Abstract
Although it is well established that tumors initiate an angiogenic switch, the molecular basis of this process remains incompletely understood. Here we show that the miRNA miR-132 acts as an angiogenic switch by targeting p120RasGAP in the endothelium and thereby inducing neovascularization. We identified miR-132 as a highly upregulated miRNA in a human embryonic stem cell model of vasculogenesis and found that miR-132 was highly expressed in the endothelium of human tumors and hemangiomas but was undetectable in normal endothelium. Ectopic expression of miR-132 in endothelial cells in vitro increased their proliferation and tube-forming capacity, whereas intraocular injection of an antagomir targeting miR-132, anti–miR-132, reduced postnatal retinal vascular development in mice. Among the top-ranking predicted targets of miR-132 was p120RasGAP, which we found to be expressed in normal but not tumor endothelium. Endothelial expression of miR-132 suppressed p120RasGAP expression and increased Ras activity, whereas a miRNA-resistant version of p120RasGAP reversed the vascular response induced by miR-132. Notably, administration of anti–miR-132 inhibited angiogenesis in wild-type mice but not in mice with an inducible deletion of Rasa1 (encoding p120RasGAP). Finally, vessel-targeted nanoparticle delivery1 of anti–miR-132 restored p120RasGAP expression in the tumor endothelium, suppressed angiogenesis and decreased tumor burden in an orthotopic xenograft mouse model of human breast carcinoma. We conclude that miR-132 acts as an angiogenic switch by suppressing endothelial p120RasGAP expression, leading to Ras activation and the induction of neovascularization, whereas the application of anti–miR-132 inhibits neovascularization by maintaining vessels in the resting state.
Endothelial cells in the adult mammal are among the least proliferative cell types, with about one in 10,000 cells entering the cell cycle at any given time2. This quiescence is rapidly reversed in response to growth factors during pathological neovascularization, particularly during tumorigenesis3. The robust proliferative switch of the quiescent endothelium is a complex process that is governed by a network of checks and balances. Small 22-nt RNAs called miRNAs are key regulators of several physiological processes, including angiogenesis4. To identify miRNAs that activate quiescent endothelium, we profiled miRNAs in both human umbilical vein endothelial cells (HUVECs) treated with the angiogenic growth factors vascular endothelial growth factor (VEGF) or basic fibroblast growth factor (bFGF) and in a human embryonic stem cell vasculogenesis model5,6 in which embryoid bodies derived from human embryonic stem cells form well defined endothelial networks after 14 d in culture (Supplementary Fig. 1). miR-132 had the highest combined rank of all miRNAs across these screens (Supplementary Fig. 2).
miR-132 is a highly conserved miRNA transcribed from an intergenic region on human chromosome 17 by the transcription factor cAMP response element binding protein (CREB)7,8. Although no studies to our knowledge have linked miR-132 to endothelial cells, miR-132 can be expressed in neuronal cells upon stimulation with brain-derived neurotropic factor (BDNF)8. Both VEGF and bFGF can rapidly induce CREB9,10, but it is not known whether this activation is sustained enough to induce expression of miR-132 in endothelial cells. To address this issue, we investigated the kinetics of CREB phosphorylation in HUVECs and found that VEGF treatment induced peak activation of CREB after 15–30 min and, more notably, induced sustained activation for up to 9 h (Supplementary Fig. 3a). Accordingly, both VEGF and bFGF upregulated miR-132 in endothelial cells 3–6 h after treatment (Supplementary Fig. 3b). By contrast, miR-132 levels did not significantly change in human aortic smooth muscle cells treated with platelet-derived growth factor-BB (PDGF-BB; data not shown), indicating that miR-132’s potential effects on neovascularization might primarily involve the endothelium.
As tumors are potent inducers of pathological neovascularization in adults, we investigated whether tumor-associated angiogenic factors can upregulate endothelial miR-132. Indeed, miR-132 was significantly upregulated in HUVECs treated with conditioned media from breast and pancreatic tumor cell lines (Supplementary Fig. 3c). In particular, conditioned medium from MDA-MB-231 human breast carcinoma cells promoted miR-132 expression to a similar degree as VEGF (Supplementary Fig. 3c). Treatment of HUVECs with MDA-MB-231–conditioned medium led to increased phosphorylation of CREB (indicating its activation) that was reversed by pretreatment with the VEGF receptor-2 (VEGFR-2) inhibitor vatalanib (Supplementary Fig. 3d). This result suggests that tumors could potentially upregulate endothelial miR-132 by activating CREB through a VEGFR-2–dependent pathway.
