Abstract
Nucleosomes impede access to DNA. Therefore, nucleosome positioning is fundamental to genome regulation. Nevertheless, the molecular nucleosome positioning mechanisms are poorly understood. This is partly because in vitro reconstitution of in vivo-like nucleosome positions from purified components is mostly lacking, barring biochemical studies. Using a yeast extract in vitro reconstitution system that generates in vivo-like nucleosome patterns at S. cerevisiae loci, we find that the RSC chromatin remodelling enzyme is necessary for nucleosome positioning. This was previously suggested by genome-wide in vivo studies and is confirmed here in vivo for individual loci. Beyond the limitations of conditional mutants, we show biochemically that RSC functions directly, can be sufficient, but mostly relies on other factors to properly position nucleosomes. Strikingly, RSC could not be replaced by either the closely related SWI/SNF or the Isw2 remodelling enzyme. Thus, we pinpoint that nucleosome positioning specifically depends on the unique properties of the RSC complex.
Keywords: in vitro reconstitution, nucleosome positioning, RSC chromatin remodelling complex, S. cerevisiae chromatin
Introduction
Eukaryotes package their nuclear DNA into a complex structure called chromatin. At the most basic level of chromatin, the DNA is wound around an octamer of histone proteins in ∼1.7 turns (Luger et al, 1997) constituting a nucleosome core particle. Nucleosome core DNA is much less accessible to DNA-binding factors than DNA in linker regions between nucleosome cores or in nucleosome-depleted regions (NDRs). Therefore, the positioning of nucleosomes with respect to the DNA sequence is a powerful lever for the regulation of DNA-templated processes, such as transcription or replication (Simpson, 1990; Venter et al, 1994; Liu et al, 2006; Field et al, 2008; Lantermann et al, 2010). This global importance of nucleosome positioning was underscored by the high degree of defined positions in recent genome-wide nucleosome mappings in organisms from yeast to man (Yuan et al, 2005; Albert et al, 2007; Lee et al, 2007; Ozsolak et al, 2007; Whitehouse et al, 2007; Field et al, 2008, 2009; Schones et al, 2008; Shivaswamy et al, 2008; Valouev et al, 2008; Mavrich et al, 2008a, 2008b; Lantermann et al, 2010). Nevertheless, the molecular mechanism for nucleosome positioning in vivo is by far not fully understood.
As nucleosomal DNA is tightly bent, it is an attractive hypothesis that intrinsic features of DNA sequences have a major role in nucleosome positioning. Some sequence features, like certain dinucleotide periodicities (Satchwell et al, 1986), intrinsically favour, and others, like poly(dA:dT) stretches (Simpson and Shindo, 1979), disfavour nucleosome formation (Travers et al, 2009). Indeed, there is a significant correlation of such features with nucleosome positioning in vivo. For example, poly(dA:dT) stretches are enriched in S. cerevisiae promoter NDRs (Iyer and Struhl, 1995; Bernstein et al, 2004; Yuan et al, 2005), and a 10 bp periodicity of AA/TT/AT dinucleotides is more prevalent in strongly positioned nucleosomes flanking NDRs (Ioshikhes et al, 2006; Segal et al, 2006; Mavrich et al, 2008a). However, such rules are not universal. S. pombe NDRs, for example, are not enriched for poly(dA:dT) stretches (Lantermann et al, 2010), and also other yeasts do not necessarily use such sequences to establish promoter NDRs (Tsankov et al, 2010).
Intrinsic DNA sequence rules of nucleosome formation may be probed by in vitro reconstitution via salt gradient dialysis, which involves only histones and DNA mixed at initially high salt concentration that is slowly diluted until nucleosomes form spontaneously (Widom, 2001). Recently, two groups reconstituted the whole S. cerevisiae genome by salt gradient dialysis and found some overall correlations of in vitro and in vivo nucleosome occupancy, particularly at the promoter NDRs (Kaplan et al, 2009; Zhang et al, 2009), but individual nucleosome positions were mostly not recapitulated (Zhang et al, 2009). Clearly, additional factors beyond just the DNA and histones determine nucleosome positions in vivo.
What are these nucleosome positioning factors? Besides a role of some abundant sequence-specific DNA-binding proteins, like budding yeast Reb1 and Abf1 (Raisner et al, 2005; Badis et al, 2008; Hartley and Madhani, 2009), ATP-dependent nucleosome remodelling enzymes are implicated in global nucleosome positioning. Such enzymes enable the assembly, disassembly or relocation of nucleosomes, and in some cases they can catalyse histone exchange events. They vary in the type of ATPase subunit and in the association with different subunits (Clapier and Cairns, 2009). The S. cerevisiae Isw2 and Isw1 remodelling enzymes were shown to move nucleosomes over intrinsically unfavourable sequences at the 5′ and 3′ ends of genes (Isw2 (Whitehouse et al, 2007)) and at mid-coding regions (Isw1 (Tirosh et al, 2010)), which in both cases were associated with suppression of erroneous transcription. Conversely, in S. pombe, which does not encode a remodelling enzyme of the ISWI family, the remodelling enzyme Mit1 appears to be involved in generating regular nucleosomal arrays (Lantermann et al, 2010). Finally, the essential remodelling enzyme RSC appears to keep NDRs nucleosome-free in S. cerevisiae (Badis et al, 2008; Hartley and Madhani, 2009). Ablation of RSC in temperature-sensitive mutants increased nucleosome occupancy at ∼55% of NDRs (Hartley and Madhani, 2009). However, such effects in conditional mutants may be indirect or confounded by cell viability issues.
Therefore, complementary to the initial identification of nucleosome positioning factors in vivo, there is an urgent need for an in vitro reconstitution system that generates in vivo-like nucleosome positioning in order to elucidate the molecular mechanism. Previously, we reported the establishment of such an in vitro system using yeast extracts that was able to successfully generate in vivo-like patterns of nucleosome positions at several yeast promoters (Korber and Horz, 2004; Hertel et al, 2005; Wippo et al, 2009). In this study, we describe the enrichment of the nucleosome positioning activity by chromatography and by fractionation of the yeast extract. We identify the RSC nucleosome remodelling complex and show directly by in vitro reconstitution that it has a specific, necessary, and in some cases even sufficient, role in nucleosome positioning at yeast promoters.
