Abstract
Expression of vascular endothelial growth factor-A (VEGFA) by tumour-associated macrophages is critical for tumour progression and metastasis. Hypoxia, a common feature of the neoplastic microenvironment, induces VEGFA expression by increased transcription, translation, and mRNA stabilization. Here, we report a new mechanism of VEGFA regulation by hypoxia that involves reversal of microRNA (miRNA)-mediated silencing of VEGFA expression. We show that the CA-rich element (CARE) in the human VEGFA 3′-UTR is targeted by at least four miRNAs. Among these miRNAs, miR-297 and -299 are endogenously expressed in monocytic cells and negatively regulate VEGFA expression. Unexpectedly, hypoxia completely reverses miRNA-mediated repression of VEGFA expression. We show that heterogeneous nuclear ribonucleoprotein L (hnRNP L), which also binds the VEGFA 3′-UTR CARE, prevents miRNA silencing activity. Hypoxia induces translocation of nuclear hnRNP L to the cytoplasm, which markedly increases hnRNP L binding to VEGFA mRNA thereby inhibiting miRNA activity. In summary, we describe a novel regulatory mechanism in which the interplay between miRNAs and RNA-binding proteins influences expression of a critical hypoxia-inducible angiogenic protein. These studies may contribute to provide miRNA-based anticancer therapeutic tools.
Keywords: CA-rich element, heterogeneous nuclear ribonucleoprotein L, hypoxia, microRNA, vascular endothelial growth factor-A
Introduction
The recruitment and infiltration of tumour-associated macrophages (TAMs) is a prominent feature of most solid tumours, and they have a principal role in tumour progression and metastasis (Lin et al, 2006; Stockmann et al, 2008). TAMs display a relatively immature phenotype but are conditioned by the hypoxic tumour microenvironment to upregulate tumourigenic genes, including vascular endothelial growth factor-A (VEGFA), a critical regulator of angiogenesis (Ferrara, 2005). TAM-derived VEGFA is primarily responsible for the ‘angiogenic switch’ that initiates tumour vascularization, and it cannot be compensated by VEGFA from other cell sources within the tumour (Lin et al, 2006; Qian and Pollard, 2010).
Hypoxia induces VEGFA expression by increased transcription, translation, and mRNA stabilization. Post-transcriptional induction of VEGFA by hypoxia is mediated by specific mRNA-binding proteins (Claffey et al, 1998; Levy, 1998; Shih and Claffey, 1999; Vumbaca et al, 2008; Ray et al, 2009). The proximal human VEGFA 3′-UTR contains a 126-nt AU-rich domain, termed the hypoxia stability region (HSR), that is critical for VEGFA mRNA stabilization under hypoxia (Claffey et al, 1998). The cytoplasmic interaction of heterogeneous nuclear ribonucleoprotein L (hnRNP L), a splicing factor with extranuclear activities (Piñol-Roma et al, 1989), with the HSR is required for VEGFA mRNA stabilization during hypoxia (Shih and Claffey, 1999). Two adjacent cis-elements have been identified in the VEGFA HSR: a 21-nt CA-rich element (CARE) that binds with high affinity to hnRNP L (Shih and Claffey, 1999) and a 29-nt GAIT (interferon-γ-activated inhibitor of translation) element that binds the heterotetrameric GAIT complex and silences inflammatory gene expression (Ray and Fox, 2007). In hypoxia, a conformational switch in the VEGFA HSR, dictated by mutually exclusive binding of the GAIT complex and hnRNP L, overrides the repressive effect of the GAIT complex and permits high-level VEGFA translation (Ray et al, 2009).
