions in plasma glucagon and insulin in TgPWS mice. Furthermore, our data also demonstrate abnormal somatostatin and PP regulation and identify a more global deficit in regulation of secretory pathways. Because the alterations in survival and function of pancreatic endocrine cells in fetal and newborn TgPWS mice precede the postnatal endocrine and metabolic abnormalities associated with failure to thrive, these findings indentify a primary mechanism. Therefore, loss of the PWS homologous imprinted genes in a mouse model leads to deregulation of molecular pathways critical for development and function of multiple islet endocrine cell types.
EXPERIMENTAL PROCEDURES
Mouse models.
The TgPWS mouse model deletion (Fig. 1A), equivalent to deletion of the homologous genes in human chromosome 15q11–q13 in most PWS and Angelman syndrome (AS) patients (48), was generated by insertion of a transgene array into mouse chromosome 7C (24, 57). The transgenic line is maintained by maternal transmission (generating TgAS mice), with the TgPWS mouse phenotype resulting after paternal transmission only; in each litter, half of the offspring are wild-type (WT) controls. Several studies characterizing the TgPWS mice were completed on the CD-1 strain background (Refs. 24, 57, 58, and 59 and the present study), but in early 2007, as part of an institutional policy, we rederived the line by embryo transfer to the Institute of Cancer Research (ICR) strain background (Taconic, Germantown, NY). CD-1 and ICR derived originally from the same line of Swiss mice (14). Subsequent breeding has occurred exclusively with ICR, and all studies were done on these mice, except where noted. Maintenance of the TgAS line was conducted at Taconic, whereas breeding to generate TgPWS litters was done at the Children's Hospital of Pittsburgh. Since hypoglycemia in TgPWS mice begins at postnatal day (P)2 and rapidly progresses (58), all studies in this report were performed on normoglycemic animals at embryonic day 17.5 (E17.5) or P1.
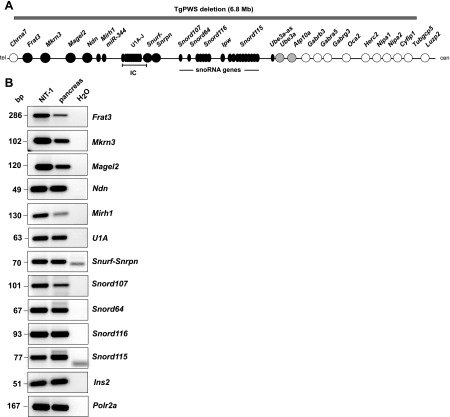
Fig. 1.
The Prader-Willi syndrome (PWS) homologous gene region in mouse is functional in the pancreas. A: genetic map and transgenic deletion mouse model of PWS (TgPWS) deletion in mouse chromosome 7C. Circles, protein-coding genes; ovals, RNA-coding genes or transcripts (Ipw and Ube3a-as have no known coding potential; not all copies of miR-344, Snord116, and Snord115 are shown); black circles and ovals, paternally expressed; gray circles, maternally expressed; white circles, biparentally expressed; IC, imprinting center; cen, centromere; tel, telomere. Gray bar indicates 6.8-Mb deletion in TgPWS mice. B: mRNA expression of imprinted, paternally expressed genes from the TgPWS deletion region in mouse pancreas and NIT-1 insulinoma cells. RT-PCR was used to assess mRNA expression, with insulin 2 (Ins2) and Polr2a used as controls. H2O controls had no RNA.
The generation of Ins-C-Timer transgenic mice that express proinsulin II with the C-peptide tagged with a fluorescent protein, Timer, has been described (5, 69). Double transgenic Ins-C-Timer;TgPWS mice were obtained by breeding TgAS males carrying the deletion on the maternal homolog with females homozygous for the Ins-C-Timer transgene. The offspring include 50% each of Ins-C-Timer;TgPWS mice and Ins-C-Timer control mice. All animals were genotyped by PCR, as described (5, 24). Approval for all experiments was obtained from the Animal Research Care Committee at the Children's Hospital of Pittsburgh of UPMC.
Glucose and hormone measurements.
Blood samples from mice at P1 were taken between 1000 and 1400. Blood glucose was determined using the FreeStyle Flash System (Abbott Diabetes Care, Alameda, CA). For hormone measurements, blood was collected from the heart cavity into capillary tubes containing EDTA, centrifuged at 4,000 rpm for 10 min at 4°C, plasma collected, and stored at −80°C. At least two litters were used for measurement of each hormone, with samples from three TgPWS or WT animals pooled for glucagon and somatostatin measurements. Pancreatic hormone contents were assayed in acid-ethanol extracts of the whole pancreas. Hormones were measured (RIA/Biomarkers Core of the Diabetes and Endocrinology Research Center, University of Pennsylvania, Philadelphia, PA) using an ultrasensitive rat insulin ELISA kit (Alpco Diagnostics, Windham, NH), a rat C-peptide RIA kit (Linco Research, St. Charles, MO), a glucagon RIA kit (Linco Research), and a rat Somatostatin-14 EIA kit (Bachem, Torrance, CA).
Islet immunohistochemical studies.
Pancreata from four TgPWS and four WT mice (CD-1) were harvested, fixed in 4% paraformaldehyde overnight at 4°C, embedded in paraffin, and cut into 6-μm sections. For staining, sections were used at regular space intervals (i.e., 1 section in 20 consecutive sections). Dewaxed, rehydrated sections were boiled in 10 mM citric acid buffer (pH 6.0) for 6 min, cooled to room temperature (RT), washed in PBS, treated with protein-blocking agent (Coulter-Immunotech, Miami, FL) for 20 min at RT, and incubated overnight at 4°C with primary antibodies (Ab). Slides were washed in PBS, incubated with secondary Ab for 1 h at RT, washed in PBS, mounted, and examined using a Nikon Microphot-FX fluorescent microscope. The following Ab were used at the indicated dilutions in PBT (PBS + 0.1% BSA + 0.2% Triton X-100): guinea pig anti-insulin (1:400; Dako, Carpinteria, CA), rabbit anti-glucagon (1:5,000; Biodesign, Saco, ME), rabbit anti-somatostatin (1:5,000; Bachem), rabbit anti-PP (1:50; Invitrogen, Carlsband, CA), Cy2-conjugated donkey anti-guinea pig IgG (1:200; Jackson Immunoresearch Laboratories, West Grove, PA), and Cy3-conjugated donkey anti-rabbit IgG (1:200; Vector Laboratories, Burlingame, CA). For measurement of β-cell area, the sections stained for insulin and glucagon were counterstained with hematoxylin, and immunopositive β-cells were counted on eight WT and 11 TgPWS sections from different mice, followed by measurement of the surface area using IP Lab Spectrum software (BD Bioscience, Rockville, MD).
For measurement of α-cell area, pancreata from three TgPWS and three WT mice were fixed in 4% paraformaldehyde, washed in PBS, and frozen embedded in Neg-50 Frozen Section Medium (Thermo Fisher Scientific, Pittsburgh, PA) for cryostat sectioning. The slides were washed with PBS, blocked with 10% fetal calf serum in PBT for 30 min at RT, and incubated with guinea pig anti-insulin (1:1,000; Linco Research) and rabbit anti-glucagon (1:1,000; Linco Research) Ab, as described above. Slides were then treated with Cy2-conjugated goat anti-guinea pig IgG and Cy3-conjugated goat anti-rabbit IgG (1:300; Jackson Laboratories) secondary Ab and examined using a Zeiss AxioImager.Z1 microscope. The α-cell-immunopositive area was estimated in seven WT and 10 TgPWS sections counterstained with hematoxylin using AxioVision Rel. 4.7 software.
For proliferation studies, a pregnant female mouse was injected intraperitoneally with 200 μg/g BrdU (Sigma-Aldrich, St. Louis, MO) 2 h before euthanization. Embryos were harvested and pancreata fixed in paraformaldehyde with frozen embedded sections processed as described above. The primary Ab were rat anti-BrdU (1:250; Novus Biological, Littleton, CO) and guinea pig anti-insulin (1:500; Linco Research), and the secondary Ab were Cy2-conjugated goat anti-guinea pig IgG and Cy3-conjugated goat anti-rat IgG. Proliferation of β-cells was expressed as the percentage of insulin-stained BrdU-positive cells to total insulin-stained β-cells in serial pancreata sections from three TgPWS and three WT embryos. Seven to 10 sections were analyzed for each animal, and total numbers of 288, 231, and 580 insulin-positive cells were counted for TgPWS and 275, 160, and 295 each for WT, respectively. For apoptosis studies, pancreas sections from embryos and mice were immunostained for cleaved caspase-3 and counterstained for insulin or glucagon, as described above. For assessment of apoptotic β- and α-cells, 19 and 23 sections from TgPWS as well as 12 and 11 sections from WT pancreas at P1 were analyzed with a total of 453 (TgPWS) and 123 (WT) insulin-positive cells as well as a total of 253 (TgPWS) and 123 (WT) glucagon-positive cells, respectively. Additional Ab were rabbit anti-cleaved caspase-3 (1:100; Cell Signaling Technology, Danvers, MA), guinea pig anti-glucagon (1:1,000; Linco Research), goat anti-Pdx1 (1:250; Abcam, Cambridge, MA), and Cy2-conjugated goat anti-guinea pig or Cy3-conjugated goat anti-rabbit IgG or Cy2-conjugated Streptavidin (each 1:300; Jackson Laboratories).
Insulin secretion.
For in vivo studies of the age of insulin stored in β-cells, pancreata from Ins-C-Timer;TgPWS and Ins-C-Timer controls at P1 were harvested and fixed in paraformaldehyde, and frozen sections were prepared as described above. Slides were analyzed using a Nikon Eclipse E800 microscope equipped with a Spot digital camera and Northern Eclipse software. The same exposure parameters and times were used for imaging islets from each genotype.
