Abstract
The endoplasmic reticulum (ER) is the cellular organelle responsible for protein folding and assembly, lipid and sterol biosynthesis, and calcium storage. The unfolded protein response (UPR) is an adaptive intracellular stress response to accumulation of unfolded or misfolded proteins in the ER. In this study, we show that the most conserved UPR sensor inositol-requiring enzyme 1 α (IRE1α), an ER transmembrane protein kinase/endoribonuclease, is required to maintain hepatic lipid homeostasis under ER stress conditions through repressing hepatic lipid accumulation and maintaining lipoprotein secretion. To elucidate physiological roles of IRE1α-mediated signalling in the liver, we generated hepatocyte-specific Ire1α-null mice by utilizing an albumin promoter-controlled Cre recombinase-mediated deletion. Deletion of Ire1α caused defective induction of genes encoding functions in ER-to-Golgi protein transport, oxidative protein folding, and ER-associated degradation (ERAD) of misfolded proteins, and led to selective induction of pro-apoptotic UPR trans-activators. We show that IRE1α is required to maintain the secretion efficiency of selective proteins. In the absence of ER stress, mice with hepatocyte-specific Ire1α deletion displayed modest hepatosteatosis that became profound after induction of ER stress. Further investigation revealed that IRE1α represses expression of key metabolic transcriptional regulators, including CCAAT/enhancer-binding protein (C/EBP) β, C/EBPδ, peroxisome proliferator-activated receptor γ (PPARγ), and enzymes involved in triglyceride biosynthesis. IRE1α was also found to be required for efficient secretion of apolipoproteins upon disruption of ER homeostasis. Consistent with a role for IRE1α in preventing intracellular lipid accumulation, mice with hepatocyte-specific deletion of Ire1α developed severe hepatic steatosis after treatment with an ER stress-inducing anti-cancer drug Bortezomib, upon expression of a misfolding-prone human blood clotting factor VIII, or after partial hepatectomy. The identification of IRE1α as a key regulator to prevent hepatic steatosis provides novel insights into ER stress mechanisms in fatty liver diseases associated with toxic liver injuries.
Keywords: ER stress, hepatic steatosis, unfolded protein response
Introduction
Hepatosteatosis, characterized by excessive accumulation of triglycerides (TGs) in hepatocytes, is considered the key metabolic component of non-alcoholic fatty liver disease (NAFLD). Hepatic lipid accumulation is regulated by a complex network that involves lipogenesis, dietary fatty acid (FA) uptake, FA oxidation, and lipid transport/secretion (Musso et al, 2009). Hepatic steatosis is the result of abnormal lipid metabolism caused by increased lipid delivery to the liver, increased de novo lipid biosynthesis, reduced lipid oxidation, and/or decreased lipid secretion (Postic and Girard, 2008; Musso et al, 2009). Although significant progress has been made in understanding the metabolic pathways associated with hepatosteatosis, the molecular signal transduction network that triggers hepatic lipid dysregulation remains to be identified. Recent findings suggest that intracellular stress response pathways from the endoplasmic reticulum (ER) have important roles in regulating expression of lipogenic factors, secretion of apolipoproteins, lipid accumulation, and lipotoxicity (Scheuner et al, 2001; Feng et al, 2003; Wei et al, 2006; Zhang et al, 2006a; Lee et al, 2008; Ota et al, 2008; Oyadomari et al, 2008; Rutkowski et al, 2008). This has raised an important notion that the ER stress response is an underlying mechanism governing the development of hepatosteatosis.
The ER is the site for folding of membrane and secreted proteins, synthesis of lipids and sterols, and storage of calcium in eukaryotic cells. A number of biological, physiological, or pathological stimuli can perturb protein folding in the ER, leading to accumulation of unfolded or misfolded proteins in the ER lumen—a condition referred to as ‘ER stress’ (Kaufman, 1999). To deal with ER stress conditions, the ER has evolved a set of signal transduction pathways collectively termed the unfolded protein response (UPR) to alter transcriptional and translational programs in the stressed cell. In metazoans, there are three basic signal transducers of the UPR, inositol-requiring 1 (IRE1), PERK (double-strand RNA-activated protein kinase-like ER kinase), and ATF6 (activating transcription factor 6). The most conserved UPR transducer is IRE1, an ER transmembrane Ser/Thr protein kinase/endoribonuclease that was originally identified as the proximal sensor for the UPR in budding yeast (Nikawa and Yamashita, 1992; Cox et al, 1993; Mori et al, 1993). The IRE1-mediated UPR signalling pathway is highly conserved in all eukaryotes. Two homologues of yeast ire1, Ire1α and Ire1β, were identified in both the murine and human genomes (Tirasophon et al, 1998; Wang et al, 1998). In mammals, IRE1α is expressed in most cells and tissues, while IRE1β expression is primarily restricted to intestinal epithelial cells. The mRNA encoding X-box-binding protein 1 (XBP1), a basic-leucine zipper (b-ZIP)-containing transcription factor, was identified as an RNA substrate for the mammalian IRE1α endoribonuclease activity (Shen et al, 2001; Yoshida et al, 2001; Calfon et al, 2002). Upon activation of the UPR, IRE1α initiates unconventional splicing of the Xbp1 mRNA to remove a 26-nucleotide intron and generate a switch in translational-reading frame. Spliced Xbp1 mRNA encodes a potent transcriptional activator that induces transcription of genes encoding functions that facilitate protein folding, secretion, and degradation in response to ER stress.
In addition to functions that are strictly involved in protein folding, trafficking, secretion, and degradation, the UPR is now recognized to have important physiological roles in metabolism, inflammation, as well as cell differentiation and survival (Ron and Walter, 2007; Todd et al, 2008; Zhang and Kaufman, 2008a). IRE1α and its endoribonuclease target Xbp1 mRNA are required for the differentiation and function of specialized cell types, and have been proposed to contribute to the pathogenesis of metabolic disease (Reimold et al, 2001; Zhang et al, 2005; Ron and Walter, 2007). Under conditions of ER stress, activated IRE1α can bind to tumour necrosis factor-receptor-associated factor 2 (TRAF2), leading to activation of c-JUN N-terminal kinase (JNK) through apoptosis signal-regulating kinase 1 (Urano et al, 2000; Nishitoh et al, 2002). This might provide stressed cells with survival or death signals by activating JNK-mediated signalling pathways. Additionally, the cytosolic domain of IRE1α can also recruit the pro-apoptotic BCL-2-associated X protein and BCL-2 antagonist, augmenting both the kinase and endoribonuclease activities of IRE1α (Hetz et al, 2006). In the liver, ER stress and UPR signalling can be triggered by metabolic factors, such as lipids, glucose, cytokines, homocysteine, and free FAs (Zhang and Kaufman, 2008a). In this study, we generated hepatocyte-specific Ire1α-null mice. Using this animal model, we show that IRE1α is essential to maintain hepatic lipid homeostasis under conditions of ER stress in the liver. Deletion of the Ire1α gene results in profound hepatosteatosis and hypolipidemia in the mice upon ER stress. The identification of a key role of IRE1α in preventing ER stress-induced hepatosteatosis provides novel insights into potential avenues to limit hepatic lipid accumulation that contribute to metabolic syndrome.
Results
Hepatocyte-specific deletion of Ire1α is efficient
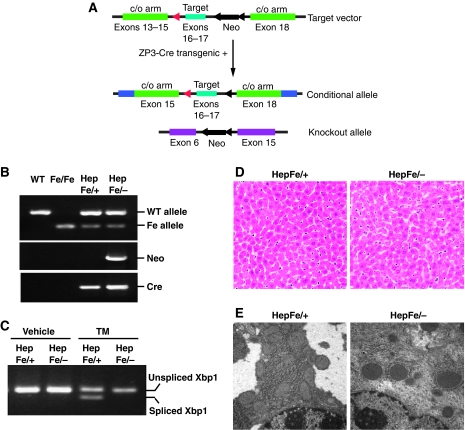
Homozygous Ire1α-null (Ire1α−/−) murine embryos die by embryonic day 12.5, while the heterozygous Ire1α+/− mice can survive as normal wild-type mice (Lee et al, 2002; Zhang et al, 2005). The most significant phenotype of whole body Ire1α deletion, as well as Xbp1 deletion, was liver hypoplasis (Reimold et al, 2000; Zhang et al, 2005). In order to study the tissue-specific and temporal roles of IRE1α in the mouse, we engineered mice with a loxP site-flanked Ire1α allele (Ire1αfe) (Figure 1A and B) (Hoess and Abremski, 1990). Mice with one or two floxed Ire1α alleles were viable, fertile, and apparently normal. Mice that harbour one floxed Ire1α allele and one Ire1α-null allele (Ire1αfe/−) were bred with transgenic mice expressing Cre recombinase under the control of the albumin promoter (Postic and Magnuson, 2000) to obtain offspring with hepatocyte-specific Ire1α deletion (Ire1αfe/−Cre+, herein called Ire1αHepfe/−) and littermate control mice (Ire1αfe/+Cre+, herein called Ire1αHepfe/+) that have a functional Ire1α allele. Functional deletion of Ire1α was evidenced by the absence of spliced Xbp1 mRNA, the target of IRE1α endoribonuclease activity, in the livers of the Ire1αHepfe/− mice challenged with a sublethal injection of tunicamycin (TM), an inhibitor of N-linked glycosylation that causes accumulation of misfolded protein in the ER (Figure 1C). In comparison, significant levels of spliced Xbp1 mRNA were detected in the control Ire1αHepfe/+ mice that harbour a wild-type Ire1α allele. We have not observed a difference in liver function between wild-type Ire1α+/+ and our experimental control Ire1αHepfe/+ mice (data not shown). Mice with hepatocyte-specific Ire1α deletion (Ire1αHepfe/−) were viable and displayed no gross developmental or morphological defects (Figure 1D). Although livers from Ire1αHepfe/− mice appeared phenotypically normal in the absence of challenge, ultrastructural analysis revealed less rough ER content in the Ire1α-null (Ire1αHepfe/−) livers, compared with the littermate control (Ire1αHepfe/+) livers (Figure 1E), suggesting a possible ER defect in the absence of IRE1α.
Figure 1.
Hepatocyte-specific deletion of Ire1α. (A) Schematic illustration of the targeting vector and the Ire1α-null alleles. The targeting vector contained loxP-flanked exons 16 and 17 of the murine Ire1α gene as well as a loxP-flanked Neomycin (Neo) cassette. The engineered mice containing the targeted allele were backcrossed with ZP3-Cre transgenic mice in order to generate mice with the loxP-flanked Ire1α allele without the Neo cassette. This mouse strain was then crossed with heterozygous Ire1α-null mice (Lee et al, 2002; Zhang et al, 2005) to produce mice containing an Ire1α conditional allele (Ire1αfe) and an Ire1α-null allele (Ire1α−). (B) Genotyping of conditional Ire1α-null mice. Mice harbouring a conditional Ire1α allele in the presence of an Ire1α-null or wild-type allele were crossed with transgenic mice that express Cre recombinase under the control of the albumin promoter to generate hepatocyte-specific Ire1α-null mice (Ire1αHepfe/−) and control mice that harbour a functional Ire1α allele (Ire1αHepfe/+). Three pairs of PCR reactions were used for genotyping: one pair amplifies the targeted allele and the wild-type allele; a second pair amplifies the Neo cassette to identify the Ire1α-null allele (Lee et al, 2002; Zhang et al, 2005); and a third set amplifies the Cre transgene. (C) Semiquantitative reverse transcription (RT)–PCR of Xbp1 mRNA. Xbp1 mRNA splicing was not detected in livers of hepatocyte-specific Ire1α-null mice (Ire1αHepfe/−) at 8 h after intraperitoneal injection of TM (2 μg/g body weight). (D, E) Liver sections from hepatocyte-specific Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice in the absence of TM challenge stained with haematoxylin and eosin (magnification × 200) (D) and transmission electron microscopy (magnification × 19 000) (E).
