This study explores the order in which individual metabolic genes are lost in an in silico evolutionary process leading from the metabolic network of Eschericia coli to that of the genome-reduced endosymbiont Buchnera aphidicola.
Keywords: constraint-based modeling, endosymbiont, evolution, metabolism
Abstract
A fundamental challenge in Systems Biology is whether a cell-scale metabolic model can predict patterns of genome evolution by realistically accounting for associated biochemical constraints. Here, we study the order in which genes are lost in an in silico evolutionary process, leading from the metabolic network of Eschericia coli to that of the endosymbiont Buchnera aphidicola. We examine how this order correlates with the order by which the genes were actually lost, as estimated from a phylogenetic reconstruction. By optimizing this correlation across the space of potential growth and biomass conditions, we compute an upper bound estimate on the model's prediction accuracy (R=0.54). The model's network-based predictive ability outperforms predictions obtained using genomic features of individual genes, reflecting the effect of selection imposed by metabolic stoichiometric constraints. Thus, while the timing of gene loss might be expected to be a completely stochastic evolutionary process, remarkably, we find that metabolic considerations, on their own, make a marked 40% contribution to determining when such losses occur.
Introduction
Symbiotic relationships include those associations in which one organism lives within the tissues of the other, either in the intracellular space or extracellularly (Douglas, 2007). One classical case of mutualism that has been a focus of numerous investigations is the symbiosis between Buchnera aphidicola and its aphid host. Evolutionary studies suggest that 160–280 million years ago (Moran et al, 1993) this aphid ancestor was infected with a free-living eubacterium in a process that led to co-speciation of the host and its symbiont. The host and the endosymbiont then became interdependent and unable to survive without each other. It is believed that Buchnera has evolved from a free-living Gram-negative ancestor quite similar to Escherichia coli. The Buchnera genome is considerably reduced compared with that of E. coli, but it has retained ancestral genes for proteins involved in DNA replication, transcription and translation, as well as chaperonins and proteins involved in secretion, energy-yielding metabolism and amino acid biosynthesis (Baumann et al, 1995; Shigenobu et al, 2000; Moran and Mira, 2001; Silva et al, 2001; Gil et al, 2002; Tamas et al, 2002; Moran et al, 2009).
The symbiosis between B. aphidicola and its host is characterized by a process in which few or no genes have been acquired as part of the transition to a symbiotic lifestyle; rather, the gene set of the ancestor has been selectively reduced so as to retain only those genes and pathways required for the symbiotic lifestyle (Dale and Moran, 2006). The symbiosis therefore has a nutritional basis. Specifically, Buchnera has retained in the genome genes for the biosyntheses of amino acids essential for the host while those for non-essential amino acids are missing, indicating complementarity and syntrophy between the host and the symbiont (Shigenobu et al, 2000). Nitrogen recycling, however, is not quantitatively important to the nutrition of aphid species studied, and there is strong evidence against bacterial involvement in the lipid and sterol nutrition of aphids (Lai et al, 1994; Douglas, 1998; Moran and Baumann, 2000). Moreover, studies have excluded the hypothesis that genome reduction in Buchnera has been accompanied by gene transfer to the host nuclear genome (Nikoh et al, 2010).
Previous work by Pál et al (2006) has addressed the problem of inferring gene content of an organism given its lifestyle, by modeling the evolution of the reduced genomes of endosymbiotic bacteria such as B. aphidicola and Wigglesworthia glossindia (Akman et al, 2002; Pál et al, 2006). Using the E. coli metabolic network (Reed et al, 2003) as a starting point, these authors developed a protocol for simulating the gradual loss, during evolution, of metabolic enzymes. This involved the random removal of genes, and hence enzymes from the network, whose contribution to the organism's growth yield (computed using a flux balance analysis (FBA) model; Fell and Small, 1986; Varma and Palsson, 1994; Kauffman et al, 2003) are vanishingly small. Starting from the E. coli model and repeating this stochastic gene removal process many times while aggregating the results, they managed to obtain end point viable minimal metabolic networks (where no genes can be further removed) that were ∼80% similar to the metabolically annotated genes of B. aphidicola. Although previous studies have considered metabolic network constraints over an evolutionary time scale (van Hoek and Hogeweg, 2009), and have studied the differential retention of metabolic genes versus non-metabolic genes following whole-genome duplication (Gout et al, 2009), Pál et al (2006) were the first to demonstrate a particular organism's evolution in silico. However, it was geared to reconstructing the final network, i.e., the end point of the evolutionary process.
Here, we aim to go significantly further and investigate whether it is possible to computationally simulate not only the network emerging at the end point of the evolutionary process, but also its dynamics. Specifically, we wish to examine whether we can predict in silico, the timing of gene loss events in a consistent manner, and study how well these predictions correspond with phylogenetic estimates of the temporal sequence of gene loss. We are interested in elucidating the relative contributions of chance and necessity in this elaborate process and learn to what extent metabolic constraints determine the observed sequence of gene loss events.
Results
Our in silico gene loss time estimations of B. aphidicola follow from a procedure similar to the evolutionary reductive simulation performed by Pál et al (2006). Briefly, the evolution of the metabolic network undergoing gene reduction is simulated in an iterative fashion as follows: In each iteration a gene is randomly chosen to be deleted from the genome. If its deletion does not reduce growth below a certain threshold, the resulting strain is considered viable, and the deleted gene is therefore considered lost and excluded from the network. If the deletion of gene reduces growth significantly (above the given threshold, i.e., it is selected against), the pertaining gene is retained. The contribution of non-essential genes to growth and their retention in the final network evolved depends on the presence of other genes backing them in the network (Deutscher et al, 2006) and on the random sequential order by which the genes are deleted in a given run. The interplay between these stochastic events and deterministic network-based constraints is elucidated via aggregating the results over many reductive evolution simulations, as described in detail in the Materials and methods section.
