Abstract
BECLIN 1 is a central player in macroautophagy. AMBRA1, a BECLIN 1-interacting protein, positively regulates the BECLIN 1-dependent programme of autophagy. In this study, we show that AMBRA1 binds preferentially the mitochondrial pool of the antiapoptotic factor BCL-2, and that this interaction is disrupted following autophagy induction. Further, AMBRA1 can compete with both mitochondrial and endoplasmic reticulum-resident BCL-2 (mito-BCL-2 and ER-BCL-2, respectively) to bind BECLIN 1. Moreover, after autophagy induction, AMBRA1 is recruited to BECLIN 1. Altogether, these results indicate that, in normal conditions, a pool of AMBRA1 binds preferentially mito-BCL-2; after autophagy induction, AMBRA1 is released from BCL-2, consistent with its ability to promote BECLIN 1 activity. In addition, we found that the binding between AMBRA1 and mito-BCL-2 is reduced during apoptosis. Thus, a dynamic interaction exists between AMBRA1 and BCL-2 at the mitochondria that could regulate both BECLIN 1-dependent autophagy and apoptosis.
Keywords: apoptosis, autophagy, endoplasmic reticulum, mitochondria
Introduction
Autophagy has a crucial role in many health and disease processes. This catabolic process in eukaryotic cells is involved in the degradation of cellular components through the lysosome machinery. During this process, portions of cytoplasm are sequestered by double-membraned vesicles, the autophagosomes, and degraded after fusion with lysosomes for subsequent recycling. BECLIN 1, the mammalian ortholog of yeast Atg6, has an evolutionarily conserved role in macroautophagy. Several BECLIN 1-interacting proteins have been reported as regulating this function, both positively (e.g., hAtg14, Bif-1, UVRAG; Takahashi et al, 2007, 2009; Itakura et al, 2008; Liang et al, 2008; Sun et al, 2009) and negatively (e.g., Rubicon, Bcl-2 family members; Pattingre et al, 2005; Levine et al, 2008; Matsunaga et al, 2009). AMBRA1 is a positive regulator of the BECLIN 1-dependent programme of autophagy (Fimia et al, 2007). Its functional deficiency in mouse embryos leads to neuroepithelial hyperplasia associated with autophagy impairment and excessive apoptotic cell death (Cecconi et al, 2007). Further, we demonstrated that autophagosome formation is primed by AMBRA1 release from the cytoskeleton: When autophagy is induced, AMBRA1 is released from dynein in an ULK1-dependent manner, and re-localizes to the endoplasmic reticulum (ER), thus enabling autophagosome nucleation. Therefore, AMBRA1 can be considered as a crucial factor in regulating autophagy initiation (Di Bartolomeo et al, 2010).
A complex relationship between autophagy and cell death exists (Levine et al, 2008; Djavaheri-Mergny et al, 2010). Currently, the well-known antiapoptotic factor BCL-2 is a key player in this context. The pool of BCL-2 resident at the ER (ER-BCL-2) is able, indeed, with the contribution of NAF-1 (nutrient-deprivation autophagy factor-1), to negatively regulate the BECLIN 1-dependent autophagic programme (Chang et al, 2010). In contrast, the mitochondrial pool of BCL-2 (mito-BCL-2) is shown to exert only an antiapoptotic function (Pattingre et al, 2005; Levine et al, 2008) even though mitochondria have been recently demonstrated to be selected sites for autophagosome formation (Hailey et al, 2010). BCL-2-regulated apoptotic cell death has been extensively studied. Overexpression of BCL-2 promotes a protective effect against a wide range of inducers of apoptosis. This antiapoptotic action derives from the fact that BCL-2 neutralizes proapoptotic BCL-2 family members, preventing mitochondrial membrane permeabilization and consequent cell death (Adams and Cory, 2007).
As previous studies have demonstrated that Beclin 1/Vps34-mediated autophagy is negatively regulated through a direct interaction between BECLIN 1 and ER-BCL-2, we investigated whether the antiapoptotic BCL-2 protein and AMBRA1 could bind each other. In this context, we found that AMBRA1 is a new partner of BCL-2 in mammalian cells and that their binding is independent of BECLIN 1. More importantly, by targeting BCL-2 either to mitochondria (mito-BCL-2) or ER (ER-BCL-2), we demonstrated that mito-BCL-2 is able to bind AMBRA1 and that this binding is disrupted after both autophagy and apoptosis induction. Our findings indicate that AMBRA1 and BCL-2 bind BECLIN 1 on the same site. AMBRA1 can thus compete with mito- and ER-BCL-2 to bind BECLIN 1. In addition, we demonstrated that, after autophagy induction, AMBRA1/BECLIN 1 interaction increases both in mitochondrial and microsomal fractions, whereas the AMBRA1–mito-BCL-2 interaction is disrupted. Altogether, these results lead us to propose a model in which, under normal conditions, a pool of AMBRA1 is docked by BCL-2 at the mitochondria, inhibiting its autophagic function; after autophagy induction, this mitochondrial pool of AMBRA1 separates from mito-BCL-2 and increases its binding to BECLIN 1 in order to favour the autophagic programme.
Results
AMBRA1 is a new partner of the antiapoptotic and antiautophagic factor BCL-2
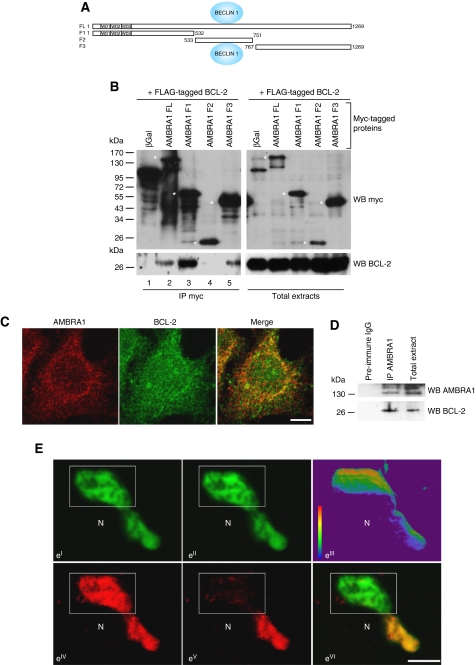
AMBRA1 interacts with BECLIN 1 and its associated kinase Vps34, and favours BECLIN 1/Vps34 functional interaction (Fimia et al, 2007). This interaction, which requires the F2 fragment of AMBRA1 (Figure 1A), is crucial in the autophagic process; taken together, AMBRA1, BECLIN 1 and Vps34 have been defined as the autophagy core-complex (He and Levine, 2010). In contrast, BECLIN 1 is negatively regulated by a direct interaction with the antiapoptotic factor BCL-2. Therefore, we investigated the existence of a putative interaction between the antiapoptotic protein BCL-2 and AMBRA1 in a co-immunoprecipitation experiment. BCL-2 and AMBRA1 were found to be associated in HEK293 cells coexpressing both AMBRA1 and BCL-2 (Figure 1B, lane 2). Next, we wanted to find the AMBRA1 domain responsible for this binding. To this end, we transfected cells with cDNAs encoding various AMBRA1 portions (Figure 1A): FL (full length), F1 (amino acids 1–532), F2 (amino acids 533-750) and F3 (amino acids 767–1269). As shown in Figure 1B (lanes 3 and 5), F1 and F3 fragments (the N-terminal and C-terminal part of the protein, respectively) are sufficient to bind BCL-2, whereas the central region (F2), which is known to bind BECLIN 1 (Fimia et al, 2007), shows no interaction. Similar results were obtained following reciprocal co-immunoprecipitation experiments (Supplementary Figure S1). BCL-2 co-immunoprecipitates with AMBRA1 (lane 2) and its F1 and F3 fragments (lanes 3 and 5). To confirm these biochemical results, we performed a confocal microscopy analysis in HeLa cells expressing detectable endogenous levels of both AMBRA1 and BCL-2. As illustrated in Figure 1C, endogenous AMBRA1 and BCL-2 showed a partial colocalization. In order to give two quantitative outputs for this colocalization, we used Pearson (rp) and Spearman (rs) statistics. Both tests produce values in the range (−1, 1), 0 indicating that there is no discernable correlation and −1 and +1 meaning strong negative and positive correlations, respectively. We obtained an rp=0.38 and an rs=0.39 confirming that AMBRA1 and BCL-2 colocalize in mammalian cells. This analysis indicates also that the interaction between the two proteins is partial. To confirm the result, we analysed the interaction between endogenous AMBRA1 and BCL-2 in HeLa cells. As illustrated in Figure 1D, we found that endogenous AMBRA1 binds specifically endogenous BCL-2. To strengthen our results on this interaction, we performed a Förster resonance energy transfer experiment to measure AMBRA1–BCL-2 proteins proximity within the cells. GFP and mCherry are two fluorescent proteins which use as an FRET pair that has been recently validated for reproducible quantitative determination of the energy transfer efficiency, both in vivo and in vitro (Albertazzi et al 2009). In order to study the interaction of BCL-2 and AMBRA1 proteins, they were fused with the GFP and mCherry fluorescent proteins and co-transfected in HEK293 cells. On a morphological ground, visual inspection of the fluorescence expression patterns in the histological material showed that BCL-2–GFP and AMBRA1–mCherry appeared to be coexpressed in structures mainly resembling mitochondria and ER. The colocalization of the two proteins was almost complete. The average FRET efficiency values measured was 11.14%±2.1 (see Figure 1E). In a previous paper, the FRET behaviour of two tandem mCherry–EGFP fusion proteins (which differed for the distance, short linker and long linker) has been investigated (Albertazzi et al 2009). A FRET efficiency value of 0.41 for the short linker and of 0.29 for the long linker was achieved for the mCherry–EGFP tandem proteins. Comparison of our AMBRA1–BCL-2 FRET value with that of the mCherry–EGFP tandem construct suggests that AMBRA1 and BCL-2 are in proximity in the interacting complex.