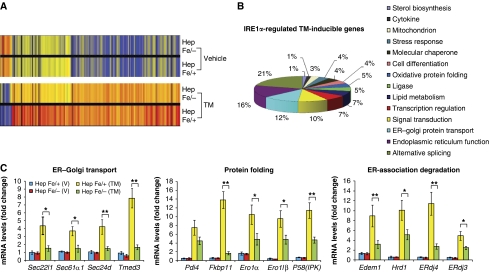
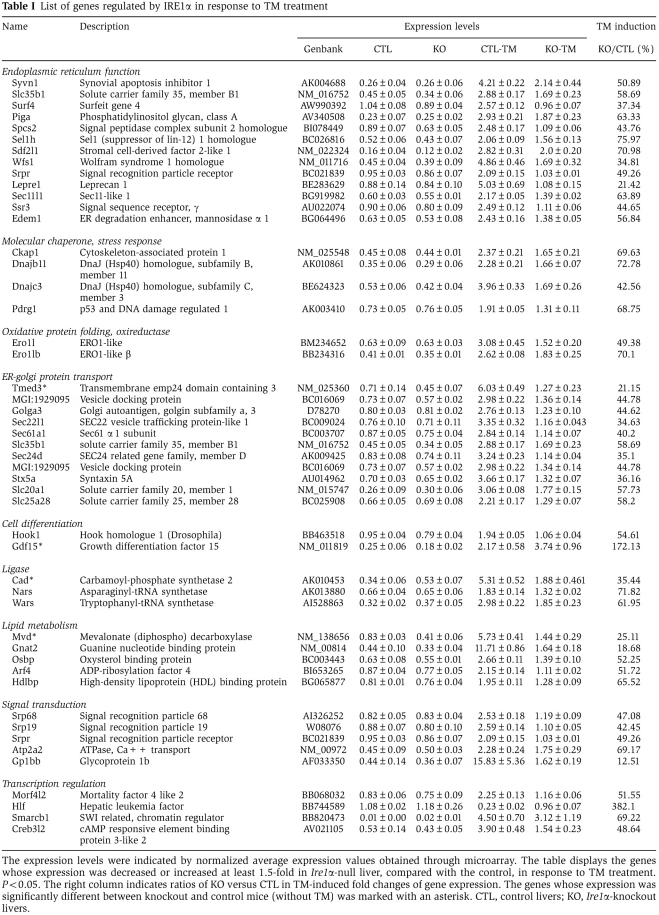
To characterize the phenotype of hepatocyte-specific Ire1α deletion as well as the molecular targets of IRE1α in the liver, we performed Affymetrix GeneChip microarray analysis using total liver mRNA from Ire1αHepfe/− and control Ire1αHepfe/+ mice injected with TM (2 μg/g body weight) induce acute ER stress in the liver (Zhang et al, 2006a). Surprisingly, deletion of Ire1α caused only a slight alteration in gene expression in the absence of stress. Upon close analysis, we identified a subset of genes with reduced or increased expression that encoded functions associated with ER signal transduction and lipid metabolism (Figure 2A) (Table I) (Supplementary Figure S1A and B). However, at 8 h after TM injection, the RNA expression profile in the Ire1αHepfe/− liver was significantly altered, compared with that characterized in the livers of control Ire1αHepfe/+ mice (Figure 2A). The microarray results indicated that IRE1α is required for ER stress-mediated induction of genes involved in ER function, intracellular stress responses, lipid metabolism, cell differentiation, alternative mRNA splicing, and signal transduction (Figure 2B) (Table I). Quantitative real-time PCR analyses confirmed that IRE1α is required for ER stress-induced expression of genes encoding functions in ER-to-Golgi protein transport (Sec22l1, Sec61α1, Sec24d, Tmed3), protein folding such as cis–trans proline isomerization and disulfide bond formation (Fkbp11, Pdi4, Ero1α, Ero1lβ, P58IPK), and ER-associated degradation (ERAD) (Edem1, Hrd1, Erdj4, Erdj3) (Figure 2C). These results support the hypothesis that IRE1α is required to maintain ER function and remodel the secretory apparatus in response to protein misfolding in the ER.
Figure 2.
IRE1α regulates expression of genes involved in protein folding, ER-to-Golgi transport, and ERAD. (A) Affymetrix microarray analysis of mRNA expression profiles in livers of Ire1αHepfe/− and Ire1αHepfe/+ mice at 3 months of age at 8 h after intraperitoneal injection of TM (2 μg/g body weight) or vehicle (150 μM dextrose). Graphic representation of ANOVA analysis is shown for the expression of 177 genes (P-value cutoff was <0.05) that were significantly regulated upon TM injection into control mice (Ire1αHepfe/+). Each vertical bar represents a single gene. Blue indicates lower expression and red indicates higher expression. (B) Percentages of IRE1α-regulated TM-inducible genes. The 177 TM-regulated genes identified by ANOVA were clustered based on their functions. The percentages of biological pathway-specific gene groups that are regulated by IRE1α are shown. (C) Quantitative real-time RT–PCR analysis of liver mRNA isolated from Ire1αHepfe/− and Ire1αHepfe/+ mice. Total liver mRNA was isolated at 8 h after injection with vehicle or TM (2 μg/g body weight) for real-time RT–PCR analysis. Expression values were normalized to β-actin mRNA levels. Fold changes of mRNA are shown in TM-treated mice compared with control mice. Each bar denotes the mean±s.e.m. (n=6 mice per group); *P<0.05, **P<0.01. P-values are shown for statistically significant differences. Edem1, ER degradation enhancing, mannosidase α-like 1; Sec22l1, SEC22 vesicle trafficking protein-like 1; Sec61α1, Sec61 α 1 subunit; Sec24d, SEC24-related gene family, member D; Tmed3, transmembrane emp24 domain containing 3; Pdi4, protein disulfide isomerase associated 4; Fkbp11, FK506-binding protein 11 of the peptidyl-prolyl cis–trans isomerase family; ERo1α, ER oxidoreductase-1 α, ERo1lβ, ERO1-like β; Hrd1, or Syvn1, synovial apoptosis inhibitor 1; ERdj4, DnaJ (Hsp40) homologue, subfamily B, member 9; ERdj3, DnaJ (Hsp40) homologue, subfamily B, member 11; P58(IPK), or Dnajc3, DnaJ (Hsp40) homologue, subfamily C, member 3.
Table 1. List of genes regulated by IRE1α in response to TM treatment.
Deletion of Ire1α causes UPR activation and defective adaptation to ER stress
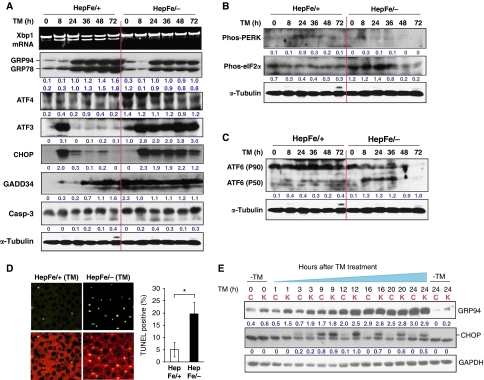
Next, we asked whether Ire1α deletion in hepatocytes alters ER homeostasis and activates the other UPR signalling pathways. To address this question, we first compared the expression of the ER chaperones GRP78/BiP and GRP94, two markers of the ER stress response, in the livers from hepatocyte-specific Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice after intraperitoneal injection of vehicle or TM. Expression of GRP78 and GRP94 in the Ire1α-deleted livers was modestly decreased compared with control livers from 8 to 72 h after TM treatment (Figure 3A). In comparison, the expression of ER stress-inducible pro-apoptotic transcription factors ATF4 (activating factor 4), CHOP (CCAAT/enhancer-binding protein (C/EBP) homologous transcription factor), and ATF3 (activating factor 3) was significantly increased in the Ire1α-null livers compared with the control livers at 8–72 h after TM treatment (Figure 3A). Because ATF4, CHOP, and ATF3 were documented to be upregulated by the PERK/eIF2α subpathway of the UPR (Harding et al, 2000; Scheuner et al, 2001; Wek et al, 2006), we thought that the PERK UPR pathway may be activated by Ire1α deletion in hepatocytes. Although we failed to detect a significant increase in phosphorylation of PERK, phosphorylation of eIF2α was increased at the early time points in the Ire1α-null livers, compared with that in the control livers (Figure 3B). Furthermore, levels of the growth arrest and DNA damage-inducible protein 34 (GADD34), a PERK/eIF2α-regulated stress protein that has a feedback regulatory role by dephosphorylating eIF2α (Novoa et al, 2001), were higher in the Ire1α-null livers at the early time points (0–24 h) after TM injection but comparable to that in the control livers at later time points (36–72 h) after TM injection (Figure 3A). It is possible that the induction of GADD34 causes dephosphorylation of eIF2α and PERK. These data suggest that Ire1α deletion in the liver leads to selective upregulation of ER stress-inducible pro-apoptotic factors ATF4, CHOP, and ATF3. This selective upregulation may reflect a defect in the ability of Ire1α-null livers to adapt to ER stress, as levels of cleaved caspase-3, the cell apoptosis executioner, and positive terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labelling (TUNEL) staining were increased in the Ire1α-null livers after TM treatment (Figure 3A and D). Analysis of ATF6 cleavage indicated that it was increased in the Ire1αHepfe/− livers compared with the Ire1αHepfe/+ livers, after TM challenge (Figure 3C), suggesting that Ire1α deletion also upregulates the ATF6 branch of the UPR in response to ER stress. Because IRE1α/XBP1 and ATF6 have redundant roles in activating expression of ER chaperones and protein-folding enzymes (Okada et al, 2002; Lee et al, 2003b; Wu et al, 2007), increased cleavage of ATF6 in Ire1α-null livers may compensate, at least partially, for the defect caused by Ire1α deletion in activating expression of UPR target genes.
Figure 3.
Ire1α deletion leads to upregulation of selective UPR genes in response to ER stress. (A–C) Xbp1 mRNA splicing and western blot analysis of livers from Ire1αHepfe/− and Ire1αHepfe/+ mice at 3 months of age after intraperitoneal injection of TM (2 μg/g body weight) for 8, 24, 36, 48, and 72 h. Mice injected with the vehicle (150 μM dextrose) were included as the control time point 0. (A) Semiquantitative RT–PCR analysis of Xbp1 mRNA splicing and western blot analysis of GRP94, GRP78/BiP, ATF4, ATF3, CHOP, GADD34, and caspase-3. Levels of α-tubulin were included as internal controls. The values below the gels represent protein signal intensities that were quantified using NIH ImageJ software and normalized to α-tubulin. (B, C) Western blot analysis of phosphorylated PERK, phosphorylated eIF2α, and ATF6. The values below the gels represent normalized protein signal intensities. (D) Immunofluorescence TUNEL staining of liver tissue sections for DNA fragmentation. Liver tissue sections were prepared from the Ire1αHepfe/− and Ire1αHepfe/+ mice after intraperitoneal injection of TM (2 μg/g body weight) for 36 h for TUNEL staining. The green fluorescence represents TUNEL-positive and red fluorescence represents propidium iodide (PI)-positive cells (magnification × 400). The right panel shows the percentages of TUNEL-positive cells determined by calculating the number of TUNEL-positive cells divided by the number of PI-positive cells from five fields of each slide. *P<0.05. (E) Western blot analysis of GRP94 and CHOP in immortalized Ire1α-null (Ire1αfe/−Cre) and control (Ire1αfe/fe) hepatocytes challenged with TM (2 μg/ml) for indicated times. Ire1α-null and control hepatocytes cultured in the absence of TM (at 0 and 24 h) were included as controls. Levels of GAPDH protein were determined as internal controls. The values below the gels represent protein signal intensities after normalization to GAPDH. C, control hepatocytes; K, Ire1α-null hepatocytes.