We first turned to search the literature for components known to exist in Buchnera's habitat, as the content of the environment has a significant effect on an organism's metabolism and hence on its gene loss time. The components found were then completed with a minimal number of metabolites that are essential for growth considering E. coli's biomass function (Supplementary Table 1.a) to form a literature-based viable media. In order to establish the robustness of this evolutionary process, we applied it on a more up to date E. coli model (Feist et al, 2007) under this literature-based viable media. The in silico deletion time of a gene in a single run of the reductive evolutionary process denotes the number of genes deleted before its own deletion occurred. To obtain a robust and consistent estimation of a gene's in silico deletion time, its mean deletion time is computed over 40 000 individual runs of the reductive evolutionary process (Materials and methods). Reassuringly, the correlation between the gene's deletion times across a pair of simulations to estimate loss times (each composed of 20 000 reductive runs as described above) is very high (Spearman's correlation of 0.92, empirical P-value <9.9e−4, Supplementary Figure 1; we chose to use empirical P-values throughout the paper as they are more strict than asymptotic P-values). This implies that, even though the genes are selected as potential candidates for deletion by chance (i.e., in a completely random manner), genes are still actually lost in a consistent and coordinated fashion, reflecting the role of necessity. Notably, even when excluding from this analysis those end point genes that are always retained in the final model, we still obtain a high mean Spearman correlation of 0.84 (empirical P-value <9.9e−4) across different runs. Many of the genes are always lost in silico and their deletion time in an individual run is random, but measured over an increasing number of reductive runs their estimated loss time converges to a deterministic value representing these genes’ global mean loss time. Focusing just on the set of 240 genes that are not always lost or always retained, we find an even higher Spearman's correlation of 0.99 between their loss time estimations obtained across different multiple runs (empirical P-value <9.9e−4, Supplementary Figure 2). All together, these results testify to the robustness and consistency of the in silico loss time estimations, and to the significant constraints that the loss of certain genes may impose on the loss times of others.
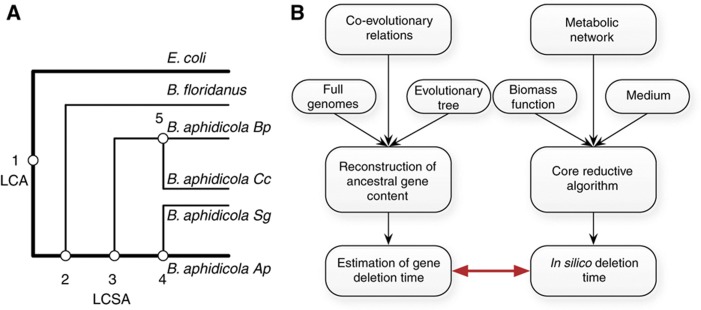
To examine how well the gene loss times predicted in silico coincide with the times these genes were actually lost during evolution, we additionally performed an ancestral gene content phylogenetic reconstruction for the sub-tree leading from several Buchnera strains (from different aphid hosts (Shigenobu et al, 2000; Tamas et al, 2002; van Ham et al, 2003; Wu et al, 2006)) to their common ancestor with E. coli (Figure 1A). Our in silico predictions of loss times (which reflect the consequences of metabolic considerations per se) were then compared with the gene loss time inferred via the phylogenetic reconstruction of all four evolutionary paths leading to the different B. aphidicola strains (while considering each path separately), overall including five states (Figure 1A; and see Figure 1B for a general description of the evolutionary reconstruction process). The latter reconstruction obviously reflects the consequences of all evolutionary forces determining gene loss.
Figure 1.
The phylogenetic tree analyzed here and a schematic overview of the computational process. (A) The phylogenetic tree of the Buchnera aphidicola strains analyzed here (Materials and methods). (B) A schematic overview of the computational process used for generating and comparing simulated and phylogenetic gene loss times.
First, it should be emphasized that the maximal mean Spearman's correlation between in silico and reconstructed gene loss times that one can possibly obtain in our setup is 0.86 and not 1 (due to numerous ties in the vector representing the phylogenetic loss time). Simulating the in silico evolutionary process described above under the literature-based viable medium and computing the correlation between the resulting in silico and reconstructed gene loss times for each of the four B. aphidicola strains, we obtain a mean Spearman's correlation of 0.46 (53% of the maximal correlation, empirical P-value <9.9e−4, see Materials and methods) averaged over these four strains (Figure 2). Notably, when excluding from the analysis those end point genes that are always retained in the pertaining species, we still obtain a significant mean Spearman's correlation of 0.37 (43% of the maximal correlation, empirical P-value <9.9e−4). Interestingly, repeating this analysis under the minimal medium (Supplementary Table 2.a) used in Pál et al (2006), we obtain a similar high mean Spearman's correlation (Supplementary Material). It should be emphasized that, in accordance with an earlier report (Pál et al, 2006), the model accurately predicts that the most preserved pathways are those involved in essential amino-acid metabolism and in central metabolism, including the pentose phosphate pathway, glycolysis and so on, while genes associated with cell envelope synthesis, lipopolysaccharides synthesis and membrane lipid metabolism are not fully retained in the final networks (Supplementary Table 1.f). It is reassuring to see that these predictions match the reports known from the literature, where it is known that B. aphidicola lacks genes for the biosynthesis of cell surface components, including lipopolysaccharides and phospholipids (Shigenobu et al, 2000). Furthermore, the extensive loss of transport capabilities and conservation of essential amino acids biosynthetic pathways are prime characteristics of the aphid symbiont (van Ham et al, 2003).
