Abstract
Spermatogonial stem cells (SSCs) provide the foundation for spermatogenesis throughout the life of a male. Because SSCs of many species can colonize the mouse testis, and glial cell line-derived neurotrophic factor (GDNF) is responsible for stimulating SSC self-renewal in rodents, we reasoned that molecular mechanisms of SSC self-renewal are similar across species. GDNF-regulated genes have been identified in mouse SSCs; however, downstream targets of GDNF are unknown in other species. The objective of this work was to identify GDNF-regulated genes in rat SSCs and to define the biological significance of these genes for rat SSC self-renewal. We conducted microarray analysis on cultured rat germ cells enriched for SSCs in the presence and absence of GDNF. Many GDNF-regulated genes were identified, most notably, Bcl6b and Etv5, which are important for mouse SSC self-renewal. Bcl6b was the most highly regulated gene in both the rat and mouse. Additionally, we identified three novel GDNF-regulated genes in rat SSCs: Bhlhe40, Hoxc4, and Tec. Small interfering RNA treatment for Bcl6b, Etv5, Bhlhe40, Hoxc4, and Tec resulted in a decrease in SSC number, as determined by transplantation, without a change in total cell number within the culture. These data indicate that, like in the mouse SSC, Bcl6b and Etv5 are important for rat SSC self-renewal, suggesting that these genes may be important for SSCs in all mammals. Furthermore, identification of three novel GDNF-regulated genes in the rat SSC extends our knowledge of SSC activity and broadens the foundation for understanding this process in higher species, including humans.
Keywords: adult stem cells, germline, growth factors, microarray, rat, self-renewal
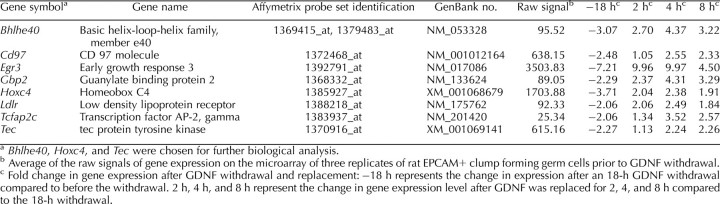
Bcl6b, Etv5, Bhlhb2, Hoxc4, and Tec are regulated by glial cell line-derived neurotrophic factor and important for spermatogonial stem cell self-renewal in the rat.
INTRODUCTION
The spermatogonial stem cell (SSC) is an adult tissue-specific stem cell that provides the foundation for spermatogenesis throughout the lifetime of a male. SSCs are members of a large group of cells termed spermatogonia, reside on the basement membrane within a cognate niche in the seminiferous epithelium of the testis, and have the capability to self-renew or differentiate into daughter cells committed to become spermatozoa. The population of spermatogonia in the testis is comprised of three types of cells: 1) stem spermatogonia (As), the true SSC capable of self-renewal divisions; 2) proliferating spermatogonia (Apr and Aal), that undergo mitotic division prior to entering the cycle of the seminiferous epithelium; and 3) differentiating spermatogonia (A1–4, B, intermediate), that have entered the cycle of the seminiferous epithelium [1]. Without the ability to undergo self-renewal, the SSC population is rapidly depleted, resulting in infertility. The process of SSC self-renewal is a multifaceted mechanism consisting of cell proliferation, prevention of apoptosis, and maintenance of the SSC phenotype. The regulation of the self-renewal vs. differentiation fate decision (which consists of some of the same mechanisms as self-renewal, such as cell proliferation) is presumed to be a highly organized process directed by factors within the stem cell niche [2]. Currently, very little is known about this important fate decision.
Direct study of SSCs within the stem cell niche has been hampered by the rarity of these cells within the testis and the lack of a definitive marker that can distinguish an SSC from a daughter proliferating spermatogonium committed to differentiation [3]. A further hindrance to study of SSCs has been the lack of methods of assay and maintenance. The development of a transplantation technique, in which labeled donor SSCs are transplanted into the testis of a sterile recipient, resulting in donor-derived spermatogenesis, has allowed for an in situ assay of SSCs [4–6]. This technique is the only definitive method to assay SSC presence and function in a given cell population, because only SSCs are capable of colonizing a recipient testis and maintaining long-term spermatogenesis. We recently developed defined, serum-free, in vitro culture systems that are ideal for studying mechanisms regulating self-renewal of mouse [7–8] and rat SSCs [9]. A drawback to the use of culture exclusively to study the biology of SSCs is that only approximately 1 in 10–15 germ cells in culture is thought to be a true SSC, with the remaining cells considered to be proliferating daughter cells [7]. Therefore, utilization of SSC culture followed by cell transplantation, to assay for the stem cell content of a given culture, is necessary to directly study SSC biology [2].
Several lines of evidence have implicated glial cell line-derived neurotrophic factor (GDNF) as the key regulator of SSC self-renewal. Using GDNF knockout and over-expression mice, Meng et al. [10] demonstrated that spermatogonial proliferation is regulated by GDNF. Additional work utilizing in vivo testis transfection of a GDNF expression vector followed by germ cell transplantation suggested that GDNF was important for SSC function [11]. Advances in the SSC culture system in our laboratory [7] provided definitive proof that mouse SSC self-renewal is dependent on GDNF. Subsequent work by us and others has demonstrated that GDNF is also essential for long-term self-renewal of rat and hamster SSCs [9, 12, 13]. Because of this conservation across rodent species, it is likely that GDNF is essential for SSC self-renewal in most, if not all, mammalian species. Furthermore, because of the ability of SSCs from a variety of species to colonize a mouse testis, it is also possible that the mechanism of GDNF-mediated SSC self-renewal is conserved among mammals [14–17].
Recently, we have begun to define the molecular regulatory process of mouse SSC self-renewal using microarray technology. Over the past decade, oligonucleotide microarray methodology has been used to characterize gene expression from whole testis tissue and individual testis cell populations [18–22]. These data sets provide a valuable tool for the discovery of differentially regulated genes across cell populations and treatments. Using microarray analysis of cultured mouse germ cells enriched for SSCs, we identified a subset of GDNF-regulated genes that are important for SSC self-renewal [23]. Furthermore, using kinase-specific inhibitors, we demonstrated that SRC family kinase (SFK) signaling mechanisms likely mediate expression of GDNF-regulated genes [24].
Although we know that GDNF supports the self-renewal of both mouse and rat SSCs, the degree of conservation of the regulation of this process between the two species is unknown. Because rat spermatogenesis occurs in mouse seminiferous tubules, and SSCs of many species are maintained and proliferate in the mouse seminiferous tubule, determining the identity of genes involved in SSC self-renewal in the rat is a critical step in extending our understanding of SSC self-renewal in higher species, including humans. Therefore, identification and subsequent analysis of GDNF-regulated genes was performed with cultured rat spermatogonia enriched for SSCs, which confirmed suspected similarities between mouse and rat SSC self-renewal, and expanded our knowledge of the regulators of this process in mammals.