For quantitative in vitro studies, pancreata from four TgPWS and four WT littermates at P1 were pooled and islets isolated as described (4) with the following modifications. Pancreata were excised and incubated in a collagenase solution at 37°C for 20 min, disrupted by manual shaking, and washed in Hanks' buffer. The islets were purified using a discontinuous Ficoll gradient, hand picked (approximately the same-sized islets were used for TgPWS and WT mice to minimize the degree of reduced β-cell mass of many TgPWS islets), and cultured overnight in RPMI containing 5 mM glucose, and then groups of 10 islets were incubated in 100 μl Krebs-Ringer medium containing 2 mM glucose for 1 h and subsequently in 20 mM glucose for 1 h. The medium was collected following each incubation and stored at −80°C, and insulin concentration was assessed as described above. The insulin content of the remaining islets was analyzed by acid-ethanol extraction.
Microarray expression profiling and data analysis.
A direct comparison between four TgPWS and four WT (CD-1) mice at P1 was performed. Total pancreatic RNA was extracted using the RNeasy Mini Kit (Qiagen) and treated with RQ1 RNase-free DNase (Promega). RNA amplification, reverse transcription, cDNA labeling, and hybridization to the mouse PancChip version 5.0 using a Cy3-Cy5 dye swap protocol (34, 71) were carried out by the Functional Genomics Core (University of Pennsylvania). To identify the set of genes significantly differentially expressed by microarray in the TgPWS and control groups, we applied two different statistical methods, PaGE (Patterns from Gene Expression) and t-test, to maximize identification of real differentially expressed genes and to minimize false-positive results. First, we used PaGE (version 5.1) to perform one-class unpaired analysis with a false discovery rate ≤20% (43). In addition, a paired t-test that uses a conventional t-test to provide the probability (P) that a difference in gene expression occurred by chance was performed. To maximize the sensitivity of the analysis, we combined both PaGE analysis (false discovery rate ≤20%) and paired t-test (P ≤ 0.05) to identify differentially expressed genes of ≥1.5-fold between TgPWS and WT pancreas. Association with gene ontology annotation data was performed using the Expression Analysis Systematic Explorer software package (http://david.abcc.ncifcrf.gov/content.jsp?file=ease/ease.htm&type=1; National Institute of Allergy and Infectious Diseases, National Institutes of Health, Frederick, MD).
Standard and quantitative RT-PCR.
cDNA was synthesized using random hexamers with the SuperScript first-strand synthesis system (Invitrogen), and either standard RT-PCR (PCR primers are in Supplemental Table S1; Supplemental Material for this article is available online at the AJP-Endocrinology and Metabolism website) or quantitative (q)RT-PCR was performed as described (59), except that the RT for snoRNAs was performed with each specific reverse primer. For qRT-PCR, primers were Ins2: 5′-GGTGGCCCGGCAGAAG-3′ [forward (F)], 5′-GCTGGTAGAGGGAGCAAATGC-3′ [reverse (R)]; Gcg: 5′-CACCAAGAGGAACCGGAACA-3′ (F), 5′-TCAGCATGCCTCTCAAATTCA-3′ (R); Sst: 5′-CCCAGACTCCGTCAGTTTCTG-3′ (F), 5′-CTTGGCCAGTTCCTGTTTCC-3′ (R); Ppy: 5′-TCAGCTCCGCAGATACATCAA-3′ (F), 5′-TCCTCGGCTCTCTTCCCATA-3′ (R); Iapp: 5′-GCTGCCTCGGACCACTGA-3′ (F), 5′-ACAGCTGGCAGTTTGGAGATG-3′ (R); Tmem27: 5′-AATTGCACTACTGGTTCTATCTGGAA-3′ (F), 5′-CCAGGTGGTCCTTTGTTGTTC-3′ (R); 7B2 (Sgne1): 5′-GCGGCCACACGGTTAAAA-3′ (F), 5′-AAGGCCAGATAGCATAGCAGAGA-3′ (R); Isl1: 5′-GGAGACATGGGCGATCCA-3′ (F), 5′-ATTGCCGCAACCAACACA-3′ (R); and Gapdh: 5′-CTCCACTCACGGCAAATTCA-3′ (F), 5′-ATGGGCTTCCCGTTGATGA-3′ (R). The ΔΔCT method was used to calculate the fold changes, and the qRT-PCR data for WT and TgPWS were expressed as means of fold changes (± SE). Statistical significance between the two groups was carried out using an independent t-test (Analyze-it, Leeds, UK).
RESULTS
TgPWS mouse model.
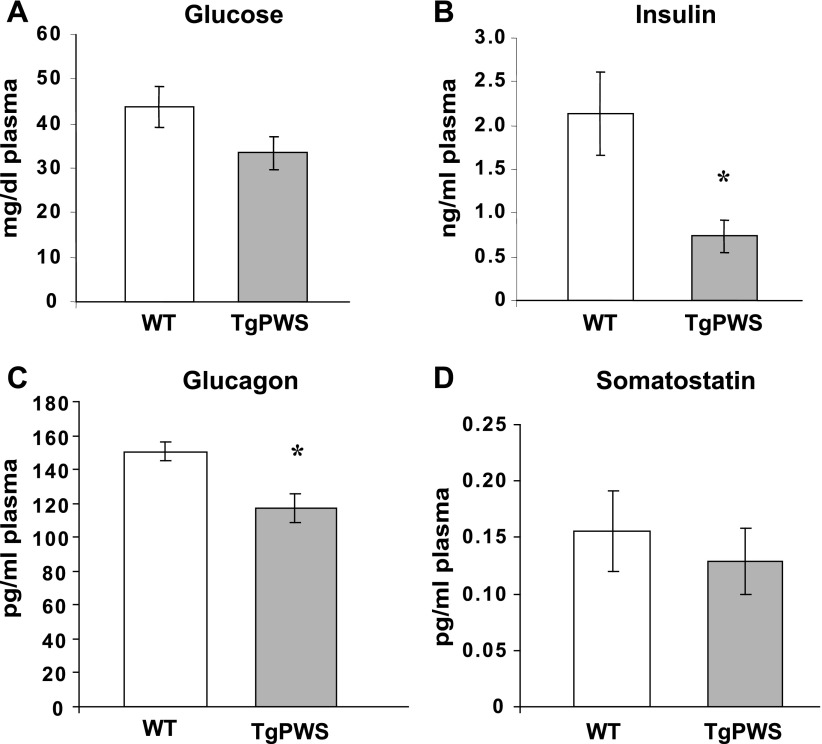
Paternal transmission of the TgPWS deletion (Fig. 1A) leads to fetal growth retardation, neonatal failure to thrive, and postnatal death within 4–7 days after birth (58). These mice also had decreased plasma levels of insulin and glucagon in fetal and postnatal life (58). Following rederivation to the ICR outbred strain (see experimental procedures), we confirmed that TgPWS (ICR) mice have an equivalent phenotype with reduced birth weights, postnatal failure to thrive, and lethality between P4 and P7 (data not shown). Endocrine abnormalities are also identical to those observed in TgPWS (CD-1) mice (58). First, there was no difference in blood glucose levels between TgPWS and WT littermates at P1 (Fig. 2A). Second, plasma insulin levels are ∼66% lower in TgPWS mice (P = 0.02) compared with their WT littermates (Fig. 2B). Third, plasma glucagon levels in TgPWS mice are ∼22% lower than in WT mice at P1 (P = 0.03; Fig. 2C). We also measured somatostatin concentrations and found that there was no difference in plasma levels between TgPWS and WT pups at P1 (Fig. 2D). We conclude that the overall phenotype is indistinguishable for TgPWS (CD-1) (58) and TgPWS (ICR) mice.
Fig. 2.
Blood glucose and plasma insulin, glucagon, and somatostatin levels in TgPWS compared with wild-type (WT) mice at postnatal day 1 (P1). A: blood glucose levels in TgPWS (n = 10) and WT littermates (n = 8). B: plasma insulin levels in TgPWS (n = 12) and WT controls (n = 17). C: glucagon levels in pooled plasma samples from TgPWS (n = 12) and WT littermates (n = 9). D: somatostatin levels in pooled plasma samples from TgPWS (n = 12) and WT littermates (n = 15). All dams were fed ad libitum. Data are presented as means ± SE. *P < 0.05, significant differences between TgPWS and WT mice (independent t-test).
The TgPWS deletion includes at least 11 paternally expressed genes, with six that encode polypeptides, one locus encoding microRNAs, and at least four sets of snoRNAs (Fig. 1A). The U1-Snurf-Snrpn-snoRNA locus is polycistronic, with at least 10 alternative functional promoters (Fig. 1A) (Refs. 10 and 48 and Stefan M and Nicholls RD, unpublished data). All these paternally expressed genes have been described as having predominant or exclusive expression in neurons (12, 16, 28, 41, 48, 70). We first determined whether or not these genes were expressed in the pancreas and in NIT-1 cells, a mouse insulinoma β-cell line. All the mouse imprinted genes homologous to the PWS domain are robustly expressed in pancreas and in NIT-1 cells (Fig. 1B). Although not a quantitative assessment, the levels detected by the same number of PCR amplification cycles for many of these genes are similar to those detected for Ins2, the major insulin gene (Fig. 1B).
Hormone content in TgPWS pancreas.