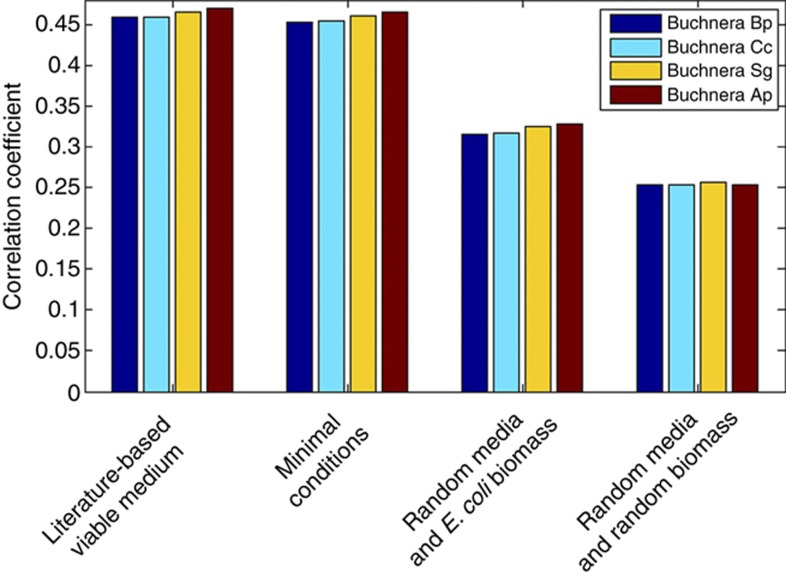
Figure 2.
Correlation results obtained by comparing in silico predicted gene loss times to the times these genes were estimated to be lost during evolution, for four different Buchnera strains. This estimation is based on a phylogenetic reconstruction of the ancestral gene content for the sub-tree, leading from different Buchnera aphidicola strains to their common ancestor with E. coli (Figure 1A). The in silico time estimations were simulated in three different situations: (1) literature-based viable medium and E. coli's biomass function (literature-based viable medium), (2) minimal medium and E. coli's biomass function as used by Pál et al (2006) (minimal conditions), and two control conditions: (3) five random media and the E. coli biomass function, and (4) five random media and random biomass functions (that together yet still form viable growth conditions, Supplementary Material).
As both the content of the environment and the composition of the biomass effect the in silico gene loss order, we examined the correlations between in silico time loss predictions obtained under random control growth/biomass conditions and the reconstructed loss times. Random media were generated by randomly selecting media components (Supplementary Material) that together allow the organism to grow considering E. coli’s biomass function. Similarly, we have generated random biomass functions by randomly selecting biomass components and searching for random media that would together obtain a viable organism. Notably, the difference in correlation values between these random condition and those described above is highly significant (Wilcoxon's P-value <1.7e−9). Moreover, Figure 2 shows that these random conditions result in markedly lower, yet positive correlation values (empirical P-value <9.9e−4, Supplementary Material). These results demonstrate that while the metabolic network topology itself already embeds some information constraining gene loss order, the model can better simulate the reductive evolution process when it is emulated under media and biomass conditions that are sufficiently close to the biological reality.
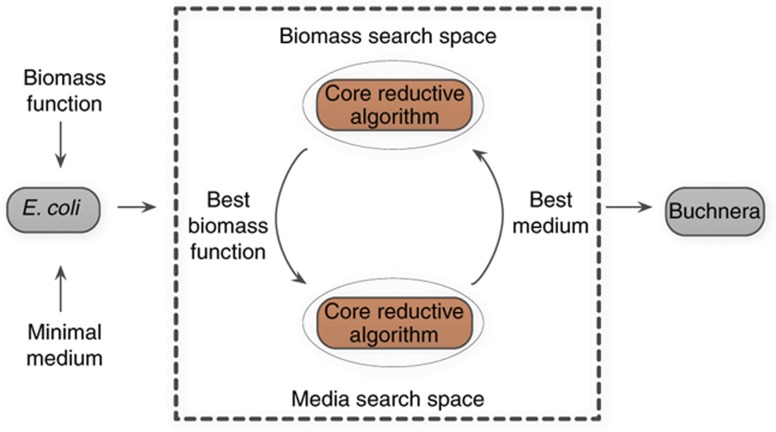
What is the upper bound on the correlation between in silico and phylogenetic gene loss times that can be achieved within our in silico framework? Estimating the maximum obtainable correlation would give an upper bound on evolutionary necessity stemming from metabolic constraints. To answer this, we next turned to search the space of potential growth media and biomass functions (Materials and methods), to study if and to what extent one can further increase the correlation between in silico and reconstructed loss times in media and biomass different from those derived from the literature or the minimal conditions that were simulated up until now. To this end, we performed the reductive evolutionary process of Pál et al (2006) as an internal kernel within a simulated annealing (SA) search algorithm, aimed at searching for the environment/biomass function that maximized our target correlation between in silico and reconstructed loss times (see Materials and methods section and Figure 3). As the search space is obviously vast, we limited the size of the media to several predefined magnitudes and found that media composed of 50 components achieve significantly higher correlation values over media of other magnitudes (hypergeometric P-value=3.06e−6 and Materials and methods). The best combination of biomass and environment found following the convergence of the SA process (Supplementary Tables 2.e and 2.f) improved the correlation over the four Buchnera strains to a mean Spearman's correlation of 0.54 (63% of the maximal correlation) considering the end point genes, and to 0.39 (44% of the maximal correlation) without the end point genes (empirical P-value <9.9e−4, Supplementary Figure 3). A list of the in silico gene loss time based on the growth condition found by the SA search process and the gene loss time based on the phylogenetic reconstruction can be found in Supplementary Table 2j. Notably, the new evolved end point network achieves a mean similarity level (area under the resulting ROC curve, AUC) of about 88% (P-value <1.41e−10) with the metabolic genes of the different Buchnera strains, akin to the similarity achieved by Pál et al (2006) (see Supplementary Figure 4 for the corresponding similarity levels to the strains that appear in the phylogenetic reconstruction used here). Interestingly, the components shared by the five media found in SA solutions obtaining the highest correlation values are enriched with components from the literature-based viable medium (hyper geometric P-value=0.03). Moreover, the five biomass functions obtaining the highest correlation values all share essential amino acids and riboflavin known to be supplied by Buchnera to its host (Douglas, 1998).
Figure 3.