MATERIALS AND METHODS
Materials and Animals
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich (St. Louis, MO). Donor cells and tissues were isolated from Sprague Dawley wild-type, Ef1-EGFP, or Mt1-lacZ transgenic rats. Immunodeficient NCr nude mice (CrTac:NCr-Foxn1nu) were purchased from Taconic Farms Inc. (Hudson, NY). All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Isolation and Culture of SSC-Enriched Testis Cell Fractions
The cell surface marker, EPCAM, is effective in isolating SSC-enriched testis cell populations from the rat [9]. Cultures of EPCAM+ clump-forming germ cells contain self-renewing SSCs capable of colonizing recipient testes after long-term culture. However, a majority of the cells within the culture are proliferating spermatogonia [9], and germ cell transplantation must be utilized to assay for the SSC. Cultures of rat EPCAM+ clump-forming germ cells were established from cells isolated from rat pups, as previously described [9] with few modifications, which are detailed in Supplemental Materials and Methods (supplemental data available online at www.biolreprod.org). Isolation of EPCAM+ cells was accomplished by magnetic-activated cell sorting and approximately 200 000 EPCAM+ cells were seeded per well of a 12-well culture dish containing 100 000 mitotically inactivated SIM mouse embryo-derived, thioguanine- and ouabain-resistant (STO) feeders. The EPCAM+ clump-forming germ cells were maintained in defined serum-free media (rSFM) with GDNF (20 ng/ml; R&D Systems, Minneapolis, MN), Gfra1 (150 ng/ml; R&D Systems), and FGF2 (1 ng/ml; BD Biosciences, San Jose, CA) [9].
Germ Cell Transplantation
The number of donor colonies of spermatogenesis in a recipient testis is a direct reflection of the SSC population in the originally injected cell population [25–27]. All germ cell transplantation experiments were conducted with long-term (cultured at least 1 mo) cultures of EPCAM+ clump-forming germ cells established from 8- or 9-day-old Sprague Dawley rats carrying the Escherichia coli LacZ gene under the control of the metallothionein 1a promoter (Mt1a-LacZ), similar to previous reports [9]. Cells (10 μl; 1 × 106/ml) were transplanted into each testis of adult busulphan-treated (50 mg/kg) nude mice. At 2–3 mo after transplantation, testes were removed and analyzed for donor-derived spermatogenesis by 5-bromo-4-chloro-3-indolyly β-d-galactosidase staining. Each experimental unit was transplanted into eight recipient testes, and each treatment was replicated two to three times. Colony number is the number of donor spermatogenic colonies in recipient testes per 105 cells originally cultured. Each colony represents the proliferation from a single SSC [25–27].
GDNF Withdrawal and RNA Isolation
Long-term cultures of wild-type Sprague Dawley EPCAM+ clump-forming germ cells were utilized for microarray analysis. Cells were isolated from cultures before and after an 18-h GDNF withdrawal, and at 2, 4, and 8 h after GDNF replacement, and frozen in Trizol. To isolate RNA for microarray, a combination of the Trizol method and RNeasy (Qiagen, Valencia, CA) columns was used, as previously described [23]. RNA was isolated using the Trizol method, as directed, for all other assays.
Microarray Processing and Analysis
After RNA isolation and cDNA production, (5 treatments, n = 3/treatment), cDNA products were hybridized to Affymetrix Rat Genome 230 2.0 Arrays by the University of Pennsylvania School of Medicine Microarray Core Facility according to the Affymetrix GeneChip Expression manual (www.affymetrix.com). Microarray analysis was conducted at the Penn Bioinformatics core. The David Bioinformatics Database (http://david.abcc.ncifcrf.gov) was utilized for functional annotation clustering.
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) was utilized to validate microarray, small interfering RNA (siRNA) knockdown, and in vivo expression of GDNF-regulated genes. After RNA isolation, samples were DNAse (Ambion, Austin, TX)-treated to remove genomic DNA and reverse transcribed using oligo(dT) priming and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). Primer sequences (Supplemental Table S1) were designed using Primer 3 software [28]. Expression levels of specific genes were assayed using SYBR green (Applied Biosystems, Foster City, CA) and an ABI 7300 sequence detection system (Applied Biosystems). Relative gene expression was determined by normalizing gene-of-interest expression levels to that of Rps2 using the formula: Relative Expression = 1/2(Ct of the gene-of-interest) − (Ct of Rps2).
Small Interfering RNA Transfection
To demonstrate that identified GDNF-regulated genes had biologic function, siRNA knockdown experiments were conducted on cultures of EPCAM+ clump-forming germ cells established from transgenic rats carrying either the GFP or mt-LacZ gene. Small interfering RNAs were purchased from Ambion (Silencer single nucleotide siRNAs; Austin, TX) or Dharmacon (siGENOME SMARTpool; cocktail-multi nucleotide siRNAs, Chicago, IL) (Supplemental Table S2). All siRNA sequences were validated using BLAST. Negative control siRNA was Ambion Silencer negative control scrambled nontargeting siRNA (product no. 4611). The transfection technique was performed as previously described [23], with few modifications, as detailed in the Supplemental Materials and Methods. Germ cells were transfected with gene-specific or negative control siRNA on Day 0 of the experimental timeline and subcultured to siRNA-free media 18 h later. The effect of siRNA knockdown on expression of specific genes was determined using qRT-PCR 18 h after transfection. The SSCs utilized for transplantation experiments were then allowed to self-renew for an additional 7 days. Because the siRNA transfection is transient in nature, cultures transfected with siRNA for genes important for SSC self-renewal would undergo a period of delay in SSC self-renewal, followed by a recovery in self-renewal ability. This delay would result in fewer SSCs per culture when compared with negative control-treated cultures after the 8-day culture period and subsequent transplantation analysis. By transfecting cells with a fluorescently labeled siRNA (Silencer FAM GAPDH siRNA; Ambion), transfection efficiency was determined to be 95 ± 0.23%. Cultures established from rats carrying a GFP transgene were used to determine cell number and viability after transfection and cultures derived from rats carrying the mt-LacZ transgene were used to determine stem cell potential via the germ cell transplantation technique. Because the SSC concentration varies between different primary cultures of EPCAM+ clump-forming germ cells, negative control siRNA (Ambion)-treated cultures were always run and analyzed in parallel to each gene-specific siRNA-treated culture, using cells from the same primary culture. By comparing the number of SSCs in negative control siRNA-treated cultures to gene-specific siRNA-treated cultures 8 days after treatment, we were able to directly evaluate the effect of specific gene knockdown on SSC activity.
Flow Cytometric Analysis of siRNA-Treated SSCs
In order to accurately determine the number of donor-derived cells in siRNA-treated cultures of EPCAM+ clump-forming germ cells, flow cytometry was used. Single-cell digests of cultured cells were assayed for GFP content using a FACSCalibur flow cytometer (BD Biosciences) on Days 0, 4, and 8 of siRNA treatment. Additionally, the percentage of apoptotic cells was determined on the same cells using a commercial nexin staining kit (Guava Technologies, Hayward, CA).
Immunofluorescence and Immunocytochemistry
Immunofluorescence was used to demonstrate the presence of the proteins for identified GDNF-regulated genes within EPCAM+ clump-forming germ cells. In order to confirm that proteins were in fact expressed in vivo, immunocytochemistry was performed on adult testis tissue. Descriptions of these techniques are provided in the Supplemental Materials and Methods.
Statistical Methods
All experiments were run in at least duplicate or triplicate using independently established cell cultures for each replicate. For determination of differences between colony numbers before and after GDNF withdrawal, ANOVA was performed using SPSS 15 (SPSS, Chicago, IL). When comparing data sets, univariate ANOVA was conducted and, if data sets contained multiple points, differences were determined using the least significant difference post hoc test. To determine colony number differences between negative control and siRNA treatments, data were first transformed using a square root transformation. No statistical differences were observed between Dharmacon and Ambion siRNAs; therefore, data from different siRNA companies were pooled. In Figures, error bars indicate the SEM.
Additional Methods
Detailed descriptions of EPCAM+ clump-forming germ cell isolation and culture, RNA isolation, microarray processing and analysis, flow cytometry, and immunoanalysis of proteins are provided in the Supplemental Materials and Methods.