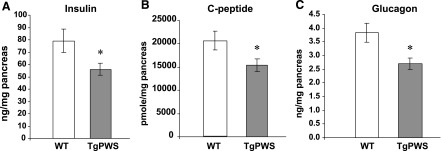
Based on the deficiencies in plasma insulin and glucagon in TgPWS mice, we next assessed pancreatic hormone levels. Total pancreatic insulin (Fig. 3A) and C-peptide (Fig. 3B) content normalized to pancreatic weight at P1 was 25–30% decreased in TgPWS compared with WT littermates (P = 0.037 for insulin and P = 0.042 for C-peptide). Similarly, glucagon content was decreased by the same extent in TgPWS islets (P = 0.009; Fig. 3C). The C-peptide/insulin content ratio was unchanged (P = 0.69) in TgPWS compared with control mice, suggesting a normal cleavage of proinsulin in TgPWS β-cells. Somatostatin content relative to total pancreatic weight did not differ between TgPWS and controls at P1 (P = 0.09; data not shown). There was no significant difference in the weight of total pancreas between TgPWS and WT mice at P1 (TgPWS: n = 22, 9.8 ± 0.7 mg; WT: n = 20, 9.9 ± 0.9 mg; P = 0.9).
Fig. 3.
Pancreatic content of insulin, C-peptide, and glucagon in TgPWS mice. Insulin (A), C-peptide (B), and glucagon (C) concentrations were estimated in total pancreas from TgPWS (n = 11) and control mice (n = 10) at P1 after acid-ethanol extraction. Results are presented as a means ± SE. *P < 0.05, significant differences between TgPWS and WT mice (independent t-test).
Abnormal islet morphology, reduced α- and β-cell mass, and increased α- and β-cell apoptosis in TgPWS pancreas.
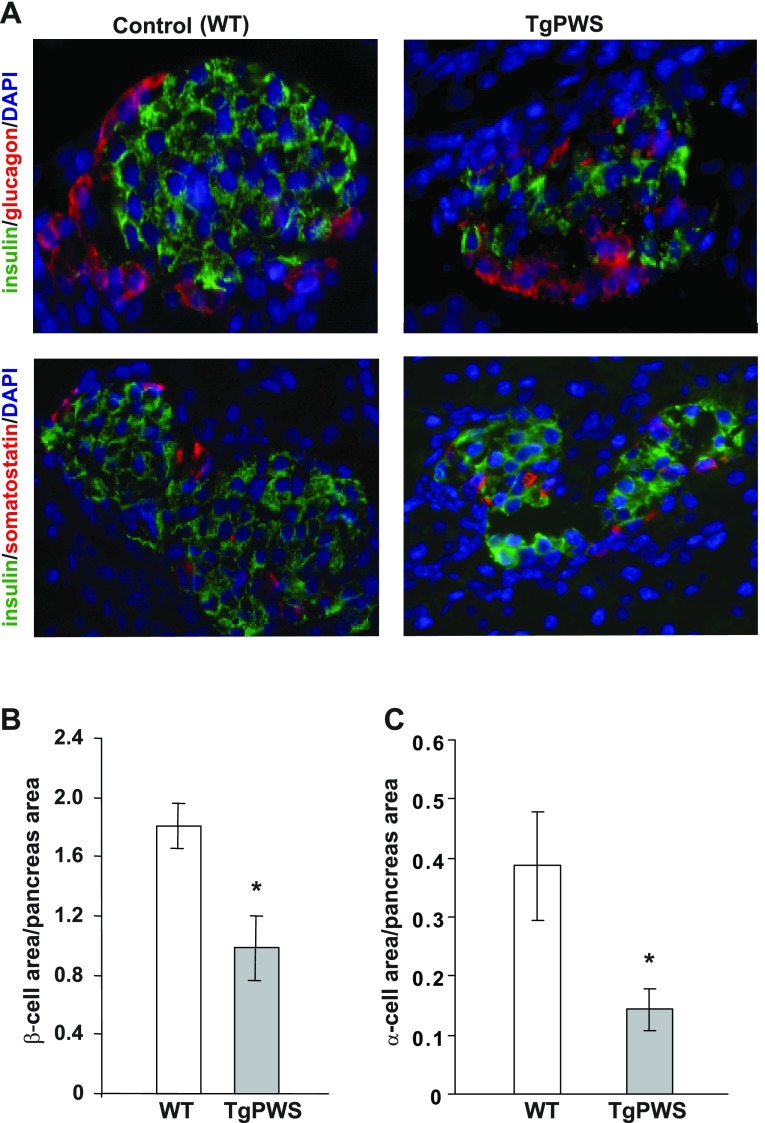
Pancreatic islet morphology was assessed by using immunostaining with markers for β-, α-, δ-, and PP cells. All four markers of islet cell lineages were identified in TgPWS pancreas (Fig. 4A and and data not shown). Nevertheless, many of the mutant islets showed a disturbed morphology with irregular shape and abnormal cellular arrangement, with α- and δ-cells present in the core of the islets (Fig. 4A). Islet cell composition was further investigated by quantitative analysis of β- and α-cell mass. The β- and α-cell areas normalized to total pancreatic area were reduced by ∼45 (P = 0.01) and ∼60% (P = 0.01), respectively, in TgPWS compared with WT mice at P1 (Fig. 4, B and C).
Fig. 4.
Altered islet morphology with reduced number of β- and α-cells in TgPWS pancreas compared with controls at P1. A: islets from TgPWS and WT pancreata were stained using antibodies against insulin (green) and glucagon or somatostatin (red) to detect β-, α-, and δ-cells, respectively. Cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; blue). B and C: β- and α-cell mass, respectively, quantified in pancreata from TgPWS and WT mice. Values represent percentage ± SE of β- and α-cell area normalized to the total tissue area of counted sections (n = 11 or 10 for TgPWS; n = 8 or 7 for WT for β- and α-cell area, respectively). *P < 0.05, significant differences between TgPWS and WT mice (independent t-test).
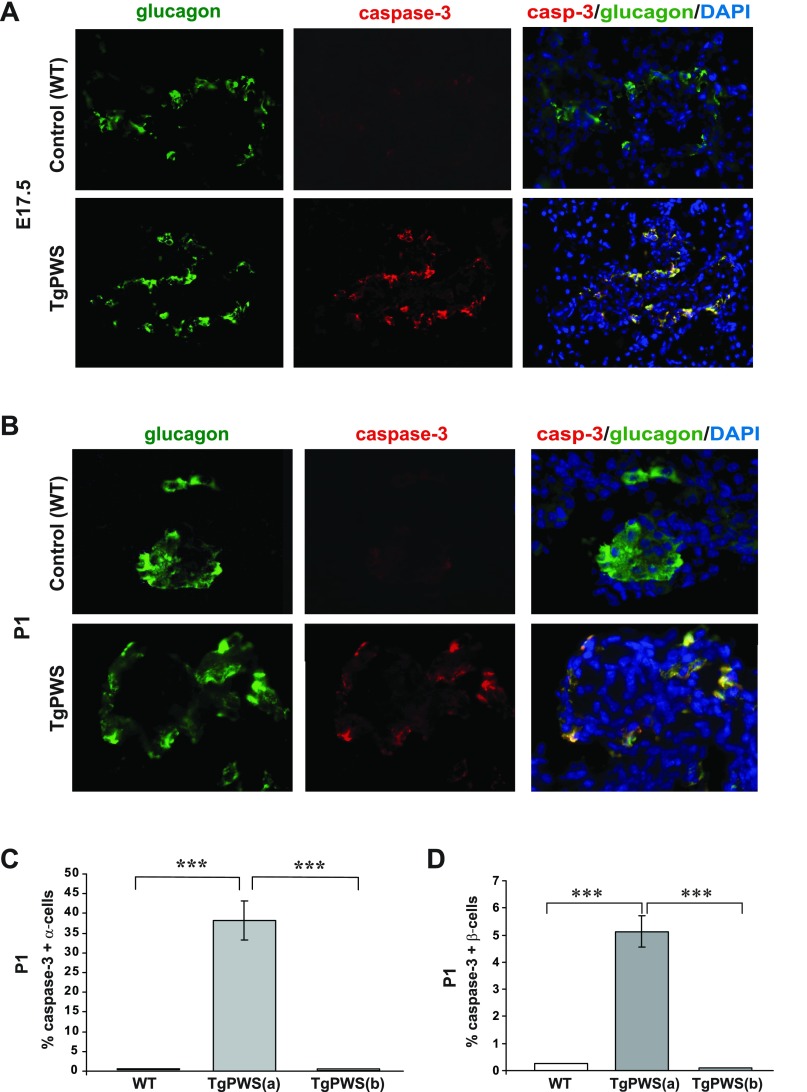
Apoptosis was examined in pancreas sections from TgPWS and WT mice at E17.5 and P1 by dual staining for activated caspase-3 and glucagon (α-cells) or insulin (β-cells). At these ages, apoptosis of α-cells was more pronounced than for β-cells in the TgPWS pancreas. Whereas essentially no apoptotic α-cells were identified in the control islets, numerous caspase-3 positive α-cells were present in TgPWS islets at E17.5 (Fig. 5A) and P1 (Fig. 5B). However, we found a range of islets in the TgPWS pancreas, with two-thirds of islets having numerous glucagon-producing cells showing evidence of apoptosis, whereas one-third of islets had few or no apoptotic α-cells (Fig. 5C). In the former group, at P1, the proportion of caspase-3-positive α-cells/islet averaged ∼38%. Similarly, in the β-cell analysis we found that one-half of the TgPWS islets had no evidence of apoptosis, whereas one-half of the islets had a small percentage of doubly stained cells for caspase-3 and insulin (Fig. 5D and Supplemental Fig. S1A). To assess β-cell replication, we performed dual staining for insulin and incorporated BrdU on pancreatic sections from mice at E17.5. At this age, there was no difference in the replication rate of TgPWS compared with WT β-cells (Supplemental Fig. S1, B and C). We conclude that the decreased number of α-cells and β-cells in TgPWS pancreas is due to increased apoptosis.