Results
The nucleosome positioning activity for the PHO8 promoter could be enriched over four sequential fractionation steps
The S. cerevisiae PHO8 promoter has promoter nucleosomes with stereotypical positioning (Yuan et al, 2005; Mavrich et al, 2008a; Jiang and Pugh, 2009), that is, an NDR of ∼120 bp that is flanked by two positioned nucleosomes with the downstream nucleosome N+1 covering the TSS (Figure 1A). Upstream of PHO8 is the divergently transcribed KRE2 gene with a similarly stereotypical promoter. In short, in the following sections, we call this entire region the ‘PHO8 promoter’.
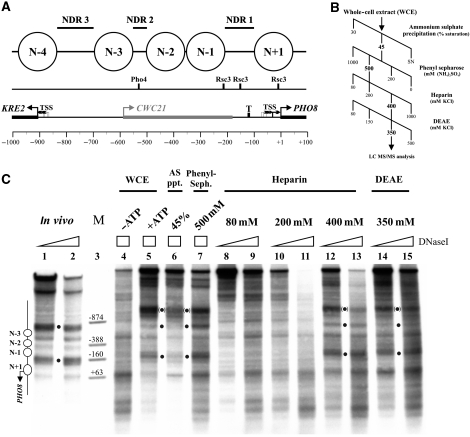
Figure 1.
The nucleosome positioning activity for the PHO8 promoter could be enriched from a yeast whole-cell extract (WCE) over four sequential fractionation steps. (A) Top panel: schematics of nucleosome positions at the KRE2-CWC21-PHO8 locus, according to Barbaric et al (1992) and Jiang and Pugh (2009). Nucleosomes are numbered relative to NDR1. Middle panel: mapped Pho4 (Barbaric et al, 1992) or predicted Rsc3 (Badis et al, 2008) binding sites (Supplementary Figure S8A). Lower panel: KRE2, CWC21 and PHO8 open reading frames (rectangular bars with large broken arrows), TATA box (T; Basehoar et al, 2004) and transcriptional start sites (TSS, small broken arrows; Miura et al, 2006). Scale bar: distance in base pairs from PHO8 ORF start. All panels drawn to scale. (B) Extract fractionation scheme. Fractions positive for the PHO8 promoter nucleosome positioning activity are labelled in bold. SN, supernatant. (C) DNaseI indirect end labelling analysis of the PHO8 promoter region in vivo or in vitro after salt gradient dialysis assembly and incubation with either WCE in the presence or absence of ATP, or with one of the indicated fractions (see B) in the presence of ATP. Black dots: diagnostic bands, which are characteristic for the in vivo pattern and seen in vitro only in the presence of ATP and the nucleosome positioning activity. Black dots in parentheses: hypersensitive site within the lacZ ORF of the pUC19 backbone specific for the in vitro pattern that always co-occurred with the in vivo-like PHO8 promoter pattern. The yeast sequence terminates close to the top marker band. Schematics on the left analogous to (A). Position of marker bands is labelled relative to the PHO8 ORF start. Ramps and boxes: relative DNaseI concentrations. All samples were electrophoresed alongside in the same gel, but the in vivo samples migrated slightly faster, probably because of different total DNA concentration.
We assembled plasmids carrying the PHO8 promoter into chromatin by salt gradient dialysis using Drosophila embryo histone octamers. As shown before (Hertel et al, 2005), this assembly by itself was unable to reconstitute the in vivo nucleosome positions (Figure 1C, lane 4, note that the pattern of salt gradient dialysis chromatin does not change in the presence of extract if no ATP is added; Hertel et al, 2005). However, incubation of such chromatin templates with a yeast whole-cell extract (WCE) and ATP shifted the nucleosomes to their in vivo positions (Figure 1C, lane 5; Hertel et al, 2005). Importantly, we analyse in vivo and in vitro chromatin samples side-by-side by using the same methodology and in the same gels. This way the nucleosome positioning patterns of different samples can be directly compared. Using this assay, we traced the nucleosome positioning activity during extract fractionation over four sequential steps (Figure 1B and C). The protein complexity was greatly reduced (Supplementary Figure S1), with only a moderate loss of the nucleosome positioning activity. As our reconstitution system could also generate in vivo-like positioning at other loci (Korber and Horz, 2004; Hertel et al, 2005; Wippo et al, 2009; Figures 2 and 3, Supplementary Figure S2, and data not shown), we tested the 500-mM ammonium sulphate phenyl sepharose fraction and the final 350-mM KCl DEAE fraction on other promoters as well. While the former fraction was as positive as the WCE for almost all loci, the latter was mainly positive for PHO8 (data not shown), indicating distinct nucleosome positioning activities for different loci.
Figure 2.
Purified RSC repositioned nucleosomes in salt gradient dialysis chromatin, but only in few cases, resulting in in vivo-like positions. DNaseI indirect end labelling analysis of the (A) PHO8, (B) RIM9, (C) CHA1 and (D) SNT1 promoter regions in vitro after assembly by salt gradient dialysis and incubation with WCE or purified RSC complex in the presence or the absence of ATP as indicated. The amount of RSC is given as the molar ratio of RSC to nucleosomes. In each panel, lanes 1 and 2 show the wt in vivo DNaseI pattern. Free DNA samples correspond to the respective non-assembled plasmids in the absence of WCE, RSC and ATP but under otherwise identical conditions. Bars in between lanes mark hypersensitive regions that correspond, at least to some degree, to NDRs of the in vivo patterns. The arrow between lanes 12 and 13 in D marks a nuclease-sensitive region that becomes inaccessible because of RSC activity. Ramps: increasing DNaseI concentrations. Position of marker bands is labelled relative to the ORF start of the respective locus. Schematics on the left are analogous to Figure 1A for the respective locus. Predicted Rsc3 binding sites (Supplementary Figure S8) are indicated by black dots.
Figure 3.
Purified RSC could rescue the nucleosome positioning activity of an extract generated from an rsc3-ts mutant grown under restrictive conditions. DNaseI indirect end labelling analysis of the (A) PHO8, (B) RIM9, (C) CHA1, and (D) SNT1 promoter regions as in Figure 2, but with WCEs generated from wild-type (BY4741) grown logarithmically at 30°C, or from rsc3-ts strain (TH8239) grown logarithmically at 25°C with or without an overnight shift to 37°C. Addition of RSC corresponded to the 1:5 ratio in Figure 2.
