A genome wide RNAi screen identifies 72 host cell genes affecting S. Typhimurium entry, including actin regulators and COPI. This study implicates COPI-dependent cholesterol and sphingolipid localization as a common mechanism of infection by bacterial and viral pathogens.
Keywords: coatomer, HeLa, Salmonella, siRNA, systems biology
Abstract
The pathogen Salmonella Typhimurium is a common cause of diarrhea and invades the gut tissue by injecting a cocktail of virulence factors into epithelial cells, triggering actin rearrangements, membrane ruffling and pathogen entry. One of these factors is SopE, a G-nucleotide exchange factor for the host cellular Rho GTPases Rac1 and Cdc42. How SopE mediates cellular invasion is incompletely understood. Using genome-scale RNAi screening we identified 72 known and novel host cell proteins affecting SopE-mediated entry. Follow-up assays assigned these ‘hits’ to particular steps of the invasion process; i.e., binding, effector injection, membrane ruffling, membrane closure and maturation of the Salmonella-containing vacuole. Depletion of the COPI complex revealed a unique effect on virulence factor injection and membrane ruffling. Both effects are attributable to mislocalization of cholesterol, sphingolipids, Rac1 and Cdc42 away from the plasma membrane into a large intracellular compartment. Equivalent results were obtained with the vesicular stomatitis virus. Therefore, COPI-facilitated maintenance of lipids may represent a novel, unifying mechanism essential for a wide range of pathogens, offering opportunities for designing new drugs.
Introduction
Salmonella enterica subspecies 1 serovar Typhimurium (S. Typhimurium or S. Tm in this paper) is a common cause of diarrhea in humans. Central to its pathogenicity is the ability to invade gut epithelial cells (Patel and Galan, 2005; Schlumberger and Hardt, 2006; McGhie et al, 2009). This process is incompletely understood and requires an intricate interplay of S. Typhimurium virulence factors and numerous host cell factors. The whole range of host cell factors involved in the invasion process has not been elucidated.
In order to invade epithelial cells, S. Typhimurium binds to the host cell surface. This process can involve reversible adhesion (e.g., via fimbriae) and irreversible docking via the Type III secretion system 1 (T1; Misselwitz et al, 2011). Then, T1 acts as a molecular syringe to inject virulence factors, so called effectors (Figure 1A). Four key effectors, SopE, SopE2, SopB and SipA, can trigger actin polymerization and mediate epithelial cell invasion in a functionally overlapping manner (Norris et al, 1998; Zhou et al, 1999; Schlumberger and Hardt, 2006). Among the key effectors, SopE, a G-nucleotide exchange factor for the Rho GTPases Rac1 and Cdc42 (Hardt et al, 1998; Rudolph et al, 1999; Friebel et al, 2001), is the most potent trigger of invasion. Both Rho GTPases signal to the Arp2/3 complex, a powerful activator for actin polymerization (Goley and Welch, 2006). Actin polymerization leads to the formation of pronounced and characteristic ruffles on the cellular surface (Finlay et al, 1991) facilitating invasion. Once inside the host cell, S. Typhimurium is enclosed in a vacuole which matures, acquires late endosome markers and positions itself close to the nucleus (Guignot et al, 2004; Marsman et al, 2004). Inside this ‘Salmonella-containing vacuole’ (SCV), S. Tm expresses a second set of virulence factors encoded on the ‘Salmonella pathogenicity island 2’ (SPI-2) (Schlumberger and Hardt, 2006).
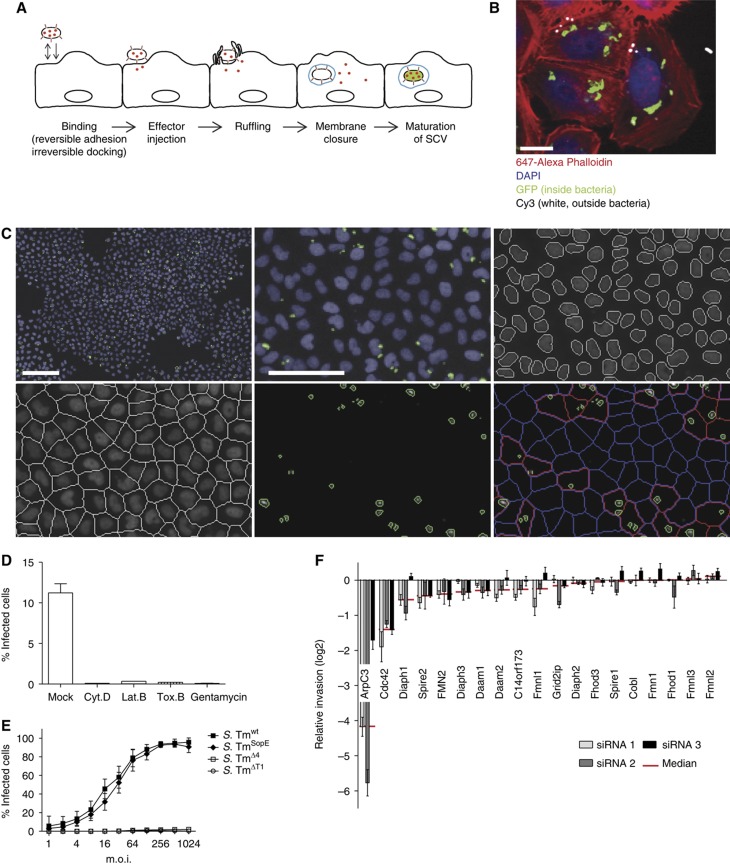
Figure 1.
Establishment of an automated assay to analyze S. Typhimurium invasion. (A) Overview showing the invasion process of S. Typhimurium divided into five major steps: (i) during the binding step, the bacteria attach to the cellular surface by reversible adhesion or irreversible docking; (ii) T1 is used as a molecular syringe to inject effectors (shown in red) into the eukaryotic cell; (iii) these effectors in turn induce membrane ruffling; (iv) subsequently the cellular membrane encloses a bacterium (membrane closure), thereby producing a Salmonella-containing vacuole (SCV, shown in blue); (v) after a maturation step, S. Tm genes important for intracellular survival are induced (green). (B) Fluorescence image showing GFP expression of S. Tmwt (pM975) only after invasion into HeLa cells (green=inside bacteria, red=actin, blue=DAPI, white=outside bacteria; scale bar=20 μm). (C) Automated image analysis strategy: S. Tmwt (pM975) infection of HeLa cells followed by the acquisition of nuclei (blue) and bacterial spots (green) using an automated microscope with a × 10 objective. Images were analyzed using CellProfiler as follows: recognition of nuclei, definition of cells, identification of bacterial spots and the allocation of these spots to cells (red outline=infected cell, blue outline=non-infected cell; scale bar whole image=100 μm, detailed image=50 μm). (D) Verification of the automated assay testing inhibitors of Salmonella invasion. HeLa cells were infected with S. Tmwt (pM975) and analyzed as described in (C). Pretreatment of HeLa cells with the inhibitors Cytochalasin D (Cyt. D), Latrunculin B (Lat. B), Toxin B (Tox. B) or the antibiotic gentamycin prevents invasion. (E) Invasion efficiencies of various Salmonella strains into HeLa cells analyzed by the automated assay showing S. TmSopE (pM975) invasion being as efficient as S. Tmwt (pM975). (F) Verification of the automated assay using siRNAs directed against different actin polymerization regulators. Depletion of ArpC3 and Cdc42 reduces S. TmSopE (pM975) invasion (red line=median of three siRNAs tested for each gene; log2 relative invasion=% infected cells with siRNA treatment divided by the median of % infected cells treated with control siRNA).
While tremendous progress has been achieved toward understanding the invasion of S. Typhimurium into host cells, the picture is far from complete. Importantly, host cell factors required for S. Typhimurium invasion have never been analyzed in a comprehensive and unbiased manner. RNAi screening has recently been introduced to study host–pathogen interactions (Ramet et al, 2002; Agaisse et al, 2005; Cheng et al, 2005; Pelkmans et al, 2005; Philips et al, 2005; Derre et al, 2007; Kuijl et al, 2007; Cherry, 2008; Elwell et al, 2008; Chong et al, 2009; Prudencio and Lehmann, 2009; Hirsch, 2010). This systematic approach has significantly advanced our understanding of the respective molecular infection processes.
To better understand the mechanism of S. Typhimurium entry, we performed a genome-scale RNAi screen for host proteins affecting SopE-mediated invasion into HeLa cells. Our experiments identified actin cytoskeletal regulators and proteins affecting host cell invasion, which were not previously implicated in this process. Systematic follow-up assays revealed a novel functional link between the coatomer I (COPI) complex, the maintenance of the membrane lipid composition and the capacity of S. Typhimurium to manipulate its host cell. This novel mechanism might be of general importance for numerous bacterial and viral pathogens.
Results
An automated assay measuring S. Typhimurium invasion into host cells
For large-scale screening of S. Typhimurium invasion, we used a modified gentamycin protection assay. In our assay, HeLa cells were infected for 20 min with wild-type S. Typhimurium (S. Tmwt (pM975); Supplementary Table SI) expressing GFP under a SPI-2 promotor (pssaG). This GFP reporter is not expressed by extracellular bacteria, but strongly induced when the pathogen resides within the SCV. After infection, medium containing gentamycin (kills all extracellular bacteria) was added, followed by a 4 h incubation step allowing for GFP expression and maturation (Schlumberger et al, 2007). Figure 1B shows specific GFP expression by intracellular S. Tmwt (pM975; ‘green’) but not by bacteria remaining extracellular (‘white’). The combination of gentamycin protection and SPI-2-driven GFP expression resulted in a specific and bright fluorescence signal of the intracellular bacteria.
To establish a genome-wide screening system, we used automated microscopy and developed an automated algorithm to identify and enumerate cells successfully invaded by S. Typhimurium. The assay determines the fraction of infected cells, as defined by the presence of at least one GFP-expressing intracellular bacterium (Figure 1C, bottom right panel, red). Preincubation of cells before the infection with medium containing gentamycin reduced the fraction of infected cells 250- to 1000-fold (Figure 1D), confirming the high specificity of the assay. In addition, toxins known to inhibit S. Typhimurium invasion showed similar effects: Cytochalasin D and Latrunculin B, which disrupt the actin cytoskeleton, and Toxin B from Clostridium difficile, which inactivates Rho GTPases, reduced the fraction of infected cells by at least 90%. These observations demonstrate that disruption of important host cell signaling pathways yields pronounced signals in this assay, validating the approach.
It should be noted that SPI-2 expression does not occur immediately after Salmonella entry but requires a maturation step of the SCV. Therefore, the modified gentamycin protection assay would be sensitive to perturbations of S. Typhimurium entry, the first steps of its intracellular life cycle, as well as SCV fusion with the lysosome and can be considered as a global assay probing Salmonella entry and SCV maturation.
Testing host cell genes for effects on S. Typhimurium invasion
While a strain lacking the four key effectors SipA, SopE, SopE2 and SopB (S. TmΔ4 (pM975)) showed only marginal invasion, an isogenic mutant lacking three key effectors with only SopE remaining (S. TmSopE (pM975)) invaded almost as efficiently as S. Tmwt (pM975) (Figure 1E). As expected, invasion of the mutant without the Type III secretion system 1 (S. TmΔT1 (pM975)) was negligible. On the basis of these observations, we decided to use S. TmSopE (pM975) for our screen. This circumvented any issues arising from the presence of SopE2, SipA or SopB, as these functionally overlapping effectors might mask phenotypes specific for SopE-induced host cell invasion.
To validate the assay, we screened a small targeted siRNA library systematically depleting known actin nucleators, including formins, the p21 subunit of the Arp2/3 complex (ArpC3) as well as the Rho GTPase Cdc42 (Cdc42). Cells were preincubated with siRNAs for 3 days and S. TmSopE (pM975) invasion was analyzed by automated microscopy. Depletion of ArpC3 reduced invasion by ∼95% (log2 median=−4.2; Figure 1F). A similar effect was observed after depletion of Cdc42 (70%, log2 median=−1.4). Depletion of Diaph1, Spire2 and Fmn2 resulted in only 32%, 26% and 25% reduced invasion, respectively, and all other formins yielded even weaker or no detectable effects on invasion at all. These data are in line with the current model of S. Typhimurium host cell invasion, which implicates that the activation of Rho GTPases by SopE and subsequent activation of the Arp2/3 complex are essential for SopE-mediated invasion (Schlumberger and Hardt, 2006). In contrast, individual formins contribute much less to S. Typhimurium entry. Overall, this experiment confirmed that the image-based invasion assay is well suited and sufficiently robust for performing a genome-scale siRNA screen.