Fig. 5.
Numerous α-cells and some β-cells undergo apoptosis in TgPWS pancreas at embryonic day 17.5 (E17.5) and P1. A and B: assessment of α-cell apoptosis in TgPWS compared with WT pancreas at E17.5 and P1, respectively, by immunostaining of activated caspase-3 (red) as a marker for apoptosis and glucagon (green) as the marker for α-cells. Cell nuclei were stained with DAPI (blue). A FITC (480/440 nm excitation) filter was used to take images for glucagon, and a tetramethylrhodamine isothiocyanate (TRITC; 535/530 nm excitation) filter was used to take images for caspase-3 immunostained cells. In the merged images, apoptotic α-cells that coexpress glucagon and caspase-3 appear yellow. C: quantification of apoptotic α-cells at P1 presented as a mean percentage of caspase-3-positive α-cells in TgPWS vs. WT islets. Islets in TgPWS were of 2 types, having an average of 38% apoptotic α-cells [17 islets in TgPWS(a)] or no apoptotic cells [8 islets in TgPWS(b)], whereas 11 islets were assessed for WT. D: quantification of apoptotic β-cells at P1 presented as mean percentage of caspase-3-positive β-cells in TgPWS vs. WT islets. Examples of immunostained TgPWS and WT pancreatic sections at P1 and E17.5 are in Supplemental Fig. S1. Islets in TgPWS pancreas at P1 were of 2 types, with 9 islets having an average of 5% apoptotic β-cells [TgPWS(a)] and 9 islets having no apoptotic cells [TgPWS(b)], whereas 12 islets were assessed for WT. ***P < 0.0001, significant differences between TgPWS(a) and WT or TgPWS(b) pancreas (independent t-test).
TgPWS islets have impaired insulin secretion.
Plasma insulin levels were significantly decreased in TgPWS mice (Fig. 2B) (58), and this could not be explained solely by the loss of β-cell mass (Fig. 4B) or the moderate decrease in pancreatic insulin content (Fig. 3A). These observations suggest that functional impairment of β-cells in the TgPWS pancreas might contribute to hypoinsulinemia. However, the small size of TgPWS fetuses and newborns, due to growth retardation and lethality, precludes the performance of glucose tolerance tests or the measurement of plasma insulin levels at multiple time points following glucose challenge in vivo. Therefore, we assessed two different parameters of insulin secretion: 1) the age of insulin stored in β-cells in vivo using a unique fluorescent reporter and 2) the amount of insulin secreted in vitro in isolated islets.
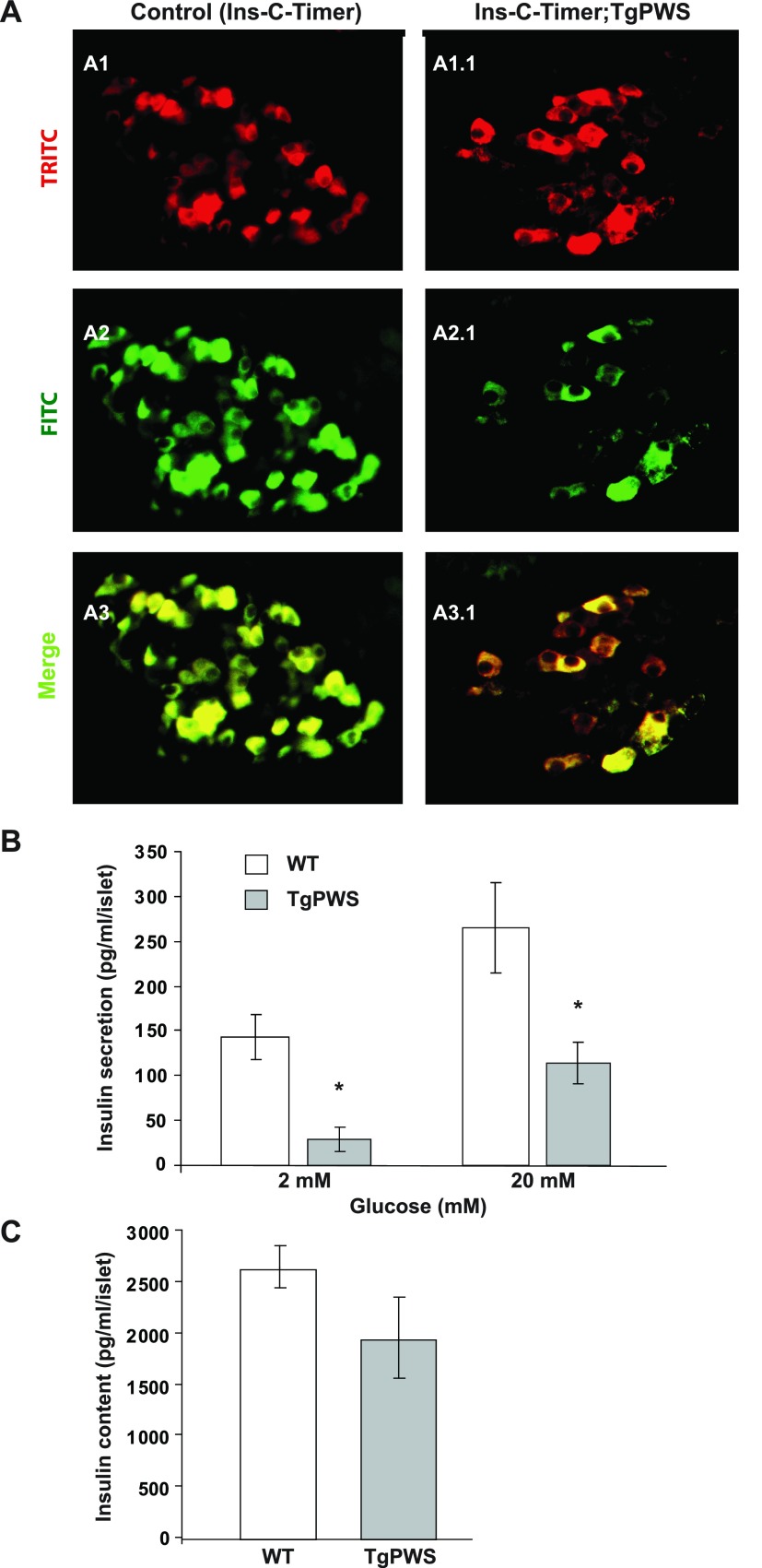
To examine the dynamics of insulin secretion in vivo, we analyzed the expression of an insulin-Timer fusion protein, Ins-C-Timer (5) in pancreas sections from TgPWS, and control mice at P1. The Ins-C-Timer reporter is unique because it changes color from green to red over 24 h after synthesis, allowing monitoring of the time taken for insulin secretion since synthesis (5); importantly, this reporter does not provide an assessment of the amount of insulin produced or secreted. In a similar manner, an Ngn3-Timer transgene was recently used to monitor temporal patterns of gene expression in pancreatic endocrine progenitor vs. differentiating cells (46), and Timer fluorescent reporters have been used to monitor exocytosis of secretory vesicles over time in neuroendocrine cells (22, 65). In our study, we found predominantly green fluorescence in control Ins-C-Timer pancreas sections (Fig. 6A), indicative of newly synthesized insulin. In contrast, TgPWS islets showed predominantly red fluorescence representing intracellular storage of insulin for a longer period of time. We conclude that newly synthesized insulin is secreted relatively rapidly in Ins-C-Timer control transgenic mice, whereas in the islets of Ins-C-Timer;TgPWS double transgenic mice, insulin is synthesized but secretion is impaired, and red fluorescence accumulates.
Fig. 6.
Altered insulin secretory dynamics in TgPWS islets. A: expression of an Ins-C-Timer reporter was analyzed as a function of age in pancreas sections from TgPWS and control littermates at P1. As is also seen in Fig. 4B, the TgPWS islet has a reduced number of β-cells. The Ins-C-Timer reporter changes color from green to red as time passes after insulin synthesis (5, 22, 46, 65). A TRITC (535/530 nm excitation) filter was used to take images from A1 and A1.1, and a FITC (480/440 nm excitation) filter was used to take images from A2 and A2.1. A3 and A3.1 represent the merged A1-A2 and A1.1-A2.1 images that report on the time since synthesis of insulin. In the control Ins-C-Timer mice, β-cells show predominantly green (newly synthesized) or yellow (intermediate age) fluorescence (A3). β-Cells from TgPWS mice show mostly red (old) and yellow (intermediate) fluorescence, indicating an age-dependent delay in insulin secretion (A1.1–A3.1). B: the amount of insulin secreted was analyzed quantitatively in isolated pooled islets from 5 TgPWS and 5 WT littermates at P1 per experiment in the presence of 2 and 20 mM glucose. C: insulin content was estimated in 20 mM glucose-stimulated islets after acid-ethanol extraction. Results in B and C are means ± SE from 3 independent experiments. *P < 0.05, significant differences between TgPWS and WT mice (independent t-test).