LC-MS/MS analysis of the final 350-mM KCl DEAE fraction
LC-MS/MS analysis of the final 350-mM KCl DEAE fraction identified 212 proteins (Supplementary Table S2), of which 95 localized outside the nucleus and 117 localized at least partially to the nucleus or had no known localization (Huh et al, 2003). These 117 proteins were, in principle, the more promising candidates, but many of them were excluded from further analysis, as yeast strains harbouring deletion or temperature-sensitive alleles of the respective genes showed the wild-type DNaseI pattern at the PHO8 promoter in vivo (Supplementary Table S2; data not shown).
Purified RSC repositioned nucleosomes in salt gradient dialysis chromatin, but only in few cases resulting in in vivo-like positions
Intriguingly, our final fraction contained 10 out of 17 subunits of the RSC complex (Supplementary Table S2), suggesting a role for this remodelling enzyme. To directly test whether the RSC complex was sufficient for proper nucleosome positioning, we chose a test set of four yeast loci in which a role of RSC in nucleosome organization had either previously been implicated (RIM9 and PHO8 (Badis et al, 2008), CHA1 (Moreira and Holmberg, 1999; Badis et al, 2008; Parnell et al, 2008)) or not (PHO8 (Parnell et al, 2008) and SNT1 (Badis et al, 2008; Hartley and Madhani, 2009)). We assembled equimolar amounts of four plasmids, each carrying one of these loci, together in the same reaction by salt gradient dialysis with purified histones. This pool of pre-assembled plasmids was the common starting material for the following experiments.
Similar to the PHO8 locus, the main NDRs and some of the positioned nucleosomes at both the CHA1 and the SNT1 locus were properly generated upon addition of WCE and ATP to salt gradient dialysis chromatin, whereas salt gradient dialysis by itself again did not recapitulate in vivo-like nucleosome positioning (Figure 2C and D, compare lanes 4–5 and 12–13 with lanes 1–2). The pattern of salt gradient dialysis chromatin and WCE without ATP was again the same as that for untreated salt gradient dialysis chromatin. So also at these loci as well, our yeast extract-based in vitro reconstitution system generated in vivo-like nucleosome organization from non-in vivo-like salt gradient dialysis chromatin. Nevertheless, salt gradient dialysis assembly alone could reconstitute the RIM9 NDR2 and ECM40 NDR3 to some extent correctly (Figure 2B, compare lanes 12–13 with lanes 1–2), while addition of WCE broadened RIM9 NDR2. The RIM9 locus turned out to be a rare example in which in vivo-like nucleosome positioning was less properly reconstituted in our yeast extract-based in vitro system. Moreover, the NDR1 at AEP1 was not met under any in vitro conditions. A strong band close to the position of NDR1 in the presence of RSC (Figure 2B, lanes 6–9) was not at the proper position as seen by indirect end labelling using a secondary cleavage with better resolution for this region (data not shown).
Strikingly, addition of purified RSC and ATP to salt gradient dialysis chromatin already generated the proper NDR at CHA1 to some degree (Figure 2C, compare lanes 6–9 with lanes 1–2) and could clearly position nucleosome N-1 at the SNT1 locus as in vivo (Figure 2D, compare lanes 6–9 with lanes 1–2). The prominent band in the salt gradient dialysis chromatin pattern (arrow between lanes 12 and 13) at the position of the SNT1 nucleosome N-1 was removed by the addition of purified RSC (or of WCE) in the presence of ATP, suggesting that RSC alone could move a nucleosome to an in vivo-like position. This was true for both tested RSC concentrations (Figure 2C and D, compare lanes 6–7 with lanes 8–9).
In contrast, addition of purified RSC was unable to reconstitute in vivo-like positioning both at the PHO8 (Figure 2A, compare lanes 6–9 with lanes 1–2) and at the RIM9 locus (Figure 2B, compare lane 6–9 with lanes 1–2), although it did change the pattern of the salt gradient dialysis chromatin (Figure 2A–D, compare lanes 6–9 with lanes 12–13) arguing for sufficient remodelling activity in the assay. Importantly, and in accordance with our earlier findings (Korber and Horz, 2004; Hertel et al, 2005; Wippo et al, 2009), the nucleosome positioning activity of both purified RSC and of the WCE was strictly dependent on the presence of ATP (Figure 2A–D, compare lanes 10–13 with lanes 4–9).
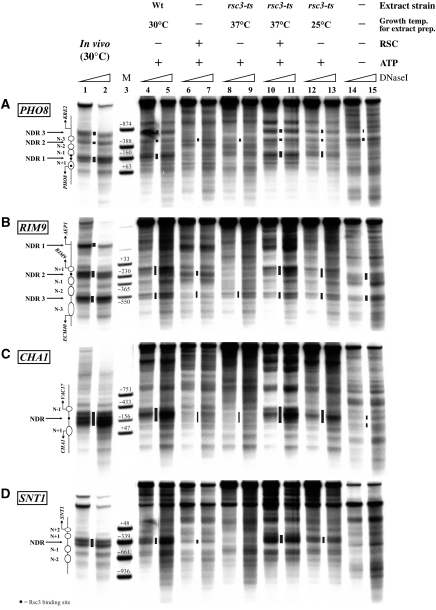
A direct and necessary role for RSC in generating in vivo-like nucleosome positions at PHO8, RIM9, CHA1 and SNT1 in vitro
As purified RSC could generate only a minor fraction of the proper nucleosome positioning in vitro, we wondered whether it was even necessary. We prepared extracts from a strain carrying a temperature-sensitive allele of the gene coding for the essential Rsc3 subunit of the RSC complex (rsc3-ts mutant (Badis et al, 2008)) that was grown at the non-permissive temperature (37°C) overnight. Such an extract (Figure 3, ‘rsc3-ts 37°C’ extract) was much less effective in positioning nucleosomes properly than the wild-type WCE (Figure 3A–D, compare lanes 8–9 with lanes 4–5), while an extract prepared from the rsc3-ts strain grown at 25°C functioned almost like the wild-type WCE (Figure 3A–D, compare lanes 12–13 with lanes 4–5). The rsc3-ts 37°C extract failed to reconstitute NDR1 and NDR3 at the PHO8 promoter, NDR2 at the RIM9 promoter, the broad NDR at the CHA1 locus and the strong NDR at the SNT1 promoter (Figure 3A–D, lanes 8–9). Nevertheless, it did change the pattern of the salt gradient dialysis chromatin starting material (Figure 3A–D, compare lanes 8–9 with 14–15), arguing for residual nucleosome remodelling activity also in this extract. In summary, the rsc3-ts 37°C extract was sufficiently impaired in its nucleosome positioning activity to confirm the necessary role of RSC and to serve as a background for rescue experiments using purified RSC complex.