General description of our computational approach. Starting from E. coli biomass and a minimal medium, we first search through the space of possible media within a given predetermined size, each time applying the core reductive algorithm to estimate the obtained similarity level to the Buchnera model, until no further improvement is achieved. Next, we fix the medium achieving the highest score and repeat this process while searching through the space of possible biomass functions. Similarly, when no further improvement is achieved, we fix the biomass resulting in the highest score for the next iteration. These two steps are repeated until we converge on a medium and biomass function where no further improvement in the correlation between in silico and reconstructed loss times can be achieved.
Overall, these results testify that an in silico evolutionary reductive process based on metabolic and stoichiometric constrains can account for about 40% (0.63^2) of the variation in the reconstructed time course of endosymbiont gene loss. The considerable temporal information that is still missed is probably a result of a combination of the action of other selection forces not considered here (e.g., regulatory or physiological constraints) and of potential stochastic components of the evolutionary process. As an additional acid test of this observation, and as the search space is obviously vast, we performed the reductive simulation in a supervised manner, strictly imposing the phylogenetic gene loss order as the actual in silico deletion order as much as possible, under the conditions obtained by the SA search (Materials and methods). This results in a mean Spearman's correlation of 0.78 (91% of the maximal correlation, empirical P-value <9.9e−4) between the in silico and phylogenetically inferred loss times. Thus, even at this extreme limit case where the simulation artificially aims to repeat the phylogenetic process exactly, even though coming close to the maximal possible value, there still remains a non-negligible unexplained component. As the conditions obtained by the SA search managed to significantly increase the correlation, we chose to perform the in-depth analysis presented in the following under these conditions.
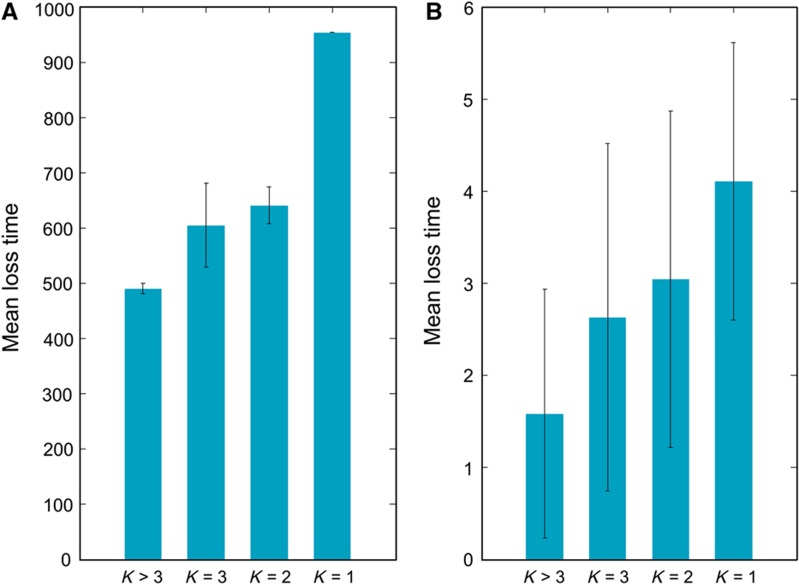
What does the metabolic model reveal about the constraints that affect the timing of gene loss? To answer this, we examined the dependency of the predicted loss time of each gene on its intrinsic network-level properties. We find that the predicted gene loss time strongly depends on the number of functional backups that the corresponding reactions of a gene have in the network under a given medium. The latter is measured by the k-robustness index introduced in Deutscher et al (2006), where k=1 denotes essential genes, k=2 denotes genes involved in synthetic lethal pairs, k=3 involves genes with at least two other functional backups and so on. Accordingly, we find a very strong inverse Spearman's correlation of −0.84 (empirical P-value <9.9e−4) between the order of gene loss predicted in silico and the k-robustness levels of the genes (Figure 4A). Notably, when excluding essential genes (k=1) from the analysis, we still obtain a high inverse Spearman's correlation of −0.65 (empirical P-value <9.9e−4). This arises as poorly backed up genes (k=2) are more likely to be retained in the final networks evolved than genes with k>2 (P-value=2.35e−8), as when their sole backup gene is lost they then become essential. An analogous association is found between the gene loss time inferred from the phylogenetic reconstruction and the k-robustness levels, with a mean Spearman's correlation of −0.51 (empirical P-value <9.9e−4, Figure 4B) over the four Buchnera strains.
Figure 4.
Mean in silico and phylogenetically loss time as a function of the k-robustness index. (A) Mean in silico gene loss time as a function of the number of backup reactions a gene has in the metabolic network (its k-robustness; Deutscher et al, 2006). (B) Phylogenetically reconstructed gene loss time, also as a function of gene backup number. Error bars in both cases represent the standard deviation.
In addition, one might expect that the relative loss time of a gene is influenced by its functional dependencies with other genes. To examine this, we performed a flux coupling analysis and identified pairs of reactions whose activities asymmetrically depend on each other, i.e., are directionally coupled (Burgard et al, 2004). Remarkably, examining these pairs, we find that genes encoding reactions whose activity is needed for activating the other reaction (and not vice versa) have a tendency to be lost later (both in silico and considering the phylogenetic loss time), as one would expect (binomial P-value <1e−14, Materials and methods). This finding complements previous reports that asymmetric coupling relationships influence the order by which new genes are acquired by horizontal transfer (i.e., an enzyme whose function depends on the presence of another enzyme tend to be acquired later) (Pál et al, 2005) and results in an asymmetric occurrence of genes across genomes (Notebaart et al, 2009).
