Abstract
Cells that have mutated their genes or are virally infected are a potential threat to a host. Consequently the immune system has evolved mechanisms for CD8 T lymphocytes to identify such cells and eliminate them. The generation of CD8 T cell responses occurs in two phases, both of which critically involve the process of antigen presentation. In the first phase sentinel cells gather antigens present in tissues and then present them to naïve CD8 T cells in ways that stimulate their maturation into effectors. In the second phase these effector cells seek out and eliminate the pathological cells. The abnormal cells are identified through their presentation of immunogenic antigens that they are producing. The antigen presentation mechanisms used by the sentinel cells can be different from those in other cells. This article will review these mechanisms with a focus in each case on how antigenic peptides are generated for presentation.
Classical (direct) pathway of MHC class I antigen presentation.
All cells transcribe a subset of their genes and translate them into proteins. In parallel they also continuously degrade these proteins in order to regulate their levels and eliminate damaged ones (1). In this process, cellular proteins are initially cleaved in the cytoplasm and nucleus into oligopeptide fragments.
To monitor what cells are producing, the immune system has evolved mechanisms to sample the library of peptides produced by cellular catabolism and display them on the cell surface for scrutiny by CD8 T cells (2). In this process a fraction of the peptides present in the cytosol are transported into the endoplasmic reticulum (ER) by the transporter of antigen presentation (TAP). In this location, peptides that are of the right length and sequence bind to newly synthesized MHC class I molecules, which then transport them to the cell surface. Through these mechanisms, cells display on their surface a sampling of their expressed gene products. In healthy cells, all of these presented peptides come from normal autologous genes and are ignored because the immune system is tolerant to them. However, if cells are synthesizing mutant proteins or ones from viruses, then peptides from these gene products will also be displayed and this allows effector CD8 T cells to recognize the abnormal cells and eliminate them.
Proteases that make the initial cleavages to generate a presented peptide.
The catabolic mechanism in cells that makes the initial cleavages to generate most presented peptides is the proteasome. Since the proteasome and its connection to antigen presentation has been well described elsewhere, we will cover this subject here relatively briefly and provide readers with references to representatives and/or recent papers and reviews.
The proteasome is a large barrel-shaped particle present in the cytosol and nucleus of all eukaryotic cells (3-5) that is responsible for degrading of a majority of cellular proteins (6). At its core is a 20S cylinder that is composed of two outer alpha rings and two inner beta rings with a central channel into which protein substrates enter and are cleaved (3, 6). The beta ring contains three active sites, each of which is formed by a different subunit: B1, B2 and B5 (3). All three of these subunits work through the same catalytic mechanism wherein an N-terminal threonine residue makes a nucleophilic attack on a peptide bond of the substrate. However the three active sites each have different specificities, cleaving preferentially on the carboxylic side of either hydrophobic residues (B5), basic residues (B1), or acidic ones (B2) (3, 6).
The strongest evidence that the proteasome plays a major role in the generation of most presented peptides comes from studies using highly specific inhibitors of the proteasome’s threonine-active sites. In living cells, these agents block completely the presentation of peptides from antigens that require proteolysis, but have no effect on ones that do not need cleavage (e.g. when epitopes are expressed directly from minigenes) and markedly limit the overall supply of peptides to MHC class I molecules (6).
While the immune system has utilized this phylogentically older pathway for a source of peptides it also evolved modifications that are thought to play a special role in antigen presentation. One modification is an alternate set of active site subunits (B1i/LMP2, B2i/MECL1, B5i/LMP7) that when expressed preferentially incorporate into newly assembling proteasomes in place of the “standard” B1, B2 and B5 subunits to form so called “immunoproteasomes” (3, 6). These are constitutively expressed in dendritic cells, lymphocytes, and thymic epithelium and are induced in all cells by proinflammatory cytokines such as IFN-γ.
The net effect of changing these beta subunits is to alter the catalytic activity of the proteasome’s active sites. This changes the cleavages made in protein substrates both quantitatively and qualitatively and thereby results in the production of a different set of peptides (7). Where it has been examined for individual antigens, immunoproteasomes sometimes make more or less of a particular epitope (3, 8). It has been predicted that immunoproteasomes generate peptides that are more favorable for antigen presentation, however, how often this is the case and how important immunoproteasomes are to MHC class I antigen presentation is not fully resolved (6, 7). Mice that lack one or two of the immunoproteasome subunits have defects in the presentation of selected epitopes and in some cases partial reductions in the generation of presented peptides overall (7). However, a more complete understanding of the contribution of immunoproteasomes to antigen presentation and maybe even to nonimmune functions awaits the generation of mice that lack all three inducible subunits.
Thymic medullary epithelium also constitutively express another B5 subunit (B5t) that preferentially incorporates with B1i plus B2i into proteasomes called “thymoproteasomes” (9). Mice that lack either B2i or especially B5t show marked reductions in the generation of CD8 T cells and defects in the T cell repertoire, presumably because changes in the peptide repertoire lead to altered T cell selection in the thymus.
Another apparent immune modification of the proteasome is the PA28 complex that is constitutively expressed in dendritic cells and is induced in other cells by IFN-γ. PA28 is a heterohexameric (α3β3) ring that binds to one or both ends of the 20S proteasome. Upon binding it increases the catalytic activity of all three of the proteasome’s active sites (3) and may do so at least in part opening the ends of the 20S particle allowing substrates to enter this particle. This change in activity can lead to altered cleavages in substrates (10) and increases or decreases in the generation of at least some MHC class I-presented peptides by purified proteasomes (11, 12) Similarly, transfection of PA28 into cells enhanced the presentation of some but not all epitopes (3) and increased the surface expression of some MHC class I molecules while decreasing the levels of other ones (12) Consistent with these findings PA28 −/− mice have a defect in presenting some antigens but not others (13).