To obtain a quantitative assessment of insulin secretion in TgPWS mice, insulin secretion was measured by static incubations of islets isolated from TgPWS and WT littermates at P1 under basal (2 mM glucose) and glucose-stimulated (20 mM) conditions. The overall amount of insulin secreted in response to 2 and 20 mM glucose was markedly decreased in isolated islets from TgPWS mice (2 mM: WT = 144 ± 25 pg·ml−1·islet−1, TgPWS = 29 ± 13 pg·ml−1·islet−1, P = 0.01; 20 mM: WT = 265 ± 50 pg·ml−1·islet−1, TgPWS = 115 ± 23 pg·ml−1·islet−1, P = 0.05; Fig. 6B). The insulin content per islet was not significantly different for TgPWS and WT islets (WT = 2,640 ± 210 pg·ml−1·islet−1, TgPWS = 1,950 ± 400 pg·ml−1·islet−1, P = 0.2; Fig. 6C). We also calculated fractional insulin release (insulin secretion normalized to insulin content). Under basal (2 mM glucose) conditions, WT islets have 5.4% of insulin content released/hour, whereas the rate for TgPWS islets is 1.5% insulin content released/hour (3.6-fold lower). Glucose-stimulated secretion (20 mM) is 10 and 5.9% insulin content released/hour for WT and TgPWS, respectively (a 1.7-fold decrease in TgPWS). We conclude that sufficient insulin is produced but that both basal and stimulatory insulin secretion are defective in the TgPWS islet.
Global patterns of gene expression in TgPWS vs. WT pancreas.
To compare the genome-wide patterns of steady-state mRNA levels in the pancreas of TgPWS and WT mice at P1, when blood glucose levels of TgPWS pups are normal (Fig. 2A) (58), we used microarrays and required two different statistical significance levels to be met (see experimental procedures). By these criteria, 65 genes were differentially expressed ≥1.5-fold in TgPWS compared with the WT pancreas (Supplemental Tables S2 and S3), with the majority (62 genes) upregulated and only three genes downregulated. The latter included Ndn, an imprinted gene that maps to the TgPWS deletion (Fig. 1A) and hence, is not detectably expressed in TgPWS tissues (59). For the upregulated genes, the fold changes ranged between 3.3- and 1.5-fold (not adjusted for the loss of islet-mass in TgPWS pancreas). To interpret the functional implications of differentially expressed genes, six non-mutually exclusive gene ontology functional groups were found to be enriched for genes upregulated in TgPWS pancreas, with the extracellular region and hormone activity gene groups scoring best (Table 1). Indeed, most upregulated genes with well-defined functions encode hormones secreted from the pancreas or other factors involved in the secretion process.
Table 1.
Overrepresented EASE gene ontology groups for mRNAs upregulated in the TgPWS pancreas
| Category | Gene Groups | Genes | %Total Genes | P Value |
|---|---|---|---|---|
| GOTERM_CC_ALL | Extracellular region | Timp2, Iapp, Sst, Ctrl, Igf2, Wfdc15b, Lamc1, Ppy, Ngfr, Igfbp6, S100a11, Col4a1, Col1a1, Scg5, Igf1r, Fbn1, Pmp22, Ins1, Ins2, Tor2a, Tmem27, Ctsf, Pyy | 37.1 | 0.00000035 |
| GOTERM_MF_ALL | Hormone activity | Iapp, Sst, Igf2, Ins1, Ins2, Ppy, Pyy | 11.3 | 0.0000029 |
| GOTERM_MF_ALL | Receptor binding | Timp2, Iapp, Sst, S100a11, Penk1, Igf2, Ins1, Ppy, Pyy, Ins2 | 16.1 | 0.00018 |
| GOTERM_CC_ALL | Extracellular matrix part | Timp2, Col1a1, Col4a1, Fbn1, Lamc1 | 8.1 | 0.00025 |
| GOTERM_BP_ALL | Cell migration | Sst, Isl1, Rras2, Rtn4, Lamc1, Ngfr | 9.7 | 0.002 |
| GOTERM_BP_ALL | Anatomical structure morphogenesis | Igfbp6, Isl1, Igf1r, Igf2, Ihpk2, Pmp22, Rtn4, Rhoj, Ndst1, Ngfr | 16.1 | 0.008 |
EASE, Expression Analysis Systematic Explorer; TgPWS, transgenic deletion mouse model of Prader-Willi syndrome. The table shows gene groups with a P value <0.01 having common cellular, metabolic, and biological processes and found to be overrepresented among the transcripts with fold changes ≥1.5-fold that were called significantly altered by both Patterns from Gene Expression and t-test. %Total genes represents the fraction of the entire set of 62 upregulated genes in TgPWS vs. wild-type pancreas. Four genes with significantly changed expression (Amotl1, Nfib, Neat1, and Camk2n1) are not included in any of the gene groups generated by EASE. Functionally related groups that include identical gene members in this analysis are represented only by the group with the most significant P value. Some genes may be represented in several groups.
Because of our stringent statistical criteria, some genes with altered expression may have been excluded from our list. For example (see Supplemental Table S3), similar to Ndn, Snurf-Snrpn is an imprinted gene in the PWS-homologous deletion and was detected as downregulated in TgPWS (1.55-fold; PaGE = 0.77, P = 0.0012). Similarly, Ube3a also maps to the TgPWS/TgAS deletion but is nonimprinted in the pancreas and, as expected, shows a 2.02-fold decrease in TgPWS (PaGE = 0.7, P = 0.036). Another gene potentially upregulated in TgPWS pancreas is glucagon (Gcg), which was spotted six times on the array with fold change ranging from 2.85 to 3.49 (PaGE from 0.75 to 0.79, P from 0.052 to 0.092). Finally, Txnip (thioredoxin-interacting protein) is potentially upregulated 1.53-fold (PaGE = 0.77, P = 0.031), a result consistent with previous observations of 1.5-fold upregulation in the TgPWS liver (59).
We assessed the chromosomal position of affected genes to identify any evidence of clustering and hence, potential coregulation in cis. The majority of the 62 upregulated genes have a random distribution except for three pairs of adjacent genes (Supplemental Table S2), including Ins2 and Igf2 in chromosome 7 (both encoding hormones), Ppy and Pyy in chromosome 11 (both encoding hormones), and Neat1 and Malat1 (Neat2) in chromosome 19 (both encoding non-protein-coding RNAs). An additional two genes (Cytb and Atp6) are encoded by the mitochondrial chromosome. Although chromosome 11 had 12 upregulated genes, other than the Ppy/Pyy pair the distribution of genes reflected the gene density on this chromosome, and each was separated by a large number of genes. Finally, of the autosomal loci, 31 upregulated genes are on the forward strand and 29 are on the reverse strand, a random distribution.
Differentially expressed endocrine genes in TgPWS vs. WT pancreas.
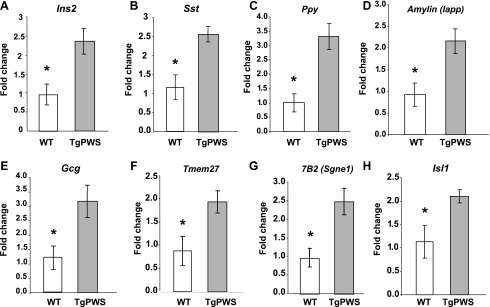
Next, we confirmed and extended the microarray results using quantitative qRT-PCR studies. The overexpression in TgPWS pancreas of the genes encoding hormones was confirmed, with insulin 2 (Ins2) expression increased 2.4-fold (P = 0.01) (Fig. 7A), expression of somatostatin (Sst) increased 2.2-fold (P = 0.01; Fig. 7B), expression of pancreatic polypeptide (Ppy) increased 3.3-fold (P = 0.005; Fig. 7C), and expression of islet amyloid polypeptide (Iapp) increased 2.3-fold (P = 0.02) (Fig. 7D). In addition, although the microarray data for Gcg suggested increased mRNA expression in TgPWS pancreas, the results did not satisfy the statistical criteria (see above). Nevertheless, we found by the more sensitive and accurate qRT-PCR method that Gcg mRNA levels are increased significantly in TgPWS pancreas by 2.6-fold (P = 0.03; Fig. 7E). It should be noted that if the reductions in β- and α-cell mass (Fig. 4, B and C) are considered, then the average magnitude in TgPWS compared with WT of upregulation of Ins2 and Gcg expression in each β- or α-cell is ∼4.4- or 6.5-fold, respectively.
Fig. 7.
Increased mRNA expression in the TgPWS pancreas of genes encoding hormones and cosecreted products (A–E), polypeptides involved in secretion (F and G), and a transcriptional regulator (H). Steady-state mRNA levels were assessed by quantitative RT-PCR using total RNA from pancreata of 4 TgPWS and 4 control mice at P1 and normalized to Gapdh mRNA levels. Results are mean fold change ± SE. *P < 0.05, significant differences between TgPWS and WT mice (independent t-test).
In addition, mRNA levels for two genes encoding components of secretory pathways were also confirmed by qRT-PCR to be increased in TgPWS deletion mice, including Tmem27 (fold change = 2.2, P = 0.03; Fig. 7F) and Sgne1 (fold change = 2.5, P = 0.01; Fig. 7G). Combined, these results show that mRNAs encoding all four major islet hormones (insulin, glucagon, somatostatin, PP), other secretory products (amylin), and secretory functions (Tmem27, 7B2/Sgne1) are upregulated in TgPWS pancreas at P1.
Interestingly, the mRNA encoding the LIM homeobox transcription factor ISL1, which plays a key role in development of the pancreas (1) and also regulates expression of several genes encoding islet hormones (21, 36, 42, 66–68, 73), was found upregulated in TgPWS pancreas by microarray (Supplemental Table S2). Confirming this, Isl1 mRNA levels were increased 1.8-fold (P = 0.04) by qRT-PCR (3.3-fold if taking into account loss of β- and α-cell mass, as described above) (Fig. 7H).