A genome-scale screen for host cell proteins affecting SopE-mediated invasion
To identify host cell genes affecting SopE-mediated invasion on a genome scale, we used the ‘druggable genome’ siRNA library (Version 2.0, Qiagen; Supplementary Table SII) covering 6978 human genes. The siRNAs were transferred into 384-well dishes. Each dish included identical sets of controls for siRNA transfection (Eg5 and Plk1 reduce cell number significantly) and positive controls, including ArpC3 (known negative effect on invasion) or gentamycin (kills bacteria before invasion). These controls allowed direct comparison of the data between the different plates and provided essential quality controls.
SiRNA-transfected HeLa cells were infected with S. TmSopE (pM975) at an m.o.i. of 64. In this screen, each gene was tested with three independent siRNAs and each test was performed in triplicates. The invasion efficiency was analyzed by the modified gentamycin protection assay. For each plate, the values were normalized to the median of all siRNAs on this plate. In addition, z-score correction was performed, yielding 361 candidate hits from the uncorrected and 190 candidate hits from the z-score corrected data (for details see Materials and methods). ‘Hit’ genes encoding central subunits of the Arp2/3 complex (Actr2, Actr3) and Cdc42 showed strong inhibitory effects, thus validating our approach (Figure 2A and B). On the basis of the lists of candidate hits and possible interaction partners not present in the original library, we assembled a new library targeting 298 genes with four siRNAs per gene (Supplementary Table SIII). This library also included siRNAs to control for transfection efficiency (Eg5, Plk1 killing cells) and effects on S. Tm invasion (ArpC3, Cdc42, Cfl1).
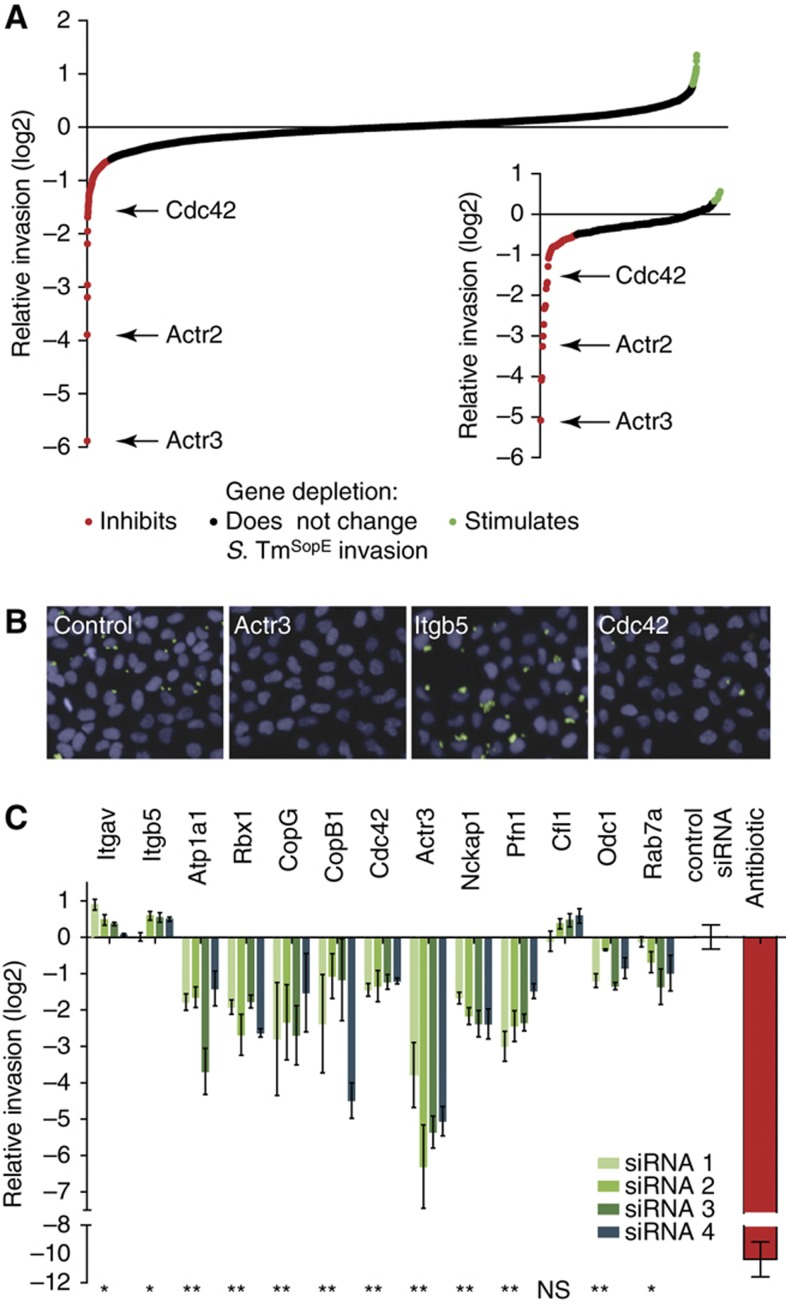
Figure 2.
Genome-scale siRNA screen reveals host cell factors required for Salmonella invasion. (A) Overview of relative invasion of S. TmSopE (pM975) into HeLa cells transfected with siRNA of the druggable genome library. Values represent the median of three siRNAs per gene. Median values above and below 1.5 times of the interquartile range were defined as positive (green) and negative (red) hits, respectively. A confirmatory screen with 298 selected genes approved 72 hits from the genome-scale screen (inserted small graph). Positive (green) and negative (red) hits were determined with median values above 0.3 and below −0.5, respectively. (B) Example images of indicated hits from the screen demonstrating reduced invasion of S. TmSopE (pM975) for Actr3- and Cdc42-depleted cells and increased invasion in the absence of Itgb5 (blue=nuclei, green=invaded bacteria; scale bar=50 μm). (C) Relative invasion of S. TmSopE (pM975) for selected hits from both screens (*P-value <0.1, **P-value <0.05, NS=not significant, Mann–Whitney U-test).
The library was tested in a confirmatory screen for SopE-mediated invasion. S. Tm invasion was normalized using the median of 12 control siRNAs targeting genes without effects on S. Tm invasion in the druggable genome screen (Materials and methods). This allowed normalization in spite of the highly biased nature of the confirmatory library (i.e., containing hits of the primary screen), which prohibited reliable threshold calculation based on the screened library, itself. Therefore, a hit was arbitrarily defined as a gene displaying a log2 of the median of the 4 siRNAs ⩽−0.5 or ⩾0.3. The confirmatory screen thus validated 72 hits (Figure 2A, insert; Supplementary Table SIII). A subset of the results is represented in Figure 2C and depletion efficiencies of these hits were verified (Supplementary Figure 1). Some hits of the primary screen could not be confirmed. This could be explained by off-target effects of the oligos used in the initial screen. Additionally, in the rescreen four instead of three oligos per gene were tested and some of the oligos were newly designed by the company. These slightly changed conditions could explain the different results obtained in the initial and the confirmatory screen.
The list of confirmed hits included important regulators of the actin cytoskeleton previously implicated in S. Typhimurium host cell invasion (Figure 2B and C), as the heptameric Arp2/3 complex of which both subunits tested in the druggable genome screen showed strong inhibitory effects. Other examples include Cdc42 (Chen et al, 1996) and the Nck-associated protein 1 (Nap1, nckp1; Shi et al, 2005; Hanisch et al, 2010), a component of the Wave complex linking Rho GTPase activation to the activation of the Arp2/3 complex (Goley and Welch, 2006). In addition, we identified Profilin 1 (Pfn1), a well-characterized actin binding protein which delivers actin monomers to sites of actin polymerization (Pollard and Cooper, 2009), that has not been studied before in the context of S. Typhimurium invasion. Hits stimulating S. TmSopE (pM975) invasion efficiency included adenylyl cyclase-associated protein 1 (Cap1; Balcer et al, 2003), which mediates the breakdown of actin fibers and can affect S. Typhimurium host cell invasion (Maciver and Hussey, 2002; McGhie et al, 2004; Paavilainen et al, 2004). Therefore, these hits confirm and extend the current model of SopE-mediated host cell invasion. The screen also identified numerous novel genes not previously linked to S. Typhimurium host cell invasion. These hits include the sodium potassium ATPase 1 (Atp1a1; Kaplan, 2002), the ring box protein 1 (Rbx1), an ubiquitin E3 ligase and essential partner for most of the proteins of the Cullin family (Petroski and Deshaies, 2005) and subunits of the heptameric COPI complex. Two subunits of the COPI complex were present in the druggable genome library of which COPB1 was missed. However, the depletion of all five subunits in the confirmatory screen reduced Salmonella invasion. Coatomer I is implicated in retrograde transport of vesicles cycling between the Golgi apparatus and the endoplasmic reticulum (Lee et al, 2002; Beck et al, 2009) and in anterograde transport of some proteins (Pepperkok et al, 1993; Orci et al, 1997). Two integrins, Itgβ5 (Itgb5) and ItgαV (Itgav; Shimaoka and Springer, 2003), were identified as strong invasion-stimulating hits (Figure 2C). To the best of our knowledge, the proteasome complex has not been implicated in Salmonella invasion before. All seven α-subunits and five of seven β-subunits were present in the genome-scale library, of which two α- and two β-subunits were identified as hits, all of which could be confirmed. The detailed composition of the mediator complex implicated in protein splicing in various cell types is still controversial (Conaway et al, 2005) but seems to include more than 20 proteins, of which seven were present in the initial library. Only Med4 was identified as a hit. It remains unclear whether the remaining subunits were missed due to experimental noise or whether an effect (if any) on Salmonella invasion is restricted to Med4. Taken together, host factors important for SopE-mediated S. Tm invasion comprise a surprising variety of cellular components and are not limited to well-established actin-regulating proteins.
Follow-up screen of candidate hits affecting host cell binding
The modified gentamycin protection assay used in the screen measures the presence of S. Tm in a mature SCV and could thus identify genes affecting any step of the invasion process. In order to assign the hits to particular steps and enable identification of functional links between the novel hits, we developed step-specific secondary assays addressing S. Tm binding, effector injection, cellular ruffling and membrane closure.
Binding to the host cell is the first step of S. Typhimurium invasion. Under the conditions used in the screen, binding is mediated mainly by the T1 system itself (Lara-Tejero and Galan, 2009; Misselwitz et al, 2011). In order to uncouple binding from the subsequent steps of the invasion process, we utilized the isogenic non-invasive strain S. TmΔ4 (Supplementary Table SI). This strain encodes a fully functional T1 system, attaches via T1 to host cells and can efficiently insert the T1 translocon conduit into the host cell membrane. However, it lacks the four key effector proteins SipA, SopE, SopE2 and SopB and is thus incapable of triggering the subsequent steps of the invasion process, i.e., membrane ruffling and invasion. To synchronize the binding process for all cells of a well, we chose a short incubation time (6 min) and an m.o.i. of 82. The unbound bacteria were washed off, the cells were fixed, and we stained the nuclei with DAPI and the bound bacteria with an anti-LPS antibody (Materials and methods). Automated microscopy and automated image analysis were adapted to enumerate cells carrying bound bacteria on the surface. Strikingly, mitotic cells always carried much higher numbers of bound bacteria than non-mitotic cells (Figure 3A and B, top panel). Cells neighboring mitotic cells often displayed this phenotype as well. The reason for this increased binding phenotype is not understood. Nevertheless, to increase the sensitivity of our assay, mitotic cells and their direct neighbors were excluded from the analysis (Misselwitz et al, 2010).
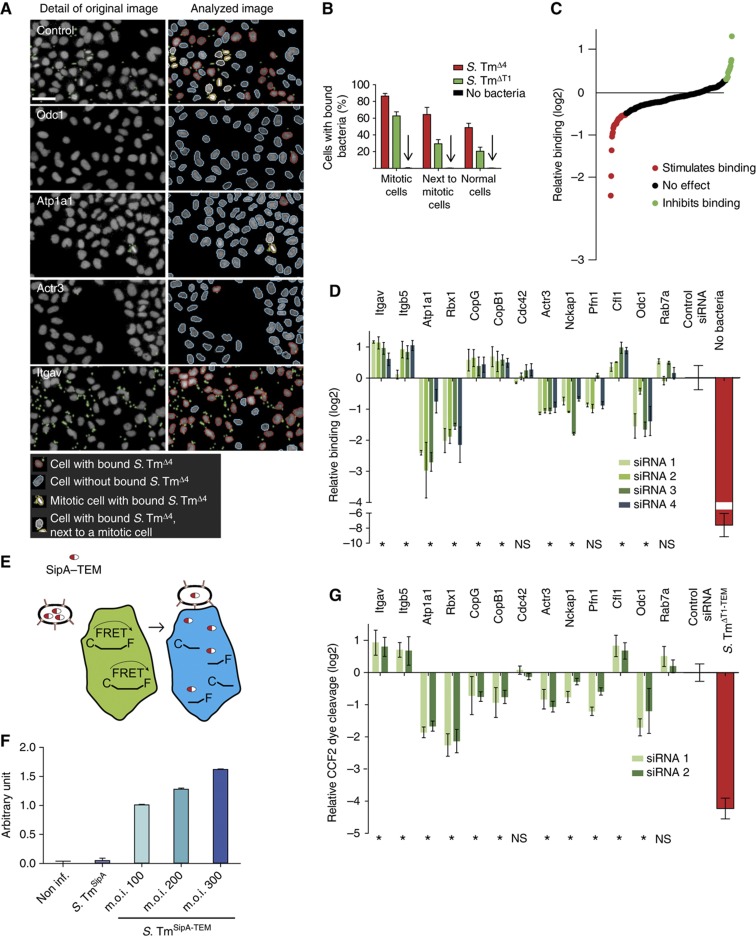
Figure 3.
