In this work, an integrated computational and synthetic biology approach is used to engineer an all-or-none response in a ligand/receptor signaling system. Rationally tuning of the resulting switch is achieved by modulating the strength of two positive feedback loops.
Keywords: cell signaling, design principles, feedback, switch-like response, synthetic biology
Abstract
The ability to engineer an all-or-none cellular response to a given signaling ligand is important in applications ranging from biosensing to tissue engineering. However, synthetic gene network ‘switches' have been limited in their applicability and tunability due to their reliance on specific components to function. Here, we present a strategy for reversible switch design that instead relies only on a robust, easily constructed network topology with two positive feedback loops and we apply the method to create highly ultrasensitive (nH>20), bistable cellular responses to a synthetic ligand/receptor complex. Independent modulation of the two feedback strengths enables rational tuning and some decoupling of steady-state (ultrasensitivity, signal amplitude, switching threshold, and bistability) and kinetic (rates of system activation and deactivation) response properties. Our integrated computational and synthetic biology approach elucidates design rules for building cellular switches with desired properties, which may be of utility in engineering signal-transduction pathways.
Introduction
In response to extracellular cues, natural biological systems generate dynamic intracellular responses that can be critical for achieving a native phenotype. A wide array of specific and modular responses can, in principle, be generated by integrating signaling elements from the cell surface, cytoplasm, and nucleus (Kiel et al, 2010). Cell-surface receptors enable ligand-specific recognition and signaling, cytoplasmic messengers provide signal processing modules, and transcriptional elements in the nucleus regulate complex changes in gene expression. Engineering these intracellular pathways for biotechnological (Alper and Stephanopoulos, 2009) and biomedical (Ro et al, 2006) applications is challenging, particularly when interfacing synthetic components with native pathways in order to elicit the desired response (Mukherji and van Oudenaarden, 2009).
A binary decision is common in certain all-or-none cellular processes such as differentiation, proliferation, survival, and apoptosis, and the ability to engineer cells with such an output can be useful in controlling fate decisions, patterning tissues, and creating intracellular logic programs (Ferrell and Machleder, 1998; Becskei et al, 2001; Bagowski et al, 2003; Melen et al, 2005; Chang et al, 2006; Anderson et al, 2007; Rinaudo et al, 2007; Win and Smolke, 2008). From the first experimental demonstration of a synthetic toggle (Gardner et al, 2000) to more recent examples of bistable switches (Ajo-Franklin et al, 2007; Deans et al, 2007), the construction of such devices (Gardner et al, 2000; Hasty et al, 2000; Yokobayashi et al, 2002; Isaacs et al, 2003; Kramer et al, 2004; Kim et al, 2006; Ajo-Franklin et al, 2007; Deans et al, 2007; Dueber et al, 2007; Friedland et al, 2009) has primarily relied on specific components and the design principles do not easily translate to a receptor-mediated signaling pathway or to a new system of interest (Kwok, 2010).
Results and discussion
We have recently reported a mathematical model of a signaling topology with two positive, linear feedback loops, inspired by lineage commitment networks in hematopoietic progenitors, which can convert graded ligand-receptor binding responses into highly ‘switch-like' transcriptional responses (Palani and Sarkar, 2008). Here, we computationally show that the two feedback loops alone are sufficient to generate bistability with strong ultrasensitivity, and we further demonstrate the approach experimentally. In this minimal topology, a ligand binds to its cognate cell-surface receptor and transmits a signal that activates a downstream transcription factor. The activated transcription factor drives its own transcription as well as that of the cognate cell-surface receptor (Figure 1A, Supplementary Table S1 and S2). Without feedback, the receptor signaling pathway has a purely hyperbolic response, with a Hill coefficient (nH) of 1.00 (Supplementary Figure S1). To identify feedback topologies that could convert this graded signal into a switch, we simulated several topological variations of a one-component system (Supplementary information). In the absence of any other sources of ultrasensitivity, we found that two interconnected, linear feedback loops—but not either loop alone unless feedback was cooperative—could generate the desired switch-like response (Supplementary Figure S1 and Supplementary information). While other sources of ultrasensitivity in an actual cellular signaling network may relax the requirement for both feedback loops, the two-feedback topology in Figure 1A should nevertheless be sufficient for creating a binary response to a signaling ligand.