DISCUSSION
The etiology of PWS is generally considered to involve hypothalamic and pituitary mechanisms (9, 10, 44); however, a new theme emerging from our study is that multiple primary deficits in the secretory components of the endocrine pancreas occur in a deletion mouse model of PWS. In particular, TgPWS newborn mice show features consistent with the clinical presentation in infants with PWS (9). Of significance is the finding that the profound decreases in plasma levels of insulin and glucagon in TgPWS mice (Ref. 58 and the present study) were present in embryonic life and preceded the development of hypoglycemia (58), indicating a primary pancreatic defect. Indeed, our results demonstrate that development (islet morphology), survival (of α- and β-cells), and function (hormone secretion) of pancreatic endocrine cells are impaired in TgPWS mice and underlie the profound hypoinsulinemia and hypoglucagonemia in this mouse model. Consequently, our studies in mice have identified a cluster of imprinted, paternally expressed genes homologous to the PWS-imprinted domain that play important roles in pancreatic endocrine cell development and function. A second theme emerging from our work is that transcriptional regulation of genes encoding hormones and secretory factors are deregulated in TgPWS pancreas. Mechanisms that may underlie this coordinate gene regulatory network response are considered below. An understanding of these novel pathways leading to profound deficits in pancreatic endocrine cells is essential for an understanding of the pathophysiology of disorders of pancreatic development and function, including the TgPWS mouse model as well as those in humans, such as type 2 diabetes.
Islet developmental and functional abnormalities in a PWS mouse model.
The first major finding is that reduced β- and α-cell mass, with concomitant decreases in pancreatic insulin, C-peptide, and glucagon levels, can partially explain the decreased plasma insulin and glucagon levels in TgPWS mice. Our findings suggest that the reduction in α- and β-cell mass resulted from increased apoptosis since β-cell replication did not differ between the two genotypes. Interestingly, apoptosis of α-cells was much more pronounced in TgPWS islets at the studied ages. Nevertheless, given the degree of loss in β-cell mass in TgPWS by birth, it is likely that β-cell death occurs earlier in fetal development and precedes α-cell apoptosis; alternatively, another cell death pathway such as autophagy may contribute to the demise of β-cells in TgPWS mice. Many genetic and environmental factors might increase the frequency of β- and α-cell death during normal development and in pathological conditions. In our study, the genetic defect in TgPWS mice may induce islet cell alterations that increase the sensitivity to apoptotic stimuli. For example, loss of function of one or more of the imprinted, paternally expressed genes in TgPWS could induce apoptotic stimuli such as deregulation of transcriptional or signaling pathways, endoplasmic reticulum stress, or mitochondrial dysfunction and augment the apoptotic process. Consistent with this hypothesis, Necdin is required to prevent apoptosis in certain types of postmitotic neurons in the mouse, likely by a p53 and caspase-3 pathway (2, 33, 40). Further studies are required to elucidate the molecular mechanisms and to identify the apoptotic pathway activated in TgPWS islet cells.
The second key finding is that the TgPWS deletion resulted in a significant impairment of insulin secretion. Despite normal levels of plasma glucose at P1, using an Ins-C-Timer reporter we found that in vivo insulin secretion is markedly delayed in TgPWS mice compared with WT littermates, contributing to the profound hypoinsulinemia. In vitro basal insulin secretion, when normalized to insulin content, was decreased 3.6-fold in TgPWS mice compared with that of WT. In addition, despite a stimulatory increase in insulin secretion in TgPWS mice similar to that in WT mice, glucose-stimulated insulin secretion was 1.7-fold lower in TgPWS compared with WT mice. It is likely that the overall in vivo insulin secretion capacity of TgPWS islets is further diminished beyond our results, given that we selected the largest islets for in vitro studies in an attempt not to a priori bias the outcome to detect reduced insulin secretion from smaller islets with lower β-cell mass. These data suggest that the TgPWS genetic defect disrupts the basal secretory apparatus to a greater degree than the glucose-stimulated secretory pathway. Indeed, we (Stefan M and Nicholls RD, unpublished data) found no significant changes between TgPWS and WT mice at P1 in mRNA pancreas expression of Glut2 (encoding the β-cell glucose transporter) or the genes encoding the ATP-sensitive potassium channel subunits Sur1 and Kir6.2, key components of the glucose-regulated pathway that increases intracellular Ca2+ to trigger insulin release (7, 52, 60). Future work is needed to determine which molecules involved in insulin secretory vesicle exocytosis are abnormally regulated in the TgPWS mouse model.
A third major finding reported here is the profound α-cell defects, including increased apoptosis, plasma hypoglucagonemia, and an abnormal insulin/glucagon ratio. Under normal circumstances, plasma glucagon levels rise in the presence of insulin deficiency, which is present in the TgPWS fetus and neonate, or during hypoglycemia (29), which begins and rapidly worsens in TgPWS pups after the first postnatal day (58). However, the expected counterregulatory increase in glucagon levels was not observed in TgPWS mice at any age (Ref. 58 and the present study), suggesting that α-cell function is impaired. These data imply that a similar signaling and/or secretory mechanism might be disturbed in both β- and α-cells in TgPWS mice. Recent studies have shown a delayed counterregulatory response to insulin-induced hypoglycemia in mice deficient for the PWS region Magel2 gene, although pancreatic hormones were not assessed (62). Loss of Magel2 in TgPWS deletion mice may contribute in part to the more severe neonatal hypoglycemia and complete lack of a counterregulatory response described here for TgPWS mice. Future studies will determine the roles of α-cell and/or hypothalamic mechanisms to impaired pancreatic hormones and hypoglycemia in PWS and mouse models.
Coordinate regulation of genes encoding secretory functions.
An intriguing fourth major observation in this study is that steady-state mRNA levels for genes encoding the major pancreatic hormones (i.e., insulin, glucagon, somatostatin, and PP) and several other secretory factors (e.g., amylin, Sgne1/7B2, and Tmem27) were each upregulated by a similar fold change in TgPWS pancreas compared with control littermates. There are several nonmutually exclusive potential causes for the increased mRNA production of genes encoding hormones in TgPWS pancreas. First, Ins2, Ins1, and Gcg upregulation might result from homeostatic feedback (either humoral or neural) secondary to detection of low hormone levels in plasma or a target tissue. Another possibility is a pancreatic feedback mechanism whereby residual endocrine cells attempt to compensate for the loss of islet cells and reduced pancreatic hormone secretion. If the upregulation of gene expression in TgPWS pancreatic islets is related to one of these physiological compensatory mechanisms, then the increases in Ins2, Ins1, Gcg, Sst, and Ppy mRNA levels would be consistent with impaired function or development of all pancreatic endocrine cells.
Second, the expression of genes encoding pancreatic hormones (and other upregulated genes) may be controlled by one or more of the PWS-imprinted genes. The paternally expressed genes that are deleted in TgPWS mice may exert their regulatory role directly such that the PWS homologous gene function(s) would normally suppress mRNA expression, splicing, or stability for genes encoding hormones. The absence of the imprinted gene function(s) in the TgPWS mouse model would thus lead to increased mRNA levels for genes encoding pancreatic hormones. An example of a potential regulatory pathway that could contribute to such a mechanism is miR-344 (see Fig. 1A). Using a novel technique (HITS-CLIP) that combines Argonaute immunoprecipitation with RNA sequencing and bioinformatics, miR-344 was identified as one of the top 20 highest expressed miRNAs in the P13 mouse brain (13). Intriguingly, of their top predicted miR-344 targets (13), five were detected in our study as upregulated in the TgPWS pancreas, Nfib, Srrm2, Rev3l, Timp2, and D4Wsu53e (see Supplemental Table S2).
Alternatively, the imprinted paternally expressed genes deleted in TgPWS mice may indirectly control the expression of genes encoding pancreatic hormones such that the absence of a PWS gene function(s) leads to upregulation of a common regulatory factor. Supporting this model, in TgPWS pancreas the steady-state levels of ISL1 mRNA were greatly increased. A number of properties associated with ISL1 further support the model. ISL1 is expressed in all hormone-producing pancreatic cells and has an essential role in the development and survival of islet cell lineages (1, 21). ISL1 also has been shown to activate expression of rat Ins1 (36, 68, 73), rat Gcg (67), rat Sst (42, 66), and mammalian Iapp/amylin (68). Recent findings further implicate ISL1 in the regulation of mouse Ins, Gcg, Sst, and Ppy in the pancreas (21) as well as Sgne1, Ngfr, and Ppp2r2b in retinal ganglion cells (47). Therefore, the upregulation of an islet-specific transcription factor(s) such as ISL1 or another factor not on our microarray in the TgPWS pancreas is likely to account for the coordinate upregulation of genes encoding hormones. Future studies are needed to confirm ISL1-binding sites in direct target genes and the mechanisms. At present, the upstream control of Isl1 is largely unknown, and whether it responds to physiological signals and/or is a direct or secondary target of a PWS region imprinted gene is still to be determined.
Implications for PWS in humans.
Interestingly, PWS patients show signs of pancreatic insufficiency with reduced insulin levels relative to the degree of obesity (23, 26, 27, 39, 49, 54, 61) and reduced PP levels in response to nutrients (63, 74, 75). Diabetes affects ∼25% of the PWS population, with both insulin-dependent and insulin-independent diabetes described (3, 51, 54). Finally, PWS subjects are well known to have GH deficiency due to deficient pituitary secretion in response to stimuli (8, 30, 31) and gonadotropin deficiency thought to be due to a hypothalamic deficiency in GnRH release. Indeed, mouse Necdin has been implicated both in GnRH gene expression and the development of GnRH neurons, providing a potential molecular basis for gonadotropin deficiency in PWS (45). Similarly, Magel2 has recently been implicated in sex-specific endocrine changes putatively of hypothalamic origin, including GH release in response to ghrelin, in Magel2-null mice (62). These observations, combined with the pancreatic data presented here for the TgPWS mouse model, are consistent with a hypothesis that similar mechanisms underlie abnormal pancreatic, pituitary, and hypothalamic hormone and peptide secretion in PWS.