Host cell factors affect Salmonella binding and effector injection. (A) Establishment of the binding assay for indicated siRNA-transfected HeLa cells (left) and corresponding image analysis (right). To analyze binding independent of cell cycle state, mitotic cells and their neighbors (showing an increased binding phenotype; indicated by yellow/white border) were excluded from the analysis using Enhanced CellClassifier. The remaining nuclei either had bound bacteria or not (nuclei with red/blue border; gray=nuclei, green=S. TmΔ4, scale bar=50 μm). (B) Differential quantification of S. TmΔ4 and S. TmΔT1 binding for mitotic cells, neighbors of mitotic cells and other cells. Each bar shows the median and standard deviation from 72 wells of two independent experiments. (C) Binding efficiency of S. TmΔ4 onto HeLa cells transfected with the confirmatory siRNA library. Data are displayed as log2 relative binding corresponding to the percentage of cells with bound bacteria of siRNA-treated cells and control siRNA-treated cells. (D) Binding efficiency of S. TmΔ4 for selected genes. The depletion of Atp1A1 and Rbx1 strongly inhibits binding of S. TmΔ4, whereas the depletion of Itgav and Itgb5 stimulates adherence to HeLa cells. (E) Scheme of the effector injection assay. HeLa cells were loaded with CCF2-AM (green) and infected with Salmonella carrying a fusion protein of SipA with β-lactamase (SipA–TEM). Translocated β-lactamase mediates cleavage of CCF2-AM inducing a shift from green to blue fluorescence. Effector translocation is defined as the ratio between blue (460 nm) and green (435 nm) fluorescence. (F) Validation of the effector injection assay: HeLa cells were infected with S. TmSipA or S. TmSipA-TEM, an increase of effector injection signal (arbitrary units) was observed with increasing m.o.i. of the bacteria. (G) Effector injection analysis of selected genes. HeLa cells transfected with siRNAs directed against copG and copB1 and infected with S. TmSipA-TEM showed a twofold reduction in effector injection efficiency. The ratio between blue and green fluorescence was normalized to control siRNAs and is displayed as log2 of the relative translocation. Two siRNAs per gene have been analyzed (*P<0.1, NS=not significant).
HeLa cells were seeded in 96-well dishes and transfected with the siRNA library for the 298 candidate hits (four siRNAs per gene), including siRNAs for quality control, i.e., Eg5, Plk1 (transfection controls), ArpC3, Cdc42, Cfl1 (strong effects on infection) and the 12 siRNAs without detectable effect for normalization. Binding was normalized to the control siRNAs and a binding hit was defined as a gene displaying a log2 of the median of four siRNAs ⩽−0.5 or ⩾0.3. By these criteria, 15 hits displayed increased binding and 38 hits displayed decreased binding (Figure 3C; Supplementary Table SIII). Several examples are shown in Figure 3D. By comparing the binding phenotypes with the invasion hits, we made two general observations:
Numerous invasion hits were also identified as binding hits (compare Figures 2C and 3D; Supplementary Table SII). These included Atp1a1 and Rbx1, as well as numerous actin regulators (Actr3, Pfn1, Nckap1) and several integrins (Itgb5, Itgav). In these cases, the invasion phenotype could for some genes partially, for others completely be assigned to the binding step. These binding hits might affect host cellular membrane stiffness, surface charges/hydrophobicity or the binding site/receptor availability on the host cell surface.
A significant number of invasion hits including Cdc42 and Rab7a, a small GTPase involved in vesicular trafficking did not show reduced binding; for components of the COPI complex (CopB1, CopG), binding was even increased. These genes must affect later steps of the invasion process.
The effector injection assay reveals a specific role of the COPI complex
Upon binding, S. Typhimurium must inject T1 effector proteins into the host cell (Figure 1A). Before host cell contact, the T1 system is preassembled, but inactive. Upon binding, a special conduit termed ‘translocon’ is inserted into the host cell membrane, thus activating the T1 system and initiating effector protein injection into the host cell. Host signals activating the T1 system are not well understood. Some hits identified in the invasion screen might affect this step of the infection process. To measure effector injection, we used an assay based on a fusion protein between β-lactamase and the T1 effector SipA (Figure 3E) (Charpentier and Oswald, 2004; Schlumberger et al, 2007). After injection into cells, the β-lactamase part of the fusion protein is able to cleave the fluorescent dye CCF2 present within the cell, thereby changing its fluorescence properties. As cellular ruffles and invasion might change the efficiency of effector translocation, we chose the strain S. TmSipA for this assay (Supplementary Table SI). This strain carried a fusion protein of SipA and β-lactamase (S. TmSipA–TEM), but lacks SopE, SopE2 and SopB, does not trigger ruffling and invades only very slowly and less efficiently into HeLa cells (Schlumberger et al, 2005 and data not shown). Infection of CCF2-loaded HeLa cells with S. TmSipA–TEM resulted in a pronounced change in fluorescence (Figure 3F). In contrast, S. TmSipA lacking β-lactamase did not induce any change in fluorescence, confirming the specificity of this assay.
Using this effector injection assay, we focused on a 90-gene subset of the 298 candidate hits and analyzed only the two siRNAs per gene, which had yielded the strongest signals in the invasion assay (Figure 2C). This subset of genes included 72 confirmed hits of the invasion assay as well as genes with effects close to our arbitrary threshold, which together could be conveniently tested on three 96-well plates. HeLa cells were seeded and transfected with siRNA as described for the confirmatory screen (compare Figure 2). The fluorescence signal of the injection assay was normalized to the 12 control siRNAs. Several examples of the injection screen are shown in Figure 3G (compare Supplementary Table SIII). A first comparison with the data of the binding screen (Figure 3A–D) allowed three global conclusions:
Depletion of subunits of the COPI complex strongly impaired effector translocation. In contrast, the depleted cells showed increased binding (Figure 3D). Thus, lack of the COPI complex seems to specifically inhibit effector translocation (Figure 3G).
All remaining binding hits also displayed injection phenotypes. Depletion of some proteins (e.g., Itgav, Itgb5, Cfl1) enhanced binding and injection, whereas depletion of others (e.g., Atp1a1, Rbx1, Actr3, Nckap1, Odc1) reduced both phenotypes. In these cases, the altered binding efficiency was sufficient to explain the altered injection efficiency.
A number of invasion hits showed neither reduced injection nor binding. These included the Rho GTPase Cdc42 and Rab7a. These genes must affect later steps of the invasion process.
The ruffling assay yielded a specific phenotype for Cdc42
Upon binding and effector protein injection, HeLa cells respond within minutes by pronounced actin rearrangements and by forming membrane ruffles. SopE is a key effector protein triggering actin rearrangements and membrane ruffles. To screen for ruffling phenotypes, HeLa cells were infected for 6 min at an m.o.i. of 250, fixed and nuclear DNA and the actin cytoskeleton were stained. By fluorescence microscopy, we observed that S. TmSopE triggered pronounced ruffles, whereas the isogenic control strain S. TmΔ4 lacking SopE, SopE2, SopB and SipA did not change the appearance of the cells (Figure 4A, top left panels).
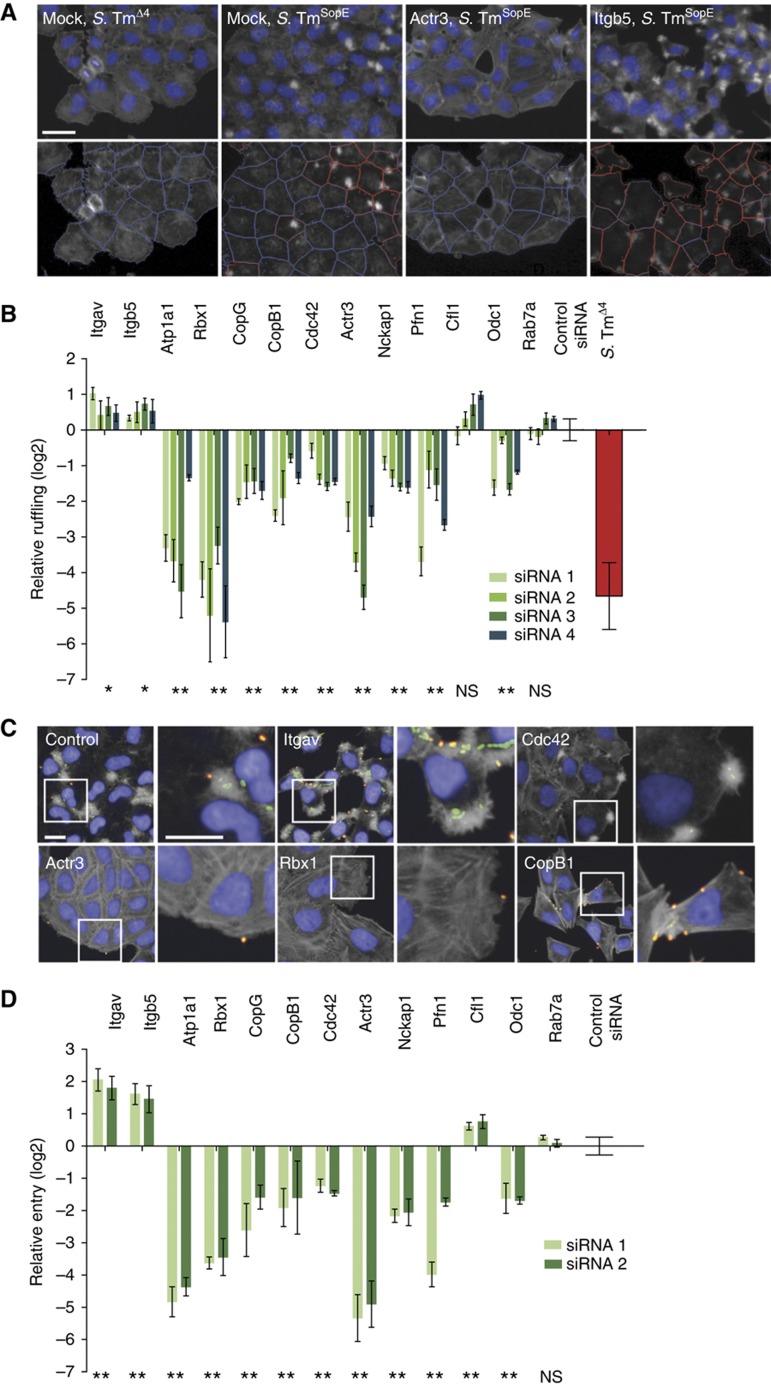
Figure 4.
Depletion of Atp1a1 and Rbx1 results in strong inhibitory effects on ruffling and membrane closure. (A) Functionality of the image analysis of ruffling cells. Upper panels: details of the original automated microscopy images (blue=nuclei, gray=actin). Lower panels: results of the automated image analysis (red outlines=ruffling cells, blue outlines=non-ruffling cells; scale bar=20 μm). (B) Examples of the hits with a positive or a negative effect on ruffling induced upon S. TmSopE invasion. Multiple test correction for 298 genes was performed to obtain the P-value (*P-value <0.1, **P-value <0.05, NS=not significant). (C) Fluorescence images showing cells treated either with control siRNAs or siRNAs directed against the indicated genes and infected with S. TmSopE (pM965) (red/green double stain=extracellular bacteria; blue=nuclei; gray=actin; green=intracellular bacteria; scale bar=20 μm). (D) Relative invasion of S. TmSopE into cells transfected with siRNAs directed against the indicated genes. Multiple test correction for 60 genes was performed on the results (**P-value <0.05; NS=not significant).