microRNAs (miRNAs) are endogenous, ∼21-nt RNA regulators of gene expression (Farh et al, 2005; Rana, 2007; Filipowicz et al, 2008; Bartel, 2009). About 30% of human genes are under the control of one or more miRNAs (Chen and Rajewsky, 2006). They are constituents of miRNA-ribonucleoprotein RNA-induced silencing complexes (miRISCs), and guide these complexes to specific mRNA targets bearing miRNA-binding sites (Rana, 2007; Bartel, 2009). In metazoans, miRISCs silence gene expression by translational repression, mRNA degradation, or a combination of both (Nilsen, 2007; Filipowicz et al, 2008; Eulalio et al, 2008a). Most studies suggest that miRNAs do not switch off their target genes completely, but rather fine-tune expression (Hobert, 2007; Karres et al, 2007; Baek et al, 2008; Bartel, 2009). miRNA expression and activity are tissue-, cell-, and developmental stage-specific (Lagos-Quintana et al, 2002; Lim et al, 2005), and aberrant function can contribute to disease (Kloosterman and Plasterk, 2006; Voorhoeve et al, 2006). VEGFA mRNA is targeted and silenced by miR-15, -16, -20a, and -20b and their downregulation by hypoxia contributes to increased VEGFA expression (Hua et al, 2006; Karaa et al, 2009; Lei et al, 2009).
An additional layer of regulatory complexity is introduced by the superimposition of crosstalk between miRNAs and RNA-binding proteins (RBPs). RBP interaction with the 3′-UTR of mRNAs can modulate activity of miRNAs, either reducing (Bhattacharyya et al, 2006; Mishima et al, 2006; Huang et al, 2007; Kedde et al, 2007) or enhancing (Hammell et al, 2009; Schwamborn et al, 2009) miRNA activity. Alternatively, RBP function can be modulated by miRNA (Eiring et al, 2010). Because RBPs and miRNAs that bind mRNA 3′-UTRs are diverse and abundant, we anticipate that many examples of these types of complex regulatory interactions are yet to be discovered (Filipowicz et al, 2008). In this report, we reveal a novel mechanism of VEGFA gene regulation involving condition-dependent crosstalk of miRNAs and an RBP. Specifically, we show that miR-297 and -299 are endogenous negative regulators of VEGFA expression in human monocytic cells, and that their function is negatively modulated by hnRNP L during hypoxia.
Results
miRNAs target the 3′-UTR CARE of VEGFA mRNA and inhibit its translation
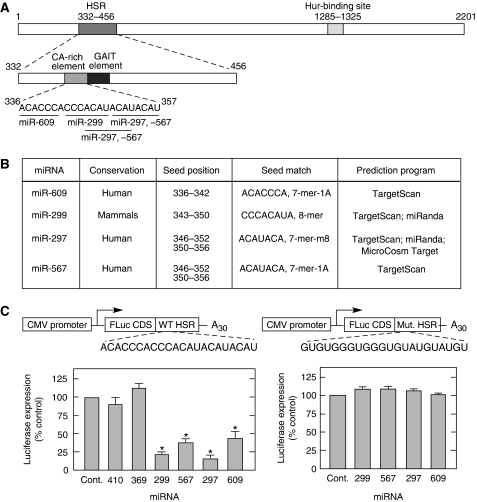
The 21-nt CARE in the HSR of the VEGFA 3′-UTR is critical for post-transcriptional regulation of VEGFA during hypoxia. To test whether miRNAs might also participate in this regulatory mechanism, we investigated potential miRNA targets in the CARE. TargetScan (Lewis et al, 2005), miRanda (John et al, 2004), and MicroCosm Targets (miRBase) (Griffiths-Jones et al, 2006) algorithms revealed miR-297, -299, -567, and -609 as candidates (Figure 1A and B). To investigate the activity of the candidate miRNAs, a luciferase reporter bearing the VEGFA HSR was cotransfected into HEK293T cells with Pre-miR™ miRNA precursors that are stable, chemically modified double-stranded RNAs that mimic endogenous, mature miRNAs. The pre-miR mimetics of the four miRNAs reduced reporter expression by about 50–80% compared with the control, a random precursor with no homology to the human genome (Figure 1C, left). Pre-miR mimetics of miR-410 and -369, predicted to target the adjacent GAIT element in the HSR, were not inhibitory, suggesting site specificity. In an additional control, a mutant HSR was generated in which ‘C’ and ‘A’ residues in the CARE were mutated to ‘G’ and ‘U’, respectively. Expression of the mutant reporter was not reduced by CARE-binding miRNAs, indicating the specific role of the CARE element in VEGFA silencing (Figure 1C, right).
Figure 1.