Figure 1.
AMBRA1 interacts with the antiapoptotic factor BCL-2 in mammalian cells. A scheme of AMBRA1 with its WD40 domains is illustrated in (A). The binding site with BECLIN 1 is also reported on AMBRA1. HEK293 cells were co-transfected with vectors encoding FLAG–BCL-2 and myc–AMBRA1 FL (full length), myc-β-galactosidase (βGal) as a negative control or myc–AMBRA1 mutants F1, F2 or F3. (B) Protein extracts were immunoprecipitated using an myc antibody. Purified complexes and corresponding total extracts were analysed by western blot (WB) using an anti-BCL-2 antibody (B). Asterisks point to the molecular weight of proteins corresponding to the original AMBRA1 fragments. (C) Partial colocalization between endogenous AMBRA1 and BCL-2 in HeLa cells. HeLa cells grown in normal media were stained by anti-AMBRA1 (red), anti-BCL-2 (green) antibodies. The merge of the two fluorescence signals is shown in the right panel. Scale bar, 6 μm. (D) Endogenous AMBRA1 co-immunoprecipitates with endogenous BCL-2 in HeLa cells. HeLa cells were lysed and protein extracts were then immunoprecipitated using an anti-AMBRA1 antibody or preimmune IgG as a negative control. Purified complexes and corresponding total extracts were analysed by WB using anti-AMBRA1 and anti-BCL-2 antibodies. (E) Confocal microscope images of FRET acceptor photobleaching assay of HEK293 cells co-transfected with BCL-2–GFP and AMBRA1–mCherry. A bleached region of the cytosol is indicated by the white rectangle. (eI) BCL-2–GFP (donor) channel before the bleach. (eII) BCL-2–GFP (donor) channel after the bleach. (eIII) Pseudo-coloured image showing an FRET efficiency values map. (eIV) AMBRA1–mCherry (acceptor) channel before the bleach. (eV) AMBRA1–mCherry (acceptor) channel after the bleach. (eVI) Merge of BCL-2–GFP and AMBRA1–mCherry channels after the bleach. N indicates a close-by nuclear area. Scale bar, 5 μm.
BECLIN 1 is not required for AMBRA1–BCL-2 interaction
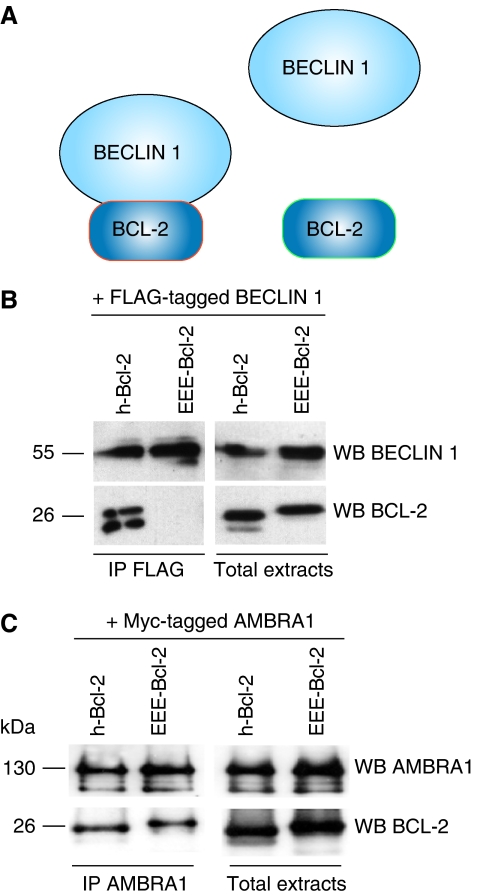
The F2 fragment of AMBRA1 is required for the interaction between AMBRA1 and BECLIN 1 (Fimia et al, 2007), but not for its binding to BCL-2. This suggests that BECLIN 1 is not necessary for this latter association. To confirm this hypothesis, we performed co-immunoprecipitation experiments using a mutant of BCL-2 unable to bind BECLIN 1. This mutant (EEE-BCL-2) possesses simultaneous glutamine substitutions at three phosphorylation sites (T69, S70 and S87), and mimics multi-phosphorylations that occur following autophagy induction (Wei et al, 2008). This impedes the interaction of BCL-2 with BECLIN 1 (Figure 2A and B). Vectors coding for AMBRA1 and for human-BCL-2 (h-BCL-2) or for the EEE-BCL-2 mutant were overexpressed in HEK293 cells; then, co-immunoprecipitations of AMBRA1 and BCL-2 were performed in normal conditions. As shown in Figure 2C, AMBRA1 is able to bind h-BCL-2 as well as the BCL-2 mutant (EEE-BCL-2) that cannot bind BECLIN 1.
Figure 2.
The interaction between AMBRA1 and BCL-2 does not require BECLIN 1. (A) Simultaneous glutamine substitutions at the three phosphorylation site on BCL-2 (EEE-BCL-2 mutant) induce a disruption of the BECLIN 1/BCL-2 complex (Pattingre et al, 2005). (B) BECLIN 1 co-immunoprecipitates with hBCL-2 but not with EEE-BCL-2 mutant in HEK293 cells. HEK293 cells were co-transfected with vectors encoding FLAG–BECLIN 1, and myc-h-BCL-2 or the EEE-BCL-2 mutant. Protein extracts were immunoprecipitated using an anti-FLAG antibody. Purified complexes and corresponding total extracts were analysed by WB using anti-BECLIN 1 and anti-BCL-2 antibodies. (C) AMBRA1 co-immunoprecipitates with both hBCL-2 and EEE-BCL-2 mutants in HEK293 cells. HEK293 cells were co-transfected with vectors encoding myc–AMBRA1, and myc-h-BCL-2 or the EEE-BCL-2 mutant. Protein extracts were immunoprecipitated using an anti-AMBRA1 antibody. Purified complexes and corresponding total extracts were analysed by WB using anti-AMBRA1 and anti-BCL-2 antibodies.