So called “hybrid” proteasomes can form with PA28 on one end and a PA700 complex on the other. PA700 confers on the proteasome complex the ability to degrade polyubiquitinated substrates (10, 14). Where examined “hybrid” proteasome particles make up ~20% of proteasomes in cells (14). Whether PA28 exerts its effects on antigen presentation through proteasomes with or without PA700 is presently not clear.
Proteasome-independent pathways of antigen processing.
Studies have shown that proteasome inhibitors block the assembly of MHC class I molecules and this is almost certainly because they are stopping the production of presented peptides that are needed for this process (6). However, the degree of inhibition of MHC class I assembly is less marked for some MHC class I molecules (15). Moreover, there are some examples of epitopes whose presentation is not blocked by proteasome inhibitors or is actually enhanced by this treatment (16, 17). These findings have been interpreted to indicate that this proteasome inhibitor-resistant presentation is being mediated by other proteases. This may indeed be the case, however caution is warranted in interpreting these results because, at the concentrations used in these studies, most proteasome inhibitors do not block all of the proteasome’s active sites (18). Therefore, in the presence of these inhibitors, proteasomes may still be making cleavages that generate the presented peptides.
One candidate for a protease that might generate presented peptides independently from the proteasome is tripeptidyl peptidase II (TPPII). TPPII was initially suggested as the major mechanism by which cells chronically exposed to proteasome inhibitors survived and generated presented peptides (19), although it is now thought that the proteasomes were not completely inhibited in these treated cells (18). However, purified TPPII can help generate an epitope (20) and the presentation of another epitope was blocked when cells were treated with TPPII inhibitors but not proteasome blockers (16, 20). However, TPPII-null mice have been generated and their cells do not show any reduction in the expression of MHC class I molecules (suggesting that peptide supply is intact) (21, 22). A role for TPPII in trimming some proteasomal products will be discussed below.
The size of MHC class I-presented peptides and the products of proteasomes.
MHC class I-presented peptides are of a remarkable uniform size, the vast majority being either 8, 9 or 10 residues, depending on the particular MHC class I molecule (2, 3, 6). This is because the ends of antigen binding groove of the class I molecule are closed and have pockets that bind the N and C-terminus of the presented peptide. Consequently only peptides of the exact right size stably bind to this complex.
When purified proteasomes were incubated with antigens the majority of peptides produced were too short (<7 residues) for antigen presentation. In contrast only a small minority (<5%) of the peptides generated were of the correct size to bind to any particular MHC class I molecule, while a substantially larger fraction (~20%) of products were too long for stable binding (23). In the one case that has been analyzed, the peptides produced by immunoproteasomes were on average longer those made from standard proteasomes (24). When such long peptides are loaded into the cytosol of cells or expressed from minigenes, they are trimmed and presented, indicating that they can serve as precursors of presented peptides.
What trims these long precursors into presented peptides? One protease involved in this process is the proteasome, which can recleave these peptides into epitopes (25). Interestingly, proteasome inhibitors block the presentation from precursors that have even a single extra C-terminal residue, indicating that these particles are the only activity in cells that can make the proper C-terminal cleavage to generate an epitope (6, 25). Consistent with these findings the cytosol of cells has little if any carboxypeptidase activity (25). In contrast, precursors that just have extra N-terminal residues are presented but in this case proteasome inhibitors do not block this process (6, 25). This suggested that there must be other activities in cells that can remove N-terminal residues. It turns out that this N-terminal trimming of peptides can occur in the cytosol, where they are first produced, or in the ER after they are transported into this location.
Peptidases and trimming of peptides in the cytosol.
N-extended peptides that are loaded or expressed in the cytosol of cells are trimmed and presented on MHC class I molecules (6, 25). Importantly this can occur to some extent in cells that are unable to cleave peptides in the ER (discussed further below) indicating that some trimming of precursors occurs in the cytosol. This trimming is mediated by aminopeptidases because precursors that are resistant to hydrolysis by these peptidases (due to a chemically blocked N terminus) fail to be trimmed and presented (26, 27). However, which specific aminopeptidases contribute to MHC class I antigen presentation is not well understood.
One aminopeptidase that may play a special role in trimming peptides is TPPII. TPPII is relatively unique because it trims longer peptides as well as shorter ones, while most other aminopeptidases only cleave peptides <14 amino acids in length. Consequently, inhibition of TPPII significantly reduces the presentation of peptides with long N-terminal extensions but not shorter ones (22, 28). How important is TPPII to antigen presentation? Although an initial report suggested that TPPII was required for the generation of most presented peptides (29), subsequent studies with inhibitors (30), RNAi-silenced cells (28) and TPPII −/− mice (21, 22) found no major defects in antigen presentation; these latter results are consistent with the finding that proteasomes generate relatively few very long peptides (see above). However, TPPII inhibitors reduce the presentation of a few individual epitopes (16), increase the presentation of others (31), and do not affect responses to many others (32), suggesting that this peptidase may play a role in the presentation of selected sequences.