CARE in human VEGFA 3′-UTR is targeted by miRNAs. (A) Schematic of human VEGFA 3′-UTR elements. The full-length VEGFA 3′-UTR (top) is expanded to show elements in the HSR (middle) and the sequence of the CARE and seed regions of predicted CARE-binding miRNAs (bottom). (B) CARE-binding miRNA candidates. (C) CARE-binding miRNAs inhibit expression of reporter bearing human VEGFA HSR. FLuc reporter constructs in pcDNA3 vector bearing either wild type (left) or mutant (right) CARE in the HSR were transfected into HEK293T cells with CARE-binding miRNA candidates (miR-297, -299, -567, and -609), miR-control (Cont.), negative controls (miR-410 and -369), and with RLuc-expressing pRL-SV40 vector as internal control. Relative Luc levels were measured after 24 h and expressed as percentage of control. Results are expressed as mean±s.d., for n=3 independent experiments. An asterisk (*) indicates a significant difference, P<0.05, two-tailed t-test.
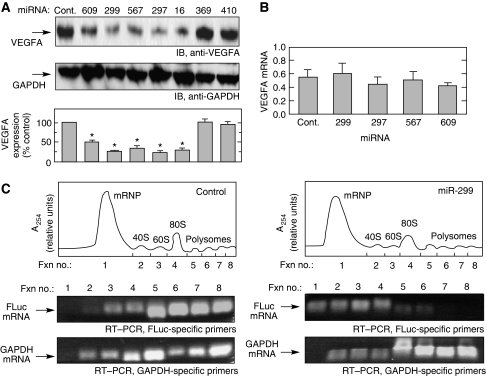
To investigate the effect of CARE-targeting miRNAs on endogenous VEGFA expression in myeloid cells, the miRNAs were overexpressed by transient transfection of U937 monocytic cells, and VEGFA was determined in cell lysates by immunoblot analysis. VEGFA expression was reduced by miR-297, -299, -567, and -609 by about 50–70% compared with controls (Figure 2A). As before, GAIT-element-targeting miRNAs were ineffective. In a positive control experiment, miR-16, which binds the distal 3′-UTR (nt 1793–1818) of human VEGFA and downregulates its expression (Karaa et al, 2009), markedly repressed VEGFA expression.
Figure 2.
The effect of CARE-binding miRNAs on VEGFA expression. (A) Ectopically expressed CARE-binding miRNAs inhibit VEGFA expression. U937 cells were transfected with CARE-binding miRNAs or controls. After 24 h, cell lysates were subjected to immunoblot analysis with anti-VEGFA (top) or anti-GAPDH (middle) antibodies. VEGFA expression was quantitated and expressed as percentage of control (B) CARE-binding miRNAs do not inhibit VEGFA mRNA expression. After transfection with miRNAs for 24 h, VEGFA mRNA amount was measured by real-time, quantitative RT–PCR and normalized to GAPDH mRNA. (C) CARE-binding miRNAs repress translation of HSR-bearing reporter RNA. HEK293T cells were cotransfected for 24 h with FLuc reporter bearing the VEGFA HSR and either control miRNA (left) or miR-299 (right). Lysates were subjected to sucrose density gradient fractionation and RNA was monitored by absorption at 254 nm (top). RNA isolated from each fraction was subjected to RT–PCR to determine FLuc (middle) and GAPDH (bottom) mRNAs. Results are expressed as mean±s.d., for n=3 independent experiments. An asterisk indicates a significant difference, P<0.05, two-tailed t-test.
miRNAs can silence gene expression by mRNA degradation or translation inhibition (Bartel, 2004; Guo et al, 2010). Quantitative real-time RT–PCR showed that CARE-binding miRNAs did not alter VEGFA mRNA expression, suggesting inhibition at the level of translation (Figure 2B). To confirm the silencing mechanism, translation efficiency was determined by polysome fractionation. A firefly luciferase (FLuc) reporter mRNA upstream of the VEGFA HSR was cotransfected with miR-299 into HEK293T cells. After 24 h, cell lysates were fractionated by sucrose gradient to separate dense, rapidly translating polysome fractions from light, non-translating ribonucleoprotein fractions (Figure 2C, top panels). RT–PCR analysis using FLuc-specific primers showed that miR-299 shifted the reporter from the rapidly translating to the non-translating fractions (Figure 2C, middle panels). As a control, GAPDH mRNA was primarily detected in the translating fractions and was unaffected by miR-299 (Figure 2C, bottom panels). Together, these results indicate translational inhibition as the mechanism by which CARE-binding miRNA reduces VEGFA expression.