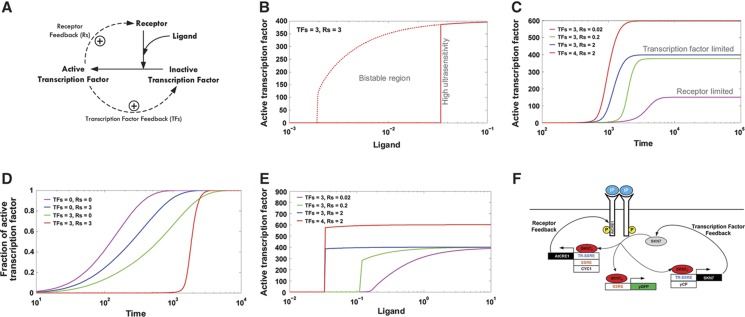
Figure 1.
Network design and computational analysis. (A) Minimal two-feedback topology. Extracellular ligand binds to its specific cell-surface receptor which signals to activate a downstream transcription factor. The activated transcription factor upregulates its own expression as well as that of the cell-surface receptor through two positive feedback loops with feedback strengths TFs and Rs, respectively. (B) Simulation showing steady-state response of active transcription factor with change in ligand concentration. Transcription factor feedback strength (TFs) and receptor feedback strength (Rs) are kept equal and constant at 3. (C) Time plot highlighting tunability in steady-state setpoint of active transcription factor in receptor-limited and transcription factor-limited regimes by varying TFs and Rs. Dimensionless ligand concentration is kept constant at 0.3. (D) Time plot of fraction of activated transcription factor levels for no feedback, receptor feedback, transcription factor feedback, and double feedback topologies showing variability in activation kinetics. Ligand concentration is kept constant at 0.3. (E) Steady-state response plot of active transcription factor with change in ligand levels for various TFs and Rs values showing independent tunability of ultrasensitivity and maximum steady-state setpoint. (F) Schematic for experimental implementation of two-feedback topology in S. cerevisiae. The initial linear signaling pathway consists of the plant cytokinin IP binding to and activating the plant receptor AtCRE1, which leads to phosphorylation and activation of the yeast endogenous transcription factor SKN7. Transcription factor feedback and receptor feedback are synthetically introduced by driving protein expression with synthetic promoters SSRE and TR-SSRE. yGFP, which is expressed through the SSRE promoter, serves as a fluorescent reporter. Source code is available for this figure at www.nature.com/msb.
Simulations of this topology exhibit high ultrasensitivity and bistability in the expression of active transcription factor (Figure 1B) for a wide range of feedback strengths at the receptor and transcription factor levels. By varying feedback strengths, the model also shows tunability in various system-level properties such as steady-state setpoint (in receptor-limited and transcription factor-limited regimes, explained below; Figure 1C) and activation kinetics (Figure 1D). Moreover, the model suggests that the topology can independently tune traditionally correlated properties such as ultrasensitivity and maximum steady-state setpoint (Figure 1E), thereby providing a flexible platform to design switches. We therefore experimentally tested whether synthetic introduction of these two feedback loops into a graded signaling pathway could convert it into a tunable, reversible switch.
While this strategy could be employed to rewire endogenous signaling pathways, a major challenge in synthetic biology is to regulate cellular behavior using non-native ligands as these molecules can ensure response specificity. Cells can be made responsive to such ligands by heterologously expressing their cognate receptors (if these receptors activate endogenous signaling proteins) or chimeric receptors (if they do not; Sadelain et al, 2009); however, further engineering of these receptor-mediated signals to generate tunable, switch-like cellular responses has remained challenging (Kwok, 2010). Here, we demonstrate our approach in Saccharomyces cerevisiae using a pathway that involves signaling of a synthetic cytokinin receptor (CRE1 from Arabidopsis thaliana (AtCRE1)) to an endogenous yeast transcription factor (SKN7; Chen and Weiss, 2005).