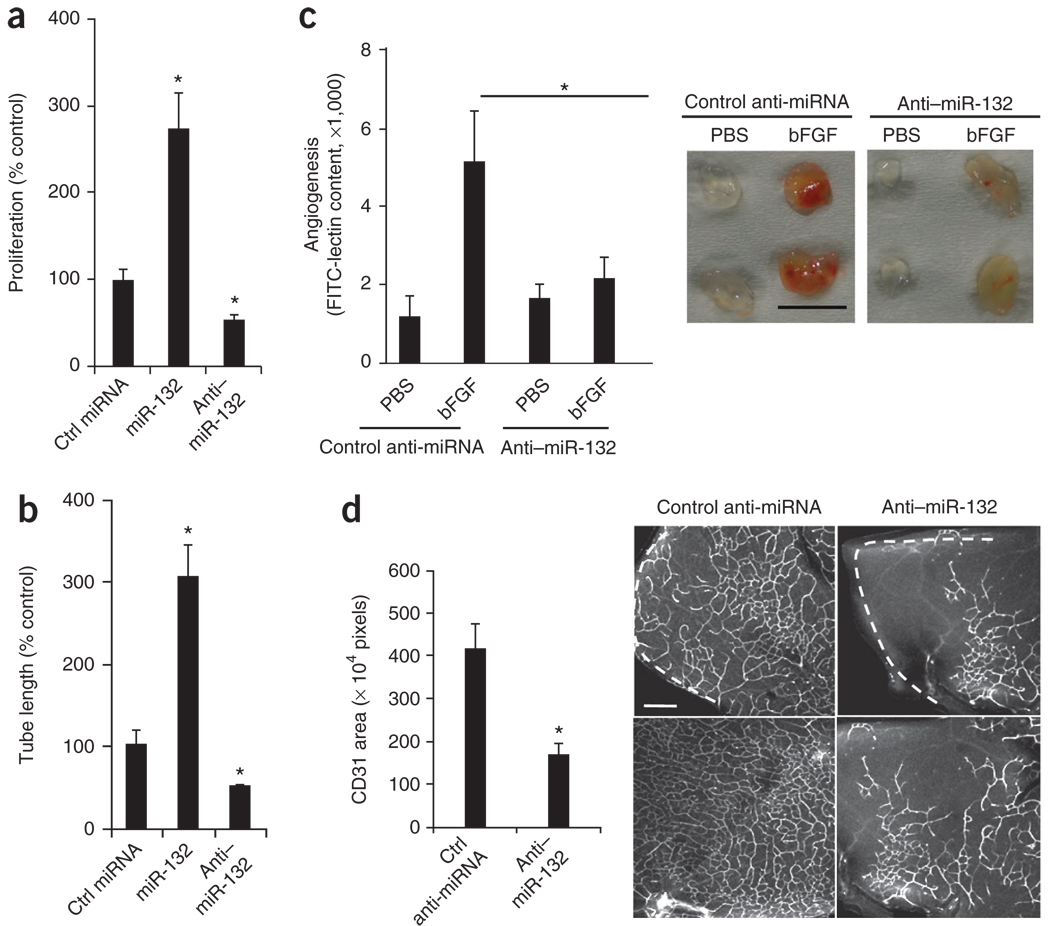
To investigate the effects of miR-132 on endothelial cells, we transfected HUVECs with mature human miR-132 or its complementary antagonist, anti–miR-132. We confirmed that these oligonucleotides were taken up by the cells (Supplementary Fig. 4a,b) and then tested their effects on cell proliferation in vitro and tube formation in a three-dimensional collagen matrix. miR-132 considerably increased cell proliferation and tube formation, whereas anti–miR-132 decreased these activities below baseline (Fig. 1a,b). Next, we investigated whether systemic administration of anti–miR-132 could inhibit angiogenesis in vivo. A single dose of anti–miR-132 significantly decreased bFGF-mediated angiogenesis in subcutaneous Matrigel implants in mice (Fig. 1c). This effect diminished over time and disappeared by day 8 (data not shown), indicating that sustained blockade of miR-132 might need a higher initial dose or continuous dosing with anti–miR-132. These data show that miR-132 can regulate growth factor-induced angiogenesis in vitro and in vivo.
Figure 1.
miR-132 regulates growth factor–mediated angiogenesis in vitro and in vivo. (a) HUVEC cell proliferation. After transfection with miR-132 or anti–miR-132 or a control (Ctrl) miRNA, HUVECs were pulsed with BrdU and cell proliferation was measured using an ELISA assay. One representative experiment of three is shown, with the average values of triplicate wells. *P < 0.01 compared to control miRNA. (b) HUVEC tube formation. 24 h after transfection as in a, HUVECs were suspended in a three-dimensional collagen matrix. Tube lengths were measured using MetaMorph software on day 4. One representative experiment of three is shown, with the average values of triplicate wells. *P < 0.01 compared to control miRNA. (c) Angiogenesis in Matrigel plugs in vivo. Growth factor–reduced Matrigel containing either PBS or bFGF was injected subcutaneously into C57BL/6 mice. Mice received 10 µg of either a control anti-miRNA or anti–miR-132 in PBS intravenously (n = 6 per group). Angiogenesis was quantified by measuring FITC-lectin content on day 5. *P < 0.05 for control bFGF plugs compared to anti–miR-132 bFGF plugs. Right micrographs show representative Matrigel plugs from each group. Scale bar, 1 cm. (d) Retinal angiogenesis. Either control anti-miRNA or anti–miR-132 (1 µg) was injected intraocularly into 6-d-old BALB/c pups (n = 5 per group). Retinas were collected and stained with CD31-specific monoclonal antibodies (mAb). Bars show mean CD31 area (n = 25 fields) calculated using MetaMorph software. *P < 0.01. Right micrographs show representative confocal images of the deep plexus retinal vasculature. White dashed lines indicate the periphery of the retinas. Scale bar, 100 µm. Bars show means ± s.e.m.