Role of endogenous CARE-binding miRNAs in regulating VEGFA expression
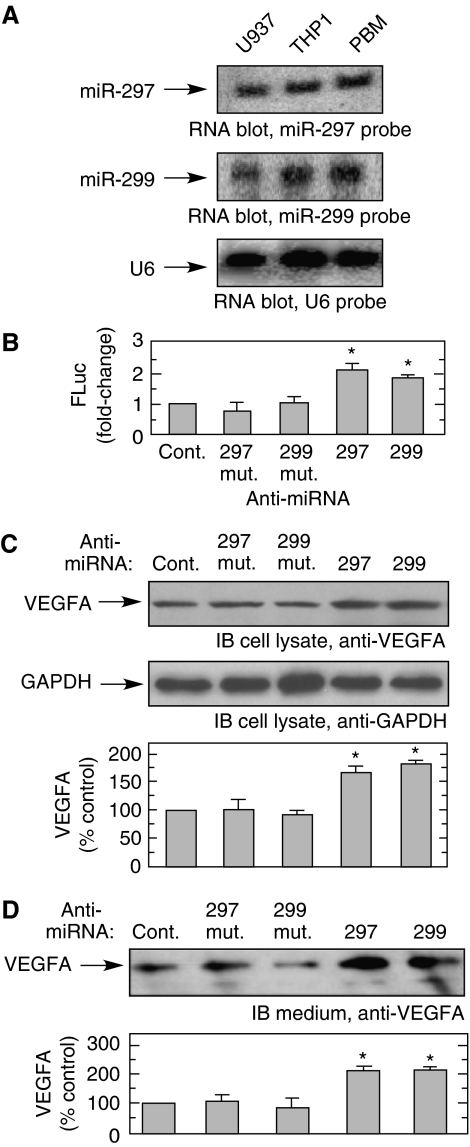
The endogenous levels of miR-297, -299, -567, and -609 were measured in monocytic cells by RNA blot using miRNA-specific probes. miR-297 and -299 were detected at comparable levels in human U937 and THP1 cell lines, and in human peripheral blood monocytes (PBMs) (Figure 3A); miR-567 and -609 were not detected in any of the cells (not shown). To determine the effect of endogenous miRNAs on VEGFA expression, U937 cells were cotransfected with FLuc reporter RNA bearing the VEGFA HSR, with chemically stabilized anti-miR-297 and -299, and with the same anti-miRNAs containing two point mutations in the seed regions as specificity controls. FLuc expression was nearly doubled by both anti-miRNAs, but not by the mutated or random control anti-miRNAs, suggesting that these endogenous miRNAs negatively regulate reporter expression (Figure 3B). To investigate the role of endogenous miRNAs on endogenous gene expression, anti-miR-297 and -299 were transfected in U937 cells and after 24 h VEGFA in cell lysates was measured by immunoblot. An ∼75% increase in VEGFA expression was observed after miRNA inhibition (Figure 3C). VEGFA is a secretory protein and transfection with either anti-miR-297 or -299 induced nearly a two-fold increase in 24 h conditioned medium (Figure 3D). These results indicate a physiological role of endogenous miR-297 and -299 in negative regulation of VEGFA in monocytic cells.
Figure 3.