This linear synthetic signaling pathway, which is graded and unimodal with respect to ligand (Chen and Weiss, 2005), has only constitutive expression of the receptor and transcription factor (cRcTF; c=constitutive, R=receptor, TF=transcription factor) and was created by integrating a single copy of AtCRE1 (Inoue et al, 2001), driven by the CYC1 promoter, into the yeast genome. For a reporter of network response, we also integrated a single copy of yEGFP3, a yeast-enhanced GFP, driven by a synthetic SSRE promoter (Chen and Weiss, 2005), which contains the binding site for the yeast endogenous transcription factor SKN7. The plant cytokinin isopentenyl adenine (IP) (Mok and Mok, 2001) was used as the ligand to activate a two-component phospho-relay pathway, in which exogenous cell-surface receptor AtCRE1 phospho-transfers and activates the cytosolic histidine kinase YPD1 (Lu et al, 2003). Phosphorylated YPD1 is shuttled to the nucleus and activates nuclear SKN7 via a second phospho-transfer. Phosphorylated SKN7 (SKN7p) binds to the SSRE promoter and activates GFP expression (Figure 1F).
Five variations of this basic network were constructed by using a single SSRE or tandem-repeat SSRE (TR-SSRE) to introduce SKN7-mediated positive feedback at moderate or strong levels, respectively. The CYC1 promoter of AtCRE1 was replaced with SSRE or TR-SSRE to create two networks with an IP-regulated positive feedback loop through the receptor only (sRcTF and tRcTF; s=SSRE, t=TR-SSRE). A network with only transcription factor feedback (cRtTF) was created by integrating a single copy of the SKN7 gene, driven by the TR-SSRE promoter, into the yeast genome. Two networks with both receptor and transcription factor feedback (sRtTF and tRtTF) were constructed by integrating a single copy of the SKN7 gene (driven by TR-SSRE) and by replacing the CYC1 promoter of AtCRE1 with either SSRE or TR-SSRE.
All six strains were grown overnight in their respective synthetic dropout (SD) media to reach steady state. The cultures were then diluted to an OD660 reading of 0.1 and induced with 1 μM IP. Aliquots from the growing cultures were extracted at different time points to quantify GFP levels by flow cytometry.
Expression of GFP is dependent on the concentration of SKN7p, which is dictated by both the expression of SKN7 and its subsequent post-translational modification via IP/AtCRE1 complex signaling. Hence, the expression of GFP can be limited by either the insufficiency of SKN7 substrate (transcription factor limited) or IP/AtCRE1 complex (receptor limited). As these two regimes generate different dynamics and could potentially be exploited to achieve greater tuning of the response, we identified the operating regime for each of our strains and characterized their consequent steady-state and dynamic operating characteristics.
Increasing the receptor feedback strength (TR-SSRE>SSRE>CYC1) (Chen and Weiss, 2005) did not change the steady-state setpoint of GFP when SKN7 was only driven by the constitutive promoter (Figure 2A). This is due to the fact that receptor expression from all tested promoters was sufficient to activate the endogenous levels of SKN7; hence, these systems are all transcription factor limited (see Figure 1C). However, when we integrated an additional copy of the SKN7 gene, driven by the strong TR-SSRE promoter, the system became receptor limited. In contrast to the transcription factor-limited regime, increasing the receptor feedback strength enabled modulation of the steady-state GFP level (Figure 2B).