We next addressed the question of how well do genomic features and network properties predict the phylogenetically reconstructed gene loss times, focusing on the genes’ mRNA levels, tAI values (tRNA adaptation index (tAI; Sharp and Li, 1987; Covert et al, 2004; Reis et al, 2004; Tuller et al, 2010a), and on the number of partners the gene products have in a protein–protein interaction (PPI) network. These variables are known to be inversely correlated with the propensity of a gene to be lost, and have previously been shown to correlate with gene loss rates in Buchnera (Delmotte et al, 2006; Tamames et al, 2007; Brinza et al, 2009). We find a Spearman's correlation of 0.43 (empirical P-value <9.9e−4) between the phylogenetically reconstructed gene loss times (our ‘gold standard’) and mRNA levels, Spearman's correlation of 0.21 (empirical P-value <9.9e−4) with the tAI values and a Spearman's correlation of 0.21 (empirical P-value <9.9e−4) with PPI degree (Supplementary Material). These correlations remain significant also after excluding the end point genes (>0.1, empirical P-value <9.9e−4). Remarkably, examining the association between in silico and reconstructed gene loss times while controlling for these genomic and PPI network variables, we still obtain a Spearman's correlation above 0.47 (empirical P-value <9.9e−4) for all four Buchnera strains. These partial correlations between in silico and reconstructed gene loss time also remain significant after excluding the end point genes of all four Buchnera strains (>0.38, empirical P-value <9.9e−4, Supplementary Material). Multiply regressing the loss times from the phylogenetic reconstruction on the in silico gene loss time predictions and the genomic and network variables, leads to improved prediction of phylogenetically reconstructed gene loss pattern compared with that obtained with the different variables alone (mean Spearman's correlation of 0.6 (empirical P-value <9.9e−4) over all four strains). Notably, the (normalized) coefficient of the in silico predictions in the regression is much higher than those of the genomic features (Supplementary Material), further testifying to the considerable independent predictive power of the model-based in silico predictions.
The most common explanation for the mechanism of gene loss occurring in the evolution of B. aphidicola is the fixation of single large deletions spanning many genes at the initial stage of genome reduction followed by single-gene deletions after the establishment of the last common symbiotic ancestor (LCSA; Moran and Mira, 2001; van Ham et al, 2003). We therefore simulated a two-phase block deletions process similar to that done in Pál et al (2006) (Materials and methods). Interestingly, we find that after a certain amount of genes are deleted from the genome, no further block deletions can occur due to the increasing density of essential genes. Notably, the maximum amount of genes that can be deleted in blocks (i.e., until no more blocks can be deleted) is in correspondence with the number of genes appearing in our phylogenetic reconstruction from the LCA (last common ancestor of Buchnera and E. coli) to the LCSA. This number is in the range of 750–850 (nodes 1–3 in Figure 1A), when q (where the probability of deleting a block with n genes is P(n)=qn, Materials and methods) is in the range of 0.5–0.9. Under the media conditions described above (minimal medium, literature-base medium and the conditions obtained by the single-gene deletion SA search), we find a mean Spearman's correlation of about 0.4 in the block deletions scenario (empirical P-value <9.9e−4). To improve the obtained correlation, we repeated the SA search under block deletion simulations. The best combination of biomass and environment found after the convergence of the SA search (Supplementary Tables 3.a, 3.b and 3.c) improved the correlation over the four Buchnera strains to a mean Spearman's correlation of 0.45 that is 53% of the maximal correlation with the end point genes, and to 0.37 (43% of the maximal correlation) without the end point genes (empirical P-value <9.9e−4). These correlations are overall lower than those obtained in the single-gene deletion simulations, however, they are higher than those obtained under random conditions and those obtained by genomic features (Supplementary Material). This may be surprising at first, but may be understood when noting that in reality, in vivo evolutionary selection process occurs on deleted blocks involving many non-metabolic genes, which are out of the scope of the metabolic in silico model studied here. That is, blocks containing non-essential metabolic genes but essential non-metabolic genes will not be deleted in vivo while they will be deleted in silico. Therefore, and although it is most likely that the evolutionary process occurred first by the loss of large blocks, we have chosen to perform the main analyses in the paper by considering only single-gene deletion simulations that better suite the confines of metabolic constraints embedded in a metabolic model, and on which we focus upon.
Conclusions
In summary, this study shows for the first time, that it is possible to go beyond capturing the end point of an evolutionary process using an in silico model, and obtain a fine-grained view of the time course of an organism's evolution toward symbiosis. A comprehensive search over numerous growth media and biomass functions reveals that strict metabolic considerations can explain about 40% of the variation observed in gene loss times, while the remaining variation is likely to be determined by other factors. The network-level functioning of a gene is found to be a stronger determinant of its time of loss than its individual genomic features. The number of functional backups the gene possesses in the network is found to be the most significant determinant of the timing of its demise.
Our simulations focus on the loss of genes reflecting the act of purifying selection. Analogous to the earlier observations of Pál et al (2006) that both necessity and chance have a significant role in reductive evolution, we find that both are likely to have part when the processes are re-examined on a more fine-grained temporal scale. ‘Necessity’ in our context refers to selection forces acting to conserve metabolic genes whose contribution to the symbiont's growth within the host is significant. However, the weight of ‘necessity’ estimated by our framework serves as a lower bound on its actual magnitude, as the true contributions of metabolic genes may go well beyond those captured in the model due to their contribution to other, non-metabolic cellular functions (as many genes may have diverse pleiotropic effects). ‘Chance’, albeit, reflects the true randomness in order of gene losses occurring in evolution that may be due to the workings of a variety of stochastic effects occurring in nature including, e.g., randomness in mutational processes driving gene loss and variations in the host's environment and so on.
Viewed from a complementary perspective, our results may have important implications for future attempts to develop more realistic phylogenetic models for gene loss: given that metabolic networks are becoming available for a wide variety of organisms, it would become possible to incorporate metabolic constraints into such reconstructions and boost their accuracy, as have been demonstrated recently for co-evolutionary constraints (Tuller et al, 2010a). In addition, the optimization approach utilized here to comprehensively search for growth media that maximize the fit between model predictions and empirical data may serve in future studies aimed at inferring the metabolic lifestyles of other, less-characterized organisms and/or their ancestors.