Mitochondrial BCL-2 preferentially binds AMBRA1 and this interaction is disrupted after autophagy induction
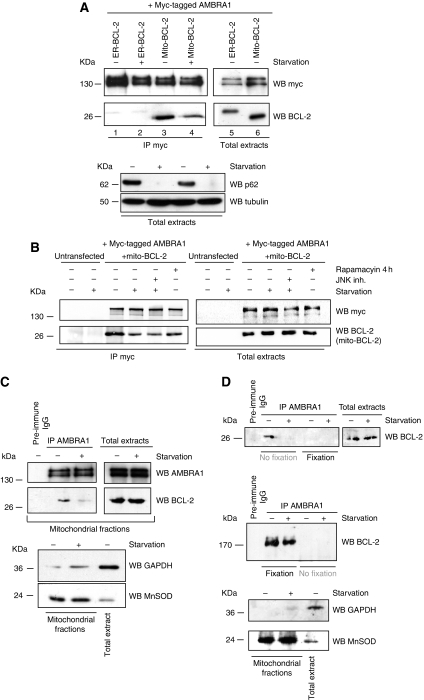
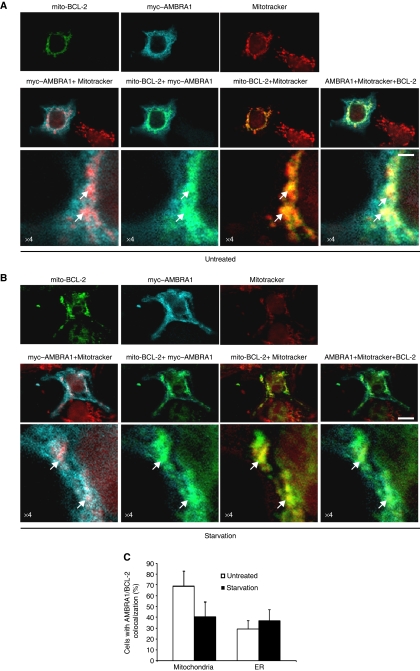
BCL-2 is predominantly found on the outer mitochondrial and ER membranes (Germain and Shore, 2003), these subcellular localizations being related to BCL-2 function (Lithgow et al, 1994). To gain an insight into the functional significance of the interaction between AMBRA1 and BCL-2, we evaluated whether AMBRA1 differentially binds the ER resident or the mitochondrial pool of BCL-2. To this end, two BCL-2 mutants with restricted subcellular localization were used in co-immunoprecipitation experiments with AMBRA1. In the first mutant, the C-terminal hydrophobic sequence of BCL-2 is exchanged for an equivalent sequence from modified ActA, which binds specifically to the cytoplasmic face of mitochondrial outer membranes (Pistor et al, 1994). The other mutant possesses a C-terminal sequence exchanged for a sequence from cytochrome b5 (DNA encoding the analogous 35 amino-acid sequence of the ER-specific isoform of rat hepatic cytochrome b5 (Mitoma and Ito, 1992), resulting in ER-specific localization (Zhu et al, 1996)). As shown in Figure 3A, myc–AMBRA1 significantly co-immunoprecipitates with mito-targeted BCL-2 (lane 3), whereas only a small amount of ER-targeted BCL-2 is found to bind AMBRA1 (lane 1). These results were confirmed by reversing the order of the co-immunoprecipitation (Supplementary Figure S2a). Knowing that the interaction between BCL-2 and BECLIN 1 is differential in normal conditions versus starvation conditions (Pattingre et al, 2005), we decided to analyse the interaction between AMBRA1 and mito-BCL-2 on autophagy induction. When cells were shifted for 4 h in an amino acid-free medium (EBSS medium), the interaction between AMBRA1 and mito-BCL-2 decreased, whereas no difference was observed in the binding between ER-BCL-2 and myc–AMBRA1. The reduction of p62/A170/SQSTM1, a ubiquitin-binding protein degraded by autophagy (Ichimura et al, 2008), was used as a marker to confirm autophagy induction (Figure 3A, lower panel). To control for the specificity of the co-immunoprecipitation between AMBRA1 and mito-BCL-2, negative controls using the myc antibody were performed in HEK293 cells, which do not overexpress myc–AMBRA1 or BCL-2 (Supplementary Figure S2B–D). The interaction between AMBRA1 and mito-BCL-2 can also be disrupted, at a lower extent, using rapamycin as an autophagy inducer (Figure 3B). During autophagy, phosphorylation of BCL-2 by JNK1 kinase abrogates the binding between BECLIN 1 and BCL-2. Thus, we also tested whether inhibition of JNK1 would be responsible for the AMBRA1–BCL-2 binding regulation. However, as illustrated in Figure 3B, the JNK1 kinase inhibitor SP600125 does not impair the release of AMBRA1 from mito-BCL-2 after autophagy induction. Next, we observed binding between endogenous AMBRA1 and mito-BCL-2. To this end, we isolated mitochondrial fractions from cells grown in normal conditions or for 4 h in EBSS. We performed an immunoprecipitation of AMBRA1 in the mitochondrial fractions, and we were able to appreciate the binding with endogenous BCL-2 in normal conditions. In contrast, this binding was disrupted after autophagy induction (Figure 3C). Of note, when checking by densitometric analysis, we can observe a binding between endogenous AMBRA1 and BECLIN 1 in these mitochondrial fractions in normal conditions, with this binding being increased following autophagy induction (Supplementary Figure S2E). To consolidate these data, we decided to carry out a mitochondrial crosslink before performing a subcellular fractionation experiment in HEK293 cells grown in normal or starvation conditions. As illustrated in Figure 3D, when cells are not fixed, BCL-2 can be detected as a 26-kDa band (appropriate molecular weight) in normal condition, and this band decreases following autophagy induction. However, after cell fixation, this band is not detectable, whereas a band of ∼170 kDa is detectable. The 170-kDa band may corresponds most likely to a complex including at least endogenous AMBRA1 and BCL-2, thus confirming the interaction. Next, we set out to establish whether this dynamic interaction could be monitored by fluorescence microscopy. As illustrated in Figure 4A, mito-BCL-2 colocalizes perfectly with the mitotracker staining, as expected, and there is a significant colocalization between myc–AMBRA1 and mito-BCL-2 in normal conditions, whereas some cells showed only a slight colocalization between ER-BCL-2 and AMBRA1 (arrows in Supplementary Figure S3A, B). We performed the same type of staining at the endogenous level to confirm this colocalization (see Supplementary Figure S4). By performing statistical colocalization analysis, we found that AMBRA1 shows a very good colocalization with mito-BCL-2, as the rp and rs values are 0.7 and 0.8, respectively (Supplementary Figure S3C). When cells are shifted to EBSS a clear decrease in AMBRA1 and mito-BCL-2 colocalization is observed (Figure 4B, starvation lane). Furthermore, the percentage of cells where myc–AMBRA1 colocalizes either with mito-BCL-2 or ER-BCL-2 was determined. As illustrated in Figure 4C, 70% cells showing colocalization between AMBRA1 and mito-BCL-2 are found in normal conditions, whereas only 40% cells show the same colocalization after autophagy induction. Such a reduction of colocalization is not observed in the case of AMBRA1–ER-BCL-2 binding, where we found ∼30% of colocalizing cells in both conditions. Of note, this colocalization seems to be due to a short ‘proximity’ between the stained compartments, rather than an overlap, as illustrated in Supplementary Figure S3B.
Figure 3.
Mitochondrial-BCL-2 preferentially binds AMBRA1, and this binding is disrupted after autophagy induction. (A) HEK293 cells were co-transfected with vectors encoding either ER-BCL-2 or mito-BCL-2 and myc-AMBRA1. Cells were grown either in normal media or in EBSS media for 4 h. Protein extracts were immunoprecipitated using an myc antibody. Purified complexes and corresponding total extracts were analysed by WB, using an anti-BCL-2 antibody or an myc antibody. Autophagy induction was controlled in total extracts by using an anti-p62 antibody. (B) Inhibition of JNK1 does not regulate the AMBRA1–mito-BCL-2 binding. HEK293 cells were co-transfected with vectors encoding myc–AMBRA1 and mito-BCL-2 or left untransfected, as a negative control. Cells were treated with rapamycin to induce autophagy or shifted in EBSS media plus the JNK1 kinase inhibitor for 4 h. Then, protein extracts were immunoprecipitated using an myc antibody. Purified complexes and corresponding total extracts were analysed by WB, by using anti-BCL-2 and myc antibodies. (C) HEK293 cells were grown in normal or in EBSS media for 4 h. Mitochondrial fractions were then isolated by centrifugation and the quality of the fractions controlled by WB by using MnSOD and GAPDH antibodies. Endogenous mitochondrial AMBRA1 was next precipitated using anti-AMBRA1 polyclonal antibody in both conditions or with the preimmune IgG as a negative control. Purified complexes were analysed by WB, by using anti-BCL-2 and anti-AMBRA1 antibodies. (D) HEK293 cells were grown in normal or in EBSS media for 4 h and then fixed 10 min with PFA 0.5% or left without fixation. Mitochondrial fractions were isolated by centrifugation and the quality of the fractions controlled by WB by using MnSOD and GAPDH antibodies. Endogenous mitochondrial AMBRA1 was next precipitated using anti-AMBRA1 polyclonal antibody in both conditions or with preimmune IgG, as a negative control. Purified complexes were analysed by WB, by using anti-BCL-2 antibody. BCL-2 was detected at 26 kDa when cells were not fixed, whereas this band disappears and a band at ∼170 kDa is detectable upon fixation.