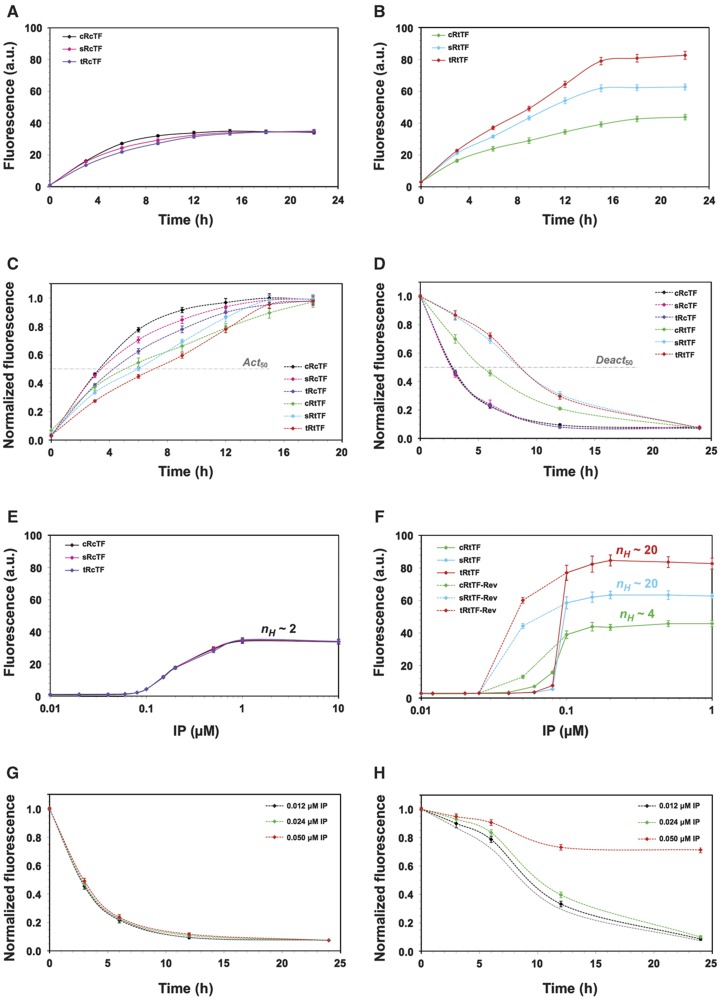
Figure 2.
Feedback modulation of kinetic and steady-state system responses. (A) Activation kinetics for cTF strains. Strains cRcTF, sRcTF, and tRcTF were induced with 1 μM IP and aliquots were taken from the cultures at different time points to determine GFP expression kinetics and steady-state levels. As these strains are all transcription factor limited, the increased receptor feedback does not enhance steady-state GFP expression. (B) Activation kinetics for tTF strains. Similarly, GFP expression kinetics and steady-state levels were quantified for strains cRtTF, sRtTF, and tRtTF after induction with 1 μM IP. The strong transcription factor feedback loop makes these strains receptor limited, so receptor feedback now has a significant effect on steady-state GFP expression. (C) Temporal activation profiles of all six strains normalized to their steady-state setpoint. All six strains reach 50% of the steady-state level at different times (Act50), with stronger feedback loops slowing the kinetics. (D) Temporal deactivation profiles of the strains normalized to the steady-state setpoint. After reaching steady-state GFP expression in 1 μM IP, the cultures were thoroughly washed and resuspended in media with no IP. All strains exhibit reversibility in the absence of IP, but the deactivation kinetics are markedly different. The time at which 50% of the initial GFP expression level is reached (Deact50) is fastest for the basic and receptor feedback strains and slowest for the double feedback strains. (E) Steady-state dose–response curves for cTF strains. For cRcTF, sRcTF, and tRcTF strains, the steady-state GFP responses to IP (0.01–10 μM) are indistinguishable and weakly ultrasensitive (nH∼2). When the strains were allowed to reach high GFP steady-state levels in 1 μM IP and were then reduced to sub-threshold concentrations of IP (0.012, 0.025, and 0.05 μM IP), the strains exhibited no memory and showed a monostable response (for clarity, reverse curves are not shown as they overlay the forward curves). (F) Steady-state dose–response curves for tTF strains. While the steady-state response for the cRtTF strain exhibits slightly greater ultrasensitivity (nH∼4), the dual-feedback strains sRtTF and tRtTF strikingly function as almost as pure binary switches (nH∼20). When the strains were allowed to reached high GFP steady-state levels in 1 μM IP and were then reduced to sub-threshold concentrations of IP (0.012, 0.025, and 0.05 μM IP), all three strains exhibited memory (reverse curves shown as dotted lines). (G) Deactivation kinetics for cRcTF strain. After reaching high GFP steady-state levels in 1 μM IP, strain cRcTF was thoroughly washed and resuspended in medium with different sub-threshold IP concentrations (0.012, 0.025, and 0.05 μM). The temporal deactivation curves indicate no temporal memory in this strain when compared with the 0 μM IP concentration curve (dotted gray line). (H) Deactivation kinetics for tRtTF strain. After reaching high GFP steady-state levels in 1 μM IP, strain tRtTF was thoroughly washed and resuspended in medium with different sub-threshold IP concentrations (0.012, 0.025, and 0.05 μM). For all sub-threshold concentrations, the strain shows temporal memory when compared with the 0 μM IP concentration curve (dotted gray line); however, the strain is only bistable at 0.05 μM IP. Source data is available for this figure at www.nature.com/msb.