Presence and activity of endogenous CARE-binding miRNAs. (A) miR-297 and -299 are endogenously expressed in monocytic cells. Total small RNAs were isolated from human U937 cells, THP1 cells, and PBM under normoxic condition. RNA was subjected to RNA blot analysis using probes against miR-297 (top), miR-299 (middle), and U6 as control (bottom). (B) Endogenous miR-297 and -299 negatively regulate expression of HSR-bearing reporter. FLuc reporter upstream of the HSR was cotransfected with anti-miR-297, -299, random sequence control (Cont.), or mutated anti-miR-297 or -299, and with RLuc-expressing vector pRL-SV40 as control for transfection efficiency. After 24 h, the relative Luc levels were determined, and expressed as fold-change compared with random control. (C) Endogenous miR-297 and -299 negatively regulate endogenous VEGFA expression. U937 cells were transfected with anti-miR-297, -299, or controls as above. Cell lysates subjected to immunoblot analysis with anti-VEGFA (top) and -GAPDH (middle) antibodies. The amount of VEGFA was expressed as percentage of random control (bottom). (D) miR-297 and -299 negatively regulate VEGFA secretion. Cells were transfected as in (C) and conditioned medium concentrated by immunoprecipitation with polyclonal anti-VEGFA antibody, immunoblotted with monoclonal anti-VEGFA antibody (top) and quantitated by densitometry (bottom). Results are expressed as mean±s.d., for n=3 independent experiments. An asterisk indicates a significant difference, P<0.05, two-tailed t-test.
Hypoxia prevents activity of miRNAs targeting the VEGFA CARE
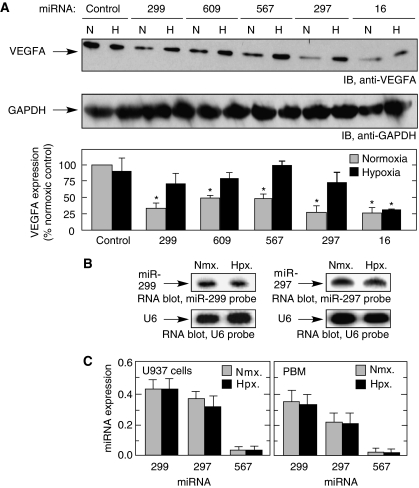
miRNA activity can be modulated in a state-specific manner, for example, by cell stress (Bhattacharyya et al, 2006; Huang et al, 2007; Kedde et al, 2007; Hammell et al, 2009), thus we investigated whether hypoxia, an established regulator of VEGFA expression, modulates the activity of CARE-binding miRNAs. The repressive activity of CARE-binding miRNAs was almost completely alleviated by hypoxia (Figure 4A). The effect of hypoxia was specific for miRNAs targeting the CARE as repression by miRNAs targeting other regions of the VEGFA 3′-UTR (Hua et al, 2006; Karaa et al, 2009; Lei et al, 2009), for example, miR-16 (Figure 4A) or miR-20a and miR-20b (data not shown) was observed in both conditions. We considered the possibility that hypoxia decreased the amount of CARE-targeting miRNAs, thereby reducing its effect on VEGFA expression. RNA blot analysis showed that hypoxia did not alter endogenous expression of CARE-binding miR-297 or -299 (Figure 4B). This result was confirmed by quantitative RT–PCR in both U937 cells and PBM (Figure 4C). These findings suggest that hypoxia might influence non-miRNA, trans-acting factors that target the VEGFA mRNA CARE.
Figure 4.
Hypoxia inhibits the activity of CARE-binding miRNAs. (A) Hypoxia overcomes miRNA-mediated inhibition of VEGFA expression. U937 cells were incubated with miRNAs under normoxic (21% pO2, N) or hypoxic (1% pO2, H) conditions for 24 h. Cell lysates were subjected to immunoblot analysis with anti-VEGFA (top) and anti-GAPDH (middle) antibodies; VEGFA was expressed as percentage of normoxic control (bottom). (B) Hypoxia does not inhibit expression of miR-297 and -299. U937 cells were incubated under conditions of normoxia (Nmx.) or hypoxia (Hpx.) for 24 h and miR-297 and -299 determined by RNA blot analysis using LNA oligomer probes and U6 probe as loading control. (C) miRNA amounts of normoxia- and hypoxia-treated U937 cells (left) or PBM (right) were determined by real-time quantitative RT–PCR after isolation of total small RNAs. RT–PCR of miR-17-5p was used for normalization. Results are expressed as mean±s.d., for n=3 independent experiments. An asterisk indicates a significant difference, P<0.05, two-tailed t-test.