Materials and methods
Constraint-based modeling of metabolic networks
A metabolic network consisting of n metabolites and m reactions can be represented by a stoichiometric matrix, denoted by S, where the entry Sij represents the stoichiometric coefficient of metabolite i in reaction j (Price et al, 2004). A constraint-based modeling (CBM) imposes mass balance, directionality and flux capacity constraints on the space of possible fluxes in the metabolic network's reactions through a set of linear equations:
where v̂ stands for the flux vector for all of the reactions in the model (i.e., the flux distribution). The exchange of metabolites with the environment is represented as a set of exchange reactions, enabling for a predefined set of metabolites to be either taken up or secreted from the growth media. The steady-state assumption represented in Equation (1) constrains the production rate of each metabolite to be equal to its consumption rate. Enzymatic directionality and flux capacity constraints define lower and upper bounds on the flux rates and are embedded in Equation (2). In this study, we used a genome-scale E. coli metabolic network of Feist et al (2007) as our starting point of the core reductive evolutionary process, accounting for 1260 metabolic genes, 2382 reactions and 1668 metabolites.
Flux balance analysis is a key computational approach within the CBM modeling framework (Fell and Small, 1986; Varma and Palsson, 1994; Kauffman et al, 2003) and is frequently used to successfully predict various phenotypes of microorganisms such as their growth yields, uptake rates (when growth rate is known), by-product secretion and knockout lethality (Price et al, 2004; Feist et al, 2009; Oberhardt et al, 2009). The objective of FBA is to maximize a biomass reaction describing the relative contribution of metabolites to the cellular biomass, while finding a steady-state flux distribution v̂ alongside additional enzymatic directionality and capacity constraints, together permitting a maximal growth yield.
Reductive evolution simulations
Reductive evolution starting from the E. coli's metabolic network was simulated using a random sequential gene deletion as previously described by Pál et al (2006). Briefly, these simulations proceed in an iterative fashion. In each iteration, we randomly choose a gene from the remaining network model and simulate its deletion by setting the flux through the corresponding reactions it encodes to zero. We next run FBA to find the maximum growth yield of the organism following this deletion. Deletions that do not reduce growth below a certain threshold are considered viable, and the gene is therefore lost and excluded from the network from here on. Otherwise, the gene is considered essential for survival and is retained in the network. This iterative process is repeated until no further genes can be deleted. As done in Pál et al (2006), the core reductive evolution procedure is repeated 2000 times, each time resulting in a different and independent evolutionary outcome. As in Pál et al (2006), an aggregative process is performed over all the resulting 2000 final-outcome network models to pick the most representative one, and its similarity to a reference set of Buchnera metabolic genes is then assessed via a standard evaluation of the AUC. Following Pál et al (2006), the cut-off for the fitness effect of simulated gene deletions was set to 0.01, based on population size and selection estimations (i.e., a gene was classified as having no fitness effect, if the biomass production rate of the knockout strain was reduced by less than a given cut-off). This threshold was used also in the k-robustness and flux coupling analyses.
Inferring in silico gene loss time in the reductive evolution simulations
The in silico evolutionary reductive model employed here was carried in a standard manner using a constraint-based metabolic modeling approach, following Gianchandani et al (2006). Applying the reductive evolutionary process, we can set each gene a value indicating its loss time. This value stands for the number of genes that were successfully deleted before its own deletion occurred. Taking the mean over these values across 2000 runs of the core evolutionary reduction process in a given medium/biomass composition provides us with an in silico estimation of genes loss time under the specified conditions (which may then be compared with the phylogenetic loss times).
Phylogenetic reconstruction and inference of Buchnera gene loss
The genomes of the analyzed Buchnera species were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/genomes/). Genes were mapped to gene family by the COG classification (Tatusov et al, 2003), and we additionally used the metabolic network of E. coli (Feist et al, 2007). The phylogenetic tree (Figure 1A) was reconstructed based on the tree of life (Ciccarelli et al, 2006); the sub-tree of the B. aphidicola was based on the distances (Maximum Parsimony (MP) score) between the genomes of the three Buchnera strains.
We used Neyman's two state model (Neyman, 1971), a version of Jukes–Cantor (Jukes and Cantor, 1969) model for inferring the edge lengths (the probability of gain/loss of a gene family) of the tree by maximum likelihood; this was done by PAML (Yang, 1997). The edge lengths correspond to the probabilities that a protein family will appear/vanish along the corresponding lineage. The ancestral reconstruction was done using three different approaches: MP, Maximum likelihood (considering the edge lengths) and Ancestral Co Evolver (ACE; Tuller et al (2010a), considering the edge lengths and additional co-evolutionary information). The results reported in the main text are those obtained using MP, as they exhibit a higher resolution of loss times, but the overall trends are similar and a detailed account of all results is provided in the Supplementary Material.
Let Pα,β denote the probability of gain/loss a gene family along the tree edge (α,β). We assume that genes cannot be gained in the evolution of endosymbionts and inferred the ancestral gene families using a generalized MP method (Fitch, 1971; Sankoff, 1975) whose penalty for the loss of a gene along the tree edge (α,β) is −log(Pα,β). In addition, our analyses were based on the ACE algorithm (Tuller et al, 2010a) with the co-evolutionary networks that appear in Tuller et al (2010a). However, similar results were obtained without the ACE or when we assume that all the edge lengths are identical.
Empirical P-value estimation of in silico gene deletion times
An empirical P-value was calculated by producing 500 random orders of gene deletions, calculating the mean loss time over these 500 orders and setting an infinity loss time (>1260) for genes that are always retained (to examine whether our simulations capture significant information on gene loss time beyond that of the end point). Next, we examined the resulting correlation between this random mean loss time and the actual phylogenetic reconstructed time. This whole process was repeated 1000 (n) times while counting the number of times a random mean loss time resulted with a correlation higher or equal to that achieved by the original vector of in silico deletion times (r). The empirical P-value is then calculated as (r+1)/(n+1). The empirical P-value reported for the correlation between the k-robustness index and the phylogenetic loss time was calculated in a similar manner by permuting the vector of k-robustness index and examining the resulting correlation to the phylogenetic loss time.