Cells contain many other cytosolic aminopeptidases that can trim peptides. Three of these, leucine aminopeptidase (LAP) (33), bleomycin hydrolase (BH) and puromycin-sensitive aminopeptidase (PSA) (34), can generate mature epitopes from precursors at least in cell free systems. However, the contribution of these peptidases to trimming in cells is less clear. Inhibitors of BH and PSA reduced the presentation of a few epitopes, however these agents were not highly specific ones (34). On the other hand BH −/−, PSA −/− and LAP −/− mutant mice or BH+LAP-double deficient animals show no defect in generating and presenting mature epitopes from N-extended precursor peptides or antigen presentation generally (33, 35, 36). These negative results may indicate that there is substantial redundancy among these enzymes, so that trimming activity is not reduced by the loss of only one or a few peptidases. Alternatively, it is possible that there are other peptidases not yet identified that play a more important role in trimming epitopes. It is also possible that LAP, BH, and/or PSA are more important for the trimming of epitopes other than the ones thus far examined.
Peptide trimming in the ER.
It is possible to target peptides directly into the ER after they are synthesized by fusing a signal sequence to the N-terminus of the peptide (6). When such constructs are expressed from minigene in cells, the signal sequence targets the peptide into the ER through the SEC61 translocon and is then removed by the signal sequence peptidase. This delivers the peptide into the ER in a manner that is independent of the TAP transporter.
When N-extended peptides were targeted into the ER via SEC61, their extra N-terminal residues were removed and the peptides were presented on MHC class I molecules (25, 27, 37). This indicated that aminopeptidases must exist in the lumen of the ER and indeed microsomes were found to have such trimming activity (38, 39). These activities were purified and identified as two members of the oxytocinase family of aminopeptidases. Although these peptidases have been given several different names, they are now most commonly referred to as ER aminopeptidase (ERAP) or ER-associated aminopeptidase (ERAAP) one and two. Mice have only ERAP1 while humans and some other mammalian species have both ERAP1 and ERAP2.
ERAP1
Most aminopeptidases continued to hydrolyze a substrate until it is converted entirely to amino acids. However ERAP1 is unique in that it trims long polypeptides rapidly until they reach a size of 9 or 8 residues and then its hydrolysis stops (40, 41). Therefore, ERAP1 seems to be specialized for generating peptides of the proper size for antigen presentation. This is not to say that ERAP1 always stops trimming when it has generated a mature peptide. Indeed it trims about 50% of 9mer peptides to 8mers, and is so doing will ultimately destroy some 9mer epitopes (40) and limit their presentation (41, 42). It has been suggested that ERAP1 might also trim the protruding ends of long peptides when they are bound to MHC class I molecules (43), however, it is not known whether this actually occurs and if ERAP1’s active site could even access such bound peptides.
ERAP1 plays an important role in MHC class I antigen presentation. Cells in which ERAP1 is eliminated through mutation or RNAi silencing (and also naturally lack ERAP2) are unable to trim N-extended peptides in the ER (41). Therefore in mouse cells and some human cells ERAP1 is the only trimming enzyme in the ER, at least for these sequences. ERAP1-deficient cells have reduced surface levels of MHC class I molecules and the peptide-MHC complexes that are made are less stable than on wild type cells (44-46). These results suggest that ERAP1 makes an important contribution both to the quantity and quality of peptides available for antigen presentation.
Interestingly, loss of ERAP1 decreases the presentation of some epitopes from full length proteins (ones presumably made as N-extended precursors), does not affect others, and increases the presentation of yet others (41, 42, 46). Consequently, ERAP1-deficiency markedly changes CD8 T cell responses to various epitopes in vivo and alters immunodominance hierarchies (46). Remarkably, ERAP1−/− T cells can recognize and respond against cells from wild type mice (44) suggesting that loss of ERAP1 may alter the overall repertoire markedly. Presumed polymorphisms in ERAP1 may similarly affect responses because ERAP1 has been linked genetically to the autoimmune disease ankylosing spondylitis (47, 48) and cancer progression (49) in humans.
Based on these findings it is clear that the antigen processing pathway generates many N-extended peptides precursors and that trimming by ERAP1 has specificity. This specificity has not yet been fully elucidated but studies with purified ERAP1 and cells expressing ERAP1 have shown that while it can remove all residues from the N-terminus of peptides, it does so at different rates (50) and is also affected by sequences in the epitope itself (40, 51, 52).
ERAP2
ERAP2 also trims residues from the N-terminus of peptides and appears to do so with different specificity from ERAP1, being more active in cleaving N-terminal basic residues from at least some substrates (53). Therefore the combined actions of ERAP1 and ERAP2 are likely to enhance the hydrolytic capacity of the ER and trim a broader set of sequences. Interestingly, ERAP2 appears to form a complex with ERAP1 and it has been suggested that this combination of 2 aminopeptidases may trim peptides in a cooperative manner (53). In contrast to ERAP1, ERAP2 does not stop trimming peptides when they are the size of mature epitopes (52).
Silencing of ERAP2 with RNAi reduced the presentation of an epitope (53). However, the contribution of ERAP2 to overall ER trimming is not yet clear. Silencing of ERAP2 only reduced overall MHC class I expression by ~10% (53). Moreover, the hierarchy of specificity in removing any of the 20 amino acids flanking an epitope was not different in cells expressing only ERAP1 or both ERAP1 and ERAP2 (50). Furthermore, ERAP2 is poorly expressed or not expressed in human brain, liver, thymus, testes, and small intestine (54); and it is not present in mice. Although additional studies are needed, these findings suggest that ERAP2 may play a more limited or specialized role in peptide trimming.
Cleavage of peptides after the ER.