Figure 4.
AMBRA1 dynamic colocalization with mito-BCL-2 and the mitochondrial network. (A, B) HEK293 cells co-transfected with vectors encoding mito-BCL-2 and myc-AMBRA1, grown either in normal media (A) or in EBSS (B) for 4 h and stained with an anti-BCL-2 antibody (green), an anti-myc–AMBRA1 antibody (blu) and Mitotracker (red). The merge of the two or three fluorescence signals are shown in the bottom panels, as indicated. Scale bar, 6 μm. White arrows point to strong triple colocalization areas. (C) AMBRA1–mito-BCL-2 colocalization decreases after autophagy induction. Quantification of cells showing colocalization between AMBRA1 and ER- or mito-BCL-2 in normal conditions and after autophagy induction is shown. Results are expressed as percentage of cells (±s.d.) showing colocalization between AMBRA1 and ER- or mito-BCL-2. Each point value represents the mean±s.d. of triplicate wells from three independent experiments. Statistical analysis was performed by analysis of variance (one-way ANOVA). *P<0.05 versus mito-BCL-2 in normal conditions.
AMBRA1 partially localizes at mitochondria in normal conditions
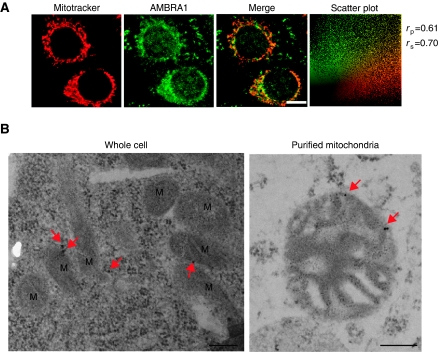
AMBRA1 is a cytoplasmic protein showing a diffuse signal in normal conditions, mostly overlapping with dynein light chains. After autophagy induction, a fraction of AMBRA1 translocates to the perinuclear region of the cell and more precisely to the ER (Di Bartolomeo et al, 2010). The binding of AMBRA1 with mito-BCL-2 led us to check whether there is a subcellular colocalization of AMBRA1 with mitochondria. As illustrated in Figure 5A, we found, indeed, that AMBRA1 presents a partial colocalization with mitochondria, as the rp and rs are 0.61 and 0.70, respectively. Of note, these quantitative results are similar to those obtained with the AMBRA1/mito-BCL-2 colocalization analysis.
Figure 5.
AMBRA1 partially colocalizes with the mitochondrial network. (A) HeLa cells grown in normal media were stained with Mitotracker (red) and an anti-AMBRA1 antibody (green). The merge of the two fluorescence signals is shown in the right panels. Scale bar, 6 μm. The Pearson correlation coefficient rp and Spearman correlation coefficient rs are indicated on the scatter plot. (B) Immunogold analysis of whole cells and purified mitochondria from HeLa cells. Fifteen nm particles label the AMBRA1 protein (red arrows). Scale bar, 0.2 μm. M, mitochondria.
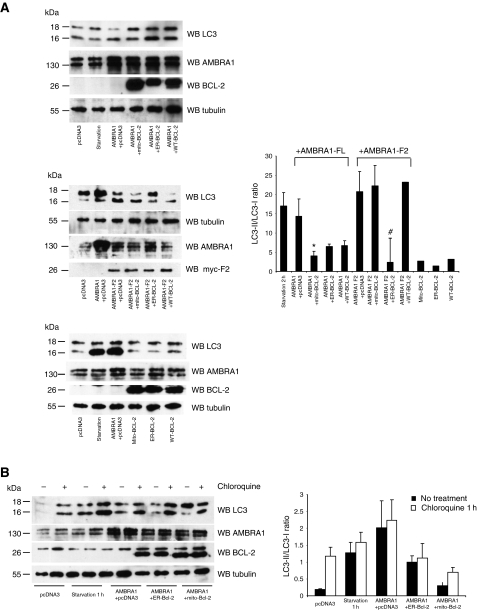
In order to confirm our observation, we performed an immunogold assay using an anti-AMBRA1 antibody on HeLa cells grown in normal conditions and on their purified mitochondria (Figure 5B and Supplementary Figure S5). Altogether, these results indicate that a pool of AMBRA1 (corresponding to 49±10% of the total protein) can localize at the mitochondria in normal conditions. Of note, after autophagy induction, this mitochondrial pool of AMBRA1 seems to be slightly reduced (see Supplementary Figure S6A). These results led us to hypothesize that mito-BCL-2 should anchor/inhibit AMBRA1 at the mitochondria in order to block its participation in the BECLIN 1-dependent autophagy programme. Thus, we decided to evaluate the effect of mito-BCL-2 on autophagy induced by AMBRA1. We overexpressed AMBRA1 in the presence of a control plasmid (pCDNA3), or either with mito-BCL-2 or WT-BCL-2. LC3 protein conversion was used as a marker of autophagy induction (Mizushima et al, 2004, 2010). As shown in Figure 6, overexpression of mito-BCL-2 is sufficient to dramatically reduce AMBRA1-induced autophagy. However, the F2 fragment of AMBRA1, which is known to bind BECLIN 1 (Fimia et al, 2007), but not BCL-2 (see Figure 1A and B), is able to evade from mito-BCL-2 inhibition but not from ER-BCL-2 effect. Finally, in order to evaluate the autophagic flux (autophagosome on-rate versus off-rate), we treated cells with the lysosome inhibitor chloroquine (20 μM) for 1 h. As shown in Figure 6B, chloroquine treatment leads to an increase of the intensity of the LC3-II bands in all the cases presented. Taken together, these results suggest that, in normal conditions, BCL-2 binds AMBRA1 at the mitochondria in order to inhibit its proautophagic function during autophagosome formation.
Figure 6.
AMBRA1 proautophagic activity is inhibited by mito-BCL-2. (A) HEK293 cells were co-transfected either with the control vector pcDNA3, vectors encoding Mito-, ER- and WT-BCL-2 as controls and a vector encoding myc–AMBRA1 or AMBRA1-F2 with or without vectors encoding mito-, ER- or WT-BCL-2. As an additional positive autophagy control, 24 h untransfected cells were shifted in EBSS medium (starvation). Total extracts were then analysed by western blot (WB) using an anti-LC3 in order to analyse autophagy induction, anti-AMBRA1, anti-BCL-2 and anti-tubulin antibodies to control protein expressions. LC3-II/LC3-I ratio was quantified by densitometric analysis using the ImageQuant software. Results are expressed as LC3-II/LC3-I ratio as a percentage of control (pcDNA3 alone). Each point value represents the mean±s.d. from three independent experiments. Statistical analysis was performed by analysis of variance (one-way ANOVA). *P<0.05 versus AMBRA1+pcDNA3. #P<0.05 versus AMBRA1 F2+pcDNA3. (B) HEK293 cells were co-transfected either with the control vector pcDNA3 and a vector encoding myc–AMBRA1 with or without vectors encoding mito-, ER- or WT-BCL-2. Twenty-four hours after transfection, cells were treated with Chloroquine A or vehicle control for 2 h. Total extracts were then analysed by western blot (WB) using an anti-LC3 antibody in order to analyse autophagy induction, anti-AMBRA1, anti-BCL-2 and anti-tubulin antibodies to control protein expressions. LC3-II/LC3-I ratio was quantified by densitometric analysis using the ImageQuant software. Results are expressed as LC3-II/LC3-I ratio. Each point value represents the mean±s.d. from three independent experiments.