To analyze the ruffling efficiency in a quantitative manner, an automated microscopy-based assay for the identification of ruffling cells was developed. Starting from images acquired by automated microscopy, nuclei and cells were identified using the image analysis software CellProfiler (Carpenter et al, 2006). With the recently released machine learning tool, Enhanced CellClassifier (Misselwitz et al, 2010), sets of ruffling and non-ruffling cells were generated to train a support vector machine (SVM) model algorithm (Cortes and Vapnik, 1995) for automatic recognition of ruffling and non-ruffling cells. Representative results of the automatic classification are shown in the lower panels of Figure 4A.
To screen for genes affecting the ruffling step, HeLa cells were seeded in 96-well plates and transfected with the siRNA library for the 298 candidate hits, or control siRNAs as described above and were infected with S. TmSopE. Ruffling was normalized using the control siRNAs and a ruffling hit was defined as a gene displaying a log2 of relative ruffling ⩽−0.5 or ⩾0.3. Using this cutoff, 29 genes displayed increased ruffling and 52 genes displayed decreased ruffling (Supplementary Table SIII). Several examples are shown in Figure 4B. We made the following observations:
Some injection hits showed equivalent phenotypes in the ruffling assay, e.g., reduced (e.g., Odc1) or enhanced ruffling (e.g., Itgb5, Itgav, Cfl1; compare Figures 3G and 4B).
For other hits (including CopB, CopG, Atp1a1, Actr3, Nckap1, Pfn1), the phenotype was even stronger in the ruffling assay than in the injection assay, suggesting an additional role for these genes at this step.
A few ruffling hits (including Cdc42) had not shown a significant effect in binding or injection assays (compare Figures 3D, G and 4B). Therefore, these hits specifically affected the ruffling step.
Other invasion hits such as Rab7a did not show a significant phenotype in the ruffling assay, suggesting a role in later steps of the invasion process.
An assay for hits affecting membrane closure
Finally, the actual entry step of S. Typhimurium into the host cell was investigated. As the modified gentamycin protection assay (Figures 1 and 2) measures S. Tm entry only after SCV maturation, an assay was developed which measures the membrane fusion event completing pathogen entry into the endocytic vacuole independently of subsequent events in the eukaryotic cell. An automated quantification of intracellular bacteria was not possible using the images acquired with the available automated epifluorescence microscope (resolution: × 20) due to the small size and spatial arrangements of cell-associated bacteria. We therefore opted for a manual quantification focusing on the 60 genes displaying the most pronounced invasion phenotypes (two siRNAs per gene). HeLa cells were infected with S. TmSopE (pM965), which constitutively express GFP. After 20 min, the cells were fixed and stained with DAPI (nuclei), TRITC-phalloidin (actin) and a S. Typhimurium LPS-specific antibody. Membrane closure was assessed by fluorescence microscopy, using differential antibody staining to distinguish extracellular bacteria (GFP+, LPS+) and internalized bacteria (GFP+, LPS−, Figure 4C).
Overall, the results obtained with this assay were similar to those obtained in the ruffling assay (compare Figure 4D and B; Supplementary Table SIII). This indicated that none of the tested genes were essential for membrane fusion or formation of the early endocytic vacuole. Furthermore, the vast majority of hits identified in our original invasion screen could be assigned to early steps of the infection process, i.e., binding, effector injection or ruffling, and did not seem to affect later maturation steps of the SCV. A notable exception was Rab7a. Rab7a depletion did not affect the membrane closure assay but had a significant phenotype in the original invasion assay. This is in line with its established role in maturation of the SCV (Meresse et al, 1999). However, besides Rab7a we were surprised to find only a few hits partially reducing SCV maturation, including the trafficking protein Vps39 and a subunit of the vacuolar ATPase (Atp6ap2, Supplementary Table SIII).
Step-by-step comparison identifies hits affecting multiple steps of the invasion process
With the large data sets available, we were able to follow each hit through the individual steps starting with binding, followed by effector injection, ruffling, membrane closure and SCV maturation, the readout of the modified gentamycin protection assay (Figure 5A). Depletion of ornithin decarboxylase 1 (Odc1; http://herkules.oulu.fi/isbn9514266315/) inhibited only the binding step. The same negative effect could be observed in all subsequent steps of the invasion process. Odc1 is therefore a bona fide binding hit. Similarly, Cdc42 meets the definition of a ruffling hit, as Cdc42 depletion did not affect binding or effector injection, but yielded similar levels of attenuation in the ruffling assay, the membrane closure assay and in SCV maturation. Rab7a represents a bona fide SCV maturation hit as the depletion influenced only the last step of the invasion process.
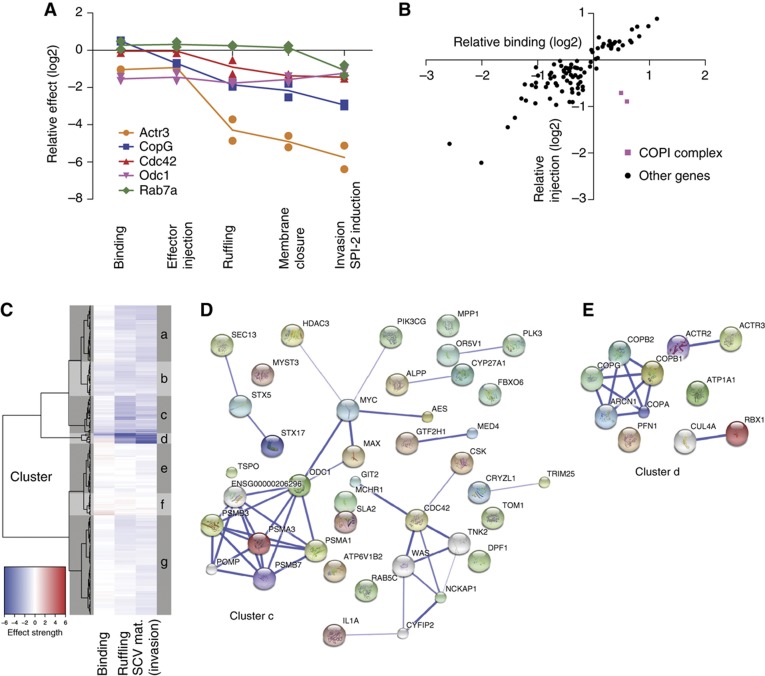
Figure 5.
Classification of hits according to their profile in the different assays and cluster analysis of the results of the whole screen. (A) Overview showing the results of the different assays describing Salmonella invasion steps for selected genes. While Odc1 shows a pure binding phenotype, Cdc42 and Rab7a are examples for pure ruffling or maturation phenotypes. Several proteins show combined phenotypes: Actr3 depletion has effects on binding and ruffling, whereas depletion of CopB1 and CopG have inhibitory effects on effector translocation and ruffling. For consistency only the results of the two strongest siRNAs are shown. (B) Scatter plot of relative binding versus relative effector injection. A strong correlation between binding and effector injection is observed for most of the hits. The outliers CopB1 and CopG are stained in purple. Each point indicates the median of the two strongest siRNAs. The results for 90 genes are shown. (C) Heatmap and clustering dendrogram based on the 300 ‘candidate hits’ obtained in the initial screen. (D, E) Functional interactions within the clusters were annotated using the STRING database (confidence cutoff=0.4, additional white nodes: five in cluster c and one in cluster d).
Importantly, many genes affected multiple steps, for instance binding and ruffling in the absence of subunits of the Arp2/3 (Actr3). Similarly, depletion of members of the COPI complex had a weak enhancing effect on binding and a specific negative effect on effector injection, and an additional negative effect on ruffling. Most likely, these effects were specific for COPI depletion, as all five subunits of the complex showed equivalent results (four oligos per gene; Supplementary Figure S2). The specific effect on injection is clearly illustrated by plotting the relative effect on binding efficiency against the relative effect on injection efficiency for each analyzed hit (Figure 5B). Thus, the COPI complex is an effector injection and a ruffling hit (Figure 5B; pink squares). Other examples of genes affecting more than one specific step of invasion include Pfn1, Actr3, Atp1a1 and Rbx1 (see Supplementary Table SIII; Supplementary Figure SIII).
Phenotypic clustering of the hits
To systematically analyze functional links between the 298 genes analyzed in detail (Supplementary Table SIII), we also used an automated clustering algorithm. We reasoned that (i) hits affecting the same cellular process should yield similar patterns of phenotypes in the step-specific assays. (ii) Hits affecting the same cellular process may interact either directly or indirectly. This could reveal cellular processes and molecular interactions, which may not have been noticed so far.
Cluster analysis of the normalized step-specific phenotypes yielded seven clusters (Figure 5C; Supplementary Table SV). Within each cluster, we automatically annotated the known functional interactions between the proteins/genes using the STRING 8.3 data base (Jensen et al, 2009).
The clusters c (Figure 5D) and d (Figure 5E) significantly enriched for known interactions, supporting the validity of our approach (P<0.003 versus random clusters of equal size). Cluster c, which harbors many binding and ruffling hits, included known regulators of membrane ruffling (Cdc42, NckAP1) as well as the ARF GTPase-activating factor Git2 (G-protein-coupled receptor kinase interactor 2; Cat-2), the proteasome, transcription factors (Myc, Max, Gtf2H1, Med4), several trafficking regulators (Sec13, Stx5, Stx17, Rab5c), proteins involved in cholesterol metabolism (Cyp27a1, TspO) and numerous genes not previously implicated in membrane ruffling (e.g., Tom1, CryZl1, Trim25). It will be of interest to determine how these factors may interact and whether they affect membrane ruffling directly or indirectly.
Cluster d harbors hits strongly affecting ruffling and vesicle maturation, including well-established actin regulators (ActR2, ActR3, Pfn1), as well as the NaK-ATPase (Atp1a1), Rbx1 and the COPI complex, which have recently been linked to regulation of the actin cytoskeleton (Valderrama et al, 2000; Wu et al, 2000; Barwe et al, 2005; Chen et al, 2009). However, their functional importance and possible interactions facilitating S. Tm invasion remain to be established. In conclusion, the cluster analysis revealed important information on established functional interactions (Figure 5D and E; lines connecting different hits) and possible novel interactions (no lines), thus providing a rich resource for future research on the host cell signaling modules driving each step of the invasion process.
The COPI complex affects the localization of cholesterol, gangliosid GM1 and the Rho GTPases Rac1 and Cdc42
Among our hits the COPI complex was unique by affecting effector injection and ruffling (Figure 5A). We therefore investigated the effects of the COPI complex depletion in detail. COPI is a central regulator of vesicular transport (Pucadyil and Schmid, 2009), suggesting that the depletion of the COPI complex might be explained by altered distribution of proteins important for S. Typhimurium invasion. Prime candidates for such proteins are the two Rho GTPases Cdc42 and Rac1 reported to have a role in SopE-mediated ruffling (Hardt et al, 1998; Friebel et al, 2001). Moreover, direct binding of Cdc42 to the γ subunit of the COPI complex has been observed (Wu et al, 2000). Redistribution of either Rho GTPase would explain the ruffling phenotype of COPI.
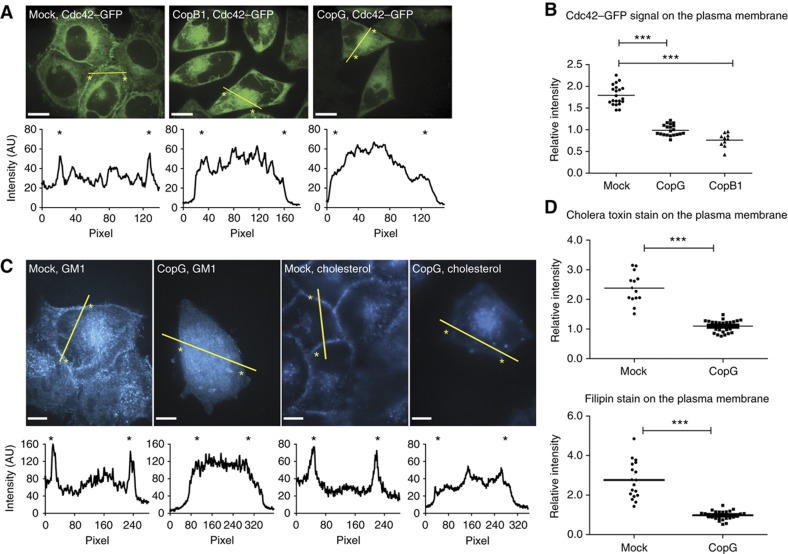
We therefore generated stable cell lines expressing Cdc42–GFP and Rac1–GFP, respectively. As shown in Figure 6A and Supplementary Figure S4, both Rho GTPases localized to the plasma membrane, to vesicles next to the nucleus and to smaller degrees to the cytoplasm. Quantification showed an enrichment of the GFP signal at the plasma membrane. Plasma membrane localization was even more pronounced for Rac1–GFP than for Cdc42–GFP (Figure 6A and B and Supplementary Figure S4A). After depletion of CopG or CopB1, both Rho GTPases were no longer found enriched at the plasma membrane, but accumulated instead in an intracellular compartment (Figure 6A and B, Supplementary Figures S4 and S7). The latter is probably equivalent to the prominent vesicular compartment accumulating markers for the trans-Golgi network (TGN), the endoplasmatic reticulum–Golgi intermediate compartment (ERGIC), the Golgi apparatus and recycling endosomes, but not for early or late endosomes or lysosomes, which had been identified recently in β-Cop (CopB1)-depleted cells (Styers et al, 2008). Depletion of several other proteins including Rbx1, Atp1A1, Actr3 did not change the localization of the Rho GTPases (data not shown), making this Rho GTPase relocalization phenotype unique among the ‘ruffling’ hits.