To further evaluate the ability of Ire1α-null hepatocytes to adapt to prolonged ER stress, we treated immortalized Ire1α-null (Ire1αfe/−Cre) and control (Ire1αfe/fe) hepatocytes with TM for up to 24 h in vitro. In response to TM, expression of ER stress-induced pro-apoptotic factor CHOP was significantly different between the Ire1α-null and control hepatocytes (Figure 3E). Although the Ire1α-null and control hepatocytes expressed similar levels of CHOP in the absence or presence of TM for up to 3 h, CHOP expression in the Ire1α-null hepatocytes was significantly greater than that in the control hepatocytes from 9 to 24 h after TM treatment (Figure 3E). CHOP expression in the control hepatocytes was induced from 3 to 9 h of TM treatment, but was subsequently attenuated, suggesting that the control hepatocytes adapted to ER stress at the later time points of TM treatment. In contrast, the Ire1α-null hepatocytes sustained high levels of CHOP expression up to 24 h after TM treatment (Figure 3E), suggesting the Ire1α-null cells are defective in their ability to adapt to prolonged ER stress.
Hepatocyte-specific Ire1α deletion increases hepatic lipid and reduces plasma lipid content upon ER stress
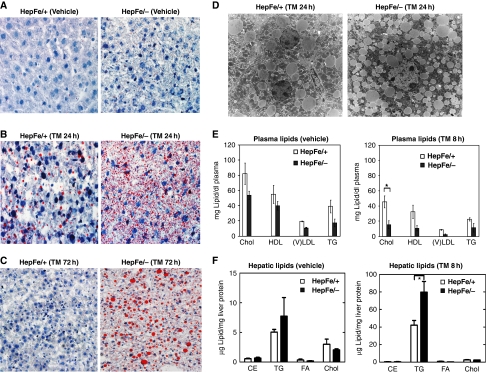
Having established a role for IRE1α in the chronic response to ER stress, we next investigated the physiological role of IRE1α in the liver. In hepatocytes, TG are synthesized in the ER and stored in the cytoplasm surrounded by a monolayer of phospholipid in distinct lipid droplet structures (Martin and Parton, 2006). Although no morphological or developmental defects were observed in the hepatocyte-specific conditional Ire1α-null (Ire1αHepfe/−) mice, Oil-red O staining of liver tissue sections revealed a slight increase in lipid content in livers from the Ire1αHepfe/− mice (Figure 4A). Under normal physiological conditions, lipid droplets maintain energy balance at the cellular and organismal levels. As a response to acute liver injury induced by sublethal doses of TM, lipid deposition is increased in the hepatocytes of wild-type mice (Yang et al, 2007; Rutkowski et al, 2008). To test the role of IRE1α in hepatic lipid accumulation in response to ER stress, we injected Ire1αHepfe/− and control Ire1αHepfe/+ mice with a dose of TM that can induce strong ER stress in the liver in vivo (Zhang et al, 2006a; Wu et al, 2007). At 24 h after TM injection, the livers from control mice displayed modest macrovesicular steatosis characterized by engorgement of the hepatocytes with larger lipid droplets (Figure 4B). In contrast, in the livers of Ire1αHepfe/− mice, TM injection induced profound microvesicular steatosis characterized by the presence of small lipid droplets that surround the nucleus (Figure 4B). At 72 h after TM injection, hepatic steatosis in the control Ire1αHepfe/+ mice was significantly resolved, as only small amounts of lipid droplets were detected in their livers (Figure 4C). In contrast, the Ire1α-null livers remained steatotic at 72 h after TM treatment, suggesting a role for IRE1α in preventing and recovering from ER stress-induced hepatic steatosis. Interestingly, at 72 h after TM treatment, Ire1α-null livers displayed macrovesicular steatosis instead of microvesicular steatosis that occurred at 24 h after TM treatment. This suggests that the formation of larger lipid droplets may be delayed in the Ire1α-null livers under conditions of ER stress. To further analyse the phenotype caused by Ire1α deletion at the subcellular level, ultrastructural analysis of liver sections was performed. In response to TM treatment, hepatocytes in the livers from Ire1αHepfe/− mice displayed significantly more lipid droplets surrounding the nucleus, compared with the livers from TM-injected control Ire1αHepfe/+ mice (Figure 4D). Taken together, these observations indicate that IRE1α is required to minimize lipid accumulation in response to acute ER stress.
Figure 4.
Hepatocyte-specific Ire1α deletion leads to hepatosteatosis and reduced plasma lipids. Ire1αHepfe/− and Ire1αHepfe/+ mice at 3 months of age were injected with TM (2 μg/g body weight) or vehicle (150 μM dextrose). At 8, 24, and 72 h after TM injection, liver tissues and plasma samples were isolated for lipid analysis. (A–C) Oil-red O staining of lipid droplets in the livers of Ire1αHepfe/− and Ire1αHepfe/+ mice after TM challenge for indicated time periods (magnification × 200). (D) Transmission electron micrographs of liver tissue sections from Ire1αHepfe/− and Ire1αHepfe/+ mice at 24 h after TM injection (magnification: × 3500). (E) Levels of plasma lipids in the Ire1αHepfe/− and Ire1αHepfe/+ mice at 8 h after injection with TM or vehicle. Chol, total plasma cholesterol; TG, triglycerides; HDL, high-density lipoproteins; (V) LDL, low and very low-density lipoproteins. Each bar denotes the mean±s.e.m. (n=6 mice per group); *P<0.05. P-values are shown for statistically significant differences. (F) Levels of liver cellular lipids in the Ire1αHepfe/− and Ire1αHepfe/+ mice at 8 h after injection with TM or vehicle. CE, cholesterol ester; TG, triglycerides; FA, fatty acids; Chol, total liver cholesterol. Each bar denotes the mean±s.e.m. (n=3 mice per group); *P<0.05.
Analysis of plasma lipid profiles demonstrated that the levels of low-density lipoproteins (LDLs) and TG were reduced by 28–55% in Ire1αHepfe/− mice, compared with that in control Ire1αHepfe/+ mice (Figure 4E). The reduction in plasma lipid contents became more profound in Ire1αHepfe/− mice at 8 h after TM injection compared with control mice (Figure 4E). Analysis of the major lipid classes in the liver demonstrated that hepatocyte-specific Ire1α deletion increased cellular TG levels modestly in the absence of TM, and more significantly at 8 h after TM injection (Figure 4F). At 24 h after TM injection, the liver TG content in Ire1αHepfe/− mice continued to increase where TG levels were approximately six times greater than those observed in the livers from control Ire1αHepfe/+ mice injected with TM (Supplementary Figure S2A). In contrast to TG, the levels of other neutral lipids, including cholesterol esters (CE), FAs, and cholesterol, were not significantly different between the livers of Ire1αHepfe/− and Ire1αHepfe/+ mice, even after TM injection (Figure 4F). Additionally, the liver content of phospholipids, including phosphatidylethanolamine (PtdEtn), phosphatidylcholine (PtdCho), and sphingomyelin, were not significantly affected by Ire1α deletion (Supplementary Figure S2B and C). Consistent with this observation, expression levels of phoshocholine cytidylyltransferase α (CCTα), the rate-limiting enzyme in PtdCho biosynthesis, were comparable in the livers from Ire1αHepfe/− and Ire1αHepfe/+ mice (Supplementary Figure S2D). These findings suggest that the increased liver TG accumulation observed upon Ire1α deletion in hepatocytes was not due to any alteration in phospholipid synthesis.
IRE1α represses expression of lipogenic transcriptional activators upon ER stress
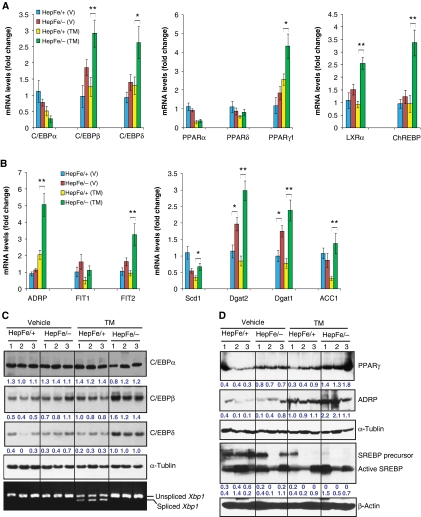
Hepatic lipid accumulation may be caused by different metabolic perturbations: increased FA delivery, increased de novo lipogenesis, decreased FA oxidation, or inadequate TG secretion as VLDL (Musso et al, 2009). To understand the molecular basis underlying the hepatic steatosis in Ire1α-null livers, we analysed expression of genes that regulate lipid biosynthesis, transport, storage, oxidation, as well as lipid droplet formation. Through quantitative gene expression analyses, we found that Ire1α deletion in hepatocytes increased expression of three classes of lipogenic regulators or enzymes that are essential for hepatic lipid deposition: (1) transcription factors of the C/EBP family, peroxisome proliferator-activated receptor γ (PPARγ), and their downstream targets; (2) adipose differentiation-related protein (ADRP) and ER-associated fat-inducing transcripts (FITs); and (3) key enzymes required for TG biosynthesis (Figure 5).
Figure 5.
IRE1α represses expression of key lipogenic regulators in the liver in response to ER stress. (A, B) Quantitative real-time RT–PCR analysis of liver mRNA in Ire1αHepfe/− and Ire1αHepfe/+ mice. Total RNAs from the livers of Ire1αHepfe/− and Ire1αHepfe/+ mice at 3 months of age at 8 h after injection with TM (2 μg/g body weight) or vehicle (V) were subjected to quantitative real-time RT–PCR analysis. Expression values were normalized to β-actin mRNA. Fold changes are shown relative to mRNA expression in one of the control (Ire1αHepfe/+) untreated mouse livers. Each bar denotes the mean±s.e.m. (n=6 mice per group); *P<0.05; **P<0.01. P-values are shown for statistically significant differences. (C, D) Western blot analysis of liver tissue from Ire1αHepfe/− and Ire1αHepfe/+ mice at 8 h after injection with TM (2 μg/g body weight) or vehicle. Semiquantitative RT–PCR analysis was performed to measure the levels of spliced and unspliced Xbp1 mRNAs as an indicator of IRE1α activity in the livers (C, bottom panel). In western blot analysis of C/EBPβ, we detected the C/EBPβ LAP (liver-enriched transcriptional activator protein) isoform of about 35 kDa in the mouse liver tissue samples (Calkhoven et al, 2000). Because PPARγ2 was not detected at the mRNA level in the liver tissue in the absence or presence of TM challenge (A, B), the PPARγ signals from the western blot analysis likely reflected PPARγ1. The experiments were repeated at least three times with consistent results, and representative data are shown. The values below the gels represent protein signal intensities after normalization to that of α-tubulin.