AMBRA1 competes with BCL-2 to bind BECLIN 1
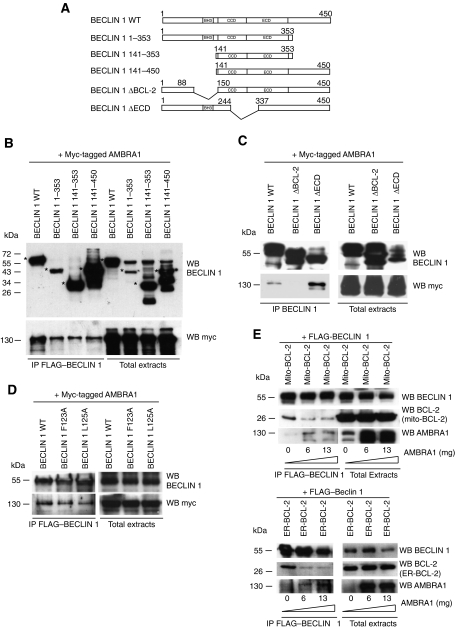
BCL-2 is known to bind BECLIN 1 on its BH3 domain. Therefore, to better understand how AMBRA1 and BCL-2 differentially regulate autophagy, we set out to determine the binding domain of AMBRA1 on BECLIN 1 by using different BECLIN 1 mutants in co-immunoprecipitation experiments with AMBRA1. In a first series of experiments, we found that three truncated BECLIN 1 mutants (aa 1–353, aa 141–353 and aa 141–450) are still able to interact with AMBRA1 (Figure 7A and B). We refined our search by using two BECLIN 1 deletion mutants: BECLIN 1 ΔBcl-2 BD (binding domain) (lacking aa 88–150) and BECLIN 1 ΔECD (lacking aa 244–337). As shown in Figure 7C, AMBRA1 can still co-immunoprecipitate with BECLIN 1 ΔECD, but not with the BECLIN 1 ΔBcl-2 BD mutant. Consequently, it appears that a part of BECLIN 1 binding domain with BCL-2 is necessary for its binding to AMBRA1. By studying more in detail the amino-acid sequence of BECLIN 1, it can be deduced that amino acids 141–150 are essential for the AMBRA1–BECLIN 1 binding. We can conclude this because the Δ141–353 mutant binds AMBRA1, whereas the Δ88–150 mutant does not. We next analysed the binding between AMBRA1 and two BH3 mutants of BECLIN 1, BECLIN 1125A and BECLIN 1F123A, which respectively interact or not with BCL-2 and BCL-XL (Pattingre et al, 2005; Maiuri et al, 2007). In fact, BECLIN 1125A mimicked the wild-type protein, while BECLIN 1F123A possesses an unfunctional BH3 domain, a mutant known to induce a strong autophagy. Figure 7D shows that AMBRA1 can bind these two BECLIN 1 mutants, indicating that an intact BH3 binding site for BCL-2 and BCL-XL is not required for the binding to AMBRA1. Even though AMBRA1 does not require a functional BH3 domain on BECLIN 1, it binds BECLIN 1 in a region similar to that used for binding BCL-2.
Figure 7.
AMBRA1 competes with BCL-2 to bind BECLIN 1. (A) Illustration of the BECLIN 1 mutants. The coiled-coiled domain (CCD) and the evolutionary conserved domain (ECD) of BECLIN 1 are indicated. (B) AMBRA1 binds all truncated mutants of BECLIN 1. Co-immunoprecipitation of AMBRA1 and different mutants of BECLIN 1 in HEK293 cells is shown. HEK293 cells were co-transfected with vectors encoding myc–AMBRA1, FLAG–BECLIN 1 1–353, FLAG–BECLIN 1 141–353 or FLAG–BECLIN 1 141–459. Protein extracts were immunoprecipitated using an anti-FLAG antibody. Purified complexes and corresponding total extracts were analysed by WB, by using anti-AMBRA1 and anti-BECLIN 1 antibodies. Asterisks point to the molecular weight of proteins corresponding to the BECLIN 1 fragments. (C) AMBRA1 cannot be co-immunoprecipitated with the Beclin 1 ΔBcl-2 mutant. HEK293 cells were co-transfected with vectors encoding myc–AMBRA1 and BECLIN 1 ΔBcl-2 BD or BECLIN 1 ΔECD. Protein extracts were immunoprecipitated using an anti-BECLIN 1 antibody. Purified complexes and corresponding total extracts were analysed by WB, by using anti-AMBRA1 and anti-BECLIN 1 antibodies. (D) Intact BH3 binding site for BCL-2 is not required for the binding to AMBRA1. HEK293 cells were co-transfected with vectors encoding myc-AMBRA1 and BECLIN 1125A and BECLIN 1F123A, which respectively interact or not with BCL-2 and BCL-XL. Protein extracts were immunoprecipitated using an myc antibody (myc–AMBRA1). Purified complexes and corresponding total extracts were analysed by WB, by using anti-myc and anti-BECLIN 1 antibodies. (E) BECLIN 1 and mito- or ER-BCL-2 binding decreases when AMBRA1 is overexpressed. HEK293 cells were transfected both with FLAG–BECLIN 1, mito- and ER-BCL-2 and increasing concentrations of myc–AMBRA1 cDNAs. Protein extracts were immunoprecipitated using an anti-FLAG antibody. Purified complexes and corresponding total extracts were analysed by WB, by using anti-BECLIN 1, anti-BCL-2 and anti-AMBRA1 antibodies. In the upper panels, bands corresponding to AMBRA1 co-immunoprecipitated with BECLIN 1 are shown at a higher exposure than the others (mito-BCL-2 and FLAG–BECLIN 1) in order to better visualize it.
In order to substantiate the hypothesis that AMBRA1 and BCL-2 may compete for binding BECLIN 1, we conducted co-immunoprecipitation experiments with both mito/ER-BCL-2 and BECLIN 1, and increasing amounts of AMBRA1 (Figure 7E). Our results show that both mito- and ER-BCL-2/BECLIN 1 interaction decrease with increasing AMBRA1 levels. This suggests that, upon induction of autophagy, AMBRA1 can compete with mito/ER-BCL-2 to bind BECLIN 1. Of note, this competition is detectable also when we perform an inverse experiment, by using BCL-2 as a competitor for the binding between AMBRA1 and BECLIN 1 (see Supplementary Figure S7).
AMBRA1–BCL-2 interaction upon apoptosis induction
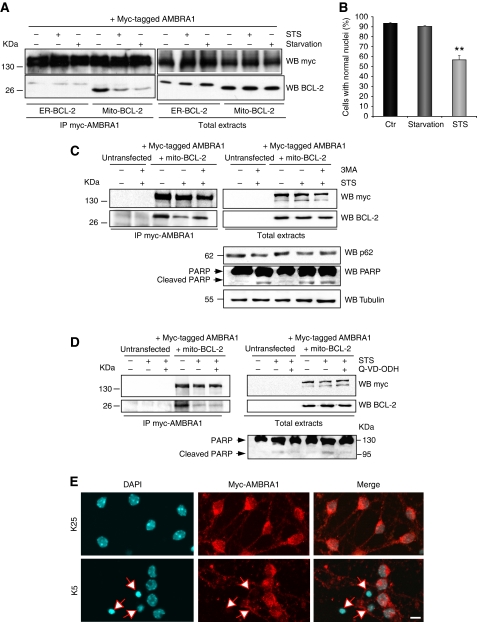
AMBRA1 functional deficiency in mouse embryos led to severe neural tube defects associated not only with autophagy impairment, but also with excessive apoptotic cell death (Fimia et al, 2007). Our observation that AMBRA1 binds the mitochondrial pool of BCL-2, which is known to exert an antiapoptotic effect, led us to examine the AMBRA1/BCL-2 interaction during apoptosis. First, we analysed AMBRA1–mito-BCL-2 interaction in HEK293 cells overexpressing both AMBRA1 and mito-BCL-2 and treated for 6 h with staurosporine (STS), an inducer of apoptosis. As illustrated in Figure 8A, we detected the same reduction of binding between AMBRA1 and mito-BCL-2 observed after induction of autophagy in the absence of cell death, as revealed by the absence of pycknotic nuclei (Figure 8B). In contrast, the interaction between AMBRA1 and ER-BCL-2 did not show any variations. In order to exclude a possible induction of autophagy by STS, cells were treated with STS in the presence of 10 mM 3-methyladenine (3-MA), an inhibitor of the kinase complex (Petiot et al, 2000). Also in this case, the disruption of AMBRA1–mito-BCL-2 complex occurs after apoptosis induction, clearly indicating that this event is independent of autophagy (Figure 8C). This decrease cannot be abrogated by addition of the general caspase inhibitor Q-VD-OPH (whose efficiency is demonstrated by PARP cleavage analysis), thus indicating that the disruption of the complex is upstream of caspase activation (Figure 8D). These results suggest that, after apoptosis induction, AMBRA1 early dissociates from mito-BCL-2.