We further examined whether this core topology could achieve kinetic tunability with respect to the rates of GFP synthesis and degradation, which correspond to the rates of SKN7 activation and deactivation, respectively. The time profiles of all strains were normalized to their steady-state values and the rate of GFP accumulation was compared among the six strains by calculating the time needed for achieving the 50% level of the final steady-state value (Act50; Figure 2C). In agreement with our model simulations (Figure 1D), the strain with the basic pathway had the fastest Act50 (3.4 h), followed by the receptor-feedback strains (3.6 h for sRcTF and 4.2 h for tRcTF). The Act50 for cRtTF and sRtTF were 5.0 and 6.0 h, respectively. The tRtTF strain was the slowest and attained the 50% mark at 6.9 h after IP induction. This observation was not unexpected, as it is well known that incorporation of feedback loops slows down the time to reach equilibrium.
To compare deactivation kinetics, the strains were grown in 1.0 μM IP for 24 h to achieve steady-state levels of GFP. The cells were then spun down, washed three times and cultured in their respective SD media without IP. All of the systems are reversible and therefore ultimately shut off; however, the rate of deactivation of SKN7 varies with feedback strength (Figure 2D). The loss of GFP signal was compared across strains by calculating the time needed for lowering the fluorescence level to 50% of the original steady-state value (Deact50). The no-feedback and receptor-feedback strains had the lowest Deact50 (∼2.5 h) followed by the transcription factor strain cRtTF (∼5.3 h). The double feedback strains had the slowest degradation rate (∼8.5 h), which could be due to the higher accumulation of SKN7p at steady state.
We then tested whether the two-feedback topology could generate a switch-like response, as predicted by the model (Figure 1B and E). All six strains were grown overnight in their respective SD media, diluted to an OD660 of 0.1, and induced with concentrations of IP spanning from 0.01 to 10 μM. The steady-state levels of GFP were plotted against IP concentration to determine the degree of ultrasensitivity in the response. The linear pathway and the networks with only receptor-mediated feedback showed a graded response (nH∼2) with respect to IP concentration (Figure 2E). The strain with only transcription factor feedback had modest ultrasensitivity (nH∼4), and most strikingly, those with both feedback loops exhibited essentially perfect binary responses (nH∼20; Figure 2F). The no-feedback and receptor-feedback strains reached setpoint saturation around 1 μM IP, whereas the other three strains reached saturation at 10-fold lower IP (Figure 2E and F).
To test the presence of memory, all six strains were induced with a high IP concentration (1 μM) and allowed to reach steady state. The cells were then spun down, washed three times, cultured in their respective SD media with sub-threshold IP concentrations (0.05, 0.025, or 0.012 μM), and allowed to reach a new steady state. The linear pathway and the pure receptor-feedback strains showed no memory of high IP induction (Figure 2E, reverse curves not shown as they overlay the forward curves) and the strain with only transcription factor feedback showed weak memory at 0.05 μM IP (∼25% of the 1.0 μM IP signal; Figure 2F). The strains with double feedback showed much stronger memory at 0.05 μM IP (∼70% of that at 1.0 μM IP; Figure 2F). The width of the bistable window (W) for each strain was estimated by the fold decrease in EC50 due to the memory of high IP. W was higher in the double feedback strain (∼2.3-fold) compared with the transcription factor feedback strain (∼1.3-fold). The presence of bistability in the transcription factor feedback strain likely arises from basal ultrasensitivity in the basic pathway and possible cooperative autoactivation of SKN7. It is important to note that if the basic pathway response were completely Michaelian and the transcription factor feedback exhibited no cooperativity, then both feedback loops would be absolutely necessary to achieve any level of bistability (Supplementary Figure S1 and Supplementary information).