Originally identified as the master regulator of adipogenesis, the nuclear receptor PPARγ has also been implicated in hepatic steastosis (Gavrilova et al, 2003; Schadinger et al, 2005; Tontonoz and Spiegelman, 2008). The PPARγ gene encodes two isoforms, PPARγ1 and PPARγ2, that are differentially expressed depending on the cell type and stress conditions (Fajas et al, 1997; Vidal-Puig et al, 1997; Dubuquoy et al, 2002; Yamazaki et al, 2010). Expression of the Pparγ2 mRNA in the liver tissues of Ire1α-null and control mice was barely detectable, either under non-stressed conditions or after TM treatment (Supplementary Figure S3). In comparison, the levels of Pparγ1 mRNA were slightly increased in the livers from Ire1αHepfe/− mice in the absence of TM challenge, and this difference became statistically significant after TM injection (Figure 5A). Additionally, other PPAR isoforms, including PPARα and PPARδ, were not significantly affected by hepatocyte-specific Ire1α deletion (Figure 5A). These results suggest that PPARγ1 is the major PPAR isoform that contributes to TM-induced hepatic steatosis. C/EBPβ and C/EBPδ act upstream of PPARγ by directly binding to the PPARγ promoter to induce its transcription (Wu et al, 1995, 1996; Tontonoz and Spiegelman, 2008). PPARγ can induce C/EBPα expression, and C/EBPα can itself bind to the C/EBP site in the Pparγ promoter, providing for a stable, self-reinforcing regulatory loop. Compared with livers from control Ire1αHepfe/+ mice, the livers from Ire1αHepfe/− mice displayed increased expression of the C/ebpβ and C/ebpδ mRNAs, and this increase became statistically significant after TM treatment (Figure 5A). However, expression levels of C/ebpα mRNA were modestly decreased in the Ire1α-null livers, compared with that in the control livers, after TM treatment for 8 h (Figure 5A). This is consistent with the previous demonstration that ER stress downregulates C/EBPα expression (Rutkowski et al, 2008; Sha et al, 2009). Furthermore, expression levels of the mRNA encoding liver X receptor α (LXRα) and carbohydrate responsive element binding protein (ChREBP) were increased in livers from Ire1αHepfe/− mice (Figure 5A), compared with that in the control livers from Ire1αHepfe/+ mice. LXRα and ChREBP are involved in the PPARγ-regulated cascade and have important roles in the induction of lipogenesis and hepatic steatosis (Tontonoz and Spiegelman, 2008; Musso et al, 2009). In addition to the upregulation of C/EBPβ, C/EBPδ, and PPARγ, hepatocyte-specific Ire1α deletion also increased expression of key factors required for hepatic lipid droplet formation, including ADRP and fat-inducing transcript 2 (FIT2) after TM injection, and other PPARγ target lipogenic genes, including aP2, Fat/Cd36, and Fsp27, in the liver tissue in response to TM treatment (Jiang and Serrero, 1992; Brasaemle et al, 1997; Kadereit et al, 2008; Matsusue et al, 2008; Musso et al, 2009) (Figure 5B and D; Supplementary Figure S4). Moreover, we also examined expression of other lipogenic genes upon hepatocyte-specific Ire1α deletion. Expression of the mRNAs encoding key enzymes required for de novo hepatic TG biosynthesis, including stearyl coenzyme A (CoA) desaturase 1 (SCD1), diacylglycerol acetyltransferase 1 (DGAT1), DGAT2, and acetyl CoA carboxylase 1, was significantly increased in the Ire1α-null livers after TM treatment (Figure 5B), which is consistent with the increased hepatic lipid accumulation in the Ire1α-null livers after TM treatment. Although transcriptional induction of the genes encoding sterol regulatory element binding protein 1c (SREBP1c), FA synthase (FASN), PPARα, PPARδ, PGC-1α, and PGC-1β was suppressed by TM treatment, there was no significant difference in the expression of these genes between Ire1αHepfe/− and Ire1αHepfe/+ mice, either in the presence or absence of TM injection (Figure 5B and D; Supplementary Figure S5). The gene expression correlated with the lipogenic phenotype in TM-induced hepatic steatosis, as the protein levels of C/EBPβ, C/EBPδ, PPARγ, and ADRP were also increased in the Ire1αHepfe/− liver, compared with the control, in response to TM challenge (Figure 5A–D). Interestingly, in response to TM treatment, levels of SREBP1c precursor, but not its cleaved and activated form, were decreased in both Ire1α-null and control livers, while the levels of cleaved/active SREBP1c were slightly increased in the Ire1α-null liver after TM treatment (Figure 5D). Apparently, under conditions of TM treatment, reduced expression of the Srebp1c mRNA reflected levels of the SREBP1c precursor, but not its mature/functional form (Supplementary Figure S5; Figure 5D). The inconsistency between transcriptional induction of specific lipogenic genes and TM-induced lipogenesis phenotype was further evidenced by induction of the genes involved in FA oxidation or uptake. Expression of the genes involved in FA oxidation, including Acox1, Cpt1α, and Cyp4a10, and the genes involved in FA uptake/transport, including Fatp2, Fat/Cd36, Fabp1, and Fabp4 (aP2), was modestly increased in the Ire1αHepfe/− liver, compared with the control, after TM treatment (Musso et al, 2009) (Supplementary Figure S4). However, our measurement of radiolabelled acetate incorporation suggested no significant difference in FA oxidation or uptake between the Ire1α-null and control primary cultured hepatocytes (data not shown), suggesting that feedback regulation may be involved in the induction of specific lipid-associated genes in TM-induced hepatic steatosis models.
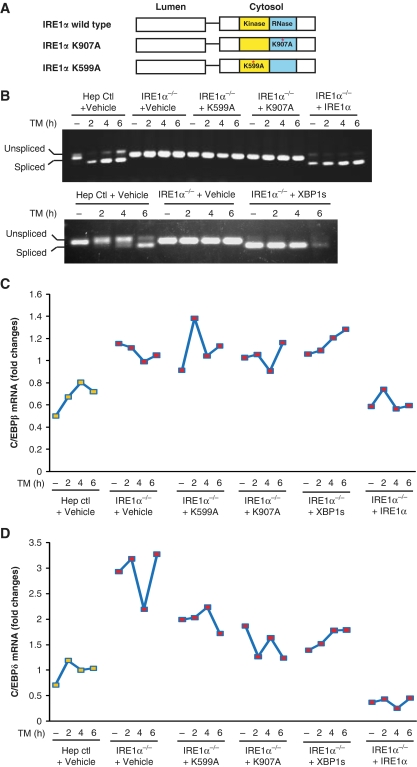
Interpretation of studies from gene-deleted organisms is always compromised by adaptive responses that may occur. To test whether IRE1α is directly involved in regulating expression of C/EBP family transcription factors under conditions of acute ER stress, we overexpressed wild-type or mutant forms of IRE1α in Ire1α-null hepatocytes. Upon accumulation of unfolded protein in the ER, IRE1α is activated through trans-auto-phosphorylation at Ser/Thr residues to elicit its endoribonuclease (RNase) activity (Schroder and Kaufman, 2005). Previously, we demonstrated that Lys to Ala mutations that destroy IRE1α kinase activity (K599A) or RNase activity (K907A) decrease Xbp1 mRNA splicing, as well as expression of downstream UPR target genes (Figure 6A) (Tirasophon et al, 1998; Zhang et al, 2005). To understand how IRE1α regulates expression of the lipogenic trans-activators C/EBPβ and C/EBPδ, we expressed wild-type IRE1α, kinase mutant IRE1α (K599A), RNase mutant (K907A) IRE1α, or spliced XBP1 by adenovirus delivery to immortalized Ire1α-null hepatocytes. To verify adenovirus-mediated expression, we examined Xbp1 mRNA splicing in the hepatocytes infected with these adenoviruses. In response to TM treatment, spliced Xbp1 mRNA was not detected in the mock-infected Ire1α-null hepatocytes or the hepatocytes infected with adenoviruses that express K599A or K907A IRE1α mutant. In contrast, spliced Xbp1 mRNA was detected in control hepatocytes (Ire1αfe/fe) that harbour two floxed Ire1α alleles and Ire1α-null hepatocytes (Ire1αfe/−Cre) infected with adenovirus that expresses wild-type IRE1α or spliced Xbp1 (Figure 6B). Analysis of expression of C/EBP family members demonstrated that Ire1α-null hepatocytes express higher levels of C/ebpβ and C/ebpδ mRNAs than the control hepatocytes (Figure 6C and D), consistent with the in vivo results (Figure 5A). Expression of the K599A or K907A IRE1α mutant in the Ire1α-null hepatocytes did not reduce C/ebpβ mRNA expression and only slightly reduced C/ebpδ mRNA expression in response to TM treatment (Figure 6C and D). In contrast, expression of wild-type IRE1α in the Ire1α-null hepatocytes reduced both C/ebpβ and C/ebpδ mRNA expression to the levels similar to those observed in the control hepatocytes (Figure 6C and D), suggesting a direct role for IRE1α in modulating expression of C/ebpβ and C/ebpδ mRNAs in hepatocytes. Moreover, overexpression of the product of IRE1α RNase, spliced XBP1, failed to decrease levels of C/ebpβ and C/ebpδ mRNAs in the Ire1α-null hepatocytes (Figure 6C and D). Taken together, these data suggest that IRE1α can decrease C/ebpβ and C/ebpδ mRNA levels in response to ER stress through mechanisms involving its kinase and RNase activities. However, overexpression of spliced Xbp1 mRNA, the target of IRE1α RNase activity, did not reduce C/ebpβ and C/ebpδ mRNA levels in the Ire1α-null hepatocytes (Figure 6C and D). This suggests that IRE1α may downregulate C/EBPβ and C/EBPδ expression through alternative mechanisms rather than splicing of Xbp1 mRNA under conditions of ER stress.
Figure 6.
Overexpression of IRE1α suppresses upregulation of CEBPβ and CEBPδ in Ire1α-null hepatocytes. (A) Depiction of domain structures for wild-type and mutant versions of IRE1α protein. K599A, IRE1α kinase mutant; K907A, IRE1α RNase mutant. Ire1α-null or control hepatocytes were infected with recombinant adenoviruses expressing different versions of IRE1α, spliced Xbp1 mRNA, or empty vector control. The infected hepatocytes were then treated with TM (2 μg/ml) for the times indicated. (B) Semiquantitative RT–PCR analysis of spliced and unspliced Xbp1 mRNAs in the Ire1α-null (Ire1αfe/−CRE) or control (Ire1αfe/fe) hepatocytes. (C, D) Quantitative real-time RT–PCR analysis of C/ebpβ and C/ebpδ mRNAs in adenovirus-infected hepatocytes. At 48 h after infection of Ire1α-null or control hepatocytes with the indicated adenoviruses, the hepatocytes were treated with TM (5 μg/ml) for the times indicated. Expression values of C/ebpβ and C/ebpδ mRNAs were normalized to β-actin mRNA. Fold changes of mRNA were measured by comparing to the expression level of mRNA in one of the empty virus-transfected control cells. Hep ctl, control (Ire1αfe/fe) hepatocytes; Ire1α−/−, Ire1α-null (Ire1αfe/−CRE) hepatocytes. A full-colour version of this figure is available at The EMBO Journal Online.