RESULTS
Identification of GDNF-Regulated Genes in Cultures of EPCAM+ Clump-Forming Germ Cells
To identify GDNF-regulated genes in cultures of EPCAM+ clump-forming germ cells, which contain the true SSC, GDNF withdrawal was conducted, followed by microarray analysis. In this culture system, SSCs grow within heterogeneous clumps of germ cells containing both the SSC and non-SSC proliferating spermatogonia (Fig. 1A). After transplantation to recipient mice, the true SSCs within the heterogeneous population of germ cells form colonies of complete spermatogenesis (Fig. 1B) [9]. In order for the microarray analysis to assess accurately gene expression in cultured SSCs, it was important to demonstrate that overnight GDNF withdrawal does not change the proportion of SSCs within the culture. Therefore, prior to microarray analysis, cultured mt-LacZ transgenic EPCAM+ clump-forming germ cells were transplanted into recipient mice before and after an 18-h GDNF withdrawal. No change in the number of colonies in recipient testes 3 mo after transplantation was observed, confirming that overnight short-term withdrawal of GDNF does not change the concentration of SSCs within the culture (Fig. 1C).
FIG. 1.
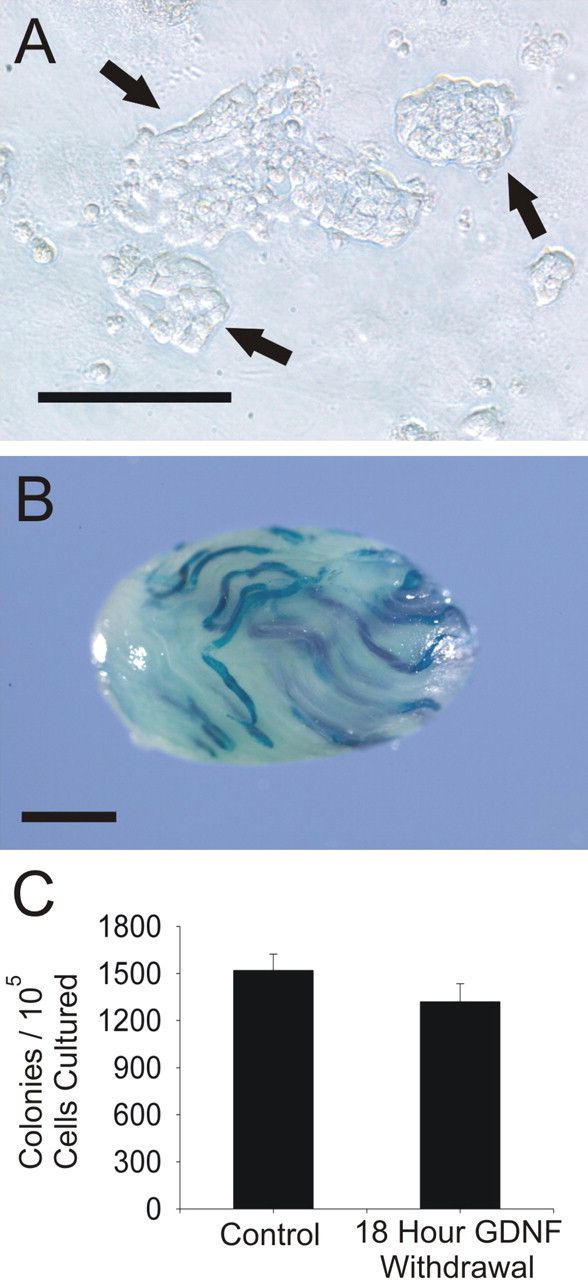
Effect of GDNF withdrawal on SSC colonization. A) Cultures of rat EPCAM+ clump-forming germ cells contain both SSCs and proliferating daughter spermatogonia that form tightly connected clumps (arrows) that loosely adhere to the feeder layer. Bar = 100 μm. B) True SSCs and their proliferating daughter spermatogonia are indistinguishable from each other unless assayed for using germ cell transplantation. After transplantation to the testes of busulphan-treated recipient nude mice, cultured rat SSCs form colonies of spermatogenesis that are easily visualized by using transgenic donor cells. In this case, the donor was transgenic for the mt-LacZ gene. Bar = 1.5 mm. C) Overnight withdrawal (18 h) of GDNF from cultured rat EPCAM+ clump-forming germ cells does not significantly influence the population of SSCs within the culture, as indicated by the lack of difference between colony numbers before and after GDNF withdrawal. Error bars represent the SEM.
Upon confirmation of SSC maintenance following GDNF withdrawal, cultured EPCAM+ clump-forming germ cells were collected before and after an 18-h GDNF withdrawal, and at 2, 4, and 8 h after GDNF replacement. Microarray analysis indicated that approximately 21 000 genes are expressed in the cultured EPCAM+ clump-forming germ cells (Supplemental Table S3), 7900 of which were GDNF-regulated. However, expression of only 77 genes changed at least 2-fold after both GDNF withdrawal and replacement. The majority of these genes were positively regulated by GDNF, and expression recovered by 4 h after GDNF replacement (Supplemental Fig. S1). Functional clustering revealed that genes up-regulated by GDNF were classified into ion binding, cell cycle, and transcriptional regulation gene families (Supplemental Table S4).
EPCAM+ clump-forming germ cells expressed several genes previously reported to be expressed and/or GDNF-regulated in SSCs ([9, 23, 24]; Supplemental Table S5). Both Bcl6b (B-cell CLL/lymphoma 6, member B [zinc finger protein]) and Etv5 (Ets variant 5), which are GDNF-regulated in the mouse SSC [23, 24], were similarly regulated in the rat EPCAM+ clump-forming germ cells. Additionally, some genes reported to be involved in SSC self-renewal in the mouse were not GDNF-regulated (Lhx1 [23, 24], Zbtb16 [previously known as Plzf] [29]), or not expressed in rat EPCAM+ clump-forming germ cells (Nanos3 [30]). The SSC markers, Epcam, Itga6, Thy1, and Cd9 [9, 31, 32], were expressed by EPCAM+ clump-forming germ cells, and, interestingly, Itga6 and Epcam were also positively GDNF-regulated. The GDNF receptors, Gfra1 and Ret, were both expressed and negatively regulated by GDNF in rat EPCAM+ clump-forming germ cells. Genes that have been suggested to be involved in SSC differentiation were either down-regulated by GDNF (Kit [33]), or not expressed (Neurog3 [34]). EPCAM+ clump-forming germ cells also expressed many genes that are important for stem cell function in hematopoietic, neural, and embryonic stem cell populations (Supplemental Table S6). Of particular interest were the expression patterns of the core embryonic stem (ES) cell pluripotency genes Pou5f1 (previously known as Oct4), Sox2, and Nanog. Both Nanog and Pou5f1 were expressed by the clump-forming germ cells; however, Sox2 was not. Furthermore, neither Nanog nor Pou5f1 were GDNF-regulated (Supplemental Fig. S2).
Conservation of GDNF-Regulated Genes Between the Mouse and Rat
Several genes (Bcl6b, Egr2, Egr3, Etv5, Lhx1, and Tspan8) were previously identified as GDNF-regulated in cultures of mouse germ cells enriched for SSCs [23]. Bcl6b, Egr2, Egr3, and Etv5 were also positively GDNF-regulated in rat EPCAM+ clump-forming germ cells; however, Lhx1 was not GDNF-regulated, and Tspan8 was not expressed (Supplemental Table S7).