Antiangiogenic agents that target the VEGFR pathway have shown considerable clinical benefit in patients with retinal diseases associated with pathological neovascularization11,12. To assess the effects of miR-132 on retinal neovascularization, we treated 6-d-old mice undergoing postnatal retinal neovascularization with a single intraocular injection of anti–miR-132 and monitored vascular growth 6 d later. Anti–miR-132 produced a 50% decrease in retinal neovascularization in the deep plexus (Fig. 1d) but had no significant effect on preestablished vessels in the superficial plexus (Supplementary Fig. 5a). Anti–miR-132 treatment did not alter perivascular coverage by smooth muscle cells in retinas (Supplementary Fig. 5b) or in established Matrigel plugs (data not shown), indicating that anti–miR-132 exerts its antiangiogenic effects by acting on endothelial cells rather than on perivascular cells. As our data show that miR-132 functions downstream of multiple angiogenic growth factors both in vitro in endothelial cells treated with VEGF and bFGF, and in vivo in bFGF Matrigel plugs, anti–miR-132 might produce a broader antivascular effect than agents that selectively target the VEGF pathway.
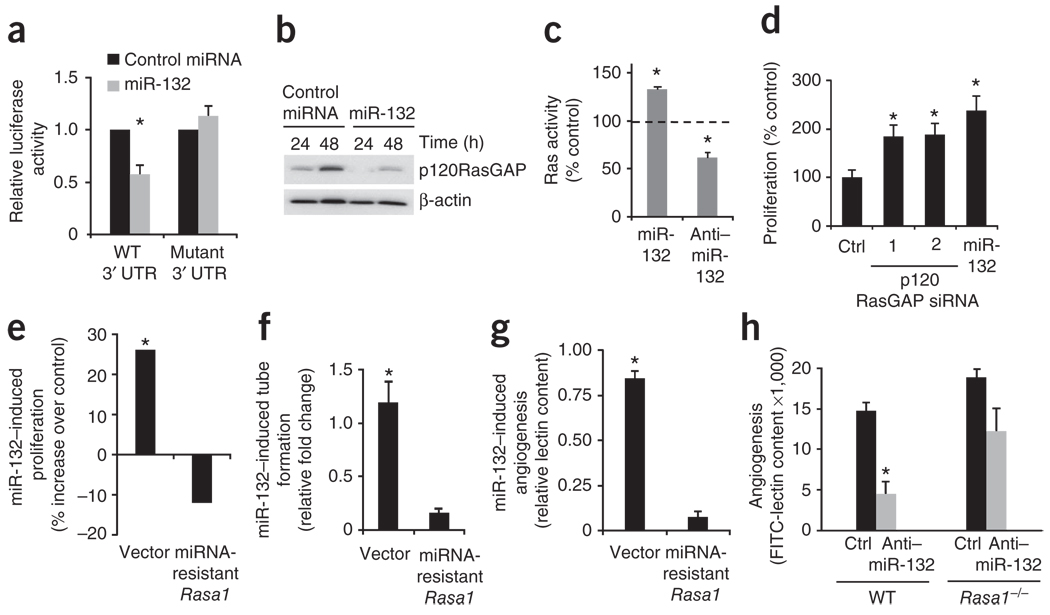
To identify the targets of miR-132 we used three algorithms, miR-base13, TargetScan14 and Pictar15, all of which predicted five potential direct targets (Supplementary Fig. 6a,b). RNAhybrid16 modeling of the 3′ untranslated regions (UTRs) of these potential targets revealed that only the top candidate—Rasa1, encoding p120RasGAP (also known as RASA1 and RasGAP)—had more than one predicted miR-132 binding site in its 3′ UTR. RNAhybrid modeling of the Rasa1 3′ UTR predicted two miR-132 binding sites, separated by 40 bases (Supplementary Fig. 6c). The presence of such cooperative miRNA binding sites has been implicated in synergistic repression of targets17. Accordingly, ectopic expression of miR-132 suppressed a luciferase reporter upstream of a 70-bp region of the Rasa1 3′ UTR (Fig. 2a). Mutagenesis of the seed sequences of the two predicted miR-132 binding sites restored luciferase expression, thereby confirming the specificity of the interaction between miR-132 and the Rasa1 3′ UTR (Fig. 2a). Contact inhibition in endothelial cells led to upregulation of p120RasGAP expression after 48 h of growth in culture (Fig. 2b), consistent with the cell density-dependent increase in GAP activity reported in other cells18. Transfection of miR-132 into HUVECs decreased endogenous p120RasGAP expression by 70% in subconfluent cells (24 h of growth) and by 50% in confluent cells (48 h of growth; Fig. 2b) but did not affect any of the other targets predicted by all three algorithms, or known regulators of angiogenesis among the targets predicted by the in silico algorithms we used (Supplementary Fig. 7). Furthermore, knockdown of miR-132 with an anti–miR-132 markedly increased p120RasGAP levels in vitro in HUVECs (Supplementary Fig. 8a) and in vivo during bFGF-induced angiogenesis in subcutaneous Matrigel implants in mice (Supplementary Fig. 8b,c).