None of the imprinted, paternally expressed loci within the TgPWS deletion region are known to be involved in regulation of pancreatic or other secretory functions, but most are of unknown function. However, many of these candidate genes encode polypeptides or RNAs that could potentially regulate the level, splicing, or modification of other RNAs (12, 37, 38, 45, 48, 59) and hence, may account for the observations. Importantly, all three PWS mouse models with a 6.8-Mb deletion (TgPWS) (Ref. 58 and the present study), an imprinting center deletion (IC-del) (72), or a maternal uniparental disomy (11) that leads to loss of function of the entire imprinted domain have an equivalent early neonatal-lethal phenotype, including the onset and decline of hypoglycemia in IC-del mice (72), suggesting that each may have equivalent pancreatic abnormalities as described here for the TgPWS deletion mice. Deletion of the Snurf-Snrpn-snoRNA polycistronic cluster in mice (64) leads to a similar albeit less severe failure to thrive phenotype, potentially implicating one or more of these loci as contributing to the pancreatic phenotype, although deletion of the two “tandemly repeated” snoRNA families in this region does not present pancreatic or neonatal abnormalities (19, 20, 55). Further studies in PWS mouse models and of these imprinted loci in pancreatic endocrine cell models will allow discovery of novel loci and pathways important for islet cell function, development, and survival and will reveal the coordinate gene regulatory network linking expression of each pancreatic hormone and other secretory factors.
GRANTS
This study was funded in part by a Children's Hospital of Pittsburgh (CHP) Innovation Award Grant and a CHP Research Advisory Committee Grant (to R. D. Nicholls), grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; DK-55704 and DK-062965 to R. A. Simmons), and an American Diabetes Association Grant 1-06-RA-39 (to P. Drain). Hormone assays were aided by the services of the RIA/Biomarkers Core of the Diabetes and Endocrinology Research Center of the University of Pennsylvania, supported by NIDDK Grant DK-19525.
DISCLOSURES
The authors do not have any financial or other conflicts of interest with the subject matter or materials discussed in this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Golbahar Houshmand, Dr. Burhan Gharaibeh, Dr. Jing He, Heather Collins, and Shanping Li for advice or technical assistance.
Present address for M. Stefan: Department of Medicine/Endocrinology, Mount Sinai School of Medicine, One Gustave L. Levy Place, Box 1118, New York, NY 10029.
REFERENCES
- 1. Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature 385: 257–260, 1997. [DOI] [PubMed] [Google Scholar]
- 2. Andrieu D, Meziane H, Marly F, Angelats C, Fernandez PA, Muscatelli F. Sensory defects in Necdin deficient mice result from a loss of sensory neurons correlated within an increase of developmental programmed cell death. BMC Dev Biol 6: 56, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bassali R, Hoffman WH, Chen H, Tuck-Muller CM. Hyperlipidemia, insulin-dependent diabetes mellitus, and rapidly progressive diabetic retinopathy and nephropathy in Prader-Willi syndrome with del(15)(q11.2q13). Am J Med Genet 71: 267–270, 1997. [PubMed] [Google Scholar]
- 4. Bertera S, Crawford ML, Alexander AM, Papworth GD, Watkins SC, Robbins PD, Trucco M. Gene transfer of manganese superoxide dismutase extends islet graft function in a mouse model of autoimmune diabetes. Diabetes 52: 387–393, 2003. [DOI] [PubMed] [Google Scholar]
- 5. Bertera S, Geng X, Tawadrous Z, Bottino R, Balamurugan AN, Rudert WA, Drain P, Watkins SC, Trucco M. Body window-enabled in vivo multicolor imaging of transplanted mouse islets expressing an insulin-Timer fusion protein. Biotechniques 35: 718–722, 2003. [DOI] [PubMed] [Google Scholar]
- 6. Bouwens L, Rooman I. Regulation of pancreatic beta-cell mass. Physiol Rev 85: 1255–1270, 2005. [DOI] [PubMed] [Google Scholar]
- 7. Bratanova-Tochkova TK, Cheng H, Daniel S, Gunawardana S, Liu YJ, Mulvaney-Musa J, Schermerhorn T, Straub SG, Yajima H, Sharp GW. Triggering and augmentation mechanisms, granule pools, and biphasic insulin secretion. Diabetes 51, Suppl 1: S83–S90, 2002. [DOI] [PubMed] [Google Scholar]
- 8. Burman P, Ritzén EM, Lindgren AC. Endocrine dysfunction in Prader-Willi syndrome: a review with special reference to GH. Endocr Rev 22: 787–799, 2001. [DOI] [PubMed] [Google Scholar]
- 9. Cassidy SB. Prader-Willi syndrome. Curr Probl Pediatr 14: 1–55, 1984. [DOI] [PubMed] [Google Scholar]
- 10. Cassidy SB, Driscoll DJ. Prader-Willi syndrome. Eur J Hum Genet 17: 3–13, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cattanach BM, Barr JA, Evans EP, Burtenshaw M, Beechey CV, Leff SE, Brannan CI, Copeland NG, Jenkins NA, Jones J. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet 2: 270–274, 1992. [DOI] [PubMed] [Google Scholar]
- 12. Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA 97: 14311–14316, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chia R, Achilli F, Festing MF, Fisher EM. The origins and uses of mouse outbred stocks. Nat Genet 37: 1181–1186, 2005. [DOI] [PubMed] [Google Scholar]
- 15. Cummings DE, Clement K, Purnell JQ, Vaisse C, Foster KE, Frayo RS, Schwartz MW, Basdevant A, Weigle DS. Elevated plasma ghrelin levels in Prader-Willi syndrome. Nat Med 8: 643–644, 2002. [DOI] [PubMed] [Google Scholar]
- 16. de los Santos T, Schweizer J, Rees CA, Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am J Hum Genet 67: 1067–1082, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Smith AJ, Purmann C, Walters RG, Ellis RJ, Holder SE, Van Haelst MM, Brady AF, Fairbrother UL, Dattani M, Keogh JM, Henning E, Yeo GS, O'Rahilly S, Froguel P, Farooqi IS, Blakemore AI. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet 18: 3257–3265, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DelParigi A, Tschöp M, Heiman ML, Salbe AD, Vozarova B, Sell SM, Bunt JC, Tataranni PA. High circulating ghrelin: a potential cause for hyperphagia and obesity in prader-willi syndrome. J Clin Endocrinol Metab 87: 5461–5464, 2002. [DOI] [PubMed] [Google Scholar]
- 19. Ding F, Prints Y, Dhar MS, Johnson DK, Garnacho-Montero C, Nicholls RD, Francke U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm Genome 16: 424–431, 2005. [DOI] [PubMed] [Google Scholar]
- 20. Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS One 3: e1709, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du A, Hunter CS, Murray J, Noble D, Cai CL, Evans SM, Stein R, May CL. Islet-1 is required for the maturation, proliferation and survival of the endocrine pancreas. Diabetes 58: 2059–2069, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duncan RR, Greaves J, Wiegand UK, Matskevich I, Bodammer G, Apps DK, Shipston MJ, Chow RH. Functional and spatial segregation of secretory vesicle pools according to vesicle age. Nature 422: 176–180, 2003. [DOI] [PubMed] [Google Scholar]
- 23. Eiholzer U, Stutz K, Weinmann C, Torresani T, Molinari L, Prader A. Low insulin, IGF-I and IGFBP-3 levels in children with Prader-Labhart-Willi syndrome. Eur J Pediatr 157: 890–893, 1998. [DOI] [PubMed] [Google Scholar]
- 24. Gabriel JM, Merchant M, Ohta T, Ji Y, Caldwell RG, Ramsey MJ, Tucker JD, Longnecker R, Nicholls RD. A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and Angelman syndromes. Proc Natl Acad Sci USA 96: 9258–9263, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldstone AP. Prader-Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol Metab 15: 12–20, 2004. [DOI] [PubMed] [Google Scholar]
- 26. Goldstone AP, Thomas EL, Brynes AE, Castroman G, Edwards R, Ghatei MA, Frost G, Holland AJ, Grossman AB, Korbonits M, Bloom SR, Bell JD. Elevated fasting plasma ghrelin in Pader-Willi syndrome adults is not solely explained by their reduced visceral adiposity and insulin resistance. J Clin Endocrinol Metab 89: 1718–1726, 2004. [DOI] [PubMed] [Google Scholar]
- 27. Goldstone AP, Patterson M, Kalingag N, Ghatei MA, Brynes AE, Bloom SR, Grossman AB, Korbonits M. Fasting and postprandial hyperghrelinemia in Prader-Willi syndrome is partially explained by hypoinsulinemia, and is not due to peptide YY3–36 deficiency or seen in hypothalamic obesity due to craniopharyngioma. J Clin Endocrinol Metab 90: 2681–2690, 2005. [DOI] [PubMed] [Google Scholar]
- 28. Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA 96: 5616–5621, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gromada J, Franklin I, Wollheim CB. Alpha-cells of the endocrine pancreas: 35 years of research but the enigma remains. Endocr Rev 28: 84–116, 2007. [DOI] [PubMed] [Google Scholar]
- 30. Grugni G, Guzzaloni G, Morabito F. Impairment of GH responsiveness to GH-releasing hexapeptide (GHRP-6) in Prader-Willi syndrome. J Endocrinol Invest 24: 340–348, 2001. [DOI] [PubMed] [Google Scholar]
- 31. Grugni G, Marzullo P, Ragusa L, Sartorio A, Trifirò G, Liuzzi A, Crinò A; Genetic Obesity Study Group of the Italian Society of Pediatric Endocrinology and Diabetology. Impairment of GH responsiveness to combined GH-releasing hormone and arginine administration in adult patients with Prader-Willi syndrome. Clin Endocrinol (Oxf) 65: 492–499, 2006. [DOI] [PubMed] [Google Scholar]