Indeed, the addition of purified RSC to the rsc3-ts 37°C extract, completely rescued the nucleosome positioning activity for all four tested loci (Figure 3A–D, compare lanes 10–11 with lanes 4–5) and yielded patterns that were even slightly more in vivo-like than those generated by the wild-type WCE. This suggests that in the wild-type WCE, RSC may even be a limiting factor for proper nucleosome positioning. The rescue by purified RSC strongly suggests that the changes observed with the rsc3-ts, arp9-ts and sth1-td strains, by us (see below, Figure 5 and Supplementary Figure S3) and by others (Supplementary Figure S4, Supplementary Table S3) (Badis et al, 2008; Parnell et al, 2008; Hartley and Madhani, 2009), were not caused by indirect effects. Moreover, purified RSC could generate much less of the proper nucleosome positioning than in combination with the rsc3-ts 37°C extract (Figure 3A–D, compare lanes 6–7 with lanes 10–11). Therefore, both RSC and the rsc3-ts 37°C extract were unable to reconstitute in vivo-like nucleosome positioning on their own, but the combination of both reconstituted the full nucleosome positioning activity. Therefore, RSC is necessary but mostly not sufficient for proper nucleosome positioning.
Interestingly, we even found an example in which purified RSC counteracted the generation of in vivo-like nucleosome positioning. We published previously that almost in vivo-like nucleosome positioning was generated at the PHO84 promoter by mere salt gradient dialysis reconstitution (Wippo et al, 2009). RSC alone disrupted this intrinsically encoded in vivo-like positioning, whereas the proper positioning was generated when RSC was added in the context of the rsc3-ts 37°C extract (Supplementary Figure S2, compare lanes 6–7 with 1–2, 4–5, 10–11 and 14–15). This further underscores the fact that additional factors from the extract are necessary to direct the role of RSC in nucleosome positioning.
The role of the RSC complex in nucleosome positioning in vitro is specific, as it could not be substituted by the SWI/SNF or Isw2 remodelling enzymes
We wondered whether the role of RSC was specific or whether other remodelling complexes could achieve similar results. The rsc3-ts 37°C extract likely still contained other remodelling enzymes. However, other remodelling enzymes might not be present in sufficient quantities to substitute for the loss of RSC function in our in vitro system. Most other remodelling enzymes are less abundant in the cell to start with (∼2000 copies of Sth1 per cell compared to ∼220 copies of Snf2; Ghaemmaghami et al, 2003), and they may be less stable during extract preparation or their concentration might have been affected indirectly because of the rsc3-ts conditions. Hence, the RSC complex might just have seemed necessary for nucleosome positioning in our in vitro system—and by extension also in previous in vivo studies—simply because it was the most abundant remodelling activity.
We added purified SWI/SNF or Isw2 remodelling enzymes in the same molar amount as previously carried out for the RSC complex to the rsc3-ts 37°C extract. These two remodelling complexes, whether alone or in combination with the rsc3-ts 37°C extract, were unable to generate in vivo-like positioning as achieved with RSC (Figure 4A–D, compare lanes 3–11 with lanes 1–2). Importantly, both remodelling enzymes individually (in the presence of ATP) altered the pattern of salt gradient dialysis chromatin (Figure 4A–D, compare lanes 8–9 and 10–11 with 12–13) to a certain extent, which confirmed sufficient activity to remodel the chromatin templates in vitro. Both remodelling enzymes did not change the pattern generated by the rsc3-ts 37°C extract (compare Figure 4A–D, lanes 3–6 with Figure 3A–D, lanes 8–9), possibly because both were already present in the rsc3-ts 37°C extract.
Figure 4.
RSC was specifically required for nucleosome positioning in vitro as both SWI/SNF and Isw2 failed to rescue the rsc3-ts 37°C extract. DNaseI indirect end labelling analysis of the (A) PHO8, (B) RIM9, (C) CHA1, and (D) SNT1 promoter regions as in Figures 2 and 3 but with addition of purified SWI/SNF or Isw2 remodelling enzymes as indicated. All remodelling enzymes were added at the same molar concentrations, corresponding to the 1:5 ratio in Figure 2.
Loss of essential subunits of the RSC remodelling complex altered chromatin structure at the PHO8, RIM9 and other promoters in vivo
Our in vitro results strongly argue for a direct role of RSC in nucleosome positioning also in vivo as suggested previously (Badis et al, 2008; Parnell et al, 2008; Hartley and Madhani, 2009). Genome-scale microarray data on changes in nucleosome occupancy upon RSC ablation were already available for the temperature-sensitive rsc3-ts allele (Badis et al, 2008) and for sth1-td degron mutants (Parnell et al, 2008; Hartley and Madhani, 2009). However, a detailed comparison between different methods is often difficult and it is usually advisable to confirm genome-wide data with locus-specific techniques for regions of interest. Therefore, we monitored the in vivo effect of RSC on chromatin patterns at selected test loci, by the same method as used for the in vitro patterns, that is, by DNaseI indirect end labelling. We used the same temperature-sensitive strains as Badis et al and Parnell et al (rsc3-ts (Badis et al, 2008), sth1-td (Parnell et al, 2008)) and included an arp9-ts mutant (Cairns et al, 1998) as Arp9 came up very prominently in our LC-MS/MS analysis (Supplementary Table S2).
At the PHO8 promoter, a broad DNaseI hypersensitive site replaced nucleosome N-3 between NDR3 and NDR2, and the short hypersensitive site at the migration position of the −160 marker band was slightly diminished, indicating increased nucleosome occupancy over NDR1. We confirmed the DNaseI indirect end labelling results by restriction enzyme accessibility. In both the arp9-ts and the rsc3-ts mutant, there was an increase in accessibility of the HpaI site located within nucleosome N-3 and a decrease for the HindIII site located within NDR1 (Supplementary Figure S3). Consistently, Badis et al (2008) observed increased nucleosome occupancy at NDR1 and a broad region of decreased occupancy at the upstream edge of nucleosome N-3 in the rsc3-ts mutant under restrictive conditions (Supplementary Figure S4A). In contrast, Parnell et al (2008) did not see significant changes in the sth1-td strain, at least for which data are available for the PHO8 promoter region (Supplementary Figure S4A), maybe because of a shorter incubation time (2 h) at the restrictive temperature (see below).