Antigens that exit the ER may be cleaved by proteases in the exocytic pathway. One such protease is furin, a member of the subtilisin family of serine proteases that is resident in the Golgi apparatus. Furin has been implicated in the generation of peptides from a few proteins that are transported via the exocytic route (55, 56). Since the presentation of these peptides does not require TAP, it is thought that furin may generate these peptides from proteins that transit the transgolgi network. However, this is unlikely to be a major source of presented peptides because TAP negative cells have very low levels of presented peptides; among which some are derived from signal sequences that are hydrolyzed in the ER, and others are not (57). Whether furin contributes to the trimming of peptides that are generated in the cytosol and transported into the ER by TAP is not known.
Peptide destruction
Proteases can also cleave within epitopes, destroying them, and this is a process that can limit antigen presentation. In fact, outside of their role in antigen presentation, most peptides are not useful in cells and their accumulation would be deleterious. As a result, cells have evolved robust catabolic activities to further degrade peptides down to amino acids, which are useful to cells. Consequently, when peptides are added to cell extracts they are rapidly degraded (58) and when injected into the cytoplasm of living cells have a half life of only seconds (26).
All of the proteases that may be involved in the generation of peptides can and probably do destroy some epitopes. Thus, the presentation of some epitopes is enhanced by proteasome inhibitors (16). Moreover, increases in the presentation of selected epitopes or in the overall surface expression of class I molecules were seen in cells in which PSA (33), BH+LAP (35), TPPII (22, 28) and ERAP1 (41, 42) were absent or inhibited. Another protease that has been implicated in this process is thimet oligopeptidase, a cytosolic metalloprotease (59). The increased antigen presentation and higher levels of MHC class I molecules after inhibiting these peptidases suggest that they normally limit the presentation of some peptides by destroying them.
The fact that cytosol is such a hostile environment for peptides has raised the question of how peptides destined for antigen presentation escape destruction. It may be that most are destroyed (it has been suggested that this is the fate of 99.9% of peptide (60)) and that the ones that do get presented are rescued by rapid transport though TAP and then binding to MHC class I molecules. However, it has also been proposed that there may be mechanisms that protect peptides from degradation. One idea is that cytosolic chaperones may bind peptides and prevent their degradation. Indeed peptides can be found bound to these proteins (61-63). However, whether binding to chaperones is important for the presentation of most peptides is not clear. In fact when thimet oligopeptidase was over expressed in cells, there was almost complete inhibition of the presentation of peptides generated in the cytosol, but no effect on the ones targeted into the ER (59). These results indicate that presented peptides are susceptible to destruction in the cytosol; this suggests either that protective mechanisms do not exist or if they do then they must be in kinetic competition with the destruction pathways. The specificity of the destructive (and protective) mechanisms has not been well characterized. This is an important issue because it may be yet another factor that influences what peptides ultimately get presented.
Cross presentation
Dendritic cells and macrophages have the unique ability to present on MHC class I not only peptides from their own endogenous antigens but also ones from antigens in their external environment through a process called cross presentation (64-67). After acquiring these antigens, dendritic cells carry this information to secondary lymphoid tissues and present it to naïve CD8 T cells in ways that initiate immune responses (64). This ability to present exogenous antigens plays a critical role in immune surveillance of tissues by allowing the immune system to monitor tissues for the presence of cancers or viruses that don’t infect dendritic cells. This pathway is also thought to allow the immune system to monitor the antigens in phagosomes of dendritic cells and macrophages and thereby allow CD8 T cells to detect phagocytes that are harboring intracellular bacteria and eliminate them (66, 67).
Antigens that are internalized by phagocytosis or macropinocytosis are cross presented several thousand more efficiently than soluble proteins (66, 67). Presumably because of these antigens that particulate (e.g. dying cells) are a major source of cross presented antigens. Although still somewhat controversial, there is accumulating evidence that the major source of cross presented peptides are intact antigens (66-68) or ones that are partially cleaved, e.g. by caspases in dying cells (69, 70). In this review we will focus on the two best characterized mechanisms by which cross presented peptides are generated from proteins in phagosomes with an emphasis on what is known about the proteases that generate these peptides. The two mechanisms we will focus on are the phagosome-to-cytosol and vacuolar pathways. It should be noted that there are also additional mechanisms by which cross presentation occurs (63, 71-73), although their role in vivo is presently unclear.
Phagosome-to-cytosol pathway.
In the phagosome-to-cytosol pathway of cross presentation, blocking proteolysis in these phagosomes does not inhibit cross presentation and may actually enhance it (66, 67). In fact, dendritic cells may promote cross presentation by raising the pH in phagosomes and thereby reducing the proteolytic destruction of antigens. Therefore, in this pathway cross presented peptides are not generated in the phagosomes themselves. Instead, the antigens are transferred from phagosomes into the cytosol where they undergo cleavage. This form of cross presentation is blocked by proteasome inhibitors indicating that these particles make the initial cleavages to generate a cross presented peptide (66, 67). The presentation is also dependent on TAP (66, 67) and at least some cases also ERAP1 (74, 75). Thus once the exogenous antigen is transferred to the cytosol, it is thought to be processed for cross presentation in much the same manner as endogenous proteins.
The peptides that are generated in the cytosol may be transported by TAP to MHC class I molecules in the ER. In addition there is evidence that many ER components, including TAP, somehow get incorporated phagosomes (73, 76). This has suggested that some peptides generated in the cytosol might be transported by TAP to MHC class I molecules in phagosomes. In this location the peptides could be further hydrolyzed by phagosomal proteases. One vacuolar peptidase that has been implicated in this process is IRAP (PLAP), which is a homologue of ERAP1 and ERAP2 (77). Like ERAP1, IRAP can remove most amino acids from the N-terminus of peptides, however unlike ERAP1 it can continue to trims peptides to a size smaller <8 residues in length (Stratikos, E. personal communication).