Figure 2.
Endothelial activation mediated by miR-132 depends on its downregulation of p120RasGAP. (a) Luciferase activity of 293T cells co-transfected with a luciferase-Rasa1 3′ UTR plasmid containing either the WT 3′ UTR or a mutated sequence, a β-galactosidase plasmid and either a control miRNA or miR-132. Bars shown mean relative luciferase activity normalized to the β-galactosidase levels (n = 3). *P < 0.05 compared to control miRNA. (b) Western blot detecting p120RasGAP in HUVECs transfected with either a control miRNA or miR-132. One representative experiment of two is shown. (c) Ras activity in HUVECs transfected with control miRNA, miR-132, control anti-miRNA or anti–miR-132. One representative experiment of two is shown, with the average value of triplicate wells. *P < 0.05 compared to control miRNA or control anti-miRNA. (d) HUVEC cell proliferation. After transfection with two different siRNAs (1 and 2) targeting p120RasGAP or with miR-132, HUVECs were pulsed with BrdU and cell proliferation was measured using an ELISA assay. Ctrl, control siRNA. One representative experiment of three is shown. *P < 0.05. Cell proliferation (e) and tube formation (f) of HUVECs. Cells were transfected with a control vector or a miRNA-resistant form of Rasa1 and, 24 h later, were transfected with miR-132 or a control miRNA. The values shown are for miR-132 transfection relative to control miRNA transfection. One representative experiment of two is shown. P < 0.05 (miR-132 compared with control miRNA). (g) Angiogenesis in Matrigel plugs in vivo. Forty-eight hours after mice were injected with bFGF Matrigel plugs, they were injected with either a control plasmid (vector) or a plasmid encoding miRNA-resistant Rasa1 in nanoparticles. After a further 24 h, mice in each group (n = 3 per group) were treated with either a control miRNA or miR-132 in RGD nanoparticles. Angiogenesis in the plugs was quantified by measuring FITC-lectin content. The values shown are for mice receiving miR-132 normalized to those receiving control miRNA. One representative experiment of two is shown. P < 0.05 (miR-132 compared with control miRNA). (h) Angiogenesis in Matrigel plugs in vivo. Tamoxifen-treated Rasa1fl/fl Ert2-ubiquitin-Cre mice (Rasa1−/−) (n = 3 per group) and wild-type mice (n = 3 per group) were injected with bFGF-containing Matrigel plugs and 2 h later treated with either a control anti-miRNA (Ctrl) or anti–miR-132. Angiogenesis was quantified by measuring FITC-lectin content. *P < 0.05 (anti–miR-132 compared with control anti-miRNA in WT mice). Bars show means ± s.e.m.
p120RasGAP is a known negative regulator of Ras that inactivates Ras by enhancing its intrinsic GTPase activity19. Accordingly, ectopic expression of miR-132 in HUVECs increased Ras activity, whereas anti–miR-132 reversed this effect, as measured in an ELISA assay using a GST-tagged, Ras-binding domain of Raf-1 to pull down active Ras (as detected with a Ras-specific antibody) (Fig. 2c). Consistent with its effects on Ras activity, anti–miR-132 substantially decreased VEGF-induced phosphorylation of mitogen-activated protein kinase extracellular related protein kinase kinase-1 (MEK-1) in HUVECs (Supplementary Fig. 9). Treatment of HUVECs with a MEK inhibitor (PD0325901) abrogated the proliferative effects of miR-132 (Supplementary Fig. 9). Moreover, knockdown of p120RasGAP using either of two different siRNAs increased endothelial proliferation to a similar extent as did miR-132 (Fig. 2d). Notably, expression of a miR-resistant Rasa1 lacking its 3′ UTR was sufficient to abrogate the miR-132–induced increase in cell proliferation (Fig. 2e) and tube formation (Fig. 2f) in vitro as well as angiogenesis in bFGF-containing Matrigel plugs in vivo (Fig. 2g).
To further elucidate the role of p120RasGAP in mediating miR-132’s effects, we crossed mice containing loxP-flanked (floxed) alleles of Rasa1 with Ert2-ubiquitin-Cre mice20 to generate mice in which Rasa1 can be deleted by tamoxifen administration. Tamoxifen binding to the estrogen receptor Ert2 leads to expression of the Cre recombinase gene downstream of the ubiquitin promoter, resulting in a deletion of the floxed allele in all cell types. Injection of tamoxifen into these mice led to a substantial decrease in p120RasGAP expression in multiple tissues (data not shown). Treatment of these mice, which lacked p120RasGAP, with anti–miR-132 did not markedly decrease angiogenesis in Matrigel plugs, as it did in control mice (Fig. 2h). Thus, the functional consequences of miR-132 in endothelial cells are predominantly, if not exclusively, mediated through its regulation of p120RasGAP expression. Moreover, we found that miR-132 could promote cell proliferation through downregulation of p120RasGAP in the mouse endothelioma cell line b.End3 (Supplementary Fig. 10), indicating that this regulatory circuit might act as a conserved angiogenic switch mechanism regulating endothelial survival, growth or both.