IRE1α is required for efficient secretion of plasma lipids
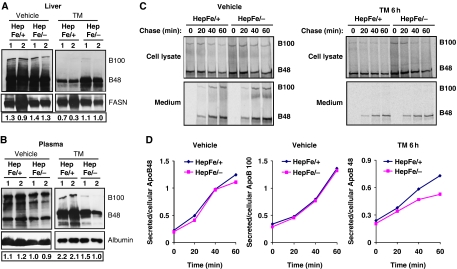
Hepatocytes secrete apolipoprotein B (ApoB)-containing lipoprotein particles (VLDL and LDL) that transport cholesterol and FAs in the form of TGs to peripheral tissues (Shelness and Sellers, 2001; Musso et al, 2009). Because Ire1αHepfe/− mice exhibit decreased plasma lipids in the absence or presence of ER stress compared with control mice (Figure 4E and F), we asked whether IRE1α is required for efficient secretion of lipoproteins into the plasma. To evaluate the involvement of IRE1α in the secretion of apolipoproteins, we first examined levels of ApoB in the liver and plasma samples from Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice in the presence or absence of TM treatment. Under the non-stressed condition, disruption of IRE1α did not significantly affect the levels of two ApoB isoforms, ApoB100 and ApoB48, in murine liver and plasma samples (Figure 7A and B). At 24 h after TM injection, more hepatic ApoB was present while less plasma ApoB was detected in Ire1α-null mice, compared with that in the control mice (Figure 7A and B). This observation suggests that IRE1α may be required for secretion of ApoB-containing lipoproteins under conditions of ER stress. To confirm this hypothesis, we analysed secretion rates of ApoB in primary hepatocytes isolated from Ire1α-null and control mice by pulse-label and pulse-chase experiments. ApoB was immunoprecipitated from cell lysates and media samples and analysed by SDS–polyacrylamide gel electrophoresis followed by fluorography. The pulse-chase experiments indicated that the secretion rates of both ApoB100 and ApoB48 were comparable between Ire1α-null and control hepatocytes under the non-stressed condition (Figure 7C and D). However, after 6 h of TM treatment, the secretion rates of ApoB48 were decreased in the Ire1α-null hepatocytes, compared with that in the control hepatocytes (Figure 7C and D). This observation is consistent with more dramatic reduction of plasma ApoB in Ire1α-null mice than the control mice when they were challenged with TM (Figure 7A and B). Moreover, in either Ire1α-null or control hepatocytes, we did not detect any significant levels of secreted ApoB100 in response to TM treatment, supporting the TM-mediated suppression of ApoB100 secretion suggested by western blot analysis with the Ire1α-null and the control mice (Figure 7A–D). Together, our observations suggest that IRE1α is required for hepatocytes to efficiently secrete ApoB-containing lipoproteins under conditions of ER stress.
Figure 7.
IRE1α is required for efficient secretion of ApoB-containing apolipoproteins. (A, B) Western blot analysis of ApoB in the liver and blood plasma samples from Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice at 24 h after injection with TM (2 μg/g body weight) or vehicle. Levels of fatty acid synthase in the liver tissues and albumin in the plasma were measured as internal controls. The values below the gels represent the normalized protein signal intensities. FASN, fatty acid synthase; Alb, albumin. (C) Primary hepatocytes from Ire1α-null (Ire1αfe/−) and control (Ire1αfe/+) mice were labelled with [35S]methionine/cysteine for 25 min and then chased for the indicated times. For TM-treated samples, the primary hepatocytes were treated with TM (10 μg/ml) for 6 h before and throughout the pulse-chase experiments. Radiolabelled ApoB-containing apolipoproteins were immunoprecipitated from the cell lysates and the media with a rabbit anti-ApoB polyclonal antibody, and resolved by 5% SDS–polyacrylamide gel electrophoresis with fluorography. The experiments were repeated at least three times with consistent results, and representative data are shown. (D) Quantification of secretion rates of ApoB48- or ApoB100-containing apolipoproteins from Ire1α-null (Ire1αfe/−) or control (Ire1αfe/+) primary hepatocytes to culture media in the absence or presence of TM treatment, as indicated in the panel (C). Secretion rates of ApoB=secreted ApoB48(100)/cellular ApoB48(100). Levels of secreted ApoB48(100) were quantified based on the signals of ApoB48(100) present in the culture media, and levels of cellular ApoB48(100) were quantified based on the signals of ApoB48(100) present in the cellular lysates of the Ire1α-null or control primary hepatocytes, as shown in the panel (C). A full-colour version of this figure is available at The EMBO Journal Online.
IRE1α protects from non-TM stress-induced hepatic steatosis
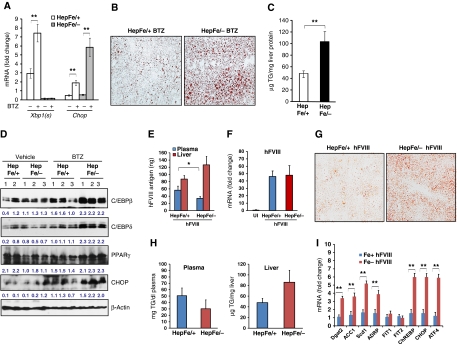
To further evaluate the role of IRE1α in preventing stress-induced hepatic steatosis in the absence of TM, we induced ER stress in the livers of Ire1α-null and control mice by alternative mechanisms. First, we treated the mice with the clinically relevant ER stress-inducing agent Bortezomib (also known as Velcade or PS-341). Bortezomib is a highly selective and reversible proteasome inhibitor that was approved for treatment of multiple myeloma and is in more extensive clinical trials for other solid tumour malignancies (Richardson et al, 2003, 2006). Although the mechanisms involved in its anti-cancer activity are still under investigation, Bortezomib was recently shown to activate the UPR and ER stress-associated apoptosis by inhibiting the 26S proteasome and ERAD (Lee et al, 2003a; Fribley et al, 2004; Nawrocki et al, 2005; Fels et al, 2008). To test whether Bortezomib can cause ER stress-associated hepatic steatosis and whether IRE1α has a protective role in Bortezomib-induced hepatic steatosis, we administered Bortezomib into Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice through tail-vein injection. Injection of Bortezomib induced ER stress in the liver, as evidenced by increased expression of spliced Xbp1 and Chop mRNAs in the liver of mice injected with Bortezomib (Figure 8A). Particularly, the expression of Chop mRNA in livers of the Ire1αHepfe/− mice was significantly greater than that of Ire1αHepfe/+ mice after Bortezomib treatment. Spliced Xbp1 mRNA in the liver from Ire1αHepfe/− mice was hardly detected (Figure 8A), reflecting the absence of IRE1α. At 36 h after injection of Bortezomib, hepatic lipid accumulation was observed in the livers of Ire1αHepfe/− and control Ire1αHepfe/+ mice (Figure 8B). However, hepatic lipid accumulation in the Ire1αHepfe/− mice was greater than that observed in the Ire1αHepfe/+ mice. Direct measurement of liver TG demonstrated that Bortezomib treatment increased TG accumulation in the Ire1αHepfe/− mice, compared with the Ire1αHepfe/+ mice (Figure 8C). The increase in TG accumulation correlated with increased expression of key lipogenic regulators, including C/EBPβ, C/EBPδ and PPARγ, in the livers of the Ire1αHepfe/− mice after Bortezomib treatment (Figure 8D). Together, these observations suggest that IRE1α is required to attenuate hepatic steatosis in response to ER stress.
Figure 8.
Mice with hepatocyte-specific Ire1α deletion exhibit severe hepatic steatosis in response to Bortezomib treatment or expression of a misfolding-prone hFVIII. (A–C) Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice at 3 months of age were injected with Bortezomib (1 μg/g body weight) into the tail-vein. At 36 h after injection, the mice were fasted for 8 h prior to euthanasia and collection of samples. (A) Quantitative real-time RT–PCR analysis for expression of Chop and spliced Xbp1 mRNAs in the liver. Expression values were normalized to β-actin mRNA. Fold changes of mRNA levels were determined by comparison to the expression level in one of the controls. Each bar denotes the mean±s.e.m. (n=6 mice per group); *P<0.05; **P<0.01. BTZ, Bortezomib. (B) Frozen liver tissue sections stained with Oil-red O for hepatic lipid contents after the Bortezomib treatment (magnification × 400). (C) Levels of intracellular liver triglycerides after the Bortezomib treatment. (D) Western blot analysis of C/EBPβ, C/EBPδ, PPARγ, and CHOP in the liver. Levels of β-actin protein were included as internal controls. The values below the gels represent normalized protein signal intensities. (E–I) Plasmid DNA vector (100 μg/mouse) expressing an hFVIII transgene encoding a misfolding-prone hFVIII (hFVIII) was transferred into the Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice at 3 months of age through hydrodynamic tail-vein injection. Liver tissues and blood plasma samples were collected at 24 h after the injection. (E) Levels of the hFVIII antigen in liver cellular lysates or plasma of Ire1α-null and control mice after the injection of the hFVIII expression vector. Each bar denotes the mean level of hFVIII antigen in plasma (ng/ml) or in liver tissue (ng/mg liver protein)±s.e.m. (n=3 mice per group). *P<0.05. (F) Quantitative real-time RT–PCR analysis for expression of hFVIII mRNA in the liver of Ire1α-null and control mice after the injection of the hFVIII expression vector. Expression values were normalized to β-actin mRNA. Fold changes of mRNA levels were determined by comparison to the expression level in the uninjected control. Each bar denotes the mean±s.e.m. (n=3 mice per group); **P<0.01. UI, uninjected control. (G) Frozen liver tissue sections stained with Oil-red O for hepatic lipid contents after the injection of the expression vector for hFVIII (magnification × 400). (H) Levels of hepatic and plasma triglycerides in the mice after the injection of hFVIII expression vector. (I) Quantitative real-time RT–PCR analysis for expression of lipogenic genes in the liver of Ire1α-null and control mice after the injection of the hFVIII expression vector. Expression values were normalized to β-actin mRNA. Fold changes of mRNA levels were determined by comparison to the expression level in one of the controls. Each bar denotes the mean±s.e.m. (n=3 mice per group); **P<0.01.
Hepatic lipid accumulation can cause lipotoxicity, represented by hepatocyte apoptosis, in the pathogenesis of fatty liver disease (Jou et al, 2008; Malhi and Gores, 2008). Consistent with this notion, increased expression of ER stress-induced pro-apoptotic factor CHOP was observed in the livers of the Ire1αHepfe/− mice (Figure 8A and D). TUNEL staining demonstrated that the percentage of apoptotic cells in the livers of Bortezomib-treated Ire1αHepfe/− mice was increased approximately five times compared with that of the Ire1αHepfe/+ mice (Supplementary Figure S6), suggesting that IRE1α has an important role in preventing ER stress-induced hepatic steatosis and apoptosis.
In addition to Bortezomib treatment, we utilized two pathophysiologically relevant stress models, expression of a human blood clotting factor VIII (hFVIII) that is prone to misfolding and partial hepatectomy, to test the involvement of IRE1α in stress-induced hepatic steatosis. Hydrodynamic DNA tail-vein injection of hFVIII expression vectors was used to direct expression of a misfolding-prone hFVIII in hepatocytes (Malhotra et al, 2008). In this study, the hFVIII is a B-domain-deleted variant of hFVIII that is secreted as a functional clotting factor. Analysis of this molecule is significant because it has previously been used in human haemophilia A gene therapy studies directing FVIII expression to the hepatocyte (Pierce et al, 2007). However, these approaches have not produced physiologically significant levels of hFVIII to date. Because newly synthesized hFVIII misfolds in the ER, the UPR is activated in mice that express hFVIII (Malhotra et al, 2008). Analysis of hFVIII secreted into the plasma demonstrated that Ire1α deletion reduces the secretion efficiency of hFVIII, with concomitant intracellular accumulation of hFVIII antigen in the liver (Figure 8E), although the mRNA expression for hFVIII was similar (Figure 8F). The decrease in hFVIII was selective as there was no difference in plasma albumin (Figure 7B). In support of the role of IRE1α in preventing ER stress-induced hepatic steatosis, FVIII expression caused greater lipid accumulation in Ire1α-null mice, compared with the control mice (Figure 8G). Moreover, Ire1α-null mice produced less plasma TGs but more hepatic TGs than the control mice after expression of hFVIII (Figure 8H). Additionally, expression of key lipogenic genes, including Dgat2, Acc1, Scd1, Adrp, and ChREBP, was increased in the Ire1α-null mice upon expression of hFVIII (Figure 8I).