Dendritic cells lacking IRAP present endogenous antigens normally but have about a 50% decrease in their ability to cross present an exogenous antigen (77). This reduction in presentation is of a similar magnitude to that observed in ERAP1-deficient cells. It is presently unknown whether these two peptidases are working together in the phagosome or in separate compartments (ERAP1 in the ER and IRAP in the phagosome).
The vacuolar pathway of cross presentation.
In the vacuolar pathway, the generation of cross presented peptides is inhibited by cysteine protease inhibitors such as leupeptin, but is resistant to proteasome inhibitors and their presentation is TAP-independent (66, 67). Therefore it is thought that the cross presented peptides are generated in the phagosome itself.
One of the vacuolar cysteine proteases that generates cross presented peptides is cathepsin S. Cathepsin S-deficient mice and their dendritic cells have a defect in vacuolar but not phagosome-to-cytosol cross presentation (78, 79). Cathepsin B, L and D-deficient cells showed no defect in cross presentation (78). In addition, IRAP might be involved in the generation of presented peptides in this compartment in DCs (77).
Why cathepsin S plays such an important role is unclear. Perhaps it can make the appropriate cleavages to generate 8-9mers peptides. In fact, recombinant cathepsin S was shown to generate a presented peptide in at least one case (78) ; however, whether it can generate other mature peptides is not known (80). Additionally cathepsin S is relatively unique in being strongly active at neutral pH and this may be important because MHC class I molecules may not stably bind peptides at acidic pH.
Since cathepsin S mice lack the vacuolar pathway and TAP-deficient animals lack the phagosome-to-cytosol pathway, it has been possible to use these genetic models to quantify the contribution of these various pathways to cross presentation. Such analyses have shown that both pathways contribute to the generation of CD8 T cell responses but that the phagosome-to-cytosol pathway contributes to a greater extent than does the vacuolar mechanism (66, 78).
Conclusions.
To supply MHC class I molecules with peptides the immune system utilized, and in some cases adapted, phylogentically older catabolic pathways that normally break down proteins. The proteasome pathway has been used and adapted with new active sites and regulators to make the initial cleavages in proteins for direct presentation and for one of the major cross presentation pathways. Aminopeptidases in the cytosol and ER trim the proteasomal products, sometimes generating the mature epitope from longer precursors and other times destroying them. Other peptidases can also cleave these peptides and destroy them. Similarly the catabolic pathways in the endocytic compartments have been used to generate peptides for one of the alternate cross presentation pathways. In this situation cathepsin S has been identified as one of the key enzymes in antigen processing. Understanding the specific proteases involved in these processes and their specificity is important because this will determine what peptides get presented and thereby the magnitude and specificity of responses.
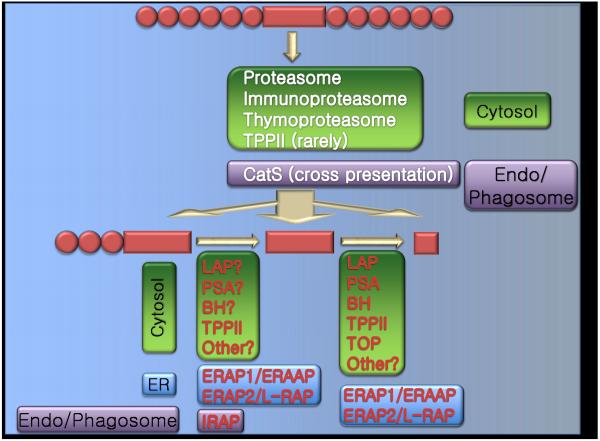
FIGURE 1.
Schematic representation of the different proteases involved in antigen processing. The proteasomal degradation step can lead to the generation of a mature epitope, a precursor peptide with an N-terminal extension, or the destruction of the epitope. Precursor peptides can be further trimmed by aminopeptidases to generate a mature epitope. Alternatively amino and/or endo peptidase activities can lead to the destruction of the epitope.
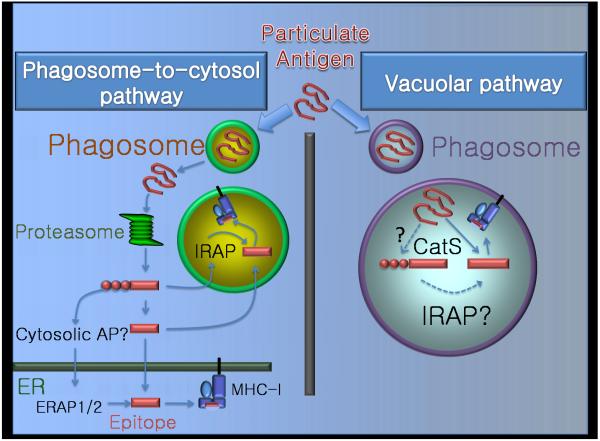
FIGURE 2.
Proteases in cross presentation. Upon uptake by dendritic cells or macrophages, exogenous Ag can be processed and presented on MHC class I molecules by two major pathways: the phagosome-to-cytosol pathway (left) and the vacuolar pathway (right). In the cytosolic pathway, proteasomes are the major proteases to generate the antigenic peptide fragments. These peptides can be further trimmed by aminopeptidases in the cytosol, ER and possibly an endosomal compartment (for details see fig.1). In the less well-understood vacuolar pathway, cathepsin S plays a key role in generating class I binding peptides. It is unclear whether other proteases (e.g. IRAP) are also involved.