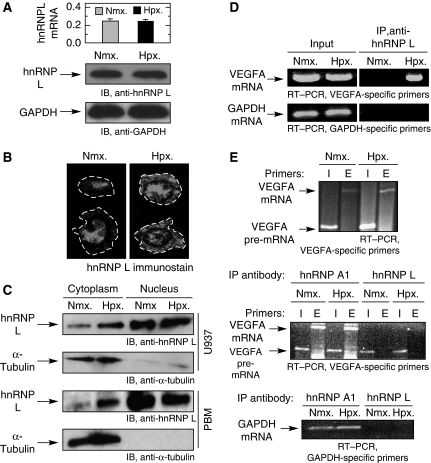
Hypoxia induces hnRNP L translocation and binding to VEGFA mRNA
hnRNP L is an abundant nucleocytoplasmic protein that binds the VEGFA CARE in human melanoma cells and increases VEGFA mRNA stability during hypoxia (Shih and Claffey, 1999). Quantitative RT–PCR (Figure 5A, top) and RNA blot analysis (not shown) showed that exposure of U937 cells to hypoxia did not significantly alter hnRNPL expression. Likewise, immunoblot analysis of cell lysates showed no effect of hypoxia on hnRNP L protein expression (Figure 5A, bottom two panels). We investigated the effect of hypoxia on hnRNP L localization. Immunofluorescence detection indicated that hnRNP L is primarily localized in the nucleus of U937 cells under normoxic conditions, but after 24 h treatment with hypoxia, substantial hnRNP L is observed in the cytoplasm (Figure 5B). To confirm the subcellular localization of hnRNP L, cell fractionation studies were done in U937 cells and PBM exposed to hypoxia or normoxia for 24 h and nuclear and cytoplasmic lysates were prepared. Hypoxia increased the cytoplasmic level of hnRNP L by three- to five-fold in both cell types (Figure 5C).
Figure 5.
Hypoxia-inducible association of cytoplasmic hnRNP L with VEGFA mRNA. (A) Hypoxia does not alter hnRNP L expression. U937 cells were incubated under normoxia (Nmx.) or hypoxia (Hpx.) for 24 h and total RNA was subjected to quantitative real-time RT–PCR using primers specific for hnRNP L; GAPDH was used as normalizer (top panel). Cell lysates were subjected to immunoblot analysis with anti-hnRNP L and -GAPDH antibodies (bottom two panels). (B) Hypoxia induces hnRNP L relocalization. U937 cells treated for 24 h under normoxia (left panel) or hypoxia (right panel) were immunostained using rabbit polyclonal anti-hnRNP L antibody. The cell boundaries are outlined (dashed lines). (C) U937 (top two panels) and PBM (bottom two panels) cells were incubated as above and nuclear and cytoplasmic fractions were immunoblotted with anti-hnRNP L and anti-α-tubulin antibodies. (D) Hypoxia induces association of hnRNP L with VEGFA mRNA. Cytosolic lysates from normoxia- and hypoxia-treated U937 cells were subjected to immunoprecipitation with anti-hnRNP L monoclonal antibody. Expression of VEGFA and GAPDH mRNAs in lysates (input, left panels) and after immunoprecipitation (right panels) was determined by RT–PCR. (E) hnRNP L binds mature VEGFA mRNA in the cytoplasm. Nuclear lysates were prepared from normoxic and hypoxic U937 cells, and VEGFA pre-mRNA and mRNA were determined by RT–PCR using intron (I)- and exon (E)-specific primers, respectively; RT–PCR of GAPDH mRNA was used as control (top). The nuclear lysates were subjected to immunoprecipitation with anti-hnRNP L and anti-hnRNP A1 monoclonal antibodies, and bound VEGFA pre-mRNA and mRNA were determined by RT–PCR (middle); RT–PCR of GAPDH mRNA was used as specificity control (bottom).