Figure 8.
AMBRA1 does not posses a proapoptotic activity but, in contrast, it may participate to cell viability. (A) AMBRA1–BCL-2 interaction is disrupted after stautosporine (STS) treatment. HEK293 cells were co-transfected with vectors encoding mito-BCL-2 and myc-AMBRA1 FL and grown either in normal conditions, in starvation condition (EBSS 4 h) or in normal media plus STS (1 μM, 6 h). Protein extracts were immunoprecipitated using an myc antibody (IP: myc-AMBRA1). Purified complexes and corresponding total extracts were analysed by WB, by using an anti-BCL-2 antibody. (B) STS but not EBSS treatment induces cell death of HEK293. HEK293 cells were co-transfected with vectors encoding mito-BCL-2 and myc–AMBRA1 and grown either in normal conditions, in starvation conditions (EBSS 4 h) or in normal media plus STS (1 μM, 6 h). Cultures were then labelled with DAPI. Cells showing condensed or fragmented nuclei were scored as dying, and the percentage of viable cells was determined 6 h after EBSS or STS treatment. For each condition, cells were counted into 10 random fields in three different wells from three independent experiments. Statistical analysis was performed by analysis of variance (one-way ANOVA). *P<0.05 versus control cells. (C) Apoptosis induced by STS treatment disrupts the complex AMBRA1/BCL-2. HEK293 cells were co-transfected or left untreated (as a negative control) with vectors encoding myc–AMBRA1 and mito-BCL-2. Cells were then treated with STS (1 μM 4 h) in presence or not with 10 mM 3-MA. Protein extracts were immunoprecipitated using an myc antibody. Purified complexes and corresponding total extracts were analysed by WB using anti-BCL-2 and myc antibodies. p62 protein levels and PARP cleavage were also analysed in total extracts to control, respectively, autophagy and apoptosis induction. (D) Caspase inhibition does not block AMBRA1/BCL-2 complex disruption following STS treatment. HEK293 cells were co-transfected or left untreated (as a negative control) with vectors encoding mito-BCL-2 and myc–AMBRA1 and grown either in normal conditions, in normal conditions plus STS (1 μM, 6 h) or plus caspase inhibitor Q-VD-ODH (10 μM, 6 h). Protein extracts were immunoprecipitated using an myc antibody (IP: myc–AMBRA1). Purified complexes and corresponding total extracts were analysed by WB, by using an anti-BCL-2 antibody. Induction of cell death by STS or the inhibitory effect of Q-VD-ODH on caspases were evaluated using an anti-PARP antibody on corresponding total extracts. (E) AMBRA1 may protect neurons from death. Overexpression of myc–AMBRA1 in CGN grown in survival medium or after induction of apoptosis is shown. After 4 DIV, CGN were transfected with an expression vector encoding myc–AMBRA1. One day later, cultures were kept in K25 medium and grown for additional 24 h or shifted in K5 medium during 15 h. Cultures were then labelled with an myc antibody and DAPI. The transfected neurons showing condensed nuclei are visible on the overlay images. Arrows point to dead neurons. Scale bar, 6 μm.
Owing to the fact that AMBRA1 is a crucial protein for the development of the nervous system, where it is expressed from the first stages of neurulation onwards (Fimia et al, 2007), we investigated a putative involvement of AMBRA1 in neuronal survival. To this end, as a model of neuronal death, we used cerebellar granule neurons (CGNs) induced to die by suppression of trophic factors. Neurons were transfected with AMBRA1 and, 24 h later, cellular viability was assessed by analysis of nuclei morphology. As shown in Figure 8E, expression of AMBRA1 does not influence membrane-depolarization-induced CGN survival (K25 medium), thus excluding a proapoptotic role of AMBRA1. However, neurons overexpressing AMBRA1 (arrows) and shifted in K5 medium (cell death condition) for 15 h seems to better survive in this condition. These results may suggest that overexpression of wild-type AMBRA1 is probably sufficient to enhance CGN survival, early after K+ depletion.
Discussion
AMBRA1 binds the mitochondrial pool of BCL-2
Our work demonstrates the existence of an interaction between AMBRA1 and BCL-2. BCL-2 has been detected in different compartments of the cell: in particular, BCL-2 is not only detected in the mitochondrial fraction (17%) but also in cytoplasmic membranes including the ER (49%) and the nuclear envelope (34%; Lithgow et al, 1994; Hoetelmans et al, 2000; Portier and Taglialatela, 2006). By looking at the different BCL-2 pools, we found that AMBRA1 binds preferentially and dynamically the mitochondrial pool of BCL-2, with the interaction being regulated by both autophagy and apoptosis induction.
Although ER-localized BCL-2, but not mitochondrial-localized BCL-2, has been shown to be important for the inhibition of autophagy (Pattingre et al, 2005), our results might imply that mitochondrial-localized BCL-2 can modulate the function of AMBRA1, so as to inhibit, in turn, the autophagic pathway. However, other studies are necessary to determine whether mito-BCL-2 is able to inhibit AMBRA1's role in the BECLIN 1/Vps34 complex formation. To this end, it will also be necessary to determine whether AMBRA1 can bind BCL-2 directly or through a cofactor. Different mechanisms have been proposed for regulating the binding of BCL-2/BCL-XL to BECLIN 1 at the ER, including JNK1-mediated BCL-2 phosphorylation, competitive disruption of BCL-2–BECLIN 1 binding by other BH3 domain proteins (Sinha and Levine, 2008) and phosphorylation of the BH3 domain of BECLIN 1 by DAP kinase (Zalckvar et al, 2009). In this study, we found that AMBRA1 can compete with BCL-2 to bind BECLIN 1 and thus may act positively in the autophagic process.
The binding between AMBRA1 and mito-BCL-2 is disrupted following autophagy induction
BCL-2 has been mainly studied in the context of cell death; yet, it is also known to carry out other functions in different cellular processes, such as cell cycle progression, glucose homeostasis, transcriptional repression by p53 and autophagy (Reed, 1998; Danial and Korsmeyer, 2004; Pattingre et al, 2005; Maiuri et al, 2007). A subtle regulation of BCL-2 at the expression level exists, due to its long half-life (Reed, 1996). However, it is known that post-translational modifications of BCL-2, and in particular phosphorylation, have crucial roles in regulating its activity (Blagosklonny, 2001; Wei et al, 2008; Pattingre et al, 2009). Recently, it has been shown that BCL-2 phosphorylation on site T69, S70 and S87 by JNK1 inhibits its antiautophagic function (Pattingre et al, 2009). How does the disruption of AMBRA1–mito-BCL-2 complex occur, following autophagy induction? We have evidence indicating that phosphorylation of BCL-2 by JNK1 does not regulate BCL-2 interaction with AMBRA1. Indeed, we found that AMBRA1 is able to co-immunoprecipitate with EEE-BCL-2, a mutant that phosphomimics the phosphorylation of BCL-2 at sites T69, S70 and S87 during autophagy induction (Wei et al, 2008). In agreement with this observation, the interaction between AMBRA1 and mito-BCL-2 remains unchanged after induction of autophagy and treatment with a JNK1 kinase inhibitor.
Another hypothesis is that the disruption of the interaction between AMBRA1 and mito-BCL-2 is regulated by post-transcriptional modifications on AMBRA1. AMBRA1 can be directly phosphorylated by the serine/threonine kinase ULK1 and, indeed, when subjected to 2D-gel electrophoresis, it shows multiple spots, corresponding to a number of modifications (Di Bartolomeo et al, 2010). In line with this evidence, mito-AMBRA1 may be subjected to similar modifications during apoptosis and autophagy.
Another hypothesis concerning the function of AMBRA1 at the mitochondria comes from new data proposed by Hailey et al (2010), in which it is demonstrated that mitochondria can serve as a source of autophagosomal membranes during starvation in mammalian cells. Although isolation of mitochondria is likely to be contaminated with ER, in line with these results, we can speculate that AMBRA1 is present at the mitochondria to participate in the production of mitochondrial autophagosomes.