Footnotes
This work was supported by the National Institutes of Health grants number 2R01AI020248-27A2 and 2R01AI043543-11A109 (to K.L.R.)
- ER
- endoplasmic reticulum
- TAP
- transporter of antigen presentation
- LMP
- MHC-linked Low Molecular Weight Protein
- MECL
- multicatalytic endopeptidase complex-like 1
- PA28
- 28-kDa proteasome activator
- PA700
- 700-kDa proteasome activator
- TPPII
- Tripeptidyl Peptidase II
- LAP
- leucine aminopeptidase
- BH
- bleomycin hydrolase
- PSA
- puromycin-sensitive aminopeptidase
- ERAP
- ER aminopeptidase
- ERAAP
- ER-associated aminopeptidase
- P-LAP
- placental leucine aminopeptidase
- L-RAP
- leukocyte-derived aminopeptidase
- TOP
- Thimet Oligopeptidase
- IRAP (PLAP)
- insulin-regulated aminopeptidase
- Cat
- Cathepsin
References.
- 1.Goldberg AL, Rock KL. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–9. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 2.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 3.Kloetzel P. The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta. 2004;1695:225–233. doi: 10.1016/j.bbamcr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Rechsteiner M, Hoffman L, Dubiel W. The multicatalytic and 26 S proteases. J. Biol. Chem. 1993;268:6065–8. [PubMed] [Google Scholar]
- 5.Goldberg AL. Functions of the proteasome: the lysis at the end of the tunnel. Science. 1995;268:522–3. doi: 10.1126/science.7725095. [DOI] [PubMed] [Google Scholar]
- 6.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv. Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 7.Van den Eynde BJ, Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol. 2001;13:147–153. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 8.Strehl B, Textoris-Taube K, Jakel S, Voigt A, Henklein P, Steinhoff U, Kloetzel PM, Kuckelkorn U. Antitopes define preferential proteasomal cleavage site usage. J. Biol. Chem. 2008;283:17891–7. doi: 10.1074/jbc.M710042200. [DOI] [PubMed] [Google Scholar]
- 9.Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 10.Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002;21:2636–45. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Textoris-Taube K, Henklein P, Pollmann S, Bergann T, Weisshoff H, Seifert U, Drung I, Mügge C, Sijts A, Kloetzel P, Kuckelkorn U. The N-terminal flanking region of the TRP2360-368 melanoma antigen determines proteasome activator PA28 requirement for epitope liberation. J. Biol. Chem. 2007;282:12749–12754. doi: 10.1074/jbc.M611644200. [DOI] [PubMed] [Google Scholar]
- 12.Yamano T, Sugahara H, Mizukami S, Murata S, Chiba T, Tanaka K, Yui K, Udono H. Allele-selective effect of PA28 in MHC class I antigen processing. J. Immunol. 2008;181:1655–1664. doi: 10.4049/jimmunol.181.3.1655. [DOI] [PubMed] [Google Scholar]
- 13.Murata S, Udono H, Tanahashi N, Hamada N, Watanabe K, Adachi K, Yamano T, Yui K, Kobayashi N, Kasahara M, Tanaka K, Chiba T. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J. 2001;20:5898–5907. doi: 10.1093/emboj/20.21.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 2000;275:14336–14345. doi: 10.1074/jbc.275.19.14336. [DOI] [PubMed] [Google Scholar]
- 15.Luckey CJ, Marto JA, Partridge M, Hall E, White FM, Lippolis JD, Shabanowitz J, Hunt DF, Engelhard VH. Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J. Immunol. 2001;167:1212–1221. doi: 10.4049/jimmunol.167.3.1212. [DOI] [PubMed] [Google Scholar]
- 16.Guil S, Rodriguez-Castro M, Aguilar F, Villasevil EM, Anton LC, Del Val M. Need for tripeptidyl-peptidase II in major histocompatibility complex class I viral antigen processing when proteasomes are detrimental. J. Biol. Chem. 2006;281:39925–34. doi: 10.1074/jbc.M608522200. [DOI] [PubMed] [Google Scholar]
- 17.Diekmann J, Adamopoulou E, Beck O, Rauser G, Lurati S, Tenzer S, Einsele H, Rammensee HG, Schild H, Topp MS. Processing of two latent membrane protein 1 MHC class I epitopes requires tripeptidyl peptidase II involvement. J. Immunol. 2009;183:1587–97. doi: 10.4049/jimmunol.0803441. [DOI] [PubMed] [Google Scholar]
- 18.Princiotta MF, Schubert U, Chen W, Bennink JR, Myung J, Crews CM, Yewdell JW. Cells adapted to the proteasome inhibitor 4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc. Natl. Acad. Sci. U.S.A. 2001;98:513–8. doi: 10.1073/pnas.021132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang EW, Kessler BM, Borodovsky A, Cravatt BF, Bogyo M, Ploegh HL, Glas R. Integration of the ubiquitin-proteasome pathway with a cytosolic oligopeptidase activity. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9990–5. doi: 10.1073/pnas.180328897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, Weinschenk T, Schild H, Laderach D, Galy A, Haas G, Kloetzel PM, Reiss Y, Hosmalin A. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat. Immunol. 2003;4:375–9. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 21.Firat E, Huai J, Saveanu L, Gaedicke S, Aichele P, Eichmann K, van Endert P, Niedermann G. Analysis of direct and cross-presentation of antigens in TPPII knockout mice. J. Immunol. 2007;179:8137–8145. doi: 10.4049/jimmunol.179.12.8137. [DOI] [PubMed] [Google Scholar]
- 22.Kawahara M, York IA, Hearn A, Farfan D, Rock KL. Analysis of the Role of Tripeptidyl Peptidase II in MHC Class I Antigen Presentation In Vivo. J. Immunol. 2009;183(10):6069–6077. doi: 10.4049/jimmunol.0803564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 24.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craiu A, Akopian T, Goldberg A, Rock KL. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide diffusion, protection, and degradation in nuclear and cytoplasmic compartments before antigen presentation by MHC class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 27.Mo XY, Cascio P, Lemerise K, Goldberg AL, Rock K. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J. Immunol. 1999;163:5851–5859. [PubMed] [Google Scholar]