Figure 6.
Cdc42, sphingolipid GM1 and cholesterol are mislocalized after depletion of the COPI complex. (A) Confocal images showing the localization of Cdc42–GFP in water-transfected cells (left), cells transfected with siRNA directed against copB1 (middle) or copG (right). The intensity plots along the lines indicated in yellow are shown below the pictures; the position of the membrane is indicated by an asterisk; scale bar=10 μm. (B) Quantification of the Cdc42–GFP signal on the membrane relative to the cytosol (***P<0.005). (C) Confocal images showing the distribution of sphingolipid GM1 (left two panels) and cholesterol (right two panels) in the cells. For each staining, water-transfected cells are shown on the left side and cells transfected with siRNA against copG on the right side; scale bar=10 μm. (D) Quantification of the relative cholera toxin β/filipin staining signal at the membrane compared with the cytosol (***P<0.005).
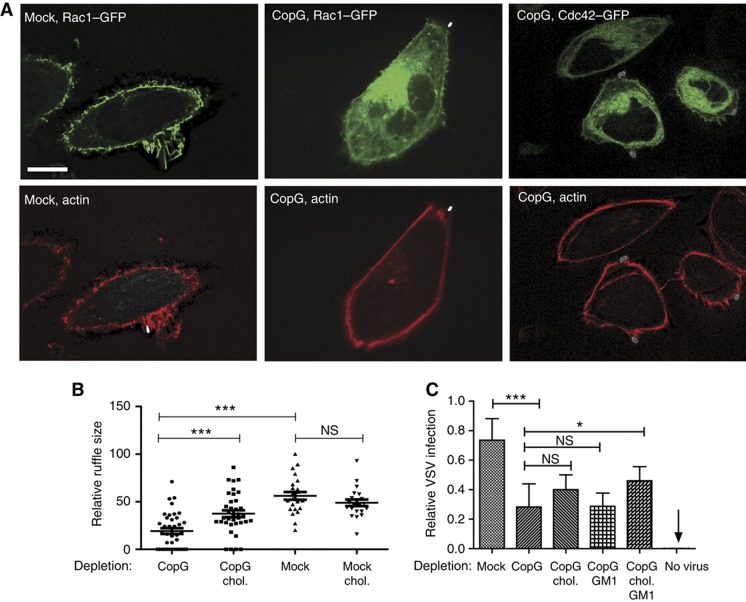
Next, it was tested whether altered lipid composition of the plasma membrane might explain Rho GTPase relocation upon COPI depletion. Cholesterol-enriched membrane domains are known to bind activated Rho GTPases via their geranylgeranyl anchors (Palazzo et al, 2004; del Pozo et al, 2004) and COPI vesicles are implicated in membrane sorting of sphingolipids and cholesterol. Membrane components such as the sphingolipid GM1 and cholesterol can be visualized by staining with the β-subunit of cholera toxin or filipin, respectively. Under normal conditions, both markers mainly localized to the plasma membrane and to some intracellular vesicles. In contrast, after depletion of CopG, the cell membrane was no longer stained by either of the dyes, but the signal accumulated in a large intracellular compartment, colocalizing with Cdc42 and Rac1 (Figure 6C and D, Supplementary Figures S5 and S6). These results suggest that cholesterol and GM1 are retained in an atypical compartment formed after depletion of different subunits of the COPI complex. This would lead to a mislocalization of Rac1 and Cdc42, effectively depleting both proteins from the cell membrane. In agreement with these data, the size of the ruffles formed at the site of Salmonella binding was significantly decreased after depletion of CopG (Figure 7A).
Figure 7.
CopG depletion abolishes actin recruitment to the site of Salmonella binding and ruffling. (A) Confocal images showing the distribution of actin and Rho GTPases upon S. TmSopE invasion for 10 min in stable Rac1- or Cdc42-GFP-expressing cell lines. The cells were transfected either with water (mock) or with siRNA directed against copG; scale bar=10 μm. (B) Size of membrane ruffles after infection with S. TmSopE for 10 min of CopG-depleted cells with or without cholesterol replenishing. The ruffle size was measured in the focal plane showing the largest diameter of the ruffle. (C) Infection of cells either transfected with water (mock) or with siRNA directed against copG by VSV for 5 h. The relative invasion was quantified using a virus construct, which leads to a green fluorescent staining of the cytoplasm upon invasion. Quantification of the amount of infected cells in untransfected cells (mock), or cells depleted of CopG, which were either complemented with cholesterol, with GM1 or with both prior to infection are shown (***P<0.005; *P<0.05; NS=not significant).
To demonstrate that the injection and/or the ruffling defects were attributable to altered membrane lipid composition, we replenished cholesterol in COPI-depleted cells by adding cholesterol complexed to methyl-β-cyclodextrin (MβCD) 5 h prior to the infection of the cells. We infected the cells and non-complemented controls for 10 min with S. TmSopE and measured the diameter of the ruffles formed by a single bacterium. Indeed, replenishing led to a significant increase in the size of the ruffle compared with the CopG-depleted cells not replenished with cholesterol (Figure 7B).
The central role of COPI in cholesterol and Rho GTPase localization was also confirmed by inhibitor experiments. Thereby, pretreatment of cells with either geranylgeranyltransferase inhibitor (GGTI, inhibits geranygeranylation of the Rho GTPases thus dislocalizing them from the plasma membrane) or MβCD (depletes cholesterol from the plasma membrane) decreased S. Typhimurium invasion (data not shown). Importantly, both inhibitors could not further diminish S. Tm invasion in CopG-depleted cells, suggesting that both cholesterol and Rho GTPases were already missing at the plasma membrane due to COPI inactivation. This confirmed that the ruffling defect of CopG-depleted cells was indeed attributable to the mislocalization of the Rho GTPases and cholesterol. Further work would be required to determine the exact molecular mechanism explaining this defect, e.g., defective Rho GTPase membrane localization in resting cells or reduced Rho GTPase recruitment after pathogen binding. Taken together, our results suggest a novel role for the COPI complex in the localization of cholesterol and Rho GTPases at the plasma membrane.
COPI-dependent membrane lipid composition also affects vesicular stomatitis virus infection
Strikingly, a large number of bacterial and viral pathogens share two phenotypes observed for SopE-mediated host cell invasion, i.e., the requirement for cholesterol-enriched microdomains in the host cell membrane and reduced infection efficiency upon COPI depletion. This includes Staphylococcus aureus (Ramet et al, 2002; Potrich et al, 2009), Escherichia coli (Ramet et al, 2002; Philips et al, 2005; Riff et al, 2005), Listeria monocytogenes (Seveau et al, 2004; Agaisse et al, 2005; Cheng et al, 2005; Gekara et al, 2005), Mycobacterium fortuitum (Philips et al, 2005), Mycobacterium tuberculosis (Munoz et al, 2009), Chlamydia caviae (Derre et al, 2007), Chlamydia trachomatis (Jutras et al, 2003; Elwell et al, 2008), influenza virus (Hao et al, 2008; Konig et al, 2010) and hepatitis c virus (Tai et al, 2009; Popescu and Dubuisson, 2010). However, a functional link between both phenotypes had never been established.
Our results suggest that the dependence on both COPI and cholesterol membrane microdomains might be functionally linked. To test this hypothesis, we analyzed the effect of COPI depletion and of replenishing cholesterol and sphingolipids such as GM1 on host cell infection by the vesicular stomatitis virus (VSV). VSV is an enveloped, single-stranded, negative-sense RNA virus of the family Rhabdoviridae. It infects a wide range of host cells and the existence of a specific receptor is still not entirely clear (Schlegel et al, 1983; Coil and Miller, 2004). Host cell entry occurs via clathrin-dependent endocytosis, and low-pH-mediated alterations in the virus glycoprotein lead to membrane fusion (Sun et al, 2005).
Indeed, CopG depletion significantly interfered with VSV host cell invasion (Figure 7C). Moreover, the infection efficiency could partially be restored by replenishing GM1; this effect was even more pronounced after replenishing both, GM1 and cholesterol. In contrast, replenishing just cholesterol without GM1 was insufficient for rescuing VSV invasion into CopG-depleted cells. Thus, some differences exist between the host cell membrane lipid requirements for VSV and SopE-dependent S. Typhimurium infection. Nevertheless, these data indicate that COPI-dependent control of the membrane lipid composition is of importance for vastly different pathogens.
Discussion
The genome-scale siRNA screen identified host cell factors mediating SopE-dependent S. Typhimurium invasion into epithelial cells. Step-specific follow-up assays detected at least one important host factor for each step, except for membrane closure. This step-specific analysis allowed the functional classification of the respective host cell factors, confirming well-established ones, revealing new ones and suggesting novel functional links between them.
Many hits identified in our screen affected the binding step. This was unexpected and suggested that binding was rate-limiting under our experimental conditions. Among the binding hits, we found numerous known regulators of actin polymerization, including Pfn1, Cofilin 1 or the Arp2/3 complex, as well as other host cell proteins implicated in cell shape or actin regulation (e.g., Itgb5, Itgav; Supplementary Tables SIII and SIV). The actin cytoskeleton determines cellular shape, membrane stiffness and the presentation of surface proteins, three parameters likely to affect pathogen binding. Therefore, genes identified as binding hits in the screen might affect these three cellular characteristics. It will be of interest to determine the underlying molecular mechanisms.
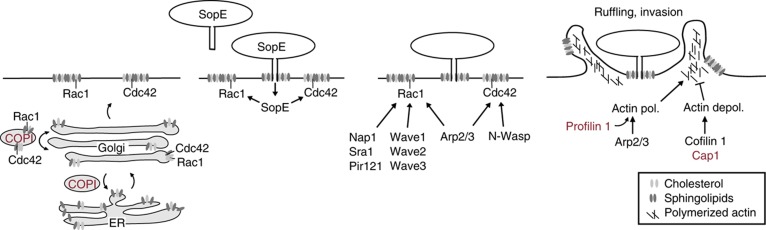
The ruffling step was affected by numerous hits and allowed us to refine the model of this key step of host cell invasion (Figure 8). In the host cell, SopE activates Cdc42 and Rac1. The presented data suggest that the Rho GTPases must be localized at the plasma membrane in order to trigger ruffling. Together with Nap1, Sra1 and Pir121 (reviewed in Schlumberger and Hardt, 2006) they lead to activation of N-Wasp or WAVE complex and subsequently to Arp2/3 complex-mediated actin polymerization. Our data suggest that this is further enhanced by Pfn1, a well-known factor delivering actin monomers to sites of actin polymerization, but not previously implicated in Salmonella infection (Pollard and Cooper, 2009). The actin polymerization step is negatively regulated by actin-depolymerizing proteins such as cofilin 1 (Dai et al, 2004; McGhie et al, 2004). Furthermore, it has been observed that Cap1 is an additional factor stimulating the breakdown of actin fibers, thus negatively regulating Salmonella invasion. This supports that the induction of membrane ruffling is the result of a complex cascade of actin polymerizing and depolymerizing events.
Figure 8.
Model for S. Typhimurium entry into HeLa cells. In order to invade, S. Typhimurium binds onto cells and injects a cocktail of effectors. This study investigates the action of the key effector SopE. SopE mediates the GTP nucleotide exchange and thereby the activation of the Rho GTPases Rac1 and Cdc42. This leads to the activation of the Arp2/3 complex, actin polymerization cellular ruffling and bacterial entry. The screen identified key regulators of this process, including Nap1, Cdc42, Profilin 1 and the Arp2/3 complex, as well as proteins mediating actin depolymerization, a process probably counteracting the activity of Rac1 and Cdc42. The correct localization seems to be as important as the activation of the Rho GTPases. Upon depletion of the COPI complex, cholesterol and Rho GTPases are mislocalized away from the cell membrane, leading to a much less efficient S. Typhimurium entry. Proteins newly identified to be implicated in S. Typhimurium entry are indicated in red. Proteins that have already been described to have a role in the infection and were found in the screen are indicated in black.