Because of the conservation of GDNF regulation of Bcl6b and Etv5, and the fact that Bcl6b was the most highly GDNF-regulated gene in both mouse [23] and rat analyses, we wanted to further characterize the expression of these genes in the rat and confirm their importance for SSC self-renewal in cultures of EPCAM+ clump-forming germ cells. The expression patterns for Bcl6b and Etv5 before and after GDNF withdrawal in EPCAM+ clump-forming germ cells were validated using qRT-PCR (Table 1 and Supplemental Fig. S3), and the presence of the protein in the germ cell clumps was demonstrated using immunofluorescence (Fig. 2A). Additionally, Bcl6b and Etv5 transcript expression was higher in cell populations enriched for SSCs, including fresh and cultured EPCAM+ cells, compared with whole testis (Fig. 2B). Spermatogonia along the basement membrane appeared to stain lightly for both BCL6B and ETV5 protein. Some differentiating stages of germ cells stained with greater intensity for these proteins (Fig. 2C).
TABLE 1.
Known GDNF-regulated genes expressed in mouse SSCs [23] identified as GDNF regulated in cultures of rat EPCAM+ clump forming germ cells enriched for SSCs that were validated by qRT-PCR.
FIG. 2.
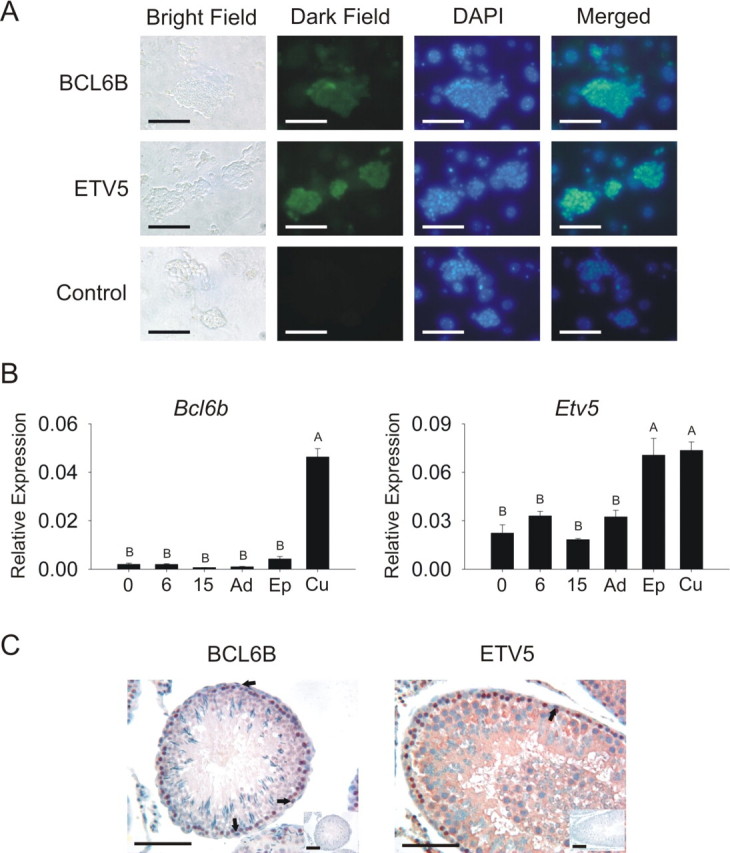
Characterization of Bcl6b and Etv5 transcript and protein expression in vivo and in vitro. A) BCL6B and ETV5 proteins were expressed in cultures of EPCAM+ clump-forming germ cells. Column 1 = bright field; column 2 = dark field, gene-specific primary antibody (GFP); column 3 = 4′,6′-diamidino-2-phenylindole (DAPI); column 4 = merged, overlay of columns 2 and 3. Control immunofluorescence images had rabbit IgG as primary antibody. Bars = 100 μm. B) Quantitative RT-PCR of gene expression in testes from Day 0 (0), Day 6 (6), Day 15 (15), and adult (Ad) testes, fresh EPCAM+ cells (Ep), and cultured EPCAM+ cells (Cu) indicated that Bcl6b and Etv5 transcripts were present in vivo and enriched in EPCAM+ clump-forming germ cells. Bars with different letters are significantly different. Error bars represent the SEM. C) BCL6B and ETV5 proteins appeared to stain at low levels in spermatogonia in adult testis tissue (arrows). Staining was more intense in other differentiated germ cells. Insets are representative images of negative control sections in which specific primary antibodies were replaced with IgG controls. Bars = 100 μm.
To unequivocally demonstrate that Bcl6b and Etv5 are required for self-renewal by the true SSC within the cultured clump-forming germ cells, siRNA-specific reduction in gene expression followed by germ cell transplantation was conducted. Cultures of EPCAM+ clump-forming germ cells were treated with siRNA for 18 h, cultured for 1 wk, and subsequently transplanted into recipient testes. Treated cultures were maintained for 1 wk prior to transplantation to evaluate potential effects of the treatment on self-renewal compared to negative control-treated cultures. Cultures transfected with siRNA for genes important for SSC self-renewal would undergo a period of delay in SSC self-renewal followed by a recovery in self-renewal ability, resulting in fewer SSCs per culture when compared with negative control-treated cultures after the 8-day culture period and subsequent transplantation analysis. The doubling time for rat SSCs in our culture system is 11 days; however, optimal growth is obtained if cells are subcultured every 8 days. Therefore, because 8 days represents nearly 73% of the self-renewal period, we felt that 8 days was an adequate period of time to determine the effects of siRNA treatment.
If Bcl6b and Etv5 are important for SSC self-renewal, siRNA treatment should result in fewer colonies of spermatogenesis upon transplantation compared to negative control-treated cultures. Small interfering RNA treatment reduced Bcl6b and Etv5 gene expression by 37.2% and 42.9%, respectively, 18 h after transfection, and expression of the constitutively active gene, Rps2, did not change after siRNA treatment, indicating that no significant off-target effects were present (Fig. 3A and Supplemental Table S2). To evaluate changes in germ cell number due to siRNA treatment, cultures of GFP+ EPCAM+ clump-forming germ cells were analyzed by flow cytometric analysis on Days 4 and 8 after siRNA treatment. A dramatic reduction in cell numbers was observed in both negative control and gene-specific siRNA-treated cultures 4 days after treatment, presumably due to lipofectamine toxicity; however, Bcl6b and Etv5 siRNA treatments did not significantly affect the number of GFP+ cells within the culture dish compared to controls at any time point examined (Fig. 3B). Additionally, no significant differences in the percentage of apoptotic cells were observed for Bcl6b or Etv5 siRNA treatment compared to controls (Fig. 3C), indicating that neither Bcl6b nor Etv5 function in apoptosis prevention within the SSC. By evaluating cells at 4 and 8 days, we were able to specifically look at the effects of gene-specific knockdown, rather than lipofectamine, on germ cell apoptosis. The number of spermatogenic colonies arising from transplanted mt-LacZ+ cultures treated with both Bcl6b and Etv5 siRNA was significantly reduced compared with negative control siRNA-treated cultures (Fig. 3, D and E). Because of the slow-growing nature of the rat EPCAM+ clump-forming germ cell cultures, and technical transplant limitations, different primary cultures were used for each gene analysis. The concentration of stem cells is highly variable in cultures of EPCAM+ clump-forming germ cells; therefore, differences were observed in the numbers of colonies from negative control-treated cultures. However, because each gene-specific siRNA-treated culture was transplanted in parallel to a negative control-treated culture from the same primary culture, a direct comparison between colonies from specific negative control siRNA and gene-specific siRNA-treated cultures can be made. These data indicate that Bcl6b and Etv5 siRNA treatments result in a decrease in SSC number without a decrease in number of total cells or an increase in the percent apoptotic cells within the germ cell clumps compared to negative control treatments. Taken together, the data demonstrate that Bcl6b and Etv5 function in SSC self-renewal in maintenance of SSC identity.
FIG. 3.