- 28.York IA, Bhutani N, Zendzian S, Goldberg AL, Rock KL. Tripeptidyl peptidase II is the major peptidase needed to trim long antigenic precursors, but is not required for most MHC class I antigen presentation. J. Immunol. 2006;177:1434–43. doi: 10.4049/jimmunol.177.3.1434. [DOI] [PubMed] [Google Scholar]
- 29.Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- 30.Marcilla M, Villasevil EM, de Castro JA. Tripeptidyl peptidase II is dispensable for the generation of both proteasome-dependent and proteasome-independent ligands of HLA-B27 and other class I molecules. Eur. J. Immunol. 2008;38:631–9. doi: 10.1002/eji.200737444. [DOI] [PubMed] [Google Scholar]
- 31.Preta G, Marescotti D, Fortini C, Carcoforo P, Castelli C, Masucci M, Gavioli R. Inhibition of serine-peptidase activity enhances the generation of a survivin-derived HLA-A2-presented CTL epitope in colon-carcinoma cells. Scand. J. Immunol. 2008;68:579–88. doi: 10.1111/j.1365-3083.2008.02175.x. [DOI] [PubMed] [Google Scholar]
- 32.Basler M, Groettrup M. No essential role for tripeptidyl peptidase II for the processing of LCMV-derived T cell epitopes. Eur. J. Immunol. 2007;37:896–904. doi: 10.1002/eji.200636372. [DOI] [PubMed] [Google Scholar]
- 33.Towne CF, York IA, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J. Immunol. 2008;180:1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 34.Stoltze L, Schirle M, Schwarz G, Schröter C, Thompson MW, Hersh LB, Kalbacher H, Stevanovic S, Rammensee HG, Schild H. Two new proteases in the MHC class I processing pathway. Nat. Immunol. 2000;1:413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 35.Towne CF, York IA, Watkin LB, Lazo JS, Rock KL. Analysis of the role of bleomycin hydrolase in antigen presentation and the generation of CD8 T cell responses. J. Immunol. 2007;178:6923–30. doi: 10.4049/jimmunol.178.11.6923. [DOI] [PubMed] [Google Scholar]
- 36.Towne CF, York IA, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J. Immunol. 2005;175:6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 37.Snyder HL, Yewdell JW, Bennink JR. Trimming of antigenic peptides in an early secretory compartment. J. Exp. Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fruci D, Niedermann G, Butler RH, van Endert PM. Efficient MHC class I-independent amino-terminal trimming of epitope precursor peptides in the endoplasmic reticulum. Immunity. 2001;15:467–476. doi: 10.1016/s1074-7613(01)00203-5. [DOI] [PubMed] [Google Scholar]
- 39.Brouwenstijn N, Serwold T, Shastri N. MHC class I molecules can direct proteolytic cleavage of antigenic precursors in the endoplasmic reticulum. Immunity. 2001;15:95–104. doi: 10.1016/s1074-7613(01)00174-1. [DOI] [PubMed] [Google Scholar]
- 40.Saric T, Chang S, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 41.York IA, Chang S, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat. Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 42.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 43.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat. Immunol. 2007;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 45.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–3. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 46.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class Ipresented peptides in vivo and plays an important role in immunodominance. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsui FWL, Haroon N, Reveille JD, Rahman P, Chiu B, Tsui HW, Inman RD. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann. Rheum. Dis. 2009 doi: 10.1136/ard.2008.103804. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 48.Maksymowych WP, Inman RD, Gladman DD, Reeve JP, Pope A, Rahman P. Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arth. Rheum. 2009;60:1317–1323. doi: 10.1002/art.24467. [DOI] [PubMed] [Google Scholar]
- 49.Fruci D, Giacomini P, Nicotra MR, Forloni M, Fraioli R, Saveanu L, van Endert P, Natali PG. Altered expression of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in transformed non-lymphoid human tissues. J. Cell Physiol. 2008;216:742–9. doi: 10.1002/jcp.21454. [DOI] [PubMed] [Google Scholar]
- 50.Hearn A, York IA, Rock KL. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J. Immunol. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evnouchidou I, Momburg F, Papakyriakou A, Chroni A, Leondiadis L, Chang S, Goldberg AL, Stratikos E. The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS ONE. 2008;3:e3658. doi: 10.1371/journal.pone.0003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang S, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17107–17112. doi: 10.1073/pnas.0500721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 54.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J. Biol. Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 55.Del-Val M, López D. Multiple proteases process viral antigens for presentation by MHC class I molecules to CD8(+) T lymphocytes. Mol. Immunol. 2002;39:235–247. doi: 10.1016/s0161-5890(02)00104-9. [DOI] [PubMed] [Google Scholar]