In most cases, the ruffling defect led to a corresponding defect in host cell invasion. However, in a few cases (e.g., proteasome complex, Rbx1), the ruffling defect was much more pronounced than the invasion defect. This might be explained by the existence of two independent modes of host cell invasion by S. Typhimurium. Indeed, a ruffling-independent mode of S. Typhimurium invasion has recently been described (Hanisch et al, 2010). The former group of ruffling hits might affect both modes of host cell invasion. In contrast, the proteasome and Rbx1 might have specifically affected the ruffling-mediated invasion, leaving the ruffling-independent invasion process untouched. This will be an interesting topic for future studies.
The COPI complex also affected membrane ruffling which was at least partially attributable to the reduced cholesterol/sphingolipid content of the plasma membrane upon CopB or CopG depletion. Depletion of COPI components resulted in cholesterol and GM1 relocalization to a large intracellular structure, probably representing the ‘common compartment’ observed in a recent study upon β-Cop-depletion harboring TGN, ERGIC, Golgi and recycling endosome membrane protein markers (Styers et al, 2008). To the best of our knowledge, the drastic effect of COPI depletion on cholesterol trafficking has not been reported previously. Nevertheless, the COPI complex has been implicated in other trafficking events. Its role for the retrograde transport from the Golgi to the endoplasmic reticulum is best understood (Bethune et al, 2006; Beck et al, 2009). However, the reason for cholesterol relocalization in CopG-depleted cells is still unclear. On the one hand, cholesterol is synthesized in the smooth endoplasmic reticulum and has a complex trafficking route, involving vesicular and non-vesicular trafficking steps (Ikonen, 2008). A deficient transport/sorting from the Golgi apparatus to other compartments might thus explain the lack of cholesterol and sphingolipids on the cell membrane and its accumulation on the common compartment. On the other hand, cholesterol is endocytosed at the cell membrane and traffics to endosomes. Subsequently, certain lipids must leave the path to the lysosome, as the cholesterol content of lysosomal membranes is low. By trapping the recycling endosomes in the common compartment, CopG depletion might interfere with cholesterol resorting to the plasma membrane. Defining the underlying mechanism of cholesterol trafficking will be an important task for future work.
Upon CopG or CopB depletion, the Rho GTPases Cdc42 and Rac1 were mislocalized in a similar manner as cholesterol and GM1. These Rho GTPases are geranylgeranylated and this modification localizes them to cholesterol-enriched lipid microdomains (del Pozo et al, 2004; Palazzo et al, 2004). In line with this hypothesis, disrupting geranylgeranylation of Rho GTPases with a chemical inhibitor released Rac1–GFP and Cdc42–GFP from the plasma membrane and prevented relocalization to the ‘common compartment’ in COPI-depleted cells (data not shown). On the basis of these observations, it is tempting to speculate that cholesterol/sphingolipid relocalization might result in the mislocalization of additional proteins, i.e., of cholesterol/sphingolipid binding membrane proteins such as Rho GTPases in COPI-depleted cells. Possibly, the cholesterol/sphingolipid relocalization phenotype might help to decipher proteins transporting these membrane lipids from proteins that are transported passively by them.
The reduced effector protein injection into COPI-depleted cells might be directly attributable to reduced cholesterol levels in the plasma membrane. The S. Typhimurium protein SipB, located at the tip of the translocon, is known to bind cholesterol with high affinity in vitro (Hayward et al, 2005), suggesting that cholesterol may directly activate bacterial effector injection by T1. In line with this hypothesis, artificial liposomes containing cholesterol and sphingolipids were shown to activate the Type III secretion system of Shigella flexneri, a closely related enteropathogenic bacterium (van der Goot et al, 2004). In addition, the Shigella effector IpaB, a homolog of SipB, could bind CD44 in cholesterol-enriched microdomains (Lafont et al, 2002), pointing to a general role of host cell membrane lipids in activating Type III secretion systems. Thus, the mislocalization of Rho GTPases, i.e., Cdc42, together with reduced effector injection efficiency into CopG-depleted cells provides satisfactory explanation for the defect in S. TmSopE-induced ruffling.
In this study, five assays were used to quantify effects on particular steps of S. Typhimurium invasion into HeLa cells. Four of these assays could be performed in a high-throughput format. A detailed discussion of the specificities of the assays is available upon request. Owing to the combination of gentamycin protection and intracellular GFP expression, the modified gentamycin protection assay (Figures 1 and 2) has a high specificity and a very robust performance, thus enabling genome-scale experiments. The binding or the ruffle assays are less robust, and require careful controls excluding the presence of staining artifacts, which may yield ‘false positives’ (i.e., over-estimation of the number of affected cells). The analysis of a large number of cells contributed to further enhancing the robustness of these assays. In the case of the ruffle assay, false positive might have occurred if the siRNA affected ruffle-like changes in the actin cytoskeleton, even in the absence of bacteria. However, the identification of step-specific genes and the high concordance of all other genes in the different subsequent assays (Supplementary Figure S2) argues that all assays were reasonably robust.
Contrary to our expectations (Schlumberger et al, 2006), the Rho GTPase Rac1 and N-WASP, linking Cdc42 and the Arp2/3 complex, have not been discovered by our initial genome-scale screen. Most likely, they belong to the ‘false negatives’, which are commonly encountered in genome-scale screens. When four new and partially different siRNAs directed against each gene were reordered, an inhibitory effect on S. Tm invasion upon depletion could be demonstrated (data not shown). Thus, due to an improved siRNA design and/or altered experimental conditions in later experiments, we could correct these false-negative genes (data not shown).
In conclusion, the presented genome-scale screen of SopE-mediated invasion revealed an array of hits enhancing the current model of triggered host cell invasion and a set of proteins not associated with S. Tm invasion so far. In addition, follow-up assays on S. Tm and VSV and published work implicate the COPI-mediated control of cholesterol and sphingolipid distribution to the host cell plasma membrane as a common mechanism affecting infection by bacterial and viral pathogens. Such host cellular functions required by phylogenetically diverse groups of pathogens are of great interest for understanding the evolution of infectious disease. Moreover, they are of practical interest, as they might offer starting points for developing novel anti-infective therapeutics with a very broad range of biological activity.
Materials and methods
Bacterial strains and plasmids
All S. Typhimurium strains used were isogenic derivatives of SL1344 (SB300) of S. enterica subspecies I serovar Typhimurium (Supplementary Table SI; Hoiseth and Stocker, 1981). Strains M701 (Muller et al, 2009), M566 (Ewen et al, 1997), SB161 (Kaniga et al, 1994) and M1114 (Schlumberger et al, 2007) have been described previously. M1128 was constructed by P22-phage transduction of the sipA∷sipA-M45-tem1 allele of strain 1104 into strain M713. M1104 has been described previously (Schlumberger et al, 2007) and strain M713 was constructed by p22-phage transduction of the sseD∷aphT allele from MvP101 (S. Typhimurium ATCC 14028 derivate; Medina et al, 1999) into strain M712 (Ehrbar et al, 2004). Plasmids pM965 (Stecher et al, 2004), pM975 (Hapfelmeier et al, 2005) and pEGFP-C3/Rac1WT (Hage et al, 2009) have been described previously. Plasmid pEGFP-C1/Cdc42hsWT was kindly provided by Dr Klaudia Giehl.
For the infection of HeLa cells, bacteria were grown in LB broth supplemented with 0.3 M NaCl and 50 mg/l Streptomycin (AppliChem) for 12 h at 37 °C and subcultured for 4 h.
Cell cultures
HeLa cells were grown in DMEM (PAA Laboratories) supplemented with 10% FCS (Omnilab) and 50 mg/l Streptomycin (AppliChem) at 37 °C and 5% CO2. HeLa cells were stably transfected with pEGFP-C3/Rac1WT or pEGFP-C1/Cdc42hsWT and clones were maintained in medium supplemented with 500 mg/l Neomycin (AppliChem).
siRNA transfection
For siRNAs a reverse transfection protocol was used. In 96-well plates (μ-clear bottom, Greiner Bio One), 2 μl of 1 μM siRNA was added to 8 μl Opti-MEM (Invitrogen) yielding a final siRNA concentration of 20 nM (after addition of cells). Lipofectamine 2000 (Invitrogen) was diluted 1:200 in Opti-MEM and incubated for 15 min at room temperature. A quantity of 10 μl per well were added and incubated for another 15 min at room temperature. These plates, henceforth referred to as cell plates, were either directly used or frozen at −80 °C. HeLa cells were seeded using 1800–2000 cells in 80 μl per well, followed by an incubation of 3 days at 37 °C and 5% CO2. For half-size plates (μ-clear bottom, half area, Greiner Bio One), all numbers were reduced to 60%. For the genome-scale screen, 384-well plates (μ-clear bottom, Greiner Bio One) were used. The volume of the different reagents was reduced to 50%. The final siRNA concentration was changed to 50 nM and Lipofectamine was used at a concentration of 1:100.
siRNA libraries
SiRNAs were ordered from Qiagen. For the genome-scale screen the ‘druggable genome’ library (Version 2.0) comprising 6978 genes and 3 siRNAs per gene were used. Data were normalized platewise to the median of all siRNAs on the respective plate. Experiments were performed in triplicates and the median for each siRNA calculated, either with or without platewise z-score correction. The median value for the three siRNAs per gene was defined as the final read-out for each gene. A hit was defined as 1.5 times the interquartile range from the median of the entire data set.
For a confirmatory screen and secondary assays, another library was assembled based on hits from the z-score corrected list (182 genes), with 80 additional hits from the uncorrected list as well as 38 additional genes with high biological probability according to our screen results, but not present in the druggable genome library. Altogether, this secondary library contained 298 genes and 4 siRNAs per gene. To enable comparison of our data with the large screen, six genes with the smallest deviation of all three siRNAs from the median of the whole screen were selected as controls for the secondary library. In total, 12 control siRNAs were used: Hs_RAB30_8, Hs_RAB30_7, Hs_P53AIP1_5, Hs_P53AIP1_6, Hs_DNAJC6_1, Hs_DNAJC6_5, Hs_ARNT2_1, Hs_ARNT2_3, Hs_FBXL2_1, Hs_FBXL2_2, Hs_ANGPTL3_2 and Hs_ANGPTL3_1. If not otherwise indicated, the median of these 12 siRNAs was used for platewise normalization of secondary assays. In all siRNA experiments, siRNAs against subunits of the Arp complex (Arc21, ArpC3_3, ArpC3_5), CDC42 (CDC42_7, CDC42_10) and CFL1 (CFL1_3, CFL1_5, CFL2_5) were used as additional controls of siRNA effects on S. Tm infection. In addition, siRNAs with known cytotoxic effects were used to control for siRNA-transfection efficiency (PLK1_2 and EG5).
Immunoblot analysis
HeLa Kyoto cells were seeded in 24-well plates and transfected as described above, using a final siRNA concentration of 20 nM. Cells of two wells transfected with siRNAs against the same proteins were lysed in 50 μl Laemmli sample buffer and incubated for 10 min at 95°C. Whole samples were subjected to 6–16% SDS–polyacrylamide gel electrophoresis. In order to confirm the depletion efficiency, the following antibodies were used: anti-actin (Santa Cruz sc-1615 and Sigma A3853), anti-calnexin (Santa Cruz sc-6465), anti-Cdc42 (BD Laboratories 610929), anti-Cfl1 (Abcam ab11062), anti-CopB1 (Abcam ab6323), anti-CopG (Gentex GTX105331), anti-Itgav (Santa Cruz sc-9969), anti-Odc1 (Abcam ab50269), anti-Pfn1 (Abcam ab50667), anti-Rab7a (Abcam ab50533) and anti-Rbx1 (anti-Roc1, Abcam ab2977). The antibody against Atp1a1 was kindly provided by Dr Jack Kaplan. Anti-Actr3 and anti-Nckap1 (Nap1) were kind gifts from Dr Klemens Rottner. Proteins were detected using either goat anti-rabbit immunoglobulin G-horseradish peroxidase (Jackson 111-035-003) or goat anti-mouse immunoglobulin G-horseradish conjugates (Sigma A4416) and using ECL substrate (Amersham), as recommended by the manufacturer. Densitometric analysis on blots was performed with the QuantityOne software (Bio-Rad). The density in siRNA-treated groups was normalized to the density of their actin or calnexin levels and to corresponding control siRNA-treated groups.