Flux-coupling analysis
Flux-coupled genes were identified by applying the previously developed flux-coupling algorithm (Burgard et al, 2004) on the E. coli metabolic reconstruction (Feist et al, 2007). Namely, we looked for directionally coupled genes where a non-zero flux for v1 implies a non-zero flux for v2, but not necessarily the reverse. In order to test whether a gene encoding reactions whose activity is needed for activating the other reaction have a tendency to be lost later, we applied a binomial test with P=0.5 testing for the significance of deviation from the theoretically expected distribution.
Searching for the most likely environment and biomass compositions via SA
As described above, we aimed to explore to what extent one can further increase the correlation between in silico and reconstructed loss times by searching over the space of potential growth media and biomass functions, to find pairs of these conditions that markedly improve the correlation between in silico gene loss time estimations to that inferred via a phylogenetic reconstruction. Our approach is based on employing SA (Kirkpatrick et al, 1983) for this search, aiming to find a fair approximation to the optimal solution.
We start our search by extending the minimal growth medium to media of size 30, 50, 80, 110 and 140 by adding randomly selected exchange metabolites and using the original biomass function (Supplementary Table 2.a, 2.b) as embedded in our starting point, the E. coli metabolic model. Our search is performed iteratively, each iteration composed of two basic steps: in the first step, we fix the biomass and search through the environments space to find the medium maximizing the mean correlation of in silico and phylogenetic loss time over the four Buchnera strains, found via repeatedly performing the core reductive evolutionary process under these conditions for a number of times (we used five repetitions in each step, limited by computational considerations due to the vast size of the search space). In the second step, we fix the resulting environment found in the previous step and now search through the biomass space, identifying biomass compositions achieving the highest mean correlation. We defined these search spaces to be a union of the uptake reactions represented in the E. coli model and the recently published metabolic network model of B. aphidicola by Thomas et al (2009): eight missing uptake reactions were added to the E. coli model for the growth media space (overall encompassing 307 potential uptake reactions). Similarly, the union of the metabolites represented in the E. coli and Buchnera biomass functions was defined as the biomass search space (overall encompassing 78 potential biomass metabolites; Supplementary Table 2.c, 2.d). These two steps are repeatedly simulated where each time the environment/biomass obtaining the highest correlation in the previous step is being fixed and a new search begins. This iterative process was carried on until no further improvement in the fitness score driving the SA process (the correlation between in silico and phylogenetic loss time) is obtained.
Searching for an upper bound on the correlation between in silico and reconstructed loss time
To evaluate an upper bound on the correlation between in silico and reconstructed loss times, we imposed the gene loss order inferred from the phylogenetic reconstruction, and repeated the evolutionary reductive simulations under the conditions obtained by the SA search (Supplementary Table 2.e and 2.f) when the genes are deleted in that order: namely, we deleted genes according to their phylogenetic loss time (i.e., genes lost in the first reductive evolution step were deleted first, then those that are lost in the second step and so on), as long as their removal did not harm the model's ability to grow. We then evaluated the correlation between the resulting in silico loss time and the reconstructed one, to obtain a bound on the maximal correlation possible.
Block deletions simulations
Similarly to the process described by Pál et al (2006), we remove a randomly chosen block of contiguous genes in the genome. Under the assumption that deletion size follows an exponential distribution, the probability of deleting a block with n genes is P(n)=qn, where q (0<q<1) specifies the exact shape of the distribution. We then calculate the impact of deleting the metabolic genes included in a deleted block. Similar to the single-gene deletion simulations, block deletions that do not reduce growth below a certain threshold are considered viable and the corresponding genes are therefore lost and excluded from the network from here on. Otherwise, the genes are considered essential for survival and are retained in the network in that specific iteration (i.e., they can still be deleted in another block deletion trial). When no further contiguous genes can be deleted, a single-gene deletion process starts until no further genes can be deleted. The results are then aggregated over 2000 simulations as in Pál et al (2006).
Various sources of information
The mRNA levels of E. coli were downloaded from Covert et al (2004), the tAI values of E. coli genes were computed as in Tuller et al (2010b) and the PPI network was taken from Tuller et al (2010a).
Supplementary Material
Supplementary Figures S1–4, Supplementary Tables S1–3
Acknowledgments
We thank Uri Gophna, Stephen G. Oliver and Tomer Shlomi for their helpful comments on the manuscript. We also thank Francois Delmotte for his kind help in the data collection process. K.Y is partially supported by a fellowship from the Edmond J. Safra Bioinformatics program at Tel-Aviv University. T.T. is a Koshland Scholar at Weizmann Institute of Science and is supported by a travel fellowship from EU grant PIRG04-GA-2008- 239317. B.P. is supported by The International Human Frontier Science Program Organization, the Hungarian Scientific Research Fund (OTKA PD 75261) and the ‘Lendület Program’ of the Hungarian Academy of Sciences. E.R's research is supported by grants from the James S. McDonnell Foundation, the EU Microme consortium, and from the Israeli Science Foundation (ISF).