The altered PHO8 promoter DNaseI pattern of the three temperature-sensitive mutants resembled the pattern of the PHO8 promoter after induction by phosphate starvation (Barbaric et al, 1992). This induced promoter pattern essentially depends on binding of the transactivator Pho4 in NDR2 (Barbaric et al, 1992; Munsterkotter et al, 2000). To control for inadvertent induction of the PHO regulon or for other Pho4-mediated effects due to ablation of essential RSC subunits, we generated pho4 rsc3-ts and pho4 arp9-ts double mutants. Importantly, the same altered chromatin structure was observed at the PHO8 promoter under restrictive conditions as in the ts single mutants (Supplementary Figure S5). Further, nucleosome positioning at the PHO84 promoter, which has a similarly low threshold of PHO induction as PHO8 (Lam et al, 2008), was largely unchanged in the rsc3-ts and arp9-ts mutants at the restrictive temperature, arguing also against inadvertent PHO regulon induction (Supplementary Figure S6).
The RIM9 NDR2 was identified as a prominent example for increased nucleosome occupancy in the rsc3-ts mutant under restrictive conditions (Badis et al, 2008; Supplementary Figure S4B). We confirmed this by DNaseI indirect end labelling and found the same effect in the arp9-ts and sth1-td strains as well. All three strains displayed significantly reduced DNaseI hypersensitivity over the RIM9 NDR2 (Figure 5B). Notably, this effect was locus specific as the nearby NDR1 at AEP1 and NDR3 at ECM40 were unaffected (Figure 5B).
Figure 5.
Loss of essential RSC subunits at elevated temperature altered chromatin structure at the PHO8, RIM9 and CHA1, but not at the SNT1 promoter. DNaseI indirect end labelling analysis of the (A) PHO8, (B) RIM9, (C) CHA1 and (D) SNT1 promoter regions in vivo. Nuclei were isolated from wild type (wt; BY4741) and strains carrying a temperature-sensitive (rsc3-ts (TH8247) and arp9-ts (YBC1536)) or temperature-sensitive degron (sth1-td (YBC2191)) allele of the indicated RSC subunits. Strains were grown logarithmically at 25°C and then shifted to the non-permissive temperature (37°C) overnight. Wt nuclei were also prepared from cells grown logarithmically at 30°C. Bars in-between lanes mark the intensity (bar width) and extent (bar length) of DNaseI hypersensitive sites. A stippled line separates samples that were not electrophoresed alongside on the same gel but combined in the figure. Asterisks indicate artefact bands. Ramps, markers and schematics as in Figure 2.
At the CHA1 locus, we did not see any effect in the sth1-td mutant, a weakly reduced NDR in the rsc3-ts mutant, although only in some experiments (Supplementary Figure S7C), and a very weak effect at the NDR in the arp9-ts mutant (Figure 5C). Hartley and Madhani (2009) also saw only small changes in a sth1-td mutant, whereas both Badis et al (2008) and Parnell et al (2008) reported clear effects (Supplementary Figure S4C and Supplementary Table S3). Therefore, in our experiment as well as in the literature, the CHA1 locus was not a clear responder to in vivo ablation of RSC subunits. This ambiguity is mirrored by two studies reporting RSC binding at CHA1 while two others did not (Supplementary Figure S4C, Supplementary Tables S3 and S4). Nevertheless, in the light of all available data we consider CHA1 as RSC target in vivo.
Finally, we observed no effects at the SNT1 locus (Figure 5D) consistent with other studies (Supplementary Figure S4D and Supplementary Table S3).
Besides the four loci that we used for our in vitro assays, we included six more loci in order to have a broader basis for the comparison of our data with published observations (Supplementary Figures S4 and S6, Supplementary Tables S3 and S4). To avoid missing any effects, we used rather harsh restrictive conditions (overnight incubation at 37°C), which compromised cell viability (47±2% for arp9-ts and <5% for rsc3-ts and sth1-td mutants). Nevertheless, it is very unlikely that this led to exaggerated or artifactual effects as our results were in excellent agreement with published data or showed even a bit weaker effects, for example, at ADH2 and CHA1 (Supplementary Table S3 and Supplementary Figure S4). Further, we tested all loci in which we saw an effect in the rsc3-ts mutant after overnight incubation at 37°C also after 6.5 h, which are the same conditions as used by Badis et al (2008) for this same strain and raised the cell viability to 31±3%. We observed the same effects as those after overnight incubation (Supplementary Figure S7). In addition, the unchanged patterns at the SNT1, ADH2 and PHO84 loci (Figure 5D and Supplementary Figure S6) argue against globally compromised chromatin structures even under the harsh overnight restrictive conditions. The only single case in which we observed more of an effect than others was the altered PHO8 promoter pattern in the sth1-td strain (see above). As this altered pattern was the same as that in the other two ts mutants (Figure 5A) and also that observed after shorter incubation times (Supplementary Figure S7A), it very likely reflects the true effect due to lack of RSC activity and could not be observed under the milder restrictive conditions used by Parnell et al (2008) (2 h at 37°C).
In summary, both our own as well as published in vivo data confirm that ablation of RSC activity in vivo interferes with nucleosome positioning. Interestingly, this was only true if essential RSC subunits were ablated as deletion of the genes encoding the non-essential RSC subunits Rtt102 or Rsc30 showed unaltered chromatin patterns at selected loci (Supplementary Figure S7 and data not shown). This clear demonstration of a role for RSC in nucleosome positioning in vivo argues that the RSC-dependent mechanism observed in vitro is not just coincidental but reflects the in vivo mechanism.
Discussion
In this study, we show for the first time by in vitro reconstitution that the RSC nucleosome remodelling complex is directly and specifically required to generate in vivo-like nucleosome positioning, especially to set up yeast promoter NDRs. There are a few cases in which RSC alone can properly determine nucleosome positioning. Nevertheless, RSC mostly requires other protein factors. Our findings provide strong evidence for the hypothesis that the in vivo nucleosome positioning machinery relies upon specific remodelling enzymes to correctly interpret nucleosome positioning cues given by the combination of DNA sequence in cis and other factors in trans. In other words, remodelling enzymes can be part of the nucleosome positioning information.