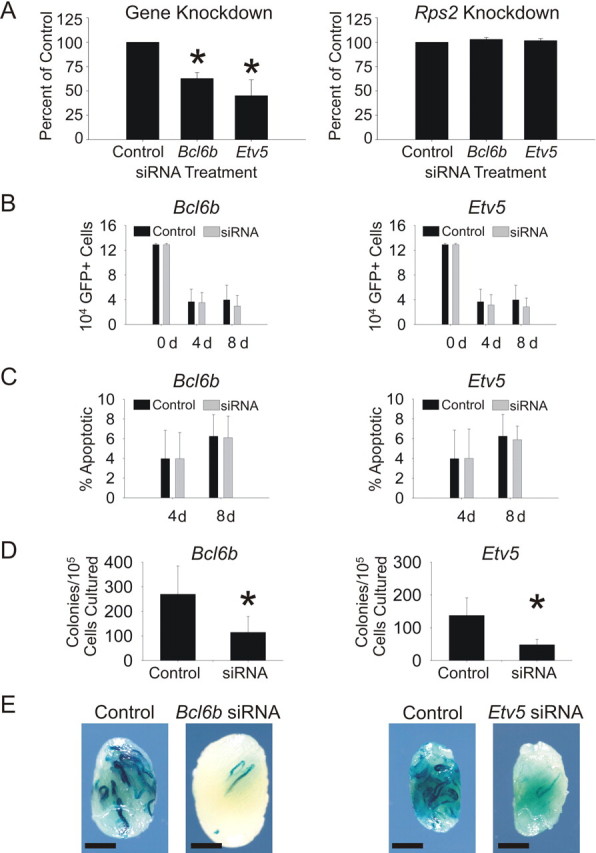
Effect of Bcl6b and Etv5 siRNA on rat SSC function. A) Bcl6b and Etv5 siRNA significantly decreased expression of Bcl6b and Etv5 in cultures of rat EPCAM+ clump-forming germ cells 18 h after transfection. Expression of Rps2 was not changed by either Bcl6b or Etv5 siRNA, indicating that no off-target effects were present. B) Bcl6b and Etv5 siRNA did not significantly change the number of donor cells in cultures of Rat GFP+ EPCAM+ clump-forming germ cells at either 4 or 8 days after transfection. C) Bcl6b and Etv5 siRNA did not significantly change the percentage of apoptotic cells (nexin positive) in cultures of Rat GFP+ EPCAM+ clump-forming germ cells at either 4 or 8 days after transfection. D) Both Bcl6b and Etv5 siRNA significantly reduced the number of SSCs in cultures of EPCAM+ clump-forming germ cells 8 days after transfection as demonstrated by the SSC transplantation assay. E) Photomicrographs of representative testes transplanted with negative control or gene-specific siRNA demonstrate the data depicted in D. Bar = 2 mm. *Statistically significant differences; error bars represent the SEM.
Identification of Bhlhe40, Hoxc4, and Tec as GDNF-Regulated and Important for Self-Renewal of Rat SSCs In Vitro
To expand our knowledge of the mechanism of GDNF-regulated SSC self-renewal, we identified, characterized, and biologically evaluated several genes previously unknown to be involved in SSC self-renewal. Seven novel GDNF-regulated genes were validated by qRT-PCR (Table 2), and three (Bhlhe40, Hoxc4, and Tec) were selected for biological validation based on their known functions and similar microarray and qRT-PCR expression patterns (Supplemental Fig. S3). Immunofluorescence analysis confirmed that basic helix-loop-helix family, member e40 (BHLHE40), homeobox C4 (HOXC4), and tec protein tyrosine kinase (TEC) proteins were expressed by EPCAM+ clump-forming germ cells (Fig. 4A). In vivo transcript expression of Bhlhe40 and Hoxc4 was higher in younger testes and cultured germ cells compared with older testes (Fig. 4B). Tec transcript expression was highest in cell populations enriched for SSCs; however, little difference in expression was observed between Day 0 and adults (Fig. 4B). Spermatogonia along the basement membrane appeared to stain lightly for HOXC4, TEC, and BHLHE40 protein. Some more advanced stages of germ cells stained with greater intensity (Fig. 4C).
TABLE 2.
Novel GDNF-regulated genes identified as GDNF regulated in cultures of rat EPCAM+ clump forming germ cells enriched for SSCs that were validated by qRT-PCR.
FIG. 4.
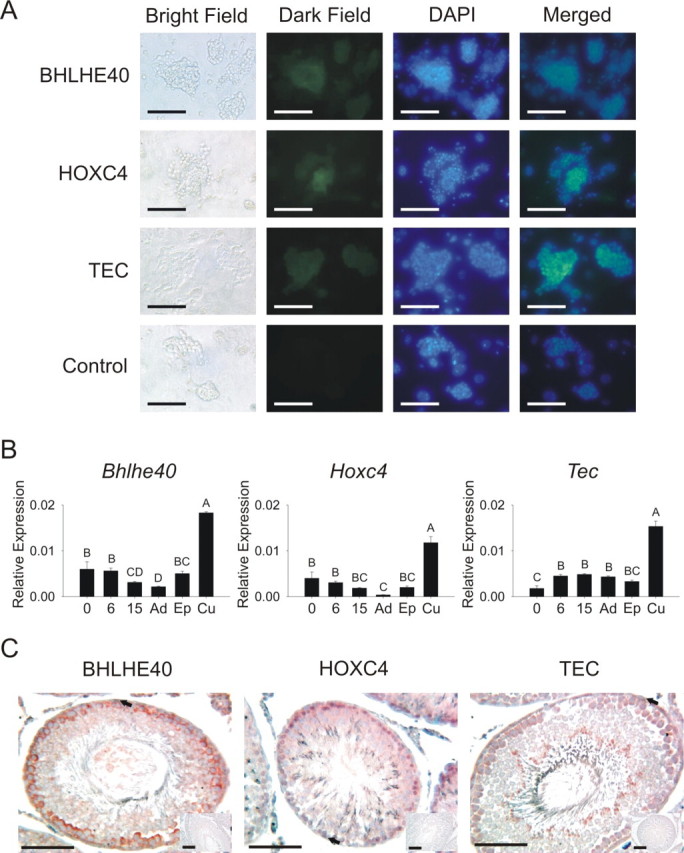
Characterization of Bhlhe40, Hoxc4, and Tec transcript and protein expression in vivo and in vitro. A) BHLHE40, HOXC4, and TEC proteins were expressed in cultures of EPCAM+ clump-forming germ cells. Column 1 = bright field; column 2 = dark field, gene-specific primary antibody (GFP); column 3 = DAPI; column 4 = merged, overlay of columns 2 and 3. Control immunofluorescence images had rabbit IgG as primary antibody. Bars = 100 μm. B) Quantitative RT-PCR of gene expression in testes from Day 0 (0), Day 6 (6), Day 15 (15), and adult (Ad) testes, fresh EPCAM+ cells (Ep), and cultured EPCAM+ cells (Cu) indicated that Bhlhe40, Hoxc4, and Tec transcripts were present in vivo and enriched in EPCAM+ clump-forming germ cells. Bars with different letters are significantly different; error bars represent the SEM. C) BHLHE40, HOXC4, and TEC proteins appeared to stain at low levels in spermatogonia in adult testis tissue (arrows). Staining was more intense in other differentiated germ cells. Insets are representative images of negative control sections in which specific primary antibodies were replaced with IgG controls. Bars = 100 μm.