Author contributions: KY, TT, BP and ER conceived and designed the research. KY, TT, BP and ER wrote the paper. KY performed the analysis and the statistical computations. TT performed the phylogenetic reconstruction.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S (2002) Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 32: 402–407 [DOI] [PubMed] [Google Scholar]
- Baumann P, Baumann L, Lai C, Rouhbakhsh D, Moran NA, Clark MA (1995) Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular Symbionts of Aphids. Annu Rev Microbiol 49: 55–94 [DOI] [PubMed] [Google Scholar]
- Brinza L, Viñuelas J, Cottret L, Calevro F, Rahbé Y, Febvay G, Duport G, Colella S, Rabatel A, Gautier C, Fayard J-M, Sagot M-F, Charles H (2009) Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. Comptes Rendus Biologies 332: 1034–1049 [DOI] [PubMed] [Google Scholar]
- Burgard AP, Nikolaev EV, Schilling CH, Maranas CD (2004) Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res 14: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO (2004) Integrating high-throughput and computational data elucidates bacterial networks. Nature 429: 92–96 [DOI] [PubMed] [Google Scholar]
- Dale C, Moran NA (2006) Molecular interactions between bacterial symbionts and their hosts. Cell 126: 453–465 [DOI] [PubMed] [Google Scholar]
- Delmotte F, Rispe C, Schaber J, Silva F, Moya A (2006) Tempo and mode of early gene loss in endosymbiotic bacteria from insects. BMC Evol Biol 6: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher D, Meilijson I, Kupiec M, Ruppin E (2006) Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat Genet 38: 993–998 [DOI] [PubMed] [Google Scholar]
- Douglas AE (1998) Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43: 17–37 [DOI] [PubMed] [Google Scholar]
- Douglas AE (2007) Symbiotic microorganisms: untapped resources for insect pest control. Trends Biotechnol 25: 338–342 [DOI] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO (2009) Reconstruction of biochemical networks in microorganisms. Nat Rev Micro 7: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA, Small JR (1986) Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem J 238: 781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20: 406–416 [Google Scholar]
- Gianchandani EP, Papin JA, Price ND, Joyce AR, Palsson BO (2006) Matrix formalism to describe functional States of transcriptional regulatory systems. PLoS Comput Biol 2: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Sabater-Muñoz B, Latorre A, Silva FJ, Moya A (2002) Extreme genome reduction in Buchnera spp.: Toward the minimal genome needed for symbiotic life. Proc Natl Acad Sci USA 99: 4454–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J-Fo, Duret L, Kahn D (2009) Differential retention of metabolic genes following whole-genome duplication. Mol Biol Evol 26: 1067–1072 [DOI] [PubMed] [Google Scholar]
- Jukes T, Cantor C (1969) Evolution of protein molecules. Munro HN (ed). Mammalian Protein Metabolism 21–123. New York: Academic Press [Google Scholar]
- Kauffman KJ, Prakash P, Edwards JS (2003) Advances in flux balance analysis. Curr Opin Biotechnol 14: 491–496 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S, Gelatt CD Jr, Vecchi MP (1983) Optimization by simulated annealing. Science 220: 671–680 [DOI] [PubMed] [Google Scholar]
- Lai CY, Baumann L, Baumann P (1994) Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA 91: 3819–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N, Mira A (2001) The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol 2: research0054.0051–research0054.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Baumann P (2000) Bacterial endosymbionts in animals. Curr Opin Microbiol 3: 270–275 [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R (2009) The Dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323: 379–382 [DOI] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H (1993) A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc Royal Soc London Series B: Biol Sci 253: 167–171 [Google Scholar]
- Neyman J (1971) Molecular studies of evolution: a source of novel statistical problems. In Statistical Decision Theory and Related Topics, Gupta S, Yackel J (eds), pp 1–27. New York: Academic Press [Google Scholar]
- Nikoh N, McCutcheon JP, Kudo T, Miyagishima S-y, Moran NA, Nakabachi A (2010) Bacterial genes in the Aphid genome: absence of functional gene transfer from buchnera to its host. PLoS Genet 6: e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notebaart R, Kensche P, Huynen M, Dutilh B (2009) Asymmetric relationships between proteins shape genome evolution. Genome Biol 10: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BØ, Papin JA (2009) Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ (2005) Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet 37: 1372–1375 [DOI] [PubMed] [Google Scholar]
- Pál C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD (2006) Chance and necessity in the evolution of minimal metabolic networks. Nature 440: 667–670 [DOI] [PubMed] [Google Scholar]
- Price N, Reed J, Palsson B (2004) Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol 2: 886–897 [DOI] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis Md, Savva R, Wernisch L (2004) Solving the riddle of codon usage preferences: a test for translational selection. Nucl Acids Res 32: 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D (1975) Minimal mutation trees of sequences. SIAM J Appl Mathematics 28: 35–42 [Google Scholar]
- Sharp PM, Li W-H (1987) The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucl Acids Res 15: 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H (2000) Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407: 81–86 [DOI] [PubMed] [Google Scholar]
- Silva FJ, Latorre A, Moya A (2001) Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet 17: 615–618 [DOI] [PubMed] [Google Scholar]
- Tamames J, Moya A, Valencia A (2007) Modular organization in the reductive evolution of protein-protein interaction networks. Genome Biol 8: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, Klasson L, Canback B, Naslund AK, Eriksson A-S, Wernegreen JJ, Sandstrom JP, Moran NA, Andersson SGE (2002) 50 Million years of genomic stasis in endosymbiotic bacteria. Science 296: 2376–2379 [DOI] [PubMed] [Google Scholar]
- Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, Rao BS, Smirnov S, Sverdlov A, Vasudevan S, Wolf Y, Yin J, Natale D (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Zucker J, Macdonald S, Sorokin A, Goryanin I, Douglas A (2009) A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst Biol 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Birin H, Gophna U, Kupiec M, Ruppin E (2010a) Reconstructing ancestral gene content by coevolution. Genome Res 20: 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Waldman YY, Kupiec M, Ruppin E (2010b) Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci 107: 3645–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RCHJ, Kamerbeek J, Palacios C, Rausell C, Abascal F, Bastolla U, Fernández JM, Jiménez L, Postigo M, Silva FJ, Tamames J, Viguera E, Latorre A, Valencia A, Morán F, Moya A (2003) Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA 100: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek MJA, Hogeweg P (2009) Metabolic adaptation after whole genome duplication. Mol Biol Evol 26: 2441–2453 [DOI] [PubMed] [Google Scholar]
- Varma A, Palsson BO (1994) Metabolic flux balancing: basic concepts, scientific and practical use. Bio Technol 12: 994–998 [Google Scholar]
- Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL, Moran NA, Eisen JA (2006) Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol 4: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood Computer Applications in BioSciences. Mol Biol Evol 13: 555–556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–4, Supplementary Tables S1–3