RSC is directly and specifically required, and in few cases even sufficient, to set up nucleosome positioning in vitro
A role of RSC in maintaining the NDRs at a large fraction of yeast promoters was suggested by three recent in vivo studies (Badis et al, 2008; Parnell et al, 2008; Hartley and Madhani, 2009). We confirmed these genome-scale results at the level of several individual promoters by DNaseI indirect end labelling using temperature-sensitive alleles of three different genes encoding essential RSC subunits. Very recently, a non-canonical RSC/nucleosome complex was suggested to reside within the NDR at the GAL1-10 promoter and to have a fine-tuning role for promoter induction (Floer et al, 2010). We confirmed that ablation of RSC activity affected this NDR, especially in the rsc3-ts mutant (Supplementary Figures S6 and S7D).
However, our own in vivo data as well as previous reports on roles for RSC in nucleosome positioning are based on conditional mutants that are compromised cells under restrictive conditions so that indirect effects may contribute to the changes at promoter NDRs. In addition, such experiments cannot distinguish whether RSC was just necessary or also sufficient for NDR formation. To answer these questions, we tested purified RSC in our in vitro reconstitution system starting from salt gradient dialysis-assembled chromatin.
In most cases, purified RSC in the presence of ATP was unable to achieve the same degree of in vivo-like nucleosome positioning as seen with the WCE. Importantly, this was not due to a lack of RSC activity in our preparation. This RSC preparation was sufficiently active to allow remodelling of the chromatin templates as the DNaseI pattern of the starting material was clearly changed by addition of RSC and ATP. Even more to the point, the successful positioning of nucleosome N-1 at the SNT1 and part of the NDR at the CHA1 locus by purified RSC in the presence of ATP is proof of principle that RSC alone can be sufficient to achieve even in vivo-like nucleosome positioning under the assay conditions. Finally, the same amount of purified RSC could completely rescue the proper nucleosome positioning activity of the rsc3-ts 37°C extract. Such an extract mimicked the in vivo phenotype in the sense that it was not able to generate the in vivo-like nucleosome positioning. As in the in vivo case, this could equally be caused by indirect effects of Rsc3 ablation on the activity of other factors. However, our rescue experiments strongly argue against indirect effects and show that RSC directly contributes to this nucleosome positioning activity. In addition, this experiment shows explicitly that additional factors from the extract are required in combination with RSC to generate the proper nucleosome positioning at most loci in vitro.
Intriguingly, both purified SWI/SNF and Isw2 remodelling enzymes failed to rescue the rsc3-ts 37°C extract, which argues that the RSC remodelling complex is specifically required for the nucleosome positioning activity and that only RSC can respond to the cues provided by the additional factors from the extract. Especially for the case of the SWI/SNF complex this specificity is somewhat surprising, as SWI/SNF and RSC are rather closely related remodelling enzymes with similar mechanistic properties in in vitro assays (Logie et al, 1999; Zhang et al, 2006), even sharing three subunits (Cairns et al, 1998). Nevertheless, both remodelling complexes may even have opposing roles; for example at the PHO8 promoter. Activation of PHO8 leads to a prominent chromatin transition at the PHO8 promoter (Barbaric et al, 1992), which essentially depends on the remodelling enzyme SWI/SNF (Gregory et al, 1998). The enrichment of RSC over the region occupied by nucleosome N-3 (Venters and Pugh, 2009; and Supplementary Figure S4A) and the strikingly similar loss of nucleosome N-3, both upon RSC inactivation and upon promoter activation, suggests that RSC ensures the proper placement of N-3 under repressive conditions, whereas SWI/SNF overrides RSC and removes nucleosome N-3 under activating conditions.
The difference in remodelling enzyme specificity may reflect differences in recruitment specificity. The RSC complex contains two subunits, Rsc3 and Rsc30, which are able to recognize a specific DNA sequence (CGCGC). The location of this motif often overlaps with the sites of nucleosome occupancy change in the rsc3-ts mutant (Badis et al, 2008). Indeed, such Rsc3 sites are present and conserved at the PHO8 promoter and other loci (Supplementary Figure 8A and B). However, recruitment of RSC through Rsc3/Rsc30 is unlikely the main or only reason for the specificity of RSC action. RSC was also necessary for proper formation of NDR3 at the PHO8 promoter, both in vivo and in vitro, even though there is no Rsc3 site nearby. Further, in vitro nucleosome positioning at the SNT1 locus, which does not contain an Rsc3 site, was strictly dependent on RSC.
Indeed, we were surprised that the rsc3-ts extract failed to reconstitute nucleosome positioning also at the SNT1 locus, although along with others (Badis et al, 2008; Hartley and Madhani, 2009) we did not see significant changes here upon RSC ablation in vivo. Again, purified RSC was able to rescue in vitro. Why this discrepancy between the RSC requirement in vivo versus in vitro at SNT1? The in vivo experiment addresses the loss of properly positioned nucleosomes upon shift to the restrictive temperature, whereas in vitro reconstitution monitors the de novo generation of correct nucleosome positioning. Therefore, other factors may maintain proper positioning in vivo even in the absence of RSC, while these factors are unable to set up proper positioning from scratch in vitro. Reb1 and Abf1 could be these factors at the SNT1 locus, as they are redundantly involved in NDR formation. Only in an abf1-td reb1-td double mutant the SNT1 NDR was compromised, but still not lost completely (Hartley and Madhani, 2009; Supplementary Figure S4D). It is also possible that nucleosome positioning at SNT1 requires a particularly low concentration of RSC activity that is still present in the ts mutants even under restrictive conditions. In vitro, this low concentration may be even further reduced because of loss of activity during extract preparation or simply by unphysiological dilution. In any case, as a locus like SNT1 was not scored in previous in vivo studies, the fraction of NDRs that depend on RSC in vivo may have been underestimated. Moreover, the presence of an Rsc3 site seems not to be a necessary indicator for a role of RSC.