The second pathophysiologic model we used to test the role of IRE1α in hepatic lipid homeostasis was partial hepatectomy, the surgical removal of liver (Fausto et al, 2006). In the clinic, partial hepatectomies are performed for liver transplantation or for surgical removal of carcinoma or hepatoma tumours (Fausto, 2001). After partial hepatectomy, the livers from Ire1α-null mice displayed profound steatosis, as reflected by accumulation of hepatic lipid droplets and increased hepatic TGs, compared with the control mice (Supplementary Figure S7A and B). Taken together, these additional non-TM pathophysiologic stress models further confirmed that IRE1α has a critical role in preventing stress-induced hepatic steatosis.
Discussion
Analysis of mice with hepatocyte-specific Ire1α deletion demonstrated that IRE1α is essential to maintain hepatic lipid homeostasis, especially under conditions of ER stress (Supplementary Figure S8). Our findings show that (1) IRE1α is required for the expression of genes that encode functions critical for oxidative protein folding, protein transport from the ER to the Golgi, and ERAD; (2) Ire1α deletion leads to selective activation of ER stress-induced pro-apoptotic transcription factors; (3) Ire1α deletion in hepatocytes causes severe hepatic steatosis and hypolipidemia after ER stress challenge; (4) IRE1α modulates lipogenesis by downregulating expression of C/EBPβ, C/EBPδ, PPARγ, and key enzymes for TG biosynthesis under conditions of ER stress; (5) IRE1α is required for efficient secretion of ApoB-containing lipoproteins; and (6) IRE1α protects the liver from hepatosteatosis in response to Bortezomib treatment, expression of the misfolding-prone hFVIII, or partial hepatectomy. These findings indicate that the most evolutionarily conserved UPR transducer IRE1α is required to repress lipid accumulation in the liver, especially when protein-folding homeostasis is disrupted in the ER.
Our studies demonstrate that deletion of Ire1α significantly alters ER function. This was reflected by defective induction of ER chaperones and enzymes that are required for protein oxidative folding, ER-to-Golgi trafficking, and ERAD (Figure 2). Importantly, Ire1α deletion led to selective upregulation of the UPR-regulated pro-apoptotic transcription factors ATF4, CHOP, and ATF3 in response to ER stress (Figure 3A). Upon ER stress, induction of ATF4, CHOP, and ATF3 is regulated by the PERK/eIF2α subpathway of the UPR (Scheuner et al, 2001; Harding et al, 2003; Wek et al, 2006). Interestingly, the Ire1α-null livers expressed much higher levels of ATF4, CHOP, and ATF3, compared with the control livers, although there was no significant activation of the PERK/eIF2α pathway at the later time points of ER stress treatment (Figure 3A–C). It is possible that elevated GADD34 expression promotes dephosphorylation of eIF2α and PERK after the longer periods of ER stress. At the later time points after TM treatment, the levels of ATF4, CHOP, and ATF3 proteins were diminished in control liver tissue and isolated hepatocytes, while their levels remained high in the Ire1α-null liver tissues and hepatocytes from 8 to 72 h after TM treatment (Figure 3A and E). Because ATF4, CHOP, and ATF3 are ER stress-induced pro-apoptotic factors, our data suggest that IRE1α is required for cells or tissues to adapt to ER stress. This was further confirmed by increased caspase-3 cleavage and positive TUNEL staining in the Ire1α-null livers after TM treatment (Figure 3A and D).
Our findings suggest that IRE1α protects from hepatosteatosis upon acute ER stress conditions through at least two regulatory mechanisms (Supplementary Figure S8): (1) IRE1α attenuates C/EBPβ, C/EBPδ, and PPARγ transcriptional control and (2) IRE1α is required for efficient secretion of apolipoprotein. PPARγ has two major isoforms, γ1 and γ2, that are generated from the same gene by alterative RNA splicing (Fajas et al, 1997; Vidal-Puig et al, 1997). PPARγ2 is highly expressed in adipose tissue and is upregulated in steatotic livers of ob/ob mice, while PPARγ1 is found at low levels in many tissues, including fat and liver tissues (Tontonoz et al, 1994; Dubuquoy et al, 2002; Schadinger et al, 2005; Zhang et al, 2006b; Musso et al, 2009; Yamazaki et al, 2010). It was proposed that both PPARγ1 and PPARγ2 contribute to formation of fatty liver, although PPARγ2 is more effective in activating the transcription of adipogenic and lipogenic genes than PPARγ1 (Vidal-Puig et al, 1997; Gavrilova et al, 2003; Matsusue et al, 2003; Zhang et al, 2004; Yamazaki et al, 2010). C/EBPβ and C/EBPδ induce transcription of PPARγ by directly binding to the PPARγ promoter (Wu et al, 1995, 1996; Tontonoz and Spiegelman, 2008). Our findings indicate that IRE1α is required to repress expression of C/EBPβ, C/EBPδ, and PPARγ1, but not PPARγ2, in hepatocytes in response to acute ER stress imposed by TM treatment. In the absence of IRE1α, ER stress significantly increased expression levels of C/EBPβ, C/EBPδ, and PPARγ1 in the liver (Figure 5). Interestingly, our data suggest that overexpression of wild-type IRE1α, but not spliced XBP1, reduced expression of C/ebpβ and C/ebpδ mRNAs in the Ire1α-null hepatocytes in response to ER stress (Figure 6C and D). Therefore, it is possible that IRE1α prevents ER stress-induced hepatic steatosis through transcriptional suppression or degradation of the C/ebpβ and C/ebpδ mRNAs upon ER stress. A recent study showed that C/EBPβ can induce expression of the Xbp1 mRNA and that spliced XBP1 directly induces expression of C/EBPα during adipogenesis (Sha et al, 2009). Consistent with a role of spliced XBP1 in expression of C/EBPα, expression of the C/ebpα mRNA was decreased in the Ire1α-null livers in the presence or absence of TM treatment (Figure 5A). Based on these observations, it is also possible that the upregulation of C/EBPβ and C/EBPδ in the Ire1α-null liver results from the feedback regulation caused by decreased expression of C/EBP1α. If that is the case, there may exist a regulatory loop in which IRE1α modulates expression of C/EBP family transcription factors under conditions of ER stress. Future studies are required to explore the mechanism by which IRE1α regulates expression of the mRNAs encoding the C/EBP transcription factor family upon ER stress.
In the absence of TM challenge, Ire1α-null mice displayed mild hepatic steatosis and decreased plasma lipids (Figure 4A, E, and F). Based on the microarray analysis, the expression of genes involved in the endomembrane protein secretion machinery, including Sec61α1, Sec24d, and Tmed3, was slightly decreased in the Ire1α-null liver under the non-stressed condition (Table I). This may lead to accumulation of hepatic lipids intracellularly, a possible cause of mild hepatic steatosis in the Ire1α-null liver. Moreover, slight increase in expression of lipogenic genes encoding C/EBPβ, C/EBPδ, PPARγ1, LXRα, ChREBP, ADRP, FIT1, FIT2, DGAT1, and DGAT2 was observed in the liver of Ire1α-null mice under the non-TM-treated condition (Figure 5A and B), which may partially account for the mild steatosis in the Ire1α-null livers under the basal condition.
Our results show that upregulation of hepatic C/EBPβ, C/EBPδ, and PPARγ1 in the absence of IRE1α correlates with increased expression of the ChREBP, Lxrα, Adrp, Fit2, Fsp27, aP2, and Fat/Cd36 genes, the targets of PPARγ and/or C/EBPβ in lipogenesis and lipid storage, in response to ER stress (Figures 5A and 8I). Consistent with the upregulation of these lipogenic trans-activators, expression of the genes encoding key enzymes required for TG biosynthesis and/or lipid droplet formation, including DGAT1, DGAT2, and SCD1, was significantly increased in the Ire1α-null livers after TM treatment (Figure 5B). Interestingly, a recent study showed that XBP1 positively regulates expression of the Scd1, Dgat2, and Acc2 mRNAs in the livers of mice fed a high-fructose diet (Lee et al, 2008). However, the requirement of IRE1α-mediated Xbp1 mRNA splicing for TG synthesis was not characterized in the study with conditional Xbp1-null mice. Alternatively, it is possible that IRE1α has different roles in regulating de novo lipogenesis under conditions of acute ER stress versus chronic metabolic stress. This raises an interesting possibility that hepatocytes may utilize the same ER stress sensor to achieve different outcomes in lipogenesis depending on the type and/or degree of the stress.
Assembly and secretion of ApoB-containing lipoproteins from hepatocytes are crucial to maintain hepatic lipid homeostasis (Musso et al, 2009; Rutledge et al, 2010). Previous studies implicated that ER stress and UPR components are critically involved in ApoB protein biosynthesis, stability, and secretion (Ota et al, 2008; Qiu et al, 2009; Rutledge et al, 2009). Our study showed that IRE1α is required for efficient secretion of ApoB-containing lipoproteins from hepatocytes to peripheral tissues (Figure 7). Secretion rates of ApoB species in the primary hepatocytes from the Ire1α-null and control mice were comparable under non-stressed conditions, although levels of ApoB48 in Ire1α-null primary hepatocytes were slightly higher than that in the control hepatocytes (Figure 7C and D). In response to TM treatment, the secretion rates of ApoB proteins were more greatly decreased in the Ire1α-null hepatocytes, compared with that in the control hepatocytes, which may partially account for the hepatic steatosis and hypolipidemia in the hepatocyte-specific Ire1α-null mice. Note that TM treatment suppressed the production of the ApoB100 species in both cultured primary hepatocytes and in mice in vivo. The mechanism by which TM suppresses production of ApoB100, but not ApoB48, requires further elucidated.
Hepatic lipid accumulation can be caused by an imbalance of any of the pathways involved in FA and TG synthesis, export, uptake, and/or oxidation. We suspect that the accumulation of liver TG in the hepatocyte-specific Ire1α-null mice might be due to metabolic alterations. For example, excess diacylglycerol can be converted to TG (Leonardi et al, 2009). Overexpression of spliced form of XBP1 was previously associated with increased level and activity of CCT, a rate-limiting enzyme in the biosynthesis of PtdCho (Sriburi et al, 2004, 2007). However, Ire1α deletion in hepatocytes did not significantly reduce production of phospholipids, including PtdCho and PtdEtn, in the presence or absence of ER stress (Supplementary Figure S2). Therefore, accumulation of hepatic TGs in Ire1α-null livers is unlikely due to impaired phospholipid metabolism. Moreover, in addition to the increased lipogenesis and defective apolipoprotein secretion in the Ire1α-null livers that we have identified, the accumulation of TGs and lipid droplets in the absence of IRE1α may result from abnormal lipid droplet assembly that occurs in the ER (Cases et al, 1998, 2001; Wolins et al, 2006). For future studies, it will be important to determine whether any ER-associated enzymes involved in lipid droplet formation are regulated by IRE1α.
Bortezomib is the only FDA-approved proteasome inhibitor that is used to treat patients with multiple myeloma (Richardson et al, 2003, 2006). Although Bortezomib was originally developed based on its ability to prevent activation of NFκB, recent studies suggest that Bortezomib causes ER stress, hepatosteatosis, and hepatoxicity (Lee et al, 2003a; Rosinol et al, 2005; Fels et al, 2008; Hernandez-Espinosa et al, 2008; Rutkowski et al, 2008). Our studies show that hepatocyte-specific Ire1α-null mice develop severe hepatic steatosis and apoptosis after Bortezomib treatment. Intravenous injection of Bortezomib caused ER stress in the liver, and IRE1α is required to attenuate Bortezomib-induced hepatoteatosis and apoptosis (Figure 8). Therefore, the regulation of hepatic lipid homeostasis by IRE1α may exist under general conditions of acute ER stress, not only TM treatment. In addition, two additional non-TM pathophysiologic stress models, in vivo expression of a misfolding-prone hFVIII and partial hepatectomy, also demonstrated that IRE1α is required to prevent ER stress-induced hepatosteatosis (Figure 8E–I; Supplementary Figure S7). Taken together, our findings provide novel insights into hepatic lipid homeostasis that requires the UPR transducer IRE1α. Therefore, interventions to modulate IRE1α-mediated signalling may provide a therapeutic avenue to treat diseases associated with hepatosteatosis and hyperlipidemia.