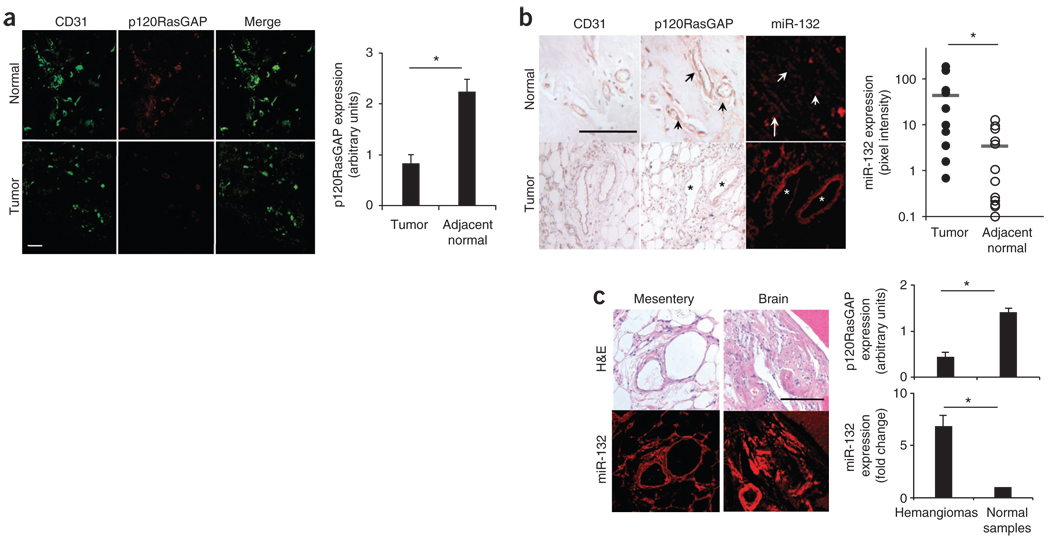
Previous studies have provided evidence that p120RasGAP acts as a crucial negative regulator of vascular development and remodeling. For instance, deletion of Rasa1 causes severe vascular remodeling and embryonic lethality in mice21. Mutations in the human gene encoding p120RasGAP have been linked to capillary and arteriovenous malformations and Parkes-Weber syndrome22–24. On the basis of these observations, we hypothesized that during tumor neovascularization, miR-132 levels would increase in angiogenic endothelial cells, thereby reducing p120RasGAP expression. Indeed, endothelial cells in human breast tumors had abundant levels of miR-132 and minimal amounts of p120RasGAP (Fig. 3a,b). By contrast, the endothelium of normal human breast tissue expressed abundant p120RasGAP but contained no detectable miR-132, as measured by in situ hybridization (Fig. 3a,b). This reciprocal expression of p120RasGAP and miR-132 was not unique to breast carcinoma but was also seen in the endothelium of orthotopic murine pancreatic tumors relative to normal mouse pancreas (Supplementary Fig. 11). In these breast tumors, p120RasGAP was readily detectable on perivascular structures associated with the tumor neovasculature (data not shown). Moreover, 75% of human hemangiomas tested (51/68) expressed miR-132 but had no detectable expression of p120RasGAP, confirming that miR-132 is a marker of hyperproliferative or activated endothelium (Fig. 3c and Supplementary Fig. 12). The reciprocal expression of miR-132 in proliferative endothelium and p120RasGAP in quiescent endothelium suggests that the regulation of p120RasGAP by miR-132 can facilitate an angiogenic switch. Our observation that PDGF-BB, a growth factor for smooth muscle and stromal cells, does not upregulate miR-132 in human aortic smooth muscle cells in vitro suggests that the miR-132–p120RasGAP regulatory mechanism is specific for the endothelium.
Figure 3.
miR-132 and p120RasGAP are expressed reciprocally in quiescent versus proliferative endothelium. (a,b) Normal human breast tissue or human breast carcinoma was analyzed for CD31 and p120RasGAP expression by immunofluorescence (a) and with miR-132 expression by in situ hybridization (b). Quantification of the results for 12 breast carcinoma tissue sections and adjacent normal tissue is also shown. Arrows and asterisks indicate blood vessels in the normal and tumor sections. Scale bar, 100 µm. *P < 0.05. (c) In situ hybridization for miR-132 and staining for p120RasGAP was performed on a human hemangioma tissue array. Representative H&E sections and miR-132 staining is shown for hemangiomas from mesentery and brain. Quantification is shown for miR-132 expression as the fold increase over normal tissue and for RasGAP expression scored on a 1–4 scale. miR-132 expression was quantified using MetaMorph software. Scale bar, 100 µm. n = 68 hemangioma samples; n = 32 normal samples. *P < 0.05, Mann-Whitney U test. Bars show means ± s.e.m.