Indeed, our results highlight the important role played by the subcellular localization of AMBRA1 for its function. Noteworthy, we found an increase of binding between AMBRA1 and BECLIN 1 in the mitochondrial fraction, after autophagy induction. Given that we have recently demonstrated that a pool of AMBRA1 and BECLIN 1 form a complex and translocate together from the dynein motor complex to the ER upon autophagy (Di Bartolomeo et al, 2010), these results, combined with the recent findings from Hailey et al lead us to propose a model in which, in normal conditions, a pool of AMBRA1 (hereafter mito-AMBRA1) is docked by BCL-2 at the mitochondria: after autophagy induction, mito-AMBRA1 dissociates from mito-BCL-2, and increases its binding with a fraction of mito-resident BECLIN 1; Mito-AMBRA1 would have therefore the potential to enhance BECLIN 1-dependent autophagy (see Supplementary Figure S8, mitochondrial model). As we observed a reduction of AMBRA1 colocalization with mitochondria in response to autophagy (see Supplementary Figure S6A), accompanied with an increase of binding between AMBRA1 and BECLIN 1 in the microsomal fraction with a reciprocal decrease of binding between BCL-2 and BECLIN 1 (see Supplementary Figure S6B), we cannot exclude that a fraction of this mitochondrial pool of AMBRA1 translocates to the ER in order to bind ER-resident BECLIN 1 and further enhances the autophagic programme (see Supplementary Figure S8, omegasome model). Although AMBRA1 has not been shown to be in a stable complex with BECLIN 1 (Zhong et al, 2009; Funderburk et al, 2010), our present results indicate that AMBRA1 and BECLIN 1 can form ‘transient’ distinct complexes that can regulate the autophagic programme.
AMBRA1 and mito-BCL-2 interaction is disrupted after both autophagy and apoptosis induction
The fact that both STS treatment and autophagy induction are able to induce the dissociation of AMBRA1 from mito-BCL-2 led us to hypothesize that these two treatments induce a common pathway. Indeed, it is known that some apoptosis inducers can trigger, at first, an autophagic response (Colell et al, 2007); this being the case, autophagy could be considered as a defensive response following stress-induced cell death. Our results, however, indicate that AMBRA1 and mito-BCL-2 interaction can be modulated by induction of both autophagy and apoptosis, thus underlying a double function of AMBRA1 both in autophagy and cell death control. Several authors have recently investigated the relationship between the proautophagic and proapoptosis activity of BECLIN 1 (Boya and Kroemer, 2009; Chang et al, 2010; Djavaheri-Mergny et al, 2010; Luo and Rubinsztein, 2010; Maiuri et al 2010). In particular, Djavaheri-Mergny et al (2010) reported that caspases can cleave BECLIN 1, thereby destroying its proautophagic activity. Thus, it will be very interesting to further investigate the relationship between AMBRA1 and caspase activities.
In conclusion, this work describes a novel interplay between the regulation of AMBRA1 function, BCL-2 and autophagy. For this reason, it opens up new areas of research in the complex relationship between apoptosis and autophagy.
Materials and methods
Antibodies
Polyclonal anti-FLAG (Sigma-Aldrich, F7425), monoclonal anti-myc 9E10 (Santa Cruz), polyclonal anti-myc A-14 (Santa Cruz), polyclonal anti-Beclin 1 (Santa Cruz), monoclonal anti-BECLIN 1 (Becton-Dickinson), mouse monoclonal anti-TUBULIN (Sigma-Aldrich), polyclonal anti-ACTIN (Sigma-Aldrich), monoclonal anti-BCL-2 (Santa Cruz, sc-7382), polyclonal anti-BCL-2 (Abcam, Ab7973), polyclonal anti-PARP antibody (Cell Signaling), polyclonal anti-MnSOD (Assay Designs), polyclonal anti-PDI (Stressgen), polyclonal anti-AMBRA1 (from SDI and Covalab), monoclonal anti-p62 (BD Transduction Laboratories) and polyclonal anti-LC3 (Cell Signaling).
Reagents and plasmid constructs
Constructs coding for AMBRA1 and its fragments were cloned in pLPCX vector (Clontech; Fimia et al, 2007). Plasmids containing either wild-type h-BCL-2-myc, EEE-BCL-2 mutant, ER-BCL-2, Mito-BCL-2, wild-type BECLIN 1 and all BECLIN 1 mutant cDNAs were present in Beth Levine's Laboratory (Howard Hughes Medical Institute, Dallas, Texas). Rapamycin and STS were purchased from Sigma-Aldrich. The Q-VD-ODH caspase inhibitor was from Kamyia Biomedical Company.
Immunoprecipitation
Cells were lysed either in HEMG buffer (25 mM HEPES (pH 8.0), 100 mM NaCl, 0.5% Nonidet P-40, 0.1 mM EDTA, 10% glycerol) plus protease and phosphatase inhibitors or in CHAPS buffer (40 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM EDTA, 0.3% of CHAPS) in order to detect endogenous AMBRA1–BCL-2 interaction. After centrifugation at 4°C for 10 min at 13 000 g, equal amounts of protein were incubated with either anti-myc, anti-FLAG, anti-Beclin 1, anti-BCL-2 or anti-AMBRA1 antibodies (2 μg) overnight at 4°C, followed by 60 min incubation with 20 μl of protein A/G sepharose beads (Amersham Bioscience). The beads were collected by centrifugation and washed four times with the HEMG buffer. Proteins bound to the beads were eluted with 30 μl of SDS–polyacrylamide gel electrophoresis sample buffer and heated to 95°C for 10 min.
Immunocytochemistry
Cells were washed in PBS and fixed with 4% paraformaldehyde in PBS for 15 min. In order to stain mitochondria, mitotracker was applied to cells, prior to fixation, 15 min at 100 nM. After permeabilization with 0.2% Triton X-100 in PBS for 5 min, cells were blocked in 2% horse serum in PBS and incubated for 1 h at RT with primary antibodies. We used the antibodies directed against AMBRA1, BCL-2 and the myc Tag. Cells were then washed in blocking buffer and incubated for 1 h with labelled anti-mouse (Alexa Fluor-488 or Alexa Fluor555, Molecular Probes, Eugene, OR) or anti-rabbit (FITC or Cy3, Jackson ImmunoResearch, West Grove, PA) secondary antibodies. Nuclei were stained with 1 μg/ml DAPI and examined under a Leica TCS SP5 confocal microscope equipped with a × 63 (NA 1.4) oil-immersion objective (Deerfield, IL). We used ‘Leica confocal’ software for analysis. In the case of AMBRA1 and mito-BCL-2 or ER-BCL-2 analysis, we used the Olympus IX70 microscope combined with SoftWorx software. Deconvolution was applied in standard condition (8 cycles). Image analysis was undertaken using the ImageJ analysis programme using the PSC colocalization plug-in (French et al, 2008) to calculate colocalization. At least 20 cells were analysed for each condition. Results are presented in terms of Pearson correlation coefficients, which represent the linear relationship of the signal intensity from the green and red channels of the analysed image.
Förster resonance energy transfer
For standard acceptor photobleaching FRET microscopy, HEK293 cells were seeded on coated glass coverslips and co-transfected with a vector encoding AMBRA1–mCherry and a vector encoding BCL-2–GFP. Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde for 10 min and, following three washes, nuclei were stained with 1 μg/ml DAPI 20 min and then placed on slides and embedded in mounting medium, as described above. FRET studies were performed on a confocal microscope (TCS SP5, Leica) using the implemented FRET acceptor photobleaching wizard. Acquisition settings were as follows: objective Plan-Apochromat × 63/1.4 NA oil immersion, pinhole 2 Airy units, image size 512 × 512. Prebleach and postbleach images were serially recorded by excitation of BCL-2–GFP at 488 nm (donor channel) with an argon laser, and mCherry–AMBRA1 at 543 nm (acceptor channel) with an helium–neon laser. Low laser intensities were used to avoid bleaching effects during acquisition, selection of the cells were made by visualizing only the donor channel to prevent premature partial bleaching of the acceptor. The acceptor was bleached with high intensity (100%) power at the 543 nm laser line for 10 iterations. This iteration time was found to be effective for bleaching mCherry–AMBRA1 in pilot experiments. The change in the fluorescence intensity between pre- and postbleach donor values (efficiency, E) was calculated using the formula E=(donor after−donor before) × 100/donor after, and was shown as a percentage; pseudo-coloured images showing FRET efficiency values were also generated. We analysed 10 cells from each of three independent experiments, where cells were maintained in basal conditions without starvation.