Modified gentamycin protection assay
HeLa cells were infected with S. TmSopE using an m.o.i of 64 for the genome-scale and 32 for the confirmatory screen. Bacteria were either added using a 384-needle head of a robot (Tecan) or a multi-channel pipette. After 22 min, the medium was replaced with medium containing gentamycin (400 μg/ml; AppliChem) followed by 4 h of incubation at 37°C, fixation using 4% paraformaldehyde (PFA) and staining with DAPI (10 μg/ml) in 0.1% Triton X-100. For the computation of S. Tm infection, an assay using automated microscopy and automated images analysis was employed: nine images per well were acquired using a cellWoRx microscope (Applied Precision) and a × 10 objective. Images were analyzed using the open source program CellProfiler (Carpenter et al, 2006) as well as customized algorithms. In brief, nuclei were analyzed using the IdentifyPrimary module of CellProfiler. Nuclei were expanded with the CellProfiler-module IdentifySecondaryObjects. Spots were identified with the IdentifyPrimary module using a fixed threshold for the intensity in the GFP channel. Finally, cells and spots were superimposed and cells containing at least one spot were counted as infected. In a preprocessing step, the optimal threshold in the GFP channel separating signal and noise best was automatically calculated by comparing gentamycin pretreated wells and mock-treated (water instead of siRNA) wells.
Binding assay
The binding assay has been described recently (Misselwitz et al, 2011). Cells were pretreated with siRNAs as described for the confirmatory screen (Figure 2). In brief, cells were infected with S. TmΔ4 (m.o.i. of 82) for 6 min using a multi-channel pipette followed by three washing steps in DMEM containing 10% FCS and PFA fixation using a microplate dispenser (WellMate, Thermo Scientific). Subsequently, bacteria were stained by indirect immunofluorescence using an anti-LPS antibody and a FITC-coated secondary antibody. Nuclei were stained using DAPI. Images were acquired on an Image Express microscope (Molecular devices) using a × 4 objective. Image analysis followed similar algorithms as described for the modified gentamycin protection assay. In a second analysis step, the Enhanced CellClassifier (Misselwitz et al, 2010) was used for recognition of mitotic nuclei and neighbors of mitotic nuclei. For the final analysis, nuclei were thus purged from mitotic nuclei and their neighbors.
Ruffling assay
Cells were infected with S. TmSopE (m.o.i. 250) for 6 min. Subsequently, cells were fixed with 4% PFA and stained with DAPI and TRITC-Phalloidin (0.5 μg/ml; Sigma). Images were subsequently acquired on a BD Pathway microscope using a × 20 objective in the DAPI and the TRITC channel. For image analysis, nuclei and cells were recognized as described for binding, except that for the definition of cell borders the information of the actin channel was also used. Subsequently, intensity and texture of cells and nuclei were measured using predefined CellProfiler modules. In addition, customized modules were designed detecting small areas with bright intensity in the actin channel. Details of our customized features and the analysis pipeline used are available upon request. After image analysis, data were imported in the program Enhanced CellClassifier (Misselwitz et al, 2010), which facilitates usage of a machine learning algorithm (SVM with a radial basis function). Thereby, the user first needs to label cells as either ruffling or non-ruffling. These labels as well as measurements for the corresponding cells are recorded by the program. Later each imaged cell can be assigned by the trained model to a particular class (i.e., ruffling or non-ruffling). It should be noted that ruffle intensity cannot be scored by the Enhanced CellClassifier. For each of the three replica of the screen, more than 10 000 cells were trained as ruffling or non-ruffling. Mitotic cells were consistently trained as non-ruffling. During training, we focused on correct recognition of non-ruffling cells. After optimization of the critical parameters C and gamma of the SVM algorithm, a fivefold cross-validation accuracy of 94% was achieved.
Membrane closure assay using differential outside staining
HeLa cells were infected with S. TmSopE carrying plasmid pM965 for constitutive GFP expression (m.o.i. of 16–33) for 20 min and immediately fixed after infection. Bacteria were stained by indirect immunofluorescence without permeabilization using an anti-LPS antibody and an FITC-coated secondary antibody. Nuclei and actin were visualized using DAPI and TRITC Phalloidin after permeabilization. For each well, cell-associated extracellular and intracellular bacteria were quantified for 100–200 nuclei. Subsequently, the average number of invaded S. TmSopE per cell was calculated. For normalization, 7 of the 12 control siRNAs were evaluated for each plate. Salmonella invasion was thus quantified for the strongest two siRNAs of our 60 top hits by two independent observers. Key hits shown in the diagrams were quantified in three independent experiments.
Effector injection assay
HeLa cells were infected with S. TmSipA–TEM (m.o.i. 125) for 6 min, followed by two washing steps in Hank's buffered salt solution (HBSS) containing gentamycin and chloramphenicol and 30 min incubation at room temperature. A CCF2 loading kit was purchased from Invitrogen and CCF2-AM was dissolved in DMSO at a 1 mM concentration and stored in aliquots at −80°C. Immediately before the assay, substrate loading solution was prepared by mixing the CCF2-AM stock solution with proprietary solutions B and C (Invitrogen) at a ratio of 1:10:156. Plates were emptied and 50 μl HBSS without antibiotics was added to the cells, followed by the addition of 10 μl substrate loading solution. Luminescence was quantified 120 min after addition of the loading kit using a Victor3 Multilabel Plate Reader (Perkin Elmer). The ratio of the blue (450 nm) and the green signal (520 nm) was calculated after platewise background correction using the average of three wells without cells.
Post-processing analysis of data
GFP invasion, ruffling and binding experiments were performed in three independent experiments. The effector injection assay was carried out in triplicates in two independent experiments; therefore, the data summarize six data points. In all experiments, siRNA controls were also conducted as well as specific controls for the various assays used. As effects mediated by S. Typhimurium (i.e., binding, ruffling, effector injection) were stable for a wide range of cell densities (data not shown), a correction for the number of nuclei or siRNA toxicity was not necessary. Statistical analysis was carried out using the Matlab implementation of the Mann–Whitney U-test, comparing the values for each siRNA tested with the control siRNAs. Multiple test correction was performed using the Matlab implementation of the Benjamini and Hochberg (1995) method.
Fluorescence staining and image acquisition for high-resolution microscopy
HeLa cells were seeded on coverslips 1 day before infection and then fixed with 4% PFA (Sigma) for 15 min and permeabilized with 0.1% Triton X-100 (Sigma) for 5 min. Cells were blocked with 3% BSA and incubated with the appropriate antibodies (TRITC Phalloidin). The coverslips were then mounted with Vectashield (Reactolab, SA) and imaged with a × 100 objective using a Zeiss Axiovert 200 m inverted microscope with an Ultraview confocal head (Perkin Elmer), a krypton argon laser (643-RYB-A01, Melles, Griot, Didam, The Netherlands) and a PLAN-Apochromat × 100 oil objective with an aperture setting of 1.3 (Zeiss). Infrared, red and green fluorescence was recorded confocally, whereas blue fluorescence (DAPI and filipin) was recorded by epifluorescence microscopy. Images were then deconvolved with Volocity 5.2.0 (Improvision, Conventry, UK) for 25 iterations.
Quantification of membrane localization of different proteins
Lines were randomly drawn through images of the different channels, and the intensity along these lines was measured by using the Intensity measurement tool of ImageJ. The data were analyzed using a MatLab program with the following algorithm: On each line the first local maximum was identified. This first maximum corresponds to the cell membrane intensity. Next, to investigate the intracellular intensity, an average over 75 pixels was calculated. To ensure a sufficient distance from the cell membrane, the starting point was defined as the first local minimum for which both adjacent local minima had a higher intensity. Finally, the ratio of both intensities was compared. All maxima and minima were visualized on the graphs and manually verified.
Treatment of cells with MβCD, GGTI and replenishment of cholesterol and GM1
Cells were seeded in DMEM supplemented with 10% FCS at an appropriate density (13 000 HeLa cells well of a 24-well plate or 2000 cells per well of a 96-well plate), subjected to siRNA treatment (20 nM siRNA, Lipofectamine in a dilution 1:200 in Opti-MEM medium) and pretreated for 24 h with GGTI at a concentration of 10 μM or with MβCD at 10 mM for 1 h. For the replenishing experiments, cells were incubated for 17 h with 3.5–14 μg/ml GM1 complexed to fatty acid-free BSA and/or for at least 5 h with 15 μg/ml cholesterol complexed to 0.37 mM MβCD.
Quantification of the ruffle size
Depletion of CopG and cholesterol replenishment was performed as described above, and cells were infected with S. TmSopE at an m.o.i. of 60 for 10 min. Only ruffles containing a single intra- or extracellular bacterium were considered. Cells were then prepared for immunofluorescence analysis as described above. Automated identification and measurements of ruffles are challenging due to a cellular variability of the signal of the actin channel, for instance, in different stages of the cell cycle. We therefore opted for a manual quantification. Stacks were acquired for all ruffles and the largest diameter of each ruffle was measured in the xy-plane using ImageJ. Future developments allowing more advanced image analysis techniques might allow for automated and unbiased ruffle quantification for large numbers of cells.
Infection with VSV
Depletion of CopG and cholesterol and GM1 replenishment was performed as described above. The cells were infected with a VSV strain overexpressing GFP (Pelkmans et al, 2005) for 5 h, fixed and analyzed by automated microscopy. Images were acquired on automated wide-field cellWoRx microscopes (Applied Precision) with a × 10 objective. Per well, 5 × 5 directly adjacent images were taken for all virus infection assays, covering over 85% of each well surface. Image-based auto-focusing was performed on the DAPI signal for each imaged site. The images were analyzed with CellProfiler combined with a software published previously for supervised classification of cellular phenotypes (Ramo et al, 2009), and image analysis methods published elsewhere (Snijder et al, 2009). The CellProfiler image analysis pipeline was used as follows: first, nuclei objects were identified based on the DAPI stain. Next, cell boundaries were estimated using nuclear expansion. Standard CellProfiler texture, intensity, size and shape features were extracted from nucleus and cell regions, as well as from complete images, for both the DAPI and virus GFP signal. We next applied supervised machine learning using the open source SVM learning tool CellClassifier (Ramo et al, 2009), to identify virus-infected cells.
Phenotypic clustering of the hits
For the clustering of the step-specific phenotypes, the R-implementation of the hierarchical method described by Ward (1963) has been applied using an Euclidean distance metric. Partitioning of the dendrogram into the clusters a–g was done manually. The number of protein–protein interactions according to the STRING database within the clusters was then compared with the number of protein–protein interactions within randomly assembled clusters of the same size, drawn from the same set of candidate genes. This step was repeated 300 times, the number of protein–protein interactions within clusters c (25) and d (11) always being higher than within the corresponding randomly assembled cluster (average 6.77 and 0.52, respectively; Jensen et al, 2009).
Supplementary Material
Supplementary Figures S1–7, Supplementary table SI
Acknowledgments
We thank Dr Klemens Rottner, Dr Axel Nohturfft, Dr Klaudia Giehl and members of the Hardt group for stimulating discussions, and Jacques Laville and the management of the Brutus cluster at ETH Zürich for excellent IT support. We also thank Dr Andreas Vonderheit for help with the MD microscope, Karin Mench and Dr Lilli Stergiou for help with the virus infections and Naomi Barrett and Silke Misselwitz for help with the figures. We are grateful to Dr Klaudia Ghiel for providing Rac1–GFP and CDC42–GFP constructs. BM was supported by a grant from the Bonizzi-Theler foundation. The project was supported by grants to WDH and BM from UBS AG on behalf of a customer and a grant (InfectX) to WDH from the Swiss SystemsX.ch initiative, evaluated by the Swiss National Science Foundation.