Remodelling enzyme-intrinsic nucleosome positioning information
Our observation of the specific role for RSC in nucleosome positioning that could not be replaced by SWI/SNF or Isw2 agrees well with several in vitro studies showing that different remodelling enzymes have distinct sequence preferences for nucleosome positioning. These may differ significantly from the DNA-intrinsically favoured positions as determined by salt gradient dialysis (Brehm et al, 2000; Flaus and Owen-Hughes, 2003; Rippe et al, 2007; Schnitzler, 2008; Pham et al, 2009). We confirm this as purified RSC, SWI/SNF and Isw2 altered the salt gradient dialysis preassembled chromatin patterns at all tested loci (Figures 2 and 4). Others stressed that different remodelling enzymes moved nucleosomes with equal efficiency irrespective of the underlying DNA sequence (Partensky and Narlikar, 2009). We note that in many in vitro studies the patterns generated by different remodelling enzymes were mainly compared with each other and not to actual in vivo positions. In contrast, our system was always gauged relative to the gold standard of in vivo nucleosome patterns. In the light of the specificity of RSC in generating such proper patterns, we suggest that RSC not only provides the ‘kinetic lubricant’ for the equilibration of nucleosomes to stable positions determined by something else but also provides part of the positioning information in itself. This interpretation would also apply if the specificity of RSC function in nucleosome positioning was due to specificity of recruitment by some factor in the extract. In the case of nucleosome N-1 at the SNT1 locus, the RSC-intrinsic information can be sufficient. Here, the combination of DNA, histones and RSC constitutes a self-organizing system yielding the exact nucleosome positioning, thus arguing against an exclusive recruitment mechanism.
Rippe et al (2007) suggested that remodelling enzyme-intrinsic preferences may be at the core of nucleosome positioning in vivo and accordingly proposed a ‘remodeller code’ for nucleosome positioning. Our SNT1 data support this hypothesis to some extent. However, we showed that in most cases other factors in addition to RSC were required for proper positioning. Therefore, we think it unlikely that there is a pure ‘remodeller code’ for nucleosome positioning but a more diverse interplay of various factors.
A model of active non-equilibrium nucleosome positioning
Segal and Widom (2009) recently suggested an ‘equilibrium model for dynamic nucleosome positioning’, which assumes that nucleosomes equilibrate in vivo to their thermodynamically favoured positions as determined by the combined effects of intrinsic DNA features, neighbouring nucleosome exclusion, transcription factor binding, histone variants/modifications and DNA methylation. In this model, remodelling enzymes would act on nucleosome positioning only as ‘kinetic lubricant’, that is, as ‘enzymes’ (in addition to their enzymatic ATPase activity) that just help nucleosomes to overcome the activation energy barrier during the equilibration process without affecting the thermodynamics of nucleosome positions. We note that a living cell is not at equilibrium, but under steady-state conditions, so that there is no need to assume equilibrium nucleosome positioning. Accordingly, Segal and Widom (2009) explicitly remark that ‘ATP-dependent chromatin remodelling complexes could actively subvert equilibrium’. As our data argue for remodelling enzyme-intrinsic nucleosome positioning in combination with other factors, we suggest that the input of energy from ATP-hydrolysis not only affects the kinetics of nucleosome positioning but also thermodynamically stabilizes positions even if they were energetically unfavourable otherwise (Korber and Becker, 2010). Indeed, we hypothesize that many if not most in vivo nucleosome architectures, as observed for example at promoter regions, are continuously and actively generated by ATP-dependent remodelling enzymes, and possibly other active processes, at the continuous expense of energy. The requirement for continuous energy input is incompatible with the assumption of equilibrium, but typical for the steady state of a living cell. It is to be noted that in our model as well, remodelling enzymes are necessary for nucleosome mobility on a physiologically relevant time scale. Therefore, once a nucleosome is positioned by a remodelling enzyme, it will stay there in a kinetically trapped state in the absence of remodelling activity. Hence, remodelling enzymes may determine nucleosome positions without remaining associated with the nucleosomes all the time.
Materials and methods
Strains and media
Yeast strains were as listed in Supplementary Table S1. Strains were grown in YPD with 0.1-g/l adenine and 1-g/l KH2PO4, except for the sth1-td strain that was grown in YP with 0.1-g/l adenine and 1-g/l KH2PO4 containing 2% raffinose and 2% galactose. Temperature-sensitive strains were grown in 400-ml medium at 25°C to an OD600 of 1.2–1.5 (spectrophotometer PMQ II, Zeiss, Germany). An equal volume of medium prewarmed to 49°C was added and the cultures were placed at 37°C for the indicated temperature overnight. Viability of temperature-sensitive mutants after overnight incubation under restrictive conditions was determined by comparing the number of single colonies after plating the same number of cells for mutant and wt (BY4741 for rsc3-ts and arp9-ts or YBC2192 for sth1-td) treated in parallel on YPDA plates at 25°C.
Yeast WCE preparation
The WCE were prepared as described (Wippo et al, 2009), with the following modifications. The extract used for the fractionation was made from commercially available baker's yeast concentrate (Deutsche Hefewerke GmbH, Nürnberg, Germany). The wild-type extract for all other experiments was made from strain BY4741grown logarithmically at 30°C. For extract preparation of TH8239 (rsc3-ts) at permissive conditions, cells were grown at 25°C and overnight at 37°C for non-permissive conditions.
Chromatin assembly and reconstitution
Chromatin was assembled by salt gradient dialysis, treated with WCE and analysed as described (Wippo et al, 2009). A measure of 0.5 μg of plasmid pUC19-PHO8-short per salt gradient assembly reaction was used for experiments in Figure 1C, and a mix of 200 ng each of plasmids pUC19-PHO8-long, pUC19-RIM9, pUC19-CHA1 and pUC19-SNT1 per assembly reaction for experiments in Figures 2, 3 and 4. For a detailed description of plasmids, see the Supplementary data.
Yeast nuclei preparation
Nuclei were prepared as described (Almer et al, 1986).
Yeast WCE fractionation
For a detailed description of the individual fractionation steps, see the Supplementary data.
Purification of remodelling enzymes
RSC2-TAP, SWI2-TAP and ISW2-FLAG were purified as described (Smith et al, 2003).
Binding site prediction
The Find Individual Motif Occurrences (FIMO) program (Version 4.1.0; available at http://meme.sdsc.edu/meme4_1/cgi-bin/fimo.cgi) was used to predict sites for Rsc3 at the PHO8, RIM9, CHA1, SNT1, RIO1, RNR3, GAL10, PHO5, PHO84 and ADH2 promoters. The position weight matrix was obtained from Supplementary Table S6 of Badis et al (2008). We note that a simple search for the Rsc3 motif CGCGC identifies the same sites as the FIMO program.