In order to validate the biological significance of Bhlhe40, Hoxc4, and Tec, siRNA treatment followed by germ cell transplantation was conducted, as described above. Bhlhe40, Hoxc4, and Tec siRNA treatments significantly reduced transcript expression in cultures of EPCAM+ clump-forming germ cells without reducing Rps2 expression 18 h after transfection (Fig. 5A). Treatment of EPCAM+ clump-forming germ cells with Bhlhe40 and Hoxc4 siRNA did not affect the number of total cells within the culture well compared to controls (Fig. 5B). Interestingly, treatment with Tec siRNA resulted in a significant increase in cell numbers 4 days after treatment compared with control (Fig. 5B); however, there was no difference at 8 days. Furthermore, no significant differences in the percentage of apoptotic cells were observed for Bhlhe40, Hoxc4, or Tec siRNA treatment compared to controls (Fig. 5C). Following transplantation, the number of spermatogenic colonies arising from gene-specific siRNA-treated cultures was reduced compared with negative control siRNA-treated cultures for all three genes (Fig. 5, D and E). These data indicate that Bhlhe40, Hoxc4, and Tec are indeed important for rat SSC self-renewal.
FIG. 5.
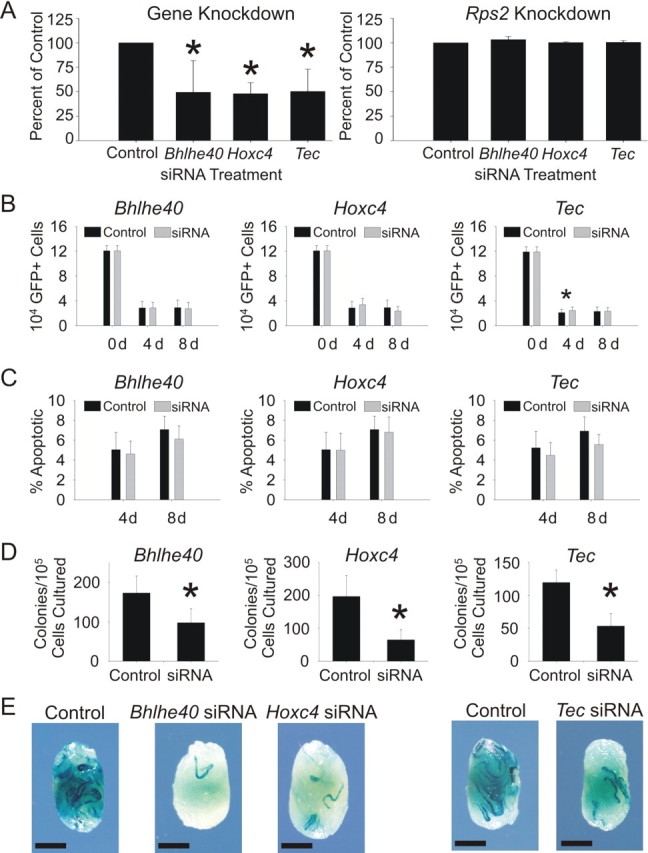
Effect of Bhlhe40, Hoxc4, and Tec siRNA on rat SSC function. A) Bhlhe40, Hoxc4, and Tec siRNA significantly decreased expression of Bhlhe40, Hoxc4, and Tec in cultures of rat EPCAM+ clump-forming germ cells 18 h after transfection. Expression of Rps2 was not changed by Bhlhe40, Hoxc4, or Tec siRNA, indicating that no off-target effects were present. B) Bhlhe40 and Hoxc4 siRNA did not significantly change the number of donor cells in cultures of Rat GFP+ EPCAM+ clump-forming germ cells at either 4 or 8 days after transfection. Cultures treated with Tec siRNA had more GFP+ cells than negative control-treated cultures at 4 days post-siRNA; however, this difference was not present after 8 days. C) Bhlhe40, Hoxc4, and Tec siRNA did not significantly change the percentage of apoptotic cells (Nexin Positive) in cultures of Rat GFP+ EPCAM+ clump-forming germ cells at either 4 or 8 days after transfection. D) Bhlhe40, Hoxc4, and Tec siRNA significantly reduced the number of SSCs in cultures of EPCAM+ clump-forming germ cells 8 days after transfection as demonstrated by the SSC transplantation assay. E) Photomicrographs of representative testes transplanted with negative control or gene-specific siRNA demonstrate the data depicted in D. Bhlhe40, Etv5 (Fig. 3), and Hoxc4 siRNA transplants were conducted at the same time with the same negative control; therefore, the same negative control image was used for all three treatments. Bar = 2 mm. *Statistically significant differences; error bars represent the SEM.
DISCUSSION
The rat SSC undergoes complete spermatogenesis within the mouse testis [6], whereas SSCs of more advanced species colonize the mouse testis, but undergo a variable degree of proliferation and little differentiation [14–17]. Additionally, GDNF has been shown to regulate the proliferation of both rat and mouse SSCs in vitro [7, 9]. Therefore, we reasoned that oligonucleotide microarray analysis of SSCs from selected species, beginning with the mouse and rat, would aid in the overall understanding of mammalian SSC self-renewal. Using this technique, we have begun to elucidate the molecular mechanisms of GDNF-mediated SSC self-renewal in the mouse [23, 24]. The objectives of this work were to identify GDNF-regulated genes in cultures of rat EPCAM+ clump-forming germ cells enriched for SSCs and to define the biological significance of a subset of these genes for rat SSC self-renewal.
To evaluate the effect of GDNF on gene expression in SSCs, it was essential to demonstrate that short-term removal of GDNF did not change the proportion of SSCs within the culture. The 18-h GDNF removal did not significantly affect the number of SSCs within the culture, indicating that this time point would be appropriate for microarray profiling of genes directly regulated by GDNF. In contrast to the rat, removal of GDNF from cultures of mouse SSCs resulted in a significant increase in stem cell number within the culture, perhaps due to hypersensitivity of GDNF following withdrawal [23]. The lack of a response in the rat may be due to a slower SSC division rate in cultures of rat germ cells (∼11 days [9]) compared with cultures of mouse germ cells (∼6 days [7]).
Microarray analysis of GDNF-regulated genes in cultures of rat EPCAM+ clump-forming germ cells revealed several thousand GDNF-regulated genes. However, only 77 genes changed 2-fold or greater after both GDNF withdrawal and replacement. Many genes that have previously been implicated as germ cell markers (Epcam, Itga6, Thy1, and Cd9; [9, 31, 32]), or important for SSC self-renewal (Bcl6b, Etv5, and Zbtb16; [23, 29]), were also expressed in the rat EPCAM+ clump-forming germ cells; however, some genes that have been implicated in SSC function, such as Nanos3 [30] and Neurog3 [34], were not expressed. Hamra et al. [21] performed a microarray on laminin-binding (SSC-enriched) vs. laminin-nonbinding rat testis cells, and several genes expressed in their SSC-enriched cell population (Bcl6b, Itga6, Egr3, and Gfra1) were also expressed by the EPCAM+ clump-forming germ cells in our study. In the work by Hamra et al. [21], functional transplantation analyses were used to confirm that the laminin-binding germ cells were indeed enriched for SSCs in order to identify germ cell genes linked to SSC function, including receptor subunits for GDNF and FGF2; however, subsequent functional transplantation analyses were not performed to determine which of the identified transcripts were required for SSC self-renewal. It is quite possible that the non-stem germ cells within our culture system also respond to GDNF withdrawal; thus, it is imperative that the importance of every gene of interest be independently validated using functional transplantation experiments.