We investigated whether hypoxia induces an interaction between hnRNPL and VEGFA mRNA in monocytic cells. Following a 24-h treatment of U937 cells with hypoxia or normoxia, cytosolic lysates were subjected to immunoprecipitation with monoclonal anti-hnRNP L antibody followed by RT–PCR using VEGFA-specific primers. No interaction of hnRNP L with VEGFA mRNA was detected in normoxic cells, but hypoxia induced a very robust and specific interaction of hnRNP L with VEGFA mRNA; RT–PCR with primers specific for GAPDH mRNA indicated specificity of the interaction (Figure 5D). Because hnRNP L is a nucleocytoplasmic protein, we examined whether the interaction between hnRNP L and VEGFA mRNA occurs in the nucleus, followed by transport to the cytoplasm, as would be expected for a shuttling factor. Both precursor and mature VEGFA mRNAs are detected in the nuclear lysates from cells treated with normoxia or hypoxia (Figure 5E, top). The same lysates were subjected to immunoprecipitation with anti-hnRNP L antibody and RT–PCR for VEGFA mRNA and pre-mRNA. hnRNP L binding to VEGFA pre-mRNA in the nucleus was detected, but binding to the mature mRNA was not (Figure 5E, middle). Immunoprecipitation using anti-hnRNP A1 was used as a positive control since it interacts with pre-mRNAs and remains mRNA-bound during transport to the cytoplasm (Mili et al, 2001). As expected, hnRNP A1 bound both VEGFA mRNA forms in the nucleus. RT–PCR with primers specific for GAPDH mRNA verified the selectivity of the interaction between VEGFA mRNA and hnRNP L (Figure 5E, bottom). We conclude that the interaction between hnRNP L and mature VEGFA mRNA occurs in the cytoplasm and is independent of nucleocytoplasmic transport.
Role of hnRNP L in overcoming miRNA-mediated repression of VEGFA
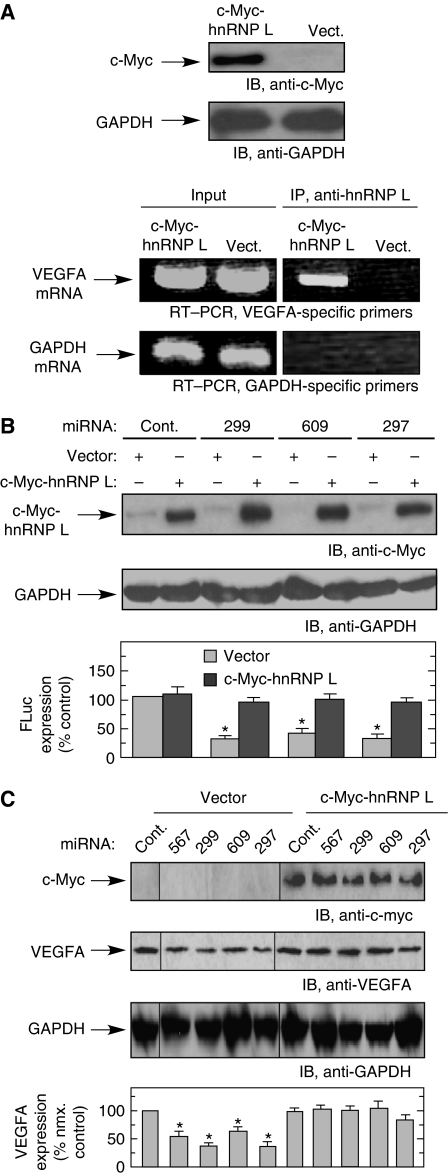
To determine whether the increase in cytoplasmic hnRNP L in hypoxia is sufficient to induce binding to VEGFA mRNA, we overexpressed hnRNP L and determined its binding to VEGFA mRNA in normoxia. U937 cells were transfected with pcDNA3-c-Myc-hnRNP L (or empty vector) for 24 h under normoxic condition and expression confirmed by immunoblot using anti-c-Myc antibody (Figure 6A, top two panels). Immunoprecipitation of cell lysate with anti-hnRNP L antibody, followed by RT–PCR with VEGFA or GAPDH-specific, primers revealed a selective interaction (Figure 6A, bottom two panels). Interestingly, the finding that ectopically expressed hnRNP L binds VEGFA mRNA suggests hypoxia or other hypoxia-inducible factors are not required, but rather the cytoplasmic concentration of hnRNP L determines its association with VEGFA mRNA. To specifically show the role of hnRNP L in overcoming miRNA-mediated inhibition, HEK293T cells were cotransfected with an expression vector encoding c-Myc-hnRNP L and FLuc reporter bearing the VEGFA HSR, in addition to CARE-binding miRNAs. Ectopic expression of hnRNP L almost completely restored expression of the reporter in the presence of the inhibitory miRNAs (Figure 6B). Similarly, the effect of hnRNP L on miRNA-mediated regulation of endogenous VEGFA expression was examined. Cotransfection of c-myc-hnRNP L with CARE-binding miRNAs blocked the repressive activity of miRNAs, completely restoring endogenous expression of VEGFA, and verifying the direct role of hnRNP L in preventing the miRNA-mediated inhibition of VEGFA expression (Figure 6C).