To test for temporal memory, cRcTF (monostable) and tRtTF (bistable) strains were grown in 1 μM IP and allowed to reached steady state. Cells were then washed three times to remove IP and then cultured in sub-threshold IP concentrations (0.05, 0.025, or 0.012 μM). The cRcTF strain did not possess any temporal memory, as its time profiles in sub-threshold IP concentrations coincided with the zero IP degradation curve (Figure 2G). However, the tRtTF strain showed significant memory for all sub-threshold concentrations when compared with the no-IP response (Figure 2H). The temporal memory led to a stable on-state only for 0.05 μM IP (Figure 2F and H).
We also analyzed the single-cell fluorescence distributions at each time point in Figures 2G and H for the 0.05 μM IP curve. The entire histogram for the cRcTF strain shifted to lower mean fluorescence at each time point, as would be expected for a monostable system (Supplementary Figure S2). By contrast, the tRtTF strain showed strong bimodality, with a large fraction of cells remaining in the high-GFP state (Supplementary Figure S2).
Thus, the ability to independently add transcription factor and receptor-feedback loops to the linear signaling pathway enables operation in either receptor-limited or transcription factor-limited regimes and confers tunability in system activation (Act50) and deactivation (Deact50) kinetics, stimulus threshold (EC50, EC90), ultrasensitivity (nH), steady-state setpoint (Ss), and bistable window (W). These strain properties are summarized in Table I.
Table 1. Properties of the strains used in this study showing tunability across various system-level properties.
| Property | cRcTF | sRcTF | tRcTF | cRtTF | sRtTF | tRtTF | Range | Relevant figures |
|---|---|---|---|---|---|---|---|---|
| NA, not applicable; ND, not determined. | ||||||||
| Feedback limitation | Transcription factor | Transcription factor | Transcription factor | Receptor | Receptor | ND | Transcription factor, receptor | Figures 2A and B |
| Steady-state setpoint, Ss (a.u.) | 34.2 | 34.6 | 35.0 | 45.8 | 63.4 | 84.6 | 2.5-fold | Figures 2E and F |
| Thresholds, EC50 and EC90 (μM) | 0.19, 0.65 | 0.20, 0.64 | 0.20, 0.70 | 0.09, 0.12 | 0.09, 0.10 | 0.09, 0.10 | 2.2, 7.0-fold | Figures 2E and F |
| Activation, Act50 (h) | 3.4 | 3.6 | 4.2 | 5.0 | 6.0 | 6.9 | 2.1-fold | Figure 2C |
| Deactivation, Deact50 (h) | 2.6 | 2.5 | 2.6 | 5.3 | 8.5 | 8.6 | 3.4-fold | Figure 2D |
| Ultrasensitivity, nH | 2.1 | 2.3 | 2.2 | 4.3 | 20.6 | 20.2 | 9.6-fold | Figures 2E and F |
| Bistable window, W | NA | NA | NA | 1.3 | 2.3 | 2.3 | Monostable, bistable | Figures 2E and F |
Our results suggest that the two-feedback topology and its topological variants may serve as a flexible platform for synthetic switch design through simple promoter engineering. To further highlight this point, we list four possible steady-state response characteristics (based on high/low combinations of ultrasensitivity and setpoint) and three possible kinetic response characteristics (based on coupled tuning of activation and deactivation rates; Table II). Examples of steady-state designs 1 and 4 as well as kinetic designs 1, 2, and 3 have been experimentally demonstrated in this study. Steady-state designs 2 and 3 could be realized by using a stronger constitutive TF promoter and a weaker TF feedback promoter, respectively (Supplementary Figure S3). Thus, this network topology, coupled with a simple set of design rules, may be used to achieve a diverse set of response behaviors.