Several genes that were identified as GDNF regulated in the mouse SSC were also GDNF regulated in cultures of rat EPCAM+ clump-forming germ cells—most notably, Bcl6b and Etv5. In fact, Bcl6b was the most highly regulated gene in both species [23]. Characterization of Bcl6b and Etv5 in vitro confirmed that both transcripts and proteins for these genes are indeed expressed in spermatogonia in cultures of EPCAM+ clump-forming germ cells enriched for SSCs. Furthermore, siRNA treatment of EPCAM+ clump-forming germ cells, followed by germ cell transplantation, confirmed the importance of both Bcl6b and Etv5 for SSC self-renewal in rats. Additionally, siRNA treatment had no influence on total cell number or apoptosis within the culture. Because siRNA treatments are transient in nature, it is unlikely that the treatment affected the ability of the SSC to home to the SSC niche. Taken together, the data indicate that these genes most likely function in maintaining the rat SSC phenotype, rather than in preventing apoptosis or stimulation of cell proliferation. In the cultured mouse SSC, Bcl6b and Etv5 siRNA resulted in delayed apoptosis and a decrease in cell numbers in vitro, in addition to a decrease in the total number of SSCs [23, 24]. These discrepancies are most likely due to the intrinsic differences between the culture systems. The proliferation rate of the SSC is much higher in the mouse culture (∼6 days) than in the rat culture (∼11 days); therefore, SSC inhibition would result in a more dramatic decrease in daughter cells within the mouse culture. Nevertheless, a direct role of these genes in the replication of the proliferating spermatogonia within the mouse, but not the rat, cannot be ruled out. To our knowledge, this is the first evidence that the intracellular mechanisms of SSC self-renewal are conserved between rats and mice, indicating that these mechanisms may be conserved in other mammals.
In an effort to increase our knowledge of GDNF-regulated genes in mammals, we identified several novel GDNF-regulated genes in cultures of rat EPCAM+ clump-forming germ cells. Three genes that were identified as GDNF regulated in the rat (Bhlhe40, Hoxc4, and Tec) were further examined for developmental expression, localization, and biological relevance. Proteins for all genes were demonstrated to be expressed in both cultured germ cells and germ cells within the testis. Protein from the identified genes (including Bcl6b and Etv5) appeared to be stained at low levels in spermatogonia of adult testes, but at higher levels by more differentiated germ cells. Because of the lack of a definitive marker for the SSC, it is impossible to demonstrate unequivocally, using histological techniques, that these proteins are indeed expressed by the SSC. However, it is possible to demonstrate functional significance of the genes, and thus their presence in SSCs, using siRNA treatments followed by quantitative transplantation experiments. Using siRNA knockdown of gene expression in cultured EPCAM+ clump-forming germ cells, followed by germ cell transplantation, we demonstrated that each gene evaluated was indeed important for SSC self-renewal. Because the siRNA transfection was transient in nature, cultures transfected with siRNA for genes important for SSC self-renewal would undergo a period of delay in SSC self-renewal, followed by a recovery in self-renewal ability. This delay resulted in fewer SSCs per culture when compared with negative control-treated cultures after the 8-day culture period and subsequent transplantation analysis. Differences in colony numbers are not due to differences in ability to colonize, but are due directly to a different number of SSCs within the siRNA-treated culture compared with the negative control. Bhlhe40, Hoxc4, and Tec were not identified as GDNF regulated in our previous mouse experiments [23]; however, array hybridizations for these genes were not adequate to determine significant differences. Thus, it is possible that Bhlhe40, Hoxc4, and Tec are also GDNF regulated in the mouse SSC, and further experimentation will clarify their roles. It is of note that Bcl6b, Etv5, Bhlhe40, Hoxc4, and Tec siRNA treatments all resulted in a decrease in SSC number within the culture dish without altering total cell number or the percent apoptotic cells. Because of this result, and the fact that all of these genes were highly regulated by GDNF, it is likely that they all function within the SSC to maintain the phenotype of the SSC, thus perpetuating the self-renewal fate decision rater than differentiation.
SSC self-renewal is a multifaceted process consisting of mechanisms such as cell proliferation, prevention of apoptosis, and maintenance of SSC phenotype. It is possible that changes in expression of evaluated genes may result in changes in expression of cell surface markers necessary for translocation of the SSC to the SSC niche after transplantation. Because the only current method to assay for the SSC is germ cell transplantation, we must assume that any change in colonization due to treatment (compared to the control treatment) is a direct downstream effect of altering one of the mechanisms that make up the process of SSC self-renewal. Based on the fact that Bcl6b, Etv5, Bhlhe40, Hoxc4, and Tec siRNA treatments did not alter cell proliferation or apoptosis, it is thus likely that these molecules are important for maintenance of the SSC phenotype. Furthermore, because the germ cells within the culture are either SSCs or their differentiating daughter cells, a change in the number of SSCs without a change in the number of differentiating daughters indicates that the ratio of SSC to differentiating germ cell in the culture has changed, and thus a shift in the ratio of self-renewal to differentiation. However, due to limitations in the rat SSC culture system and transplantation, more detailed experiments must be conducted to elucidate the exact role of these genes in the process of SSC self-renewal. Additionally, even though Lipofectamine reduced cell numbers, the effect was the same for negative control and gene specific siRNA treated cultures, therefore we are confident that the results from the siRNA experiments do in fact reflect an importance of these genes for efficient SSC self-renewal, presumably by maintaining the SSC phenotype.
Biological roles for Bcl6b and Etv5 have been explored using mouse knockout models [23, 35, 36]. Unfortunately, generation of knockout rats is challenging, and specific functional roles for these genes in rat SSC self-renewal cannot be confirmed in vivo. Bcl6b is a POZ family transcription factor, and experiments with mouse SSCs indicate that BCL6B is important for maintenance of the undifferentiated state, apoptosis, or proliferation of SSCs [23]. The fact that Bcl6b siRNA results in a decrease in proliferation of rat SSCs without directly affecting the proliferating daughter cells in the rat SSC culture indicates that, in the rat, Bcl6b is most likely important for maintenance of the undifferentiated state or influences proliferation of SSCs, but not for more differentiated germ cells. Etv5 is a transcription factor that has been clearly demonstrated to be necessary for mouse SSC self-renewal [23, 24, 36]. Additionally, a role of Etv5 in SSC niche function has also been proposed [35]. Bhlhe40 is a basic helix-loop-helix transcription factor that, in the mouse, is expressed in activated CD34+ T cells. Similar to Bcl6b, which has a role in T-cell proliferation [37], loss of Bhlhe40 in the mouse results in spontaneous activation of B and T cells, and it has been proposed that Bhlhe40 is important for the maintenance of inactivated immune cells [38]. In the testis, Bhlhe40 may have a similar function in maintaining the undifferentiated state of SSCs. Hoxc4 is a transcription factor that is important for embryonic patterning of the thoracic vertebra and proliferation of cells in the esophageal musculature [39]. The mechanistic role of Hoxc4 in SSCs is unclear, but results of the current study indicate that it is important for SSC self-renewal in the rat. Tec is a cytoplasmic protein tyrosine kinase that has been implicated in hematopoiesis, primarily in the development of the peripheral B-cell pool [40]. SFKs are important for GDNF signal transduction in the mouse SSC [24], and the structure of TEC is similar to that of SFKs [40]. It is possible that TEC kinase functions in the rat SSC similar to SFKs in the mouse SSC by mediating the GDNF signal. Further exploration of the function of these genes in SSCs will lead to a better understanding of their roles in GDNF-mediated SSC self-renewal.
To better understand the mechanism of pluripotency, we examined the levels of Sox2, Nanog, and Pou5f1 in the cultures of rat EPCAM+ germ cells. Both Nanog and Pou5f1 were expressed by the cultured cells; however, Sox2 was not. This result is in contrast to cultures of Thy1+ mouse germ cells [23] in which Sox2 and Pou5f1 were expressed and Nanog was not. In ES cells, Nanog expression and its apparent role in maintaining self-renewal and pluripotency is regulated by the continued action of Pou5f1 and Sox2. Lack of Sox2 expression by cultures of rat EPCAM+ germ cells, combined with the observation that GDNF does not regulate Pou5f1, indicates that self-renewal of ES cells and rat SSCs are regulated in different manners.