Author contributions: BM, SD, PV, LP and WDH designed the experiments; BM, SD, PV, RS, SR and MS performed the experiments; BM, SD, PV, BS, MS, CvM and WDH analyzed the results; and BM, SD, PV and WDH wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE (2005) Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309: 1248–1251 [DOI] [PubMed] [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL (2003) Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol 13: 2159–2169 [DOI] [PubMed] [Google Scholar]
- Barwe SP, Anilkumar G, Moon SY, Zheng Y, Whitelegge JP, Rajasekaran SA, Rajasekaran AK (2005) Novel role for Na,K-ATPase in phosphatidylinositol 3-kinase signaling and suppression of cell motility. Mol Biol Cell 16: 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Rawet M, Wieland FT, Cassel D (2009) The COPI system: molecular mechanisms and function. FEBS Lett 583: 2701–2709 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg J (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 57: 289–300 [Google Scholar]
- Bethune J, Wieland F, Moelleken J (2006) COPI-mediated transport. J Membr Biol 211: 65–79 [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X, Oswald E (2004) Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol 186: 5486–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Hobbie S, Galan JE (1996) Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 274: 2115–2118 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35: 841–855 [DOI] [PubMed] [Google Scholar]
- Cheng LW, Viala JP, Stuurman N, Wiedemann U, Vale RD, Portnoy DA (2005) Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci USA 102: 13646–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S (2008) Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr Opin Microbiol 11: 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong R, Swiss R, Briones G, Stone KL, Gulcicek EE, Agaisse H (2009) Regulatory mimicry in Listeria monocytogenes actin-based motility. Cell Host Microbe 6: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coil DA, Miller AD (2004) Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J Virol 78: 10920–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW (2005) The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem Sci 30: 250–255 [DOI] [PubMed] [Google Scholar]
- Cortes C, Vapnik V (1995) Support-vector networks. AT&T Labs Res 20: 273–297 [Google Scholar]
- Dai S, Sarmiere PD, Wiggan O, Bamburg JR, Zhou D (2004) Efficient Salmonella entry requires activity cycles of host ADF and cofilin. Cell Microbiol 6: 459–471 [DOI] [PubMed] [Google Scholar]
- del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA (2004) Integrins regulate Rac targeting by internalization of membrane domains. Science 303: 839–842 [DOI] [PubMed] [Google Scholar]
- Derre I, Pypaert M, Dautry-Varsat A, Agaisse H (2007) RNAi screen in Drosophila cells reveals the involvement of the Tom complex in Chlamydia infection. PLoS Pathog 3: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrbar K, Hapfelmeier S, Stecher B, Hardt WD (2004) InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J Bacteriol 186: 1215–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN (2008) RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog 4: e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen SW, Naughton PJ, Grant G, Sojka M, Allen-Vercoe E, Bardocz S, Thorns CJ, Pusztai A (1997) Salmonella enterica var Typhimurium and Salmonella enterica var Enteritidis express type 1 fimbriae in the rat in vivo. FEMS Immunol Med Microbiol 18: 185–192 [DOI] [PubMed] [Google Scholar]
- Finlay BB, Ruschkowski S, Dedhar S (1991) Cytoskeletal rearrangements accompanying salmonella entry into epithelial cells. J Cell Sci 99 (Part 2): 283–296 [DOI] [PubMed] [Google Scholar]
- Friebel A, Ilchmann H, Aelpfelbacher M, Ehrbar K, Machleidt W, Hardt WD (2001) SopE and SopE2 from Salmonella Typhimurium activate different sets of RhoGTPases of the host cell. J Biol Chem 276: 34035–34040 [DOI] [PubMed] [Google Scholar]
- Gekara NO, Jacobs T, Chakraborty T, Weiss S (2005) The cholesterol-dependent cytolysin listeriolysin O aggregates rafts via oligomerization. Cell Microbiol 7: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD (2006) The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 7: 713–726 [DOI] [PubMed] [Google Scholar]
- Guignot J, Caron E, Beuzon C, Bucci C, Kagan J, Roy C, Holden DW (2004) Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J Cell Sci 117: 1033–1045 [DOI] [PubMed] [Google Scholar]
- Hage B, Meinel K, Baum I, Giehl K, Menke A (2009) Rac1 activation inhibits E-cadherin-mediated adherens junctions via binding to IQGAP1 in pancreatic carcinoma cells. Cell Commun Signal 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch J, Ehinger J, Ladwein M, Rohde M, Derivery E, Bosse T, Steffen A, Bumann D, Misselwitz B, Hardt WD, Gautreau A, Stradal TE, Rottner K (2010) Molecular dissection of Salmonella-induced membrane ruffling versus invasion. Cell Microbiol 12: 84–98 [DOI] [PubMed] [Google Scholar]
- Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y (2008) Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454: 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller A, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt WD (2005) The Salmonella Pathogenicity Island (SPI)-1 and SPI-2 Type III secretion systems allow Salmonella Serovar Typhimurium to trigger Colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174: 1675–1685 [DOI] [PubMed] [Google Scholar]
- Hardt WD, Chen LM, Schuebel KE, Bustelo XR, Galan JE (1998) S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93: 815–826 [DOI] [PubMed] [Google Scholar]
- Hayward RD, Cain RJ, McGhie EJ, Phillips N, Garner MJ, Koronakis V (2005) Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol Microbiol 56: 590–603 [DOI] [PubMed] [Google Scholar]
- Hirsch AJ (2010) The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol 5: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291: 238–239 [DOI] [PubMed] [Google Scholar]
- Ikonen E (2008) Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol 9: 125–138 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras I, Abrami L, Dautry-Varsat A (2003) Entry of the lymphogranuloma venereum strain of Chlamydia trachomatis into host cells involves cholesterol-rich membrane domains. Infect Immun 71: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Bossio JC, Galan JE (1994) The Salmonella Typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol 13: 555–568 [DOI] [PubMed] [Google Scholar]
- Kaplan JH (2002) Biochemistry of Na,K-ATPase. Annu Rev Biochem 71: 511–535 [DOI] [PubMed] [Google Scholar]
- Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L et al. (2010) Human host factors required for influenza virus replication. Nature 463: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van den Eeden SJ, Geluk A, Poot A, van der Marel G, Beijersbergen RL, Overkleeft H, Ottenhoff TH, Neefjes J (2007) Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 450: 725–730 [DOI] [PubMed] [Google Scholar]
- Lafont F, Tran Van Nhieu G, Hanada K, Sansonetti P, van der Goot FG (2002) Initial steps of Shigella infection depend on the cholesterol/sphingolipid raft-mediated CD44-IpaB interaction. EMBO J 21: 4449–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Galan JE (2009) The Salmonella Typhimurium SPI-1 type III secretion translocases mediate intimate attachment to non-phagocytic cells. Infect Immun 77: 2635–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Park DS, Razani B, Russell RG, Pestell RG, Lisanti MP (2002) Caveolin-1 mutations (P132L and null) and the pathogenesis of breast cancer: caveolin-1 (P132L) behaves in a dominant-negative manner and caveolin-1 (−/−) null mice show mammary epithelial cell hyperplasia. Am J Pathol 161: 1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Hussey PJ (2002) The ADF/cofilin family: actin-remodeling proteins. Genome Biol 3: reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsman M, Jordens I, Kuijl C, Janssen L, Neefjes J (2004) Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell 15: 2954–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V (2009) Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12: 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V (2004) Control of actin turnover by a salmonella invasion protein. Mol Cell 13: 497–510 [DOI] [PubMed] [Google Scholar]
- Medina E, Paglia P, Nikolaus T, Muller A, Hensel M, Guzman CA (1999) Pathogenicity island 2 mutants of Salmonella Typhimurium are efficient carriers for heterologous antigens and enable modulation of immune responses. Infect Immun 67: 1093–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meresse S, Steele-Mortimer O, Finlay BB, Gorvel JP (1999) The rab7 GTPase controls the maturation of Salmonella Typhimurium-containing vacuoles in HeLa cells. EMBO J 18: 4394–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Kreibich SK, Rout S, Stecher B, Periaswamy B, Hardt WD (2011) Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect Immun 79: 330–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Strittmatter G, Periaswamy B, Schlumberger MC, Rout S, Horvath P, Kozak K, Hardt WD (2010) Enhanced CellClassifier: a multi-class classification tool for microscopy images. BMC Bioinform 11: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AJ, Hoffmann C, Galle M, Van Den Broeke A, Heikenwalder M, Falter L, Misselwitz B, Kremer M, Beyaert R, Hardt WD (2009) The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6: 125–136 [DOI] [PubMed] [Google Scholar]
- Munoz S, Rivas-Santiago B, Enciso JA (2009) Mycobacterium tuberculosis entry into mast cells through cholesterol-rich membrane microdomains. Scand J Immunol 70: 256–263 [DOI] [PubMed] [Google Scholar]
- Norris FA, Wilson MP, Wallis TS, Galyov EE, Majerus PW (1998) SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci USA 95: 14057–14059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sollner TH, Rothman JE (1997) Bidirectional transport by distinct populations of COPI-coated vesicles. Cell 90: 335–349 [DOI] [PubMed] [Google Scholar]
- Paavilainen VO, Bertling E, Falck S, Lappalainen P (2004) Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol 14: 386–394 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG (2004) Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science 303: 836–839 [DOI] [PubMed] [Google Scholar]
- Patel JC, Galan JE (2005) Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr Opin Microbiol 8: 10–15 [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M (2005) Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436: 78–86 [DOI] [PubMed] [Google Scholar]
- Pepperkok R, Scheel J, Horstmann H, Hauri HP, Griffiths G, Kreis TE (1993) Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell 74: 71–82 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Philips JA, Rubin EJ, Perrimon N (2005) Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309: 1251–1253 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA (2009) Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu CI, Dubuisson J (2010) Role of lipid metabolism in hepatitis C virus assembly and entry. Biol Cell 102: 63–74 [DOI] [PubMed] [Google Scholar]
- Potrich C, Bastiani H, Colin DA, Huck S, Prevost G, Dalla Serra M (2009) The influence of membrane lipids in Staphylococcus aureus gamma-hemolysins pore formation. J Membr Biol 227: 13–24 [DOI] [PubMed] [Google Scholar]
- Prudencio M, Lehmann MJ (2009) Illuminating the host—how RNAi screens shed light on host-pathogen interactions. Biotechnol J 4: 826–837 [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL (2009) Conserved functions of membrane active GTPases in coated vesicle formation. Science 325: 1217–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA (2002) Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416: 644–648 [DOI] [PubMed] [Google Scholar]
- Ramo P, Sacher R, Snijder B, Begemann B, Pelkmans L (2009) CellClassifier: supervised learning of cellular phenotypes. Bioinformatics 25: 3028–3030 [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A (1992) The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70: 401–410 [DOI] [PubMed] [Google Scholar]
- Riff JD, Callahan JW, Sherman PM (2005) Cholesterol-enriched membrane microdomains are required for inducing host cell cytoskeleton rearrangements in response to attaching-effacing Escherichia coli. Infect Immun 73: 7113–7125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Weise C, Mirold S, Hillenbrand B, Bader B, Wittinghofer A, Hardt WD (1999) Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for RhoGTPases. J Biol Chem 274: 30501–30509 [DOI] [PubMed] [Google Scholar]
- Schlegel R, Tralka TS, Willingham MC, Pastan I (1983) Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell 32: 639–646 [DOI] [PubMed] [Google Scholar]
- Schlumberger MC, Hardt WD (2006) Salmonella type III secretion effectors: pulling the host cell's strings. Curr Opin Microbiol 9: 46–54 [DOI] [PubMed] [Google Scholar]
- Schlumberger MC, Kappeli R, Wetter M, Muller AJ, Misselwitz B, Dilling S, Kremer M, Hardt WD (2007) Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol Microbiol 65: 741–760 [DOI] [PubMed] [Google Scholar]
- Schlumberger MC, Muller AJ, Ehrbar K, Winnen B, Duss I, Stecher B, Hardt WD (2005) Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc Natl Acad Sci USA 102: 12548–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seveau S, Bierne H, Giroux S, Prevost MC, Cossart P (2004) Role of lipid rafts in E-cadherin—and HGF-R/Met—mediated entry of Listeria monocytogenes into host cells. J Cell Biol 166: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Scita G, Casanova JE (2005) WAVE2 signaling mediates invasion of polarized epithelial cells by Salmonella Typhimurium. J Biol Chem 280: 29849–29855 [DOI] [PubMed] [Google Scholar]
- Shimaoka M, Springer TA (2003) Therapeutic antagonists and conformational regulation of integrin function. Nat Rev Drug Discov 2: 703–716 [DOI] [PubMed] [Google Scholar]
- Snijder B, Sacher R, Rämö P, Damm E, Liberali P, Pelkmans L (2009) Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461: 520–523 [DOI] [PubMed] [Google Scholar]
- Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD (2004) Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72: 4138–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styers ML, O’Connor AK, Grabski R, Cormet-Boyaka E, Sztul E (2008) Depletion of beta-COP reveals a role for COP-I in compartmentalization of secretory compartments and in biosynthetic transport of caveolin-1. Am J Physiol Cell Physiol 294: C1485–C1498 [DOI] [PubMed] [Google Scholar]
- Sun X, Yau VK, Briggs BJ, Whittaker GR (2005) Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338: 53–60 [DOI] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT (2009) A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama F, Luna A, Babia T, Martinez-Menarguez JA, Ballesta J, Barth H, Chaponnier C, Renau-Piqueras J, Egea G (2000) The golgi-associated COPI-coated buds and vesicles contain beta/gamma-actin. Proc Natl Acad Sci USA 97: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot FG, Tran van Nhieu G, Allaoui A, Sansonetti P, Lafont F (2004) Rafts can trigger contact-mediated secretion of bacterial effectors via a lipid-based mechanism. J Biol Chem 279: 47792–47798 [DOI] [PubMed] [Google Scholar]
- Ward JH (1963) Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association 58: 236–244 [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ (1997) The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WJ, Erickson JW, Lin R, Cerione RA (2000) The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature 405: 800–804 [DOI] [PubMed] [Google Scholar]
- Zhou D, Mooseker MS, Galan JE (1999) Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283: 2092–2095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–7, Supplementary table SI