Rsc3 site alignment
The orthologous sequences for PHO8, RIM9, CHA1, GAL10 and RIO1 from S. paradoxus, S. mikatae, S. bayanus, S. kudriavzevii, S. castelli and S. kluyveri were taken from Kellis et al (2003) and Cliften et al (2003). The ORF sequence plus 1000 bp upstream from each yeast species were aligned with the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using the default settings.
Supplementary Material
Acknowledgments
We thank Harm van Bakel, Tim Hughes, Charlie Boone (University of Toronto), Timothy Parnell and Brad Cairns (University of Utah) for sharing data and yeast strains. We also thank Paul Hartley and Hiten Madhani (UCSF) for sharing data, Christina Bech Hertel (University of Munich) for technical advice, Dorothea Blaschke (University of Munich) for technical assistance, Gözde Güçlüler (Izmir Technical Institute) for work on this project during her summer stay as an Amgen Scholar in the group of PK, Mark Ptashne (Sloan Kettering Institute) for sharing data before publication, Axel Imhof (University of Munich) for invaluable advice on mass spectrometry, Gernot Längst for technical and scientific advice and discussions and Peter Becker (University of Munich) for his continuous interest and support. This work was funded by the German Research Community (Deutsche Forschungsgemeinschaft (DFG), Transregio 05 to PK), by the European Community (NET grant within the Network of Excellence The Epigenome to PK) and by the Amgen Foundation (Amgen Scholarship to Gözde Güçlüler). CLP is supported by a grant from NIGMS (GM49650).
Author contributions: CJW designed and performed the vast majority of all experimental work. LI performed LC-MS/MS analysis. SW purified RSC, SWI/SNF and Isw2 complexes in the group of CLP. AH helped with the extract fractionation. PK conceived and supervised the entire study. CJW, CLP and PK wrote the manuscript. This paper is dedicated by CJW to his parents.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 [DOI] [PubMed] [Google Scholar]
- Almer A, Rudolph H, Hinnen A, Horz W (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J 5: 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S, Fascher KD, Horz W (1992) Activation of the weakly regulated Ph08 promoter in Saccharomyces cerevisiae—chromatin transition and binding-sites for the positive regulatory protein Ph04. Nucleic Acids Res 20: 1031–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF (2004) Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL (2004) Global nucleosome occupancy in yeast. Genome Biol 5: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J 19: 4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD (1998) Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell 2: 639–651 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76 [DOI] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E (2009) Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet 41: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E (2008) Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2003) Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol Cell Biol 23: 7767–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, Hicks J, Bryant GO, Ptashne M (2010) A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gregory PD, Barbaric S, Horz W (1998) Analyzing chromatin structure and transcription factor binding in yeast. Methods 15: 295–302 [DOI] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel CB, Langst G, Horz W, Korber P (2005) Nucleosome stability at the yeast PHO5 and PHO8 promoters correlates with differential cofactor requirements for chromatin opening. Mol Cell Biolo 25: 10755–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF (2006) Nucleosome positions predicted through comparative genomics. Nature Genet 38: 1210–1215 [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K (1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF (2009) A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Korber P, Becker PB (2010) Nucleosome dynamics and epigenetic stability. Essays Biochem 48: 63–74 [DOI] [PubMed] [Google Scholar]
- Korber P, Horz W (2004) In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J Biol Chem 279: 35113–35120 [DOI] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O’Shea EK (2008) Chromatin decouples promoter threshold from dynamic range. Nature 453: 246 -U216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C (2007) A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD (2006) Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res 16: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie C, Tse C, Hansen JC, Peterson CL (1999) The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry 38: 2514–2522 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF (2008a) A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF (2008b) Nucleosome organization in the Drosophila genome. Nature 453: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T (2006) A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci USA 103: 17846–17851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JMA, Holmberg S (1999) Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J 18: 2836–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsterkotter M, Barbaric S, Horz M (2000) Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J Biol Chem 275: 22678–22685 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Song JS, Liu XS, Fisher DE (2007) High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol 25: 244–248 [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR (2008) RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky PD, Narlikar GJ (2009) Chromatin remodelers act globally, sequence positions nucleosomes locally. J Mol Biol 391: 12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CD, He X, Schnitzler GR (2009) Divergent human remodeling complexes remove nucleosomes from strong positioning sequences. Nucleic Acids Res 38: 400–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Langst G (2007) DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci USA 104: 15635–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA (1986) Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191: 659–675 [DOI] [PubMed] [Google Scholar]
- Schnitzler GR (2008) Control of nucleosome positions by DNA sequence and remodeling machines. Cell Biochem Biophys 51: 67–80 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui KR, Cuddapah S, Roh TY, Barski A, Wang ZB, Wei G, Zhao KJ (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen LY, Thastrom A, Field Y, Moore IK, Wang JPZ, Widom J (2006) A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J (2009) What controls nucleosome positions? Trends Genet 25: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR (2008) Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol 6: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT (1990) Nucleosome positioning can affect the function of a Cis-acting DNA element invivo. Nature 343: 387–389 [DOI] [PubMed] [Google Scholar]
- Simpson RT, Shindo H (1979) Conformation of DNA in chromatin core particles containing poly(dAdT)-poly(dAdT) studied by 31 P NMR spectroscopy. Nucleic Acids Res 7: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL (2003) Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol 10: 141–145 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N (2010) Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol 11: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A, Caserta M, Churcher M, Hiriart E, Di Mauro E (2009) Nucleosome positioning-what do we really know? Mol Biosyst 5: 1582–1592 [DOI] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ (2010) The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8: e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM (2008) A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res 18: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter U, Svaren J, Schmitz J, Schmid A, Horz W (1994) A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J 13: 4848–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF (2009) A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T (2007) Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–U1033 [DOI] [PubMed] [Google Scholar]
- Widom J (2001) Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys 34: 269–324 [DOI] [PubMed] [Google Scholar]
- Wippo CJ, Krstulovic BS, Ertel F, Musladin S, Blaschke D, Sturzl S, Yuan GC, Horz W, Korber P, Barbaric S (2009) Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol Cell Biol 29: 2960–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ (2005) Genome-scale identification of nucleosome positions in S.cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K (2009) Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C (2006) DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell 24: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.