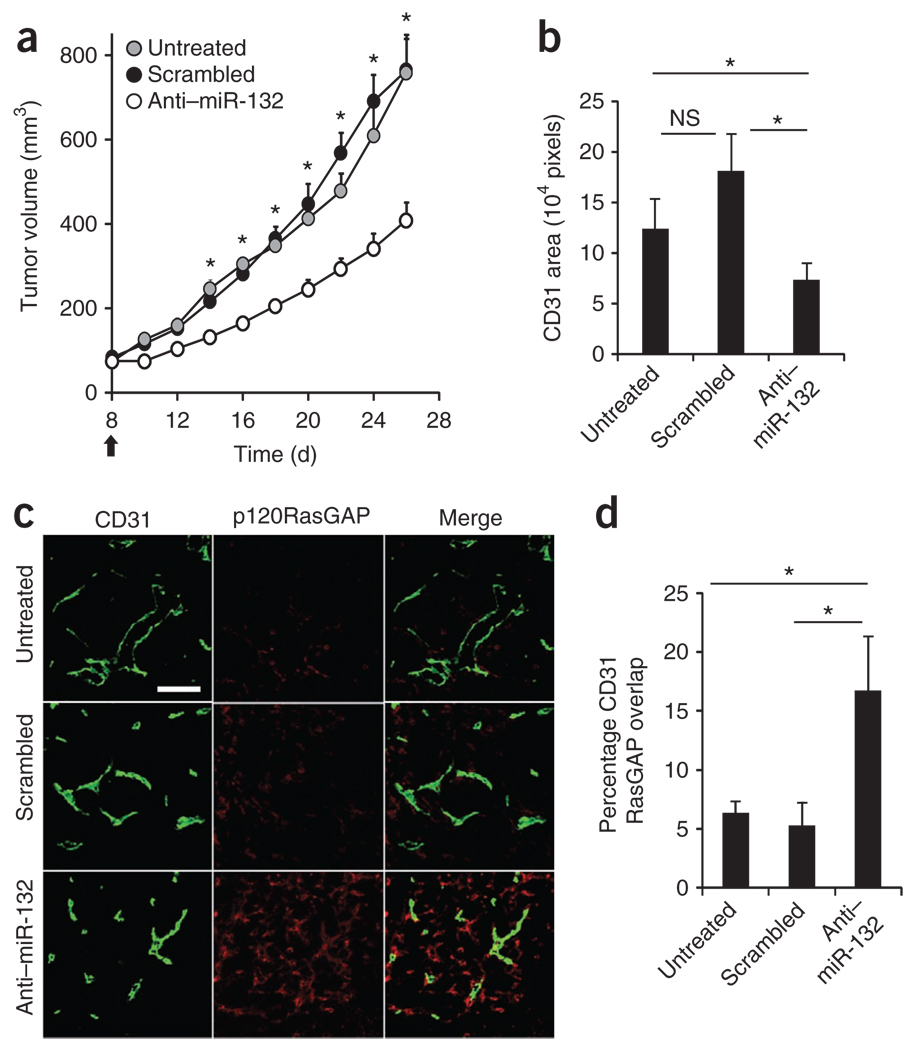
On the basis of these observations, we hypothesized that inhibition of miR-132 in the tumor vasculature might result in decreased angiogenesis and tumor burden by restoring the expression of p120RasGAP. To test this hypothesis, we used an integrin αvβ3-targeted nanoparticle that can deliver nucleic acids25 or drugs1 to the tumor neovasculature for selective delivery of anti–miR-132 to the tumor endothelium of mice. We confirmed that these nanoparticles were able to deliver a fluorescently labeled anti–miR-132 to tumor vasculature (Supplementary Fig. 13). Systemic administration of anti–miR-132 nanoparticles not only blocked angiogenesis induced by a VEGF-secreting ovarian carcinoma in mice (Supplementary Fig. 14) but also significantly decreased tumor burden and angiogenesis in an orthotopic xenograft mouse model of human breast carcinoma, MDA-MB 231 (Fig. 4a,b and Supplementary Fig. 14). After treatment with anti–miR-132 nanoparticles, the vasculature of these mice showed increased endothelial p120RasGAP (Fig. 4c,d), thereby confirming that antagonism of miR-132 can restore p120RasGAP expression during neovascularization in vivo. Although anti–miR-132 treatment did not affect the expression p120RasGAP in myeloid cells, tumor cells or stromal cells (data not shown), we did observe a twofold increase in p120RasGAP in lymphatic endothelial cells identified by positive staining for lymphatic vessel endothelial hyaluronan receptor (LYVE-1; Supplementary Fig. 15). However, unlike the CD31+ endothelial area, which was reduced upon anti–miR-132 treatment (Fig. 4b), the area of LYVE-1+ cells was not markedly different between the groups (data not shown), suggesting that anti–miR-132 does not affect lymphangiogenesis in these tumors. Moreover, anti–miR-132 did not affect the proliferation of the MDA-MB-231 cells in vitro and had no effect on perivascular cells as measured by the area of smooth muscle actin staining colocalizing with CD31 in the MDA-MB-231 tumors (data not shown).