- 32. Gunay-Aygun M, Schwartz S, Heeger S, O'Riordan MA, Cassidy SB. The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics 108: E92, 2001. [DOI] [PubMed] [Google Scholar]
- 33. Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci 28: 8772–8784, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaestner KH, Lee CS, Scearce LM, Brestelli JE, Arsenlis A, Le PP, Lantz KA, Crabtree J, Pizarro A, Mazzarelli J, Pinney D, Fischer S, Manduchi E, Stoeckert CJ, Jr, Gradwohl G, Clifton SW, Brown JR, Inoue H, Cras-Méneur C, Permutt MA. Transcriptional program of the endocrine pancreas in mice and humans. Diabetes 52: 1604–1610, 2003. [DOI] [PubMed] [Google Scholar]
- 35. Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, Caliebe A, Kuechler A, Horsthemke B, Buiting K. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet 17: 582–590, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature 344: 879–882, 1990. [DOI] [PubMed] [Google Scholar]
- 37. Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311: 230–232, 2006. [DOI] [PubMed] [Google Scholar]
- 38. Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 19: 1153–1164, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krochik AG, Ozuna B, Torrado M, Chertkoff L, Mazza C. Characterization of alterations in carbohydrate metabolism in children with Prader-Willi syndrome. J Pediatr Endocrinol Metab 19: 911–918, 2006. [DOI] [PubMed] [Google Scholar]
- 40. Kurita M, Kuwajima T, Nishimura I, Yoshikawa K. Necdin downregulates CDC2 expression to attenuate neuronal apoptosis. J Neurosci 26: 12003–12013, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee S, Walker CL, Wevrick R. Prader-Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr Patterns 3: 599–609, 2003. [DOI] [PubMed] [Google Scholar]
- 42. Leonard J, Serup P, Gonzalez G, Edlund T, Montminy M. The LIM family transcription factor Isl-1 requires cAMP response element binding protein to promote somatostatin expression in pancreatic islet cells. Proc Natl Acad Sci USA 89: 6247–6251, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manduchi E, Grant GR, McKenzie SE, Overton GC, Surrey S, Stoeckert CJ., Jr Generation of patterns from gene expression data by assigning confidence to differentially expressed genes. Bioinformatics 16: 685–698, 2000. [DOI] [PubMed] [Google Scholar]
- 44. Miller JL, Goldstone AP, Couch JA, Shuster J, He G, Driscoll DJ, Liu Y, Schmalfuss IM. Pituitary abnormalities in Prader-Willi syndrome and early onset morbid obesity. Am J Med Genet A 146A: 570–577, 2008. [DOI] [PubMed] [Google Scholar]
- 45. Miller NL, Wevrick R, Mellon PL. Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet 18: 248–260, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyatsuka T, Li Z, German MS. Chronology of islet differentiation revealed by temporal cell labeling. Diabetes 58: 1863–1868, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mu X, Fu X, Beremand PD, Thomas TL, Klein WH. Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci USA 105: 6942–6947, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2: 153–175, 2001. [DOI] [PubMed] [Google Scholar]
- 49. Paik KH, Jin DK, Lee KH, Armstrong L, Lee JE, Oh YJ, Kim S, Kwon EK, Choe YH. Peptide YY, cholecystokinin, insulin and ghrelin response to meal did not change, but mean serum levels of insulin is reduced in children with Prader-Willi syndrome. J Korean Med Sci 22: 436–441, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Partsch CJ, Lämmer C, Gillessen-Kaesbach G, Pankau R. Adult patients with Prader-Willi syndrome: clinical characteristics, life circumstances and growth hormone secretion. Growth Horm IGF Res 10, Suppl B: S81–S85, 2000. [DOI] [PubMed] [Google Scholar]
- 51. Ristow M. Neurodegenerative disorders associated with diabetes mellitus. J Mol Med 82: 510–529, 2004. [DOI] [PubMed] [Google Scholar]
- 52. Rorsman P, Renstrom E. Insulin granule dynamics in pancreatic beta cells. Diabetologia 46: 1029–1045, 2003. [DOI] [PubMed] [Google Scholar]
- 53. Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet 40: 719–721, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schuster DP, Osei K, Zipf WB. Characterization of alterations in glucose and insulin metabolism in Prader-Willi subjects. Metabolism 45: 1514–1520, 1996. [DOI] [PubMed] [Google Scholar]
- 55. Skryabin BV, Gubar LV, Seeger B, Pfeiffer J, Handel S, Robeck T, Karpova E, Rozhdestvensky TS, Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet 3: e235, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stefan M, Nicholls RD. What have rare genetic syndromes taught us about the pathophysiology of the common forms of obesity? Curr Diab Rep 4: 143–150, 2004. [DOI] [PubMed] [Google Scholar]
- 57. Stefan M, Claiborn KC, Stasiek E, Chai JH, Ohta T, Longnecker R, Greally JM, Nicholls RD. Genetic mapping of putative Chrna7 and Luzp2 neuronal transcriptional enhancers due to impact of a transgene-insertion and 6.8 Mb deletion in a mouse model of Prader-Willi and Angelman syndromes. BMC Genomics 6: 157, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stefan M, Ji H, Simmons RA, Cummings DE, Ahima RS, Friedman MI, Nicholls RD. Hormonal and metabolic defects in a Prader-Willi syndrome mouse model with neonatal failure to thrive. Endocrinology 146: 4377–4385, 2005. [DOI] [PubMed] [Google Scholar]
- 59. Stefan M, Portis T, Longnecker R, Nicholls RD. A nonimprinted Prader-Willi Syndrome (PWS)-region gene regulates a different chromosomal domain in trans but the imprinted PWS loci do not alter genome-wide mRNA levels. Genomics 85: 630–640, 2005. [DOI] [PubMed] [Google Scholar]
- 60. Straub SG, Sharp GW. Glucose-stimulated signaling pathways in biphasic insulin secretion. Diabetes Metab Res Rev 18: 451–463, 2002. [DOI] [PubMed] [Google Scholar]
- 61. Talebizadeh Z, Butler MG. Insulin resistance and obesity-related factors in Prader-Willi syndrome: comparison with obese subjects. Clin Genet 67: 230–239, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tennese AA, Wevrick R. Impaired hypothalamic regulation of endocrine function and delayed counterregulatory response to hypoglycemia in magel2-null mice. Endocrinology 152: 967–978, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tomita T, Greeley G, Jr, Watt L, Doull V, Chance R. Protein meal-stimulated pancreatic polypeptide secretion in Prader-Willi syndrome of adults. Pancreas 4: 395–400, 1989. [DOI] [PubMed] [Google Scholar]
- 64. Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet 8: 1357–1364, 1999. [DOI] [PubMed] [Google Scholar]
- 65. Tsuboi T, Kitaguchi T, Karasawa S, Fukuda M, Miyawaki A. Age-dependent preferential dense-core vesicle exocytosis in neuroendocrine cells revealed by newly developed monomeric fluorescent timer protein. Mol Biol Cell 21: 87–94, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vallejo M, Penchuk L, Habener JF. Somatostatin gene upstream enhancer element activated by a protein complex consisting of CREB, Isl-1-like, and alpha-CBF-like transcription factors. J Biol Chem 267: 12876–12884, 1992. [PubMed] [Google Scholar]
- 67. Wang M, Drucker DJ. The LIM domain homeobox gene isl-1 is a positive regulator of islet cell-specific proglucagon gene transcription. J Biol Chem 270: 12646–12652, 1995. [DOI] [PubMed] [Google Scholar]
- 68. Wang M, Drucker DJ. Activation of amylin gene transcription by LIM domain homeobox gene isl-1. Mol Endocrinol 10: 243–251, 1996. [DOI] [PubMed] [Google Scholar]
- 69. Watkins S, Geng X, Li L, Papworth G, Robbins PD, Drain P. Imaging secretory vesicles by fluorescent protein insertion in propeptide rather than mature secreted peptide. Traffic 3: 461–471, 2002. [DOI] [PubMed] [Google Scholar]
- 70. Watrin F, Le Meur E, Roeckel N, Ripoche MA, Dandolo L, Muscatelli F. The Prader-Willi syndrome murine imprinting center is not involved in the spatio-temporal transcriptional regulation of the Necdin gene. BMC Genet 6: 1, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. White P, Kaestner KH. Gene expression analysis in diabetes research. Methods Mol Biol 560: 239–261, 2009. [DOI] [PubMed] [Google Scholar]
- 72. Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19: 25–31, 1998. [DOI] [PubMed] [Google Scholar]
- 73. Zhang H, Wang WP, Guo T, Yang JC, Chen P, Ma KT, Guan YF, Zhou CY. The LIM-homeodomain protein ISL1 activates the insulin gene promoter directly through synergy with BETA2. J Mol Biol 392: 566–577, 2009. [DOI] [PubMed] [Google Scholar]
- 74. Zipf WB, O'Dorisio TM, Cataland S, Sotos J. Blunted pancreatic polypeptide responses in children with obesity of Prader-Willi syndrome. J Clin Endocrinol Metab 52: 1264–1266, 1981. [DOI] [PubMed] [Google Scholar]
- 75. Zipf WB, O'Dorisio TM, Cataland S, Dixon K. Pancreatic polypeptide responses to protein meal challenges in obese but otherwise normal children and obese children with Prader-Willi syndrome. J Clin Endocrinol Metab 57: 1074–1080, 1983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.