Table 2. Design rules for generating systems with desired steady-state or kinetic properties.
| Steady-state design | Ultrasensitivity (nH) | Steady-state setpoint (Ss) | Example strain(s) | Relevant figures (see also Table I) |
|---|---|---|---|---|
| 1 | Low | Low | cRcTF, sRcTF, tRcTF, cRtTF | Figures 2E and F |
| 2 | Low | High | cRcTF with stronger constitutive TF promoter | Supplementary Figure S3 |
| 3 | High | Low | tRtTF with weaker TF feedback promoter | Supplementary Figure S3 |
| 4 | High | High | sRtTF, tRtTF | Figure 2F |
| Kinetic design |
Activation (Act50) |
Deactivation (Deact50) |
Example strain(s) |
Relevant figures (see also Table I) |
| 1 | Slow | Slow | sRtTF, tRtTF | Figures 2C and D |
| 2 | Intermediate | Intermediate | cRtTF | Figures 2C and D |
| 3 | Fast | Fast | cRcTF, sRcTF, tRcTF | Figures 2C and D |
It is also noteworthy that the ultrasensitivity and bistability of this two-feedback topology are quite robust with respect to parameter variation (NA Shah and CA Sarkar, under review), so the generation of switch-like responses should not be heavily dependent on specific rate constants associated with the chosen molecular components or host cell. This robust and tunable design strategy may therefore enable rational manipulation of steady-state and kinetic responses of a variety of receptor signals.
Materials and methods
Computational modeling
An ordinary differential equation-based model of the two-feedback topology (Figure 1A) was constructed using mass-action kinetics for all explicitly modeled reactions and the rapid-equilibrium approximation for transcriptional feedback. The complete set of differential equations, initial conditions, and parameter values for the deterministic model are given in Supplementary Tables S1 and S2. The steady-state response plots and time profiles for various feedback strengths in Figure 1B–E were generated using custom scripts in MATLAB (The Mathworks). The model is also available in SBML format (see Supplementary information) and in the BioModels Database (accession number MODEL1102160000).
Plasmid construction
Plasmids and their properties are provided in Supplementary Table S3. E. coli strain XL1-Blue, which was used for all plasmid construction, and yeast integration plasmids pRS403 and pRS405 were purchased from Stratagene. Yeast centromere plasmids DQ232595, DQ232596, and DQ232600 were kindly provided by Ron Weiss (Massachusetts Institute of Technology; Chen and Weiss, 2005). The SKN7 gene was PCR amplified from the S. cerevisiae genome. Plasmids were constructed as follows. For pSP001, DQ232600 was cut with PvuI to extract CEN6/ARSH4, which was ligated into PvuI-digested pRS405. For pSP002 and pSP003, DQ232600 was digested with BamHI to excise AtCRE1a-PSSRE, which was ligated into DQ232595 and DQ232596, respectively; then, CEN6/ARSH4 and HIS3 were excised from the modified DQ232595 and DQ232596 plasmids (PvuI site) and ligated into pRS405. For pSP004, the SKN7 gene was cloned to replace yEGFP3 in DQ232596 using the BamHI/AscI restriction sites; CEN6/ARSH4 in the modified DQ232596 vector was removed (PvuI site) and ligated into pRS403 plasmid. All plasmid constructs were verified by sequencing.
Yeast strains, genomic screens, and culture
Yeast strains and their properties are listed in Supplementary Table S4. The background yeast strain, TM182, lacks the endogenous osmosensor receptor SLN1, which enables selective activation of SKN7 only via AtCRE1; to rescue SLN1 deletion-induced lethality due to HOG1 activation, this strain also overexpresses the HOG1 phosphatase PTP2 through a galactose-inducible promoter (Chen and Weiss, 2005). Yeast transformations were performed using the high-efficiency LiAc/SS carrier DNA/PEG method with standard SD media. The cRcTF, sRcTF, and tRcTF strains were constructed by integrating a single copy of pSP001, pSP002, and pSP003, respectively, into the LEU2 locus of the TM182 strain. The feedback strain cRtTF was constructed by integrating a single copy of pSP004 into the HIS3 locus of the cRcTF strain. Double feedback strains sRtTF and tRtTF were built by integrating a single copy of pSP004 into the HIS3 locus of the sRcTF and tRcTF strains, respectively. Yeast genomic DNA isolation was performed using the phenol/chloroform/isoamyl alcohol method. The isolated genomic DNA was probed for stable single integration of the transformed gene at the desired locus through PCR. The wild-type TM182 strain was used as a negative control in all experiments. Yeast strains were grown at 30°C in their respective SD Ura−/His−/Leu− medium with 2% galactose. IP was purchased from Sigma-Aldrich.