Figure 4.
Targeted delivery of anti–miR-132 decreases tumor burden by restoring endothelial p120RasGAP. Nude mice were implanted with 2 × 106 MDA-MB 231 cells in the mammary fat pad. The tumors were measured 8 d later, and mice with palpable tumors of similar volumes were randomly assigned to three groups. Mice were left untreated or received 50 µg of scrambled anti-miRNA or anti–miR-132 every 48 h until day 24. (a) Tumor volumes (mean ± s.e.m) of at least seven mice per group, starting from the day of treatment (black arrow). *P < 0.05 compared to anti–miR-132. (b–d) Immunofluorescent staining of CD31 and p120RasGAP in tumor sections from at least three mice per group. (b) Microvessel density measured by CD31 area. (c) Representative images of p120RasGAP expression in CD31-positive cells. Scale bar, 100 µm. (d) Quantification of the overlap of CD31 and p120RasGAP expression (percentage of CD31 cells that are also positive for RasGAP expression). Pixel density was quantified using MetaMorph software. Bars are mean ± s.e.m of multiple images from at least three mice per group. *P < 0.05 (anti–miR-132 compared to untreated or scrambled). NS, not significant.
Angiogenesis is a complex process that depends on the balance of pro- and antiangiogenic factors that influence the quiescence or proliferative state of the endothelium in a wide variety of disease states. We show here that miR-132 is expressed in activated endothelial cells, where it suppresses p120RasGAP expression, permitting activation of Ras and leading to neovascularization. Moreover, neutralization of miR-132 with an antagomir suppresses retinal and tumor angiogenesis, pointing toward a new therapeutic strategy for diseases associated with pathological angiogenesis. miR-126 has recently been described to affect developmental neovascularization by targeting several well known angiogenic pathways and transcription factors such as Sprouty-related EVH-1 domain containing-1 (SPRED-1)26–28, but the role of miR-126 in pathological angiogenesis has yet to be determined.
Other miRNAs, notably miR-296 (ref. 29) and miR-92a (ref. 30), have been described as positive or negative regulators of angiogenesis. miR-296 was identified in a screen for endothelial miRNAs that were dysregulated during co-culture with a human glioma cell line and miR-296 was shown to target HGS (hepatocyte growth factor–regulated tyrosine kinase substrate), an endosome-associated protein that affects sorting and trafficking of growth factor receptors31. Although blockade of miR-296 decreases angiogenesis in tumor xenografts29, it is unclear whether such blockade produces a substantial decrease in tumor burden in vivo. In our screens, miR-296 was upregulated in growth factor–treated endothelial cells but not in the developmental angiogenesis screen using human embryonic stem cells. By contrast, miR-132 was upregulated in endothelial activation during both developmental and pathological neovascularization. Although these two models have different kinetics and growth factor requirements and recapitulate distinct physiological responses, they share common features: activation of the proliferation, migration and tube formation programs of endothelial cells. Accordingly, anti–miR-132 blocked not only growth factor–induced angiogenesis in vitro and in vivo but also developmental angiogenesis in neonatal retinas, highlighting the conserved nature of this angiogenic switch mechanism.
Although p120RasGAP has been reported to have Ras-dependent and Ras-independent functions32, growth factor–mediated activation of Ras is sufficient to induce a proangiogenic phenotype in primary endothelial cells33. In fact, the loss of p120RasGAP alone was sufficient to mediate an enhanced angiogenic response to bFGF in Matrigel plugs (Supplementary Fig. 16). Suppression of p120RasGAP expression seems to be crucial in facilitating angiogenesis in vivo, because anti–miR-132 can both restore p120RasGAP expression and decrease neovascularization in several tumor models. On the basis of these observations, we believe it is likely that miR-132 can regulate various endothelial cell activities, including proliferation, survival, migration and tube formation, to exert its effects on angiogenesis.