Isolation of microsomal fraction
HEK293 cells were plated in ten 10-cm Petri dishes and co-transfected with vectors encoding both FLAG–Beclin 1 and myc–AMBRA1. Five dishes were then shifted in EBSS media during 4 h and the other five dishes grown in normal media. Cells were washed twice with 5 ml of a solution of 0.137 M NaCl/2.7 mM KCl/8 mM Na2HPO4/1.5 mM KH2PO4 (PBS), harvested in a solution of 5 mM EDTA in PBS and washed with 5 ml of PBS. The cells were swollen at 0°C for 10 min in 2 ml of hypotonic solution of 10 mM Tris–HCl, pH 7.5/0.5 mM MgCl2, and then protease inhibitor cocktail was added at 1 mM. Cells were homogenized with 30 strokes in a glass Dounce homogenizer and the homogenate was diluted in a equal volume of a solution of 0.5 M sucrose/6 mM 2-mercaptoethanol/40 μM CaCl2/300 mM KCl/10 mM Tris–HCl, pH 7.5. The suspension was centrifugated at 10 000 g for 20 min to pellet nuclei and mitochondria. The supernatant was made at 0.6 M KCl. The suspension was centrifugated at 100 000 g for 60 min to sediment the microsomal fraction. The pellet was suspended in a solution containing 0.25 M sucrose, 0.15 M KCl, 3 mM 2-mercaptoethanol, 20 μM CaCl2, 10 mM Tris–HCl (pH 7.5), and centrifugated again at 100 000 g for 60 min. The pellet of protein was suspended in the same solution (Maruyama and MacLennan, 1988).
Transmission electron microscopy and immunogold staining
Mitochondria were isolated from HeLa cells by standard differential centrifugation, and suspended in isolation buffer (0.2 M sucrose, 10 mM Tris-MOPS (pH 7.4), 0.1 mM EGTA-Tris, 0.1% delipidated bovine serum albumin, BSA). They were then fixed in 2% freshly depolymerized paraformaldehyde and 0.2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 2 h at 4°C. Samples were rinsed in buffer partially dehydrated and embedded in Epon resin (Fluka), sectioned and collected on uncoated nickel grids. Ultrathin sections were processed for immunogold technique. Grids were preincubated with 10% normal goat serum in 10 mM PBS containing 1% BSA and 0.13% NaN3 (medium A), for 15 min at RT; sections were then incubated with primary antibody, rabbit polyclonal anti-AMBRA1 (ProSci Inc.) diluted 1:50 in medium A, overnight at 4°C. After rinsing in medium containing 0.01% Tween-20 (Merck), a second incubation with primary antibody mouse anti-BCL-2 (Santa Cruz) diluted 1:50 in medium A was performed, for 2 h at RT. After incubation in medium A for 15 min at RT, sections were incubated for 1 h at RT in a solution of goat anti-rabbit IgG conjugated to 15 nm colloidal gold (British BioCell Int.) and goat anti-mouse IgG conjugated to 5 nm colloidal gold (British BioCell Int.) both diluted 1:30 in medium A containing fish gelatine 0.01%. Finally, grids were thoroughly rinsed in distilled water, contrasted with aqueous 2% uranyl acetate for 20 min, and Reynolds solution (3.5% tri-sodium citrate, 2.6% lead nitrate, Fluka) for 10 min, and then photographed in a Technai 20 (FEI Company, Eindhoven, the Netherlands) electron microscope.
Mitochondrial crosslinking
HEK293 cells were grown in normal medium or in EBSS medium during 4 h and fixed with PFA 0.5% during 10 min. After three washes with PBS, mitochondrial fractions were purified as described before. We also performed an immunoprecipitation of endogenous AMBRA1 with an antibody anti-AMBRA1 or with preimmune IgG as a negative control.
Supplementary Material
Acknowledgments
This study was partly supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Telethon Foundation, the Italian Ministry of Health and the Italian Ministry of University and Research. Beth Levine lab is partly supported by the National Institute of Health (NIH ROI CA109618). Flavie Strappazzon is the recipient of a post-doctoral Marie Curie grant ‘TRACKS’ ITN. Silvia Campello is supported by a Fp-pEoplE-iEF-2008 grant (grant agreement 235595). We thank Dr Palma Mattioli for confocal microscopy analysis, and Martin W Bennett and Magdalena Acuna Villa for excellent secretarial and editorial work.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams JM, Cory S (2007) Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr Opin Immunol 19: 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertazzi L, Arosio D, Marchetti L, Ricci F, Beltram F (2009) Quantitative FRET analysis with the EGFP-mCherry fluorescent protein pair. Photochem Photobiol 85: 287–297 [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV (2001) Unwinding the loop of Bcl-2 phosphorylation. Leukemia 15: 869–874 [DOI] [PubMed] [Google Scholar]
- Boya P, Kroemer G (2009) Beclin 1: a BH3-only protein that fails to induce apoptosis. Oncogene 28: 2125–2127 [DOI] [PubMed] [Google Scholar]
- Cecconi F, Di Bartolomeo S, Nardacci R, Fuoco C, Corazzari M, Giunta L, Romagnoli A, Stoykova A, Chowdhury K, Fimia GM, Piacentini M (2007) A novel role for autophagy in neuro development. Autophagy 3: 506–508 [DOI] [PubMed] [Google Scholar]
- Chang NC, Nguyen M, Germain M, Shore GC (2010) Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J 29: 606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR (2007) GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 5: 983–997 [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116: 205–219 [DOI] [PubMed] [Google Scholar]
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antoniolo M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, D′Amelio M, Nardacci R, Romagnoli A, Piacentini M, Cecconi F, Fimia GM (2010) The dynamic interaction of Ambra1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 191: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M, Maiuri MC, Kroemer G (2010) Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 12: 1717–1719 [DOI] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3: 619–628 [DOI] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z (2010) The Beclin 1-VPS34 complex-at the crossroads of autophagy and beyond. Trends Cell Biol 6: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Shore GC (2003) Cellular distribution of Bcl-2 family proteins. Sci STKE 173: pe10. [DOI] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 4: 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B (2010) The Beclin 1 interactome. Curr Opin Cell Biol 2: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoetelmans R, van Slooten HJ, Keijzer R, Erkeland S, van de Velde CJ, Dierendonck JH (2000) Bcl-2 and Bax are present in interphase nuclei of mammalian cells. Cell Death Differ 4: 384–392 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 283: 22847–22857 [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 19: 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Sinha S, Kroemer G (2008) Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 4: 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10: 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T, van Driel R, Bertram JF, Strasser A (1994) The protein product of the oncogene bcl-2 is a component of the nuclear envelope, the endoplasmic reticulum, and the outer mitochondrial membrane. Cell Growth Differ 5: 411–417 [PubMed] [Google Scholar]
- Luo S, Rubinsztein DC (2010) Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 17: 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Criollo A, Kroemer G (2010) Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J 29: 515–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G (2007) Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 26: 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, MacLennan DH (1988) Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc Natl Acad Sci 85: 3314–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11: 385–396 [DOI] [PubMed] [Google Scholar]
- Mitoma J, Ito A (1992) The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J 11: 4197–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2: 132–139 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P (2009) Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem 284: 2719–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939 [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P (2000) Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2: 992–998 [DOI] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Niebuhr K, Domann E, Wehland J (1994) The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J 13: 758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier BP, Taglialatela G (2006) Bcl-2 localized at the nuclear compartment induces apoptosis after transient overexpression. J Biol Chem 281: 40493–40502 [DOI] [PubMed] [Google Scholar]
- Reed JC (1996) Mechanisms of Bcl-2 family protein function and dysfunction in health and disease. Behring Inst Mitt 97: 72–100 [PubMed] [Google Scholar]
- Reed JC (1998) Bcl-2 family proteins. Oncogene 17: 3225–3236 [DOI] [PubMed] [Google Scholar]
- Sinha S, Levine B (2008) The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 27: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Zhong Q (2009) Regulation of Beclin 1 in autophagy. Autophagy 5: 713–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 9: 1142–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Meyerkord CL, Wang HG (2009) Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ 16: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B (2008) JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 30: 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A (2009) DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 10: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait B, Heintz N, Yue Z (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol 3-kinase complex. Nat Cell Biol 4: 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW (1996) Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J 15: 4130–4141 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.