- 56.Tiwari N, Garbi N, Reinheckel T, Moldenhauer G, Hammerling GJ, Momburg F. A transporter associated with antigen-processing independent vacuolar pathway for the MHC class I-mediated presentation of endogenous transmembrane proteins. J. Immunol. 2007;178:7932–42. doi: 10.4049/jimmunol.178.12.7932. [DOI] [PubMed] [Google Scholar]
- 57.Weinzierl AO, Rudolf D, Hillen N, Tenzer S, van Endert P, Schild H, Rammensee HG, Stevanovic S. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur. J. Immunol. 2008;38:1503–10. doi: 10.1002/eji.200838136. [DOI] [PubMed] [Google Scholar]
- 58.Beninga J, Rock KL, Goldberg AL. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 59.York IA, Mo AXY, Lemerise K, Zeng W, Shen Y, Abraham CR, Saric T, Goldberg AL, Rock KL. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18:429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 60.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 61.Kunisawa J, Shastri N. The group II chaperonin TRiC protects proteolytic intermediates from degradation in the MHC class I antigen processing pathway. Mol. Cell. 2003;12:565–576. doi: 10.1016/j.molcel.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 62.Kunisawa J, Shastri N. Hsp90alpha chaperones large C-terminally extended proteolytic intermediates in the MHC class I antigen processing pathway. Immunity. 2006;24:523–534. doi: 10.1016/j.immuni.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 63.Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu. Rev. Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 64.Heath WR, Belz GT, Behrens GMN, Smith CM, Forehan SP, Parish IA, Davey GM, Wilson NS, Carbone FR, Villadangos JA. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol. Rev. 2004;199:9–26. doi: 10.1111/j.0105-2896.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 65.Savina A, Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 2007;219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 66.Rock KL, Shen L. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol. Rev. 2005;207:166–83. doi: 10.1111/j.0105-2896.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- 67.Shen L, Rock KL. Priming of T cells by exogenous antigen cross-presented on MHC class I molecules. Curr. Opin. Immunol. 2006;18:85–91. doi: 10.1016/j.coi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, Gibbs J, Bennink JR, Yewdell JW. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 2004;304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- 69.Rawson PM, Molette C, Videtta M, Altieri L, Franceschini D, Donato T, Finocchi L, Propato A, Paroli M, Meloni F, Mastroianni CM, d’Ettorre G, Sidney J, Sette A, Barnaba V. Cross-presentation of caspase-cleaved apoptotic self antigens in HIV infection. Nat. Med. 2007;13:1431–9. doi: 10.1038/nm1679. [DOI] [PubMed] [Google Scholar]
- 70.Pang B, Neijssen J, Qiao X, Janssen L, Janssen H, Lippuner C, Neefjes J. Direct antigen presentation and gap junction mediated cross-presentation during apoptosis. J. Immunol. 2009;183:1083–90. doi: 10.4049/jimmunol.0900861. [DOI] [PubMed] [Google Scholar]
- 71.Ackerman AL, Kyritsis C, Tampe R, Cresswell P. Access of soluble antigens to the endoplasmic reticulum can explain cross-presentation by dendritic cells. Nat. Immunol. 2005;6:107–13. doi: 10.1038/ni1147. [DOI] [PubMed] [Google Scholar]
- 72.Neijssen J, Herberts C, Drijfhout JW, Reits E, Janssen L, Neefjes J. Cross-presentation by intercellular peptide transfer through gap junctions. Nature. 2005;434:83–8. doi: 10.1038/nature03290. [DOI] [PubMed] [Google Scholar]
- 73.Desjardins M, Houde M, Gagnon E. Phagocytosis: the convoluted way from nutrition to adaptive immunity. Immunol. Rev. 2005;207:158–165. doi: 10.1111/j.0105-2896.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 74.Firat E, Saveanu L, Aichele P, Staeheli P, Huai J, Gaedicke S, Nil A, Besin G, Kanzler B, van Endert P, Niedermann G. The role of endoplasmic reticulum-associated aminopeptidase 1 in immunity to infection and in cross-presentation. J. Immunol. 2007;178:2241–8. doi: 10.4049/jimmunol.178.4.2241. [DOI] [PubMed] [Google Scholar]
- 75.Blanchard N, Gonzalez F, Schaeffer M, Joncker NT, Cheng T, Shastri AJ, Robey EA, Shastri N. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat. Immunol. 2008;9:937–44. doi: 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burgdorf S, Schölz C, Kautz A, Tampé R, Kurts C. Spatial and mechanistic separation of cross-presentation and endogenous antigen presentation. Nat. Immunol. 2008;9:558–566. doi: 10.1038/ni.1601. [DOI] [PubMed] [Google Scholar]
- 77.Saveanu L, Carroll O, Weimershaus M, Guermonprez P, Firat E, Lindo V, Greer F, Davoust J, Kratzer R, Keller SR, Niedermann G, van Endert P. IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science. 2009;325:213–7. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- 78.Shen L, Sigal LJ, Boes M, Rock KL. Important role of cathepsin S in generating peptides for TAP-independent MHC class I crosspresentation in vivo. Immunity. 2004;21:155–65. doi: 10.1016/j.immuni.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 79.Chen L, Jondal M. Brefeldin A inhibits vesicular MHC class I processing in resting but not in CpG- and disruption-activated DC. Mol. Immunol. 2008;46:158–65. doi: 10.1016/j.molimm.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 80.Belizaire R, Unanue ER. Targeting proteins to distinct subcellular compartments reveals unique requirements for MHC class I and II presentation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17463–17468. doi: 10.1073/pnas.0908583106. [DOI] [PMC free article] [PubMed] [Google Scholar]