Materials and methods
Materials
Chemicals were purchased from Sigma unless indicated otherwise. Synthetic oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Antibodies against phosphorylated eIF2α, phosphorylated PERK, and phosphorylated JNK proteins were purchased from Cell Signaling Technologies, Inc. (Danvers, MA). Antibodies against ATF4, CHOP, ATF3, GADD34, C/EBPα, C/EBPβ, C/EBPδ, and PPARγ were purchased from Santa Cruz Biotechnologies, Inc. (Santa Cruz, CA). Antibody against ATF6 was purchased from Imgenex Corp (San diego, CA). Antibodies against GRP78/BiP and GRP94 were purchased from Stressgen (Ann Arbor, MI). Antibody against α-tubulin was purchased from Sigma. Antibody against SREBP1c was purchased from Thermo Scientific (Rockford, IL). Bortezomib reagent of clinical quality was kindly provided by Dr Joanne Wang at the Wayne State University School of Medicine.
Generation of IRE1α conditional knockout mice
A targeting vector for conditional deletion that spans exons 16 and 17 of the Ire1α gene was constructed in pFlox vector (Figure 1A). The region for deletion was 0.65 kb sequence that spans exons 16–17 with flanking intron sequence of the Ire1α gene. After production of chimeric mice and F1 progeny from selected targeted clones, ZP3-Cre-recombinase expression in vivo was utilized to select progeny with deletion of neomycin (Neo) gene cassette flanked by loxP sites, but not the loxP-flanked Ire1α gene sequence. This yielded a strain of mice with a loxP-flanked Ire1α gene allele (Ire1αfe/fe) with apparently undisrupted expression of the Ire1α gene. This strain of mice allows deletion of the functional Ire1α gene allele through crossing with transgenic mice expressing temporally controlled or tissue-specific Cre recombinase. To increase the deletion efficiency for the Ire1α gene allele, we cross the Ire1αfe/fe mice with previously established heterozygous Ire1α-null (Ire1α+/−) mice (Lee et al, 2002) to generate a mouse strain with a loxP-flanked Ire1α allele and an Ire1α-null allele (Ire1αfe/−) (Figure 1A). Three sets of PCR amplifications were used for genotyping of Ire1α conditional null mice: (1) for PCR amplification of the Ire1α floxed and wild-type alleles, we used primer sequences: 5′-CAGAGATGCTGAGTGAAGAC-3′, and 5′-ACAGTGGTTCCTGTGAAGGT-3′. The floxed allele generates a 241-bp band and the wild-type allele produces a 377-bp fragment; (2) for PCR amplification of Neo gene cassette to identify the Ire1α-null allele, we used primer sequences as previously described (Lee et al, 2002); (3) for PCR amplification of the Cre transgene we used primer sequences: 5′-GCGGTCTGGCAGTAAAAACTATC-3′, and 5′-GTGAAACAGCATTGCTGT CAC TT-3′.
GeneChip microarray and bioinformatics analysis
IRE1α conditional null (Ire1αHepfe/−) or the control (Ire1αHepfe/+) mice of 3 months old were injected intraperitoneally with TM (2 μg/g body weight) or vehicle (150 μM dextrose), and liver tissue was isolated at 8 h after the injection. Total RNA was purified from liver tissues and subjected to Affymetrix microarray analysis. For each microarray replicate, 10 μg RNA was fragmented and hybridized to Murine Genome 430.2 gene chips following Affymetrix instructions. The microarray data were analysed using several software packages. D-Chip (2008 Version) (http://www.dchip.org) was used to normalize the data set (Li and Wong, 2001). The median intensity from all unnormalized array chips varies from 93 to 179 with P call between 52.5–58.5%. The original signals were scaled towards the median intensity array in the group at the probe intensity level. Normalization was performed using a normalization curve with invariant probe sets. GeneSpring GX (Silicon Genetics, Redwood City, CA) was used to identify differentially expressed genes and hierarchical clustering. ANOVA was used to analyse the difference. The P-value cutoff was 0.05. Since there are 45 101 probes in the Affymetrix Mouse 430.2 chip, the Benjamini and Hochberg false discovery rate was used for multiple testing corrections. All the significant genes between two groups were filtered with expression level based on 1.5-fold changes. DAVID 2008 version software was used to functionally analyse the differentially expressed gene annotations and their related biological pathways (Dennis et al, 2003).
Transmission electron microscopy
Liver tissues were fixed with 1.25% formaldehyde, 2.5% glutaraldehyde, 0.03% picric acid in 100 mM sodium cacodylate buffer. After washing with 100 mM sodium cacodylate buffer, tissues were treated for 1 h with 1% osmium tetroxide and 1.5% potassium ferrocyanide, and then 30 min with 0.5% uranyl acetate in 50 mM maleate buffer, pH 5.2. After dehydration in ethanol, tissues were treated for 1 h in propylenoxide and then embedded in Epon/Araldite resin. Ultrathin sections were collected on EM grids and observed by using a JEOL 1200EX transmission electron microscope at an operating voltage of 60 kV.
Mouse plasma lipid profiling
Mice were fasted for 8 h prior to euthanization and collection of blood samples. Levels of plasma lipids in the mice were determined enzymatically using commercial kits (Roche Diagnostics Corporation). Approximately 120 μl of mouse blood plasma was collected and subjected to analyses using commercial kits for total cholesterol (Roche cat #: 3 313 018), HDL (Roche cat #: 3 034 569), and Triglyceride (Roche cat #: 3 034 658) (Allain et al, 1974; Stein, 1994). Levels of plasma LDL were calculated based on the formula: LDL=total cholesterol–HDL–(TGs/5).
Measurement of liver cellular lipids
Lipids were extracted from ∼50 mg of liver using a modification of the Bligh and Dyer procedure as previously described (Leonardi et al, 2009). Mice were fasted for 8 h prior to euthanization and sample collection. The amount of each major lipid class was measured by flame-ionization thin layer chromatography. TGs, cholesterol, and CEs were separated using hexane:ether (90/10, v/v). Phospholipids were resolved using chloroform:methanol:acetic acid:water (50/25/8/2, v/v/v/v). The lipids were identified by co-migration with authentic standards and quantified by comparison with known amounts of standard lipids. Additionally, measurement of levels of liver cellular TGs was performed using a TG assay kit according to the manufacturer's instructions (BioAssay Systems, ETGA-200). Briefly, ∼100 mg of fresh liver was homogenized in 500 μl PBS and then centrifuged to remove the tissue debris. The supernatant, including the fatty debris on top, was transferred into a new tube, and then add another 500 μl of 10% Triton-X100 in PBS was added for TG measurement using the kit.
Generation of immortalized Ire1α-null hepatocytes and recombinant adenoviral infection
Primary murine hepatocytes were isolated from mice harbouring floxed homozygous Ire1α alleles using a standard protocol (Berry et al, 1991), and were immortalized by SV40 T antigen. Immortalized hepatocytes were treated with adenovirus-Cre in order to delete the floxed Ire1α exons. Recombinant adenoviruses for the overexpression of flag-tagged IRE1α (wild type and its mutant forms K599A and K907A) (Qiu et al, 2010) were kindly provided by Dr Yong Liu (Institute for Nutritional Sciences, Shanghai, China) and amplified using the AdEasy System (Stratagene). Adenovirus expressing spliced XBP1 was kindly provided by Dr Umut Ozcan (Harvard University) (Park et al, 2010). Recombinant adenovirus expressing GFP was kindly provided by Dr Jiande Lin (University of Michigan). The Ire1α-null and control hepatocytes at about 60% confluency were infected with adenoviruses expressing IRE1α wild-type, its mutant forms, or spliced XBP1 at an MOI of 100 for 48 h before they were treated with TM at the indicated concentrations.
ApoB western blot analysis with mouse liver and plasma samples and ApoB pulse-chase experiments with mouse primary hepatocytes
Liver tissue and blood plasma samples were isolated from Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) mice in the absence or presence of TM treatment (2 μg/g body weight) for 8 h. To prepare the samples for ApoB western blot analysis, the liver tissue samples were diluted three times and the plasma samples were diluted five times before they were denatured. The denatured samples were resolved by 5% SDS–polyacrylamide gel electrophoresis, transferred onto 0.45 μm PVDF membranes, and followed by the incubation with a rabbit anti-ApoB polyclonal antibody (a kind gift from Dr Zhouji Chen, Washington University). For the pulse-chase experiments with mouse primary hepatocytes, livers from both Ire1α-null (Ire1αHepfe/−) and control (Ire1αHepfe/+) were first perfused by HBSS supplemented with 8 mM HEPES (pH 7.35) and 1 mM sodium pyruvate followed by collagenase digestion. Hepatocytes were pelleted by centrifugation at 50 g for 2 min, washed three times with DMEM media, and then seeded in the collagen-coated plates with Williams E media. Cells were attached in the plates for 48 h before the pulse-chase experiments. Hepatocytes were labelled with [35S]methionine/cysteine (140 μCi/ml) for 25 min and chased at the indicated times. For TM-treated samples, the cells were treated with TM (10 μg/ml) for 6 h before and throughout the pulse-chase experiments. Labelled ApoB apolipoprotein complexes were immunoprecipitated from the cell lysates and the media with the anti-ApoB antibody, resolved by 5% SDS–PAGE gel, and exposed to a phosphoimager screen.
Oil-red O staining
Frozen liver tissue sections were stained with Oil-red O for lipid content according to the standard protocol. Briefly, frozen liver tissue sections of 8 μm were air dried, and then fixed in formalin. The fixed sections were rinsed with 60% isopropanol before they were stained with freshly prepared Oil-red O solution for 15 min. After Oil-red O staining, liver sections were rinsed again with 60% isopropanol. Nuclei were lightly stained with alum haematoxylin 5.
TUNEL for apoptosis
To visualize the apoptotic events in the liver tissue sections of Ire1α-null and control mice, we used a fluorescent TUNEL staining kit (Clontech) to detect DNA fragmentation with the paraffin-embedded tissue section slides according to the manufacture's instruction (Zhang and Kaufman, 2008b). The apoptotic cells exhibited green fluorescence using a standard fluorescein filter set (520±20 nm). The cells were also stained with propidium iodide (PI), to identify all cells by strong red cytoplasmic fluorescence. The percentage of apoptotic cells was determined by calculating the number of TUNEL-positive cells divided by the number of PI-positive cells from five fields of each slide.
Hydrodynamic tail-vein injection and quantification of hFVIII antigen
We expressed a misfolding-prone hFVIII (B domain-deleted of human FVIII) that has a secretion defect like the wild-type hFVIII in murine liver using hydrodynamic tail-vein injection (Malhotra et al, 2008). Plasmid DNA samples (100 μg) were diluted in 2.5 ml lactated Ringer buffer and infused over 10 s into the tail-vein as previously described (Miao et al, 2004). Retro-orbital blood collection was performed at 24 h after injection for measurement of hFVIII activity and antigen in the plasma (Miao et al, 2004). The hFVIII antigen was quantified by using an anti-human FVIII light-chain sandwich ELISA from Affinity Biologicals Inc. (Ancaster, Canada).
Statistics
Experimental results are shown as mean±s.e.m. (for variation between animals or experiments). The mean values for biochemical data from the experimental groups were compared by a paired or unpaired, two-tailed Student’s t-test. Statistical tests with P<0.05 were considered significant.