There is emerging evidence that some of the current antiangiogenic therapies, which target single pathways such as the VEGFR pathway, can lead to the development of ‘evasive resistance’ in tumors through the upregulation of alternative growth factors34. In this context, antagonism of endogenous regulators such as miR-132 that are induced under pathological conditions and target ‘gatekeepers’ of endothelial activation such as p120RasGAP may represent a powerful strategy to inhibit angiogenesis in a wide range of pathological conditions. To our knowledge, the findings reported here provide the first description of a miRNA-regulated angiogenic switch. Our studies show that this switch can be regulated to both disrupt and facilitate neovascularization. The possibility of delivering miRNAs or antagomirs to activated endothelium, as demonstrated here using miR-132–containing nanoparticles targeted to integrin αVβ3, suggests opportunities for manipulating miRNA levels in the endothelium to control pathological neovascularization.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Barnes, E. Goka, B. Walsh and D. Wu for technical support. We thank S. Weng and J. Desgrosellier for discussions. We thank E. Brown (University of Pennsylvania) for the Ert2-ubiquitin-Cre mice. We thank R. Kerbel (University of Toronto) for the fast-growing variant of MDA-MB-231 breast carcinoma cells. We thank S. Kajiji (Scripps Research Institute) for FG human pancreatic adenocarcinoma cells. This work was supported by US National Institutes of Health grants HL078912, CA104898 and CA050286 to D.A.C. and HL096498 to P.D.K. S.A. is supported in part by an American Heart Association postdoctoral fellowship 09POST2040038.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
S.A. and D.A.C. designed the study. E.A.M., B.K.M. and R.M. designed the nanoparticles. J.N.L. established the human ES cell vasculogenesis model. D.J.S. helped with the TaqMan microRNA panel experiments and analysis. P.E.L. and P.D.K. generated and characterized the Rasa1fl/fl mice. S.A., L.S., L.M.A. and M.H. performed experiments and analyzed data. S.A., S.M.W. and D.A.C. analyzed data and wrote the manuscript. D.A.C. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Murphy EA, et al. Nanoparticle-mediated drug delivery to tumor vasculature suppresses metastasis. Proc. Natl. Acad. Sci. USA. 2008;105:9343–9348. doi: 10.1073/pnas.0803728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobson B, Denekamp J. Endothelial proliferation in tumours and normal tissues: continuous labelling studies. Br. J. Cancer. 1984;49:405–413. doi: 10.1038/bjc.1984.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkman J. The role of angiogenesis in tumor growth. Semin. Cancer Biol. 1992;3:65–71. [PubMed] [Google Scholar]
- 4.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci. Signal. 2009;2:pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindquist JN, Cheresh DA, Snyder EY. Derivation of vasculature from embryonic stem cells. Curr. Protoc. Stem Cell Biol. 2010;12:1.1F.9.1–1.1F.9.6. doi: 10.1002/9780470151808.sc01f09s12. [DOI] [PubMed] [Google Scholar]
- 6.Kelly MA, Hirschi KK. Signaling hierarchy regulating human endothelial cell development. Arterioscler. Thromb. Vasc. Biol. 2009;29:718–724. doi: 10.1161/ATVBAHA.109.184200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nudelman AS, et al. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vo N, et al. A cAMP-response element binding protein–induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayo LD, Kessler KM, Pincheira R, Warren RS, Donner DB. Vascular endothelial cell growth factor activates CRE-binding protein by signaling through the KDR receptor tyrosine kinase. J. Biol. Chem. 2001;276:25184–25189. doi: 10.1074/jbc.M102932200. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y, et al. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 11.Kenneth TE, Kertes PJ. Ranibizumab in neovascular age-related macular degeneration. Clin. Interv. Aging. 2006;1:451–466. doi: 10.2147/ciia.2006.1.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gragoudas ES, Adamis AP, Cunningham ET, Jr, Feinsod M, Guyer DR. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 16.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino M, Kawakita M, Hattori S. Characterization of a factor that stimulates hydrolysis of GTP bound to ras gene product p21 (GTPase-activating protein) and correlation of its activity to cell density. Mol. Cell. Biol. 1988;8:4169–4173. doi: 10.1128/mcb.8.10.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5–8. doi: 10.1016/0092-8674(89)90976-8. [DOI] [PubMed] [Google Scholar]
- 20.Lapinski PE, et al. Generation of mice with a conditional allele of the p120 Ras GTPase-activating protein. Genesis. 2007;45:762–767. doi: 10.1002/dvg.20354. [DOI] [PubMed] [Google Scholar]
- 21.Henkemeyer M, et al. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- 22.Boon LM, Mulliken JB, Vikkula M. RASA1: variable phenotype with capillary and arteriovenous malformations. Curr. Opin. Genet. Dev. 2005;15:265–269. doi: 10.1016/j.gde.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Eerola I, et al. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am. J. Hum. Genet. 2003;73:1240–1249. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershkovitz D, Bercovich D, Sprecher E, Lapidot M. RASA1 mutations may cause hereditary capillary malformations without arteriovenous malformations. Br. J. Dermatol. 2008;158:1035–1040. doi: 10.1111/j.1365-2133.2008.08493.x. [DOI] [PubMed] [Google Scholar]
- 25.Hood JD, et al. Tumor regression by targeted gene delivery to the neovasculature. Science. 2002;296:2404–2407. doi: 10.1126/science.1070200. [DOI] [PubMed] [Google Scholar]
- 26.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Würdinger T, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonauer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 31.Komada M, Kitamura N. The Hrs/STAM complex in the downregulation of receptor tyrosine kinases. J. Biochem. 2005;137:1–8. doi: 10.1093/jb/mvi001. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni SV, Gish G, van der Geer P, Henkemeyer M, Pawson T. Role of p120 Ras-GAP in directed cell movement. J. Cell Biol. 2000;149:457–470. doi: 10.1083/jcb.149.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meadows KN, Bryant P, Vincent PA, Pumiglia KM. Activated Ras induces a proangiogenic phenotype in primary endothelial cells. Oncogene. 2004;23:192–200. doi: 10.1038/sj.onc.1206921. [DOI] [PubMed] [Google Scholar]
- 34.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.