Analysis of GFP expression
For all IP-induction experiments, GFP levels were quantified using a Guava flow cytometer (Millipore) with a 488-nm argon excitation laser and a 525/30 nm emission filter. For each sample, 10 000 events were acquired and the fluorescence data were analyzed using FlowJo 7.5 (Tree Star).
Supplementary Material
Supplementary Figures S1–3, Supplementary Tables S1–4
Acknowledgments
We thank Christine J Li, Najaf A Shah, Ellen C O'Shaughnessy, Pamela A Barendt, and Ming-Tang Chen for helpful discussions. This work was supported by a grant from the American Heart Association (#0835132N), a National Science Foundation CAREER award (#1055231), and startup funds from the University of Pennsylvania to CAS.
Author Contributions: SP and CAS conceived and designed the experiments. SP performed the experiments. SP and CAS analyzed the data. SP and CAS wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA (2007) Rational design of memory in eukaryotic cells. Genes Dev 21: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Stephanopoulos G (2009) Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol 7: 715–723 [DOI] [PubMed] [Google Scholar]
- Anderson JC, Voigt CA, Arkin AP (2007) Environmental signal integration by a modular AND gate. Mol Syst Biol 3: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagowski CP, Besser J, Frey CR, Ferrell JE Jr (2003) The JNK cascade as a biochemical switch in mammalian cells: ultrasensitive and all-or-none responses. Curr Biol 13: 315–320 [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L (2001) Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J 20: 2528–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Oh PY, Ingber DE, Huang S (2006) Multistable and multistep dynamics in neutrophil differentiation. BMC Cell Biol 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MT, Weiss R (2005) Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol 23: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Deans TL, Cantor CR, Collins JJ (2007) A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell 130: 363–372 [DOI] [PubMed] [Google Scholar]
- Dueber JE, Mirsky EA, Lim WA (2007) Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol 25: 660–662 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr, Machleder EM (1998) The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 280: 895–898 [DOI] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ (2009) Synthetic gene networks that count. Science 324: 1199–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342 [DOI] [PubMed] [Google Scholar]
- Hasty J, Pradines J, Dolnik M, Collins JJ (2000) Noise-based switches and amplifiers for gene expression. Proc Natl Acad Sci USA 97: 2075–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Hasty J, Cantor CR, Collins JJ (2003) Prediction and measurement of an autoregulatory genetic module. Proc Natl Acad Sci USA 100: 7714–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, Yus E, Serrano L (2010) Engineering signal transduction pathways. Cell 140: 33–47 [DOI] [PubMed] [Google Scholar]
- Kim J, White KS, Winfree E (2006) Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol Syst Biol 2: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M (2004) An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol 22: 867–870 [DOI] [PubMed] [Google Scholar]
- Kwok R (2010) Five hard truths for synthetic biology. Nature 463: 288–290 [DOI] [PubMed] [Google Scholar]
- Lu JM, Deschenes RJ, Fassler JS (2003) Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot Cell 2: 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen GJ, Levy S, Barkai N, Shilo BZ (2005) Threshold responses to morphogen gradients by zero-order ultrasensitivity. Mol Syst Biol 1: 2005.002810.1038/msb4100036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Ann Rev Plant Phys 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Mukherji S, van Oudenaarden A (2009) Synthetic biology: understanding biological design from synthetic circuits. Nat Rev Genet 10: 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S, Sarkar CA (2008) Positive receptor feedback during lineage commitment can generate ultrasensitivity to ligand and confer robustness to a bistable switch. Biophys J 95: 1575–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y (2007) A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol 25: 795–801 [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943 [DOI] [PubMed] [Google Scholar]
- Sadelain M, Brentjens R, Rivière I (2009) The promise and potential pitfalls of chimeric antigen receptors. Curr Opin Immunol 21: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD (2008) Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold FH (2002) Directed evolution of a genetic circuit. Proc Natl Acad Sci USA 99: 16587–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–3, Supplementary Tables S1–4