Previously, we identified three factors (Bcl6b, Etv5, and Lhx1) that are GDNF regulated and important for SSC self-renewal through SFK signaling mechanisms in the mouse [23, 24]. The present work demonstrates that the function of Bcl6b and Etv5 are likely similar between the mouse and the rat. In fact, Bcl6b was the most highly GDNF-regulated gene in both the rat and mouse SSC. These data indicate that GDNF-regulated SSC self-renewal is at least partially conserved between these two species. Furthermore, we have identified and biologically validated three more factors (Bhlhe40, Hoxc4, and Tec) that are GDNF regulated and important for SSC self-renewal in the rat, which further extends our knowledge of the mechanism of SSC self-renewal in mammals (Supplemental Fig. S4). The dramatic regulation of Bcl6b in both the mouse and rat, as well as the GDNF regulation of Etv5, suggests other exciting areas for continued investigation of SSC regulation. Using these two genes as an entry point should allow for discoveries about the downstream regulation of SSC self-renewal in the mouse and rat. This knowledge is extremely valuable for the continued development of SSC applications in more advancing species, including livestock and humans.
Supplementary Material
Acknowledgments
We thank Drs. J. Oatley, X. Wu, and Z. Niu for critical evaluation of the manuscript, Dr. Rex Hess for advice on immunohistochemistry, C. Freeman and R. Naroznowski for assistance with animal maintenance, Dr. J. Maltzman and M. Schmidt for assistance with flow cytometry, J. Hayden for photography, and the University of Pennsylvania Cell Morphology Core for histological preparations. We also thank Dr. Fearon for Bcl6b antibody used for immunofluorescence.
Footnotes
1Supported by the National Institutes of Health (National Institute of Child Health and Human Development, HD044445 and HD052728) and the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation. Microarray data has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under the accession number GSE15652.
REFERENCES
- Russell LD, Ettlin RA, Hakim AP, Clegg ED. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River Press; 1990: 1 40 [Google Scholar]
- Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol 2006; 419: 259 282 [DOI] [PubMed] [Google Scholar]
- Tegelenbosch RAJ, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res 1993; 290: 193 200 [DOI] [PubMed] [Google Scholar]
- Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A 1994; 91: 11303 11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmerman JW. Spermatogenesis following male germ cell transplantation. Proc Natl Acad Sci U S A 1994; 91: 11298 112302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Avarbock MR, Maika SD, Hammer RE, Brinster RL. Rat spermatogenesis in mouse testis. Nature 1996; 381: 418 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A 2004; 101: 16489 16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol Reprod 2004; 71: 722 731 [DOI] [PubMed] [Google Scholar]
- Ryu B-Y, Kubota H, Avarbock MR, Brinster RL. Conservation of spermatogonial stem cell renewal signaling between mouse and rat. Proc Natl Acad Sci U S A 2005; 102: 14302 14307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287: 1489 1493 [DOI] [PubMed] [Google Scholar]
- Yomogida K, Yagura Y, Tadokoro Y, Nishimune Y. Dramatic expansion of germinal stem cells by ectopically expressed human glial cell line-derived neurotrophic factor in mouse Sertoli cells. Biol Reprod 2003; 69: 1303 1307 [DOI] [PubMed] [Google Scholar]
- Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, Garbers DL. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc Natl Acad Sci U S A 2005; 102: 17430 17435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Muneto T, Lee J, Takenaka M, Chuma S, Nakatsuji N, Horiuchi T, Shinohara T. Long-term culture of male germline stem cells from hamster testes. Biol Reprod 2008; 78: 611 617 [DOI] [PubMed] [Google Scholar]
- Dobrinski I, Avarbock MR, Brinster RL. Germ cell transplantation from large domestic animals into mouse testes. Mol Reprod Dev 2000; 57: 270 279 [DOI] [PubMed] [Google Scholar]
- Oatley JM, Reeves JJ, McLean DJ. Biological Activity of cryopreserved bovine spermatogonial stem cells during in vitro culture. Biol Reprod 2004; 71: 942 947 [DOI] [PubMed] [Google Scholar]
- Nagano M, McCarrey JR, Brinster RL. Primate spermatogonial stem cells colonize mouse testes. Biol Reprod 2001; 64: 1409 1416 [DOI] [PubMed] [Google Scholar]
- Nagano M, Patrizio P, Brinster RL. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril 2002; 78: 1225 1233 [DOI] [PubMed] [Google Scholar]
- McLean DJ, Friel PJ, Pouchnik D, Griswold MD. Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone treated rat Sertoli cells. Mol Endocrinol 2002; 16: 2780 2792 [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod 2004; 72: 492 501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319 330 [DOI] [PubMed] [Google Scholar]
- Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, Garbers DL. Defining the spermatogonial stem cell. Dev Biol 2004; 269: 393 410 [DOI] [PubMed] [Google Scholar]
- Schmidt JA, de Avila JM, McLean DJ. Analysis of gene expression in bovine testis tissue prior to ectopic testis tissue xenografting and during the grafting period. Biol Reprod 2007; 76: 1071 1080 [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Teleranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A 2006; 103: 9524 9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem 2007; 282: 25842 25851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrinski I, Ogawa T, Avarbock MR, Brinster RL. Computer assisted image analysis to assess colonization of recipient seminiferous tubules by spermatogonial stem cells from transgenic donor mice. Mol Reprod Dev 1999; 53: 142 148 [DOI] [PubMed] [Google Scholar]
- Nagano M, Avarbock MR, Brinster RL. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol Reprod 1999; 60: 1429 1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Miki H, Ogonuki N, Takehashi M, Morimoto T, Ogura A, Shinohara T. Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol Reprod 2006; 75: 68 74 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. Krawetz S, Misener S. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000: 365 386 [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36: 647 652 [DOI] [PubMed] [Google Scholar]
- Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, Grimaldi P. Potential role of Nanos3 in maintaining the undifferentiated spermatogonia population. Dev Biol 2008; 313: 725 738 [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A 2003; 100: 6487 6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Toyokuni S, Shinohara T. CD9 is a surface marker on mouse and rat male germline stem cells. Biol Reprod 2004; 70: 70 75 [DOI] [PubMed] [Google Scholar]
- Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T, Nishikawa S. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 1991; 113: 689 699 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, Suda T, Nabeshima Y. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol 2004; 269: 447 458 [DOI] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao G-Q, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 2005; 436: 1030 1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow CM, Hostetler CE, Griswold MD, Hofmann MC, Murphy KM, Cooke PS, Hess RA. ETV5 is required for continuous spermatogenesis in adult mice and may mediate blood testes barrier function and testicular immune privilege. Ann N Y Acad Sci 2007; 1120: 144 151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders PM, Hunter PJ, Telaranta AI, Carr JM, Marshall JL, Carrasco M, Murakami Y, Palmowski MJ, Cerundolo V, Kaech SM, Ahmed R, Fearon DT. BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proc Natl Acad Sci U S A 2004; 102: 7418 7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lu B, Li RQ, Flavell RA, Taneja R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol 2001; 2: 1040 1047 [DOI] [PubMed] [Google Scholar]
- Bouletand M, Capecchi MR. Targeted disruption of hoxc-4 causes esophageal defects and vertebral transformations. Dev Biol 1996; 177: 232 249 [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Jung S, Sunshine MJ, Hatam F, Xu Y, Baltimore D, Mano H, Littman DR. Severe B cell deficiency in mice lacking the tec kinase family members Tec and Btk. J Exp Med 2000; 192: 1611 1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.