Supplementary Material
Acknowledgments
Portions of this work were supported by NIH Grants DK042394, HL052173, and HL057346 (RJK) and also by the American Heart Association Grants 0635423Z and 09GRNT2280479 (KZ), and a NIH Grant 1R21ES017829-01A1 (KZ). We thank Dr Yong Liu for providing adenovirus expressing flag-tagged IRE1α, Dr Joanne Wang for providing Bortezomib reagent, Dr Jaeseok Han for technical support, Dr Harmeet Malhi for insightful comments, and Ms Jan Mitchell for her effort in preparing this paper. This work used the Chemistry Laboratory of the Michigan Diabetes Research and Training Center funded by a NIH Grant 5P60DK20572. We acknowledge Dr Thom Saunders for preparation of transgenic mice and the Transgenic Animal Model Core of the University of Michigan's Biomedical Research Core Facilities. The microarray data for gene expression in the livers of Ire1α-null and control mice in the absence or presence of ER stress has been submitted to the public database: NCBI GEO. The access number: GSE27038; GEO link: http://www.ncbi.nlm.nih.gov/geo/info/linking.html.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allain CC, Poon LS, Chan CS, Richmond W, Fu PC (1974) Enzymatic determination of total serum cholesterol. Clin Chem 20: 470–475 [PubMed] [Google Scholar]
- Berry MN, Edwards AM, Barritt GJ (1991) Isolated hepatocytes preparation, properties and applications. In Laboratory Techniques in Biochemistry and Molecular Biology, pp 59–81. New York: Elsevier Science Publishing Co., Inc [Google Scholar]
- Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C (1997) Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res 38: 2249–2263 [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A (2000) Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev 14: 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV Jr (1998) Identification of a gene encoding an acyl CoA: diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci USA 95: 13018–13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV Jr (2001) Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem 276: 38870–38876 [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- Dubuquoy L, Dharancy S, Nutten S, Pettersson S, Auwerx J, Desreumaux P (2002) Role of peroxisome proliferator-activated receptor gamma and retinoid X receptor heterodimer in hepatogastroenterological diseases. Lancet 360: 1410–1418 [DOI] [PubMed] [Google Scholar]
- Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J (1997) The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem 272: 18779–18789 [DOI] [PubMed] [Google Scholar]
- Fausto N (2001) Liver regeneration: from laboratory to clinic. Liver Transpl 7: 835–844 [DOI] [PubMed] [Google Scholar]
- Fausto N, Campbell JS, Riehle KJ (2006) Liver regeneration. Hepatology 43: S45–S53 [DOI] [PubMed] [Google Scholar]
- Fels DR, Ye J, Segan AT, Kridel SJ, Spiotto M, Olson M, Koong AC, Koumenis C (2008) Preferential cytotoxicity of Bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res 68: 9323–9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, Ron D, Tabas I (2003) The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol 5: 781–792 [DOI] [PubMed] [Google Scholar]
- Fribley A, Zeng Q, Wang CY (2004) Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol 24: 9695–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, Reitman ML (2003) Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem 278: 34268–34276 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Hernandez-Espinosa D, Minano A, Martinez C, Ordonez A, Perez-Ceballos E, de Arriba F, Mota RA, Ferrer F, Gonzalez M, Vicente V, Corral J (2008) Inhibition of proteasome by Bortezomib causes intracellular aggregation of hepatic serpins and increases the latent circulating form of antithrombin. Lab Invest 88: 306–317 [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312: 572–576 [DOI] [PubMed] [Google Scholar]
- Hoess RH, Abremski K (1990) The Cre-lox recombination system. In Nucleic Acids and Molecular Biology, Eckstein F, Lilley DM (eds) Vol. 4, pp 99–109. Berlin and Heidelberg: Springer-Verlag [Google Scholar]
- Jiang HP, Serrero G (1992) Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc Natl Acad Sci USA 89: 7856–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou J, Choi SS, Diehl AM (2008) Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis 28: 370–379 [DOI] [PubMed] [Google Scholar]
- Kadereit B, Kumar P, Wang WJ, Miranda D, Snapp EL, Severina N, Torregroza I, Evans T, Silver DL (2008) Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci USA 105: 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ (1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13: 1211–1233 [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH (2003a) Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci USA 100: 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH (2003b) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH (2008) Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16: 452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi R, Frank MW, Jackson PD, Rock CO, Jackowski S (2009) Elimination of the CDP-ethanolamine pathway disrupts hepatic lipid homeostasis. J Biol Chem 284: 27077–27089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2: research0032.1–0032.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Gores GJ (2008) Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin Liver Dis 28: 360–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ (2008) Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci USA 105: 18525–18530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG (2006) Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7: 373–378 [DOI] [PubMed] [Google Scholar]
- Matsusue K, Haluzik M, Lambert G, Yim SH, Gavrilova O, Ward JM, Brewer B Jr, Reitman ML, Gonzalez FJ (2003) Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest 111: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S, Gonzalez FJ (2008) Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab 7: 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao HZ, Sirachainan N, Palmer L, Kucab P, Cunningham MA, Kaufman RJ, Pipe SW (2004) Bioengineering of coagulation factor VIII for improved secretion. Blood 103: 3412–3419 [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J (1993) A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74: 743–756 [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M (2009) Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Prog Lipid Res 48: 1–26 [DOI] [PubMed] [Google Scholar]
- Nawrocki ST, Carew JS, Dunner K Jr, Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ (2005) Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res 65: 11510–11519 [DOI] [PubMed] [Google Scholar]
- Nikawa JI, Yamashita S (1992) IRE1 enclodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol Microbiol 6: 1441–1446 [DOI] [PubMed] [Google Scholar]
- Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H (2002) ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev 16: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K (2002) Distinct roles of ATF6 and PERK in transcription during the mammalian unfolded protein response. Biochem J 366: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Gayet C, Ginsberg HN (2008) Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J Clin Invest 118: 316–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D (2008) Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab 7: 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K, Ozcan U (2010) The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med 16: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce GF, Lillicrap D, Pipe SW, Vandendriessche T (2007) Gene therapy, bioengineered clotting factors and novel technologies for hemophilia treatment. J Thromb Haemost 5: 901–906 [DOI] [PubMed] [Google Scholar]
- Postic C, Girard J (2008) Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest 118: 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Magnuson MA (2000) DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26: 149–150 [DOI] [PubMed] [Google Scholar]
- Qiu W, Su Q, Rutledge AC, Zhang J, Adeli K (2009) Glucosamine-induced endoplasmic reticulum stress attenuates apolipoprotein B100 synthesis via PERK signaling. J Lipid Res 50: 1814–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Mao T, Zhang Y, Shao M, You J, Ding Q, Chen Y, Wu D, Xie D, Lin X, Gao X, Kaufman RJ, Li W, Liu Y (2010) A crucial role for RACK1 in the regulation of glucose-stimulated IRE1alpha activation in pancreatic beta cells. Sci Signal 3: ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH (2000) An essential role in liver development for transcription factor XBP-1. Genes Dev 14: 152–157 [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature 412: 300–307 [DOI] [PubMed] [Google Scholar]
- Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, Rajkumar SV, Srkalovic G, Alsina M, Alexanian R, Siegel D, Orlowski RZ, Kuter D, Limentani SA, Lee S, Hideshima T, Esseltine DL, Kauffman M, Adams J, Schenkein DP et al. (2003) A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med 348: 2609–2617 [DOI] [PubMed] [Google Scholar]
- Richardson PG, Mitsiades C, Hideshima T, Anderson KC (2006) Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med 57: 33–47 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Rosinol L, Montoto S, Cibeira MT, Blade J (2005) Bortezomib-induced severe hepatitis in multiple myeloma: a case report. Arch Intern Med 165: 464–465 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ (2008) UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell 15: 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge AC, Qiu W, Zhang R, Kohen-Avramoglu R, Nemat-Gorgani N, Adeli K (2009) Mechanisms targeting apolipoprotein B100 to proteasomal degradation: evidence that degradation is initiated by BiP binding at the N terminus and the formation of a p97 complex at the C terminus. Arterioscler Thromb Vasc Biol 29: 579–585 [DOI] [PubMed] [Google Scholar]
- Rutledge AC, Su Q, Adeli K (2010) Apolipoprotein B100 biogenesis: a complex array of intracellular mechanisms regulating folding, stability, and lipoprotein assembly. Biochem Cell Biol 88: 251–267 [DOI] [PubMed] [Google Scholar]
- Schadinger SE, Bucher NL, Schreiber BM, Farmer SR (2005) PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol Endocrinol Metab 288: E1195–E1205 [DOI] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ (2005) The Mammalian unfolded protein response. Annu Rev Biochem 74: 739–789 [DOI] [PubMed] [Google Scholar]
- Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L (2009) The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab 9: 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelness GS, Sellers JA (2001) Very-low-density lipoprotein assembly and secretion. Curr Opin Lipidol 12: 151–157 [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107: 893–903 [DOI] [PubMed] [Google Scholar]
- Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW (2007) Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem 282: 7024–7034 [DOI] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW (2004) XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167: 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein EA, M G (1994) Lipids, lipoproteins, and apolipoproteins. In Tietz Textbook of Clinical Chemistry, Burtis CA AE (ed), 2nd edn, pp 1002–1093. Philadelphia: W.B. Saunders Co [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ (1998) A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12: 1812–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH (2008) The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol 8: 663–674 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Graves RA, Budavari AI, Erdjument-Bromage H, Lui M, Hu E, Tempst P, Spiegelman BM (1994) Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR gamma and RXR alpha. Nucleic Acids Res 22: 5628–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM (2008) Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 77: 289–312 [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Vidal-Puig AJ, Considine RV, Jimenez-Linan M, Werman A, Pories WJ, Caro JF, Flier JS (1997) Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J Clin Invest 99: 2416–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D (1998) Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J 17: 5708–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wang D, Topczewski F, Pagliassotti MJ (2006) Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab 291: E275–E281 [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG (2006) Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11 [DOI] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE (2006) A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett 580: 5484–5491 [DOI] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ (2007) ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell 13: 351–364 [DOI] [PubMed] [Google Scholar]
- Wu Z, Bucher NL, Farmer SR (1996) Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPbeta, C/EBPdelta, and glucocorticoids. Mol Cell Biol 16: 4128–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xie Y, Bucher NL, Farmer SR (1995) Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev 9: 2350–2363 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Shiraishi S, Kishimoto K, Miura S, Ezaki O (2010) An increase in liver PPARgamma2 is an initial event to induce fatty liver in response to a diet high in butter: PPARgamma2 knockdown improves fatty liver induced by high-saturated fat. J Nutr Biochem. PMID: 20801631 (in press) [DOI] [PubMed] [Google Scholar]
- Yang L, Jhaveri R, Huang J, Qi Y, Diehl AM (2007) Endoplasmic reticulum stress, hepatocyte CD1d and NKT cell abnormalities in murine fatty livers. Lab Invest 87: 927–937 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Zhang J, Fu M, Cui T, Xiong C, Xu K, Zhong W, Xiao Y, Floyd D, Liang J, Li E, Song Q, Chen YE (2004) Selective disruption of PPARgamma 2 impairs the development of adipose tissue and insulin sensitivity. Proc Natl Acad Sci USA 101: 10703–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ (2008a) From endoplasmic-reticulum stress to the inflammatory response. Nature 454: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ (2008b) Identification and characterization of endoplasmic reticulum stress-induced apoptosis in vivo. Methods Enzymol 442: 395–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ (2006a) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124: 587–599 [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ (2005) The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest 115: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Hernandez-Ono A, Siri P, Weisberg S, Conlon D, Graham MJ, Crooke RM, Huang LS, Ginsberg HN (2006b) Aberrant hepatic expression of PPARgamma2 stimulates hepatic lipogenesis in a mouse model of obesity, insulin resistance, dyslipidemia, and hepatic steatosis. J Biol Chem 281: 37603–37615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.