Figure 6.
hnRNP L overexpression increases VEGFA expression in normoxia by inhibiting activity of CARE-binding miRNAs. (A) Overexpressed hnRNP L binds VEGFA mRNA in normoxia. U937 cells were transfected with pcDNA3-c-Myc-hnRNP L or empty vector (Vect.) under normoxia for 24 h. Cytosolic lysates were immunoblotted with anti-c-Myc tag and anti-GAPDH antibodies (top two panels). Cytosolic lysates were subjected to immunoprecipitation with anti-hnRNP L monoclonal antibody. Expression of VEGFA and GAPDH mRNAs in lysates (input, left panels) and after immunoprecipitation (right panels) was determined by RT–PCR (bottom two panels). (B) Overexpression of hnRNP L in normoxia prevents miRNA-mediated repression of HSR-bearing reporter. FLuc reporter bearing the VEGFA HSR was cotransfected into HEK293T cells with CARE-binding miRNAs (or control miRNA, Cont.), pcDNA3-c-Myc-hnRNP L or vector, and RLuc as transfection efficiency control. Lysates were subjected to immunoblot analysis with anti-c-Myc tag (top) and -GAPDH antibodies (middle). The relative level of FLuc was normalized by RLuc expression and expressed as percentage of control (bottom). (C) Overexpressed hnRNP L alleviates miRNA-mediated repression of endogenous VEGFA in normoxia. U937 cells were cotransfected with pcDNA3-c-Myc-hnRNP L or vector and CARE-binding miRNAs for 24 h under normoxia. Lysates were subjected to immunoblot analysis with anti-c-Myc tag, VEGFA, and GAPDH antibodies (the samples were run on the same gel but the lanes were rearranged for clarity). VEGFA expression was quantitated by densitometry and expressed as percent of normoxic control. Results are expressed as mean±s.d., for n=3 independent experiments. An asterisk indicates a significant difference, P<0.05, two-tailed t-test.
Discussion
Induction of VEGFA during hypoxia is a critical pathophysiological response facilitated by diverse transcriptional and post-transcriptional mechanisms. The unusually long 3′-UTR of human VEGFA mRNA appears to be a ‘hot-spot’ for post-transcriptional control by hypoxia. Hypoxia stabilizes VEGFA mRNA by interaction of the RBP Hur to AU-rich elements in the 3′-UTR (Brennan and Steitz, 2001; Goldberg-Cohen et al, 2002), or by an independent mechanism in which hnRNP L binds the CARE in the HSR of the 3′-UTR (Claffey et al, 1998; Shih and Claffey, 1999). Hypoxia, via an HSR-mediated mechanism, can increase translation of VEGFA in monocytic cells exposed to interferon-γ. In this case, hypoxia induces a conformational switch in the VEGFA HSR by increasing hnRNP L binding to the CARE, which in turn reduces binding of the GAIT complex to the adjacent GAIT element, and overrides translational repression (Ray et al, 2009). Previous reports have shown that the VEGFA 3′-UTR is targeted by multiple miRNAs. For example, miR-15b, -16, -20a, and -20b downregulate VEGFA expression, but the levels of the miRNAs themselves are downregulated during hypoxia, thus enabling VEGFA expression (Hua et al, 2006). Likewise, miR-126 downregulates VEGFA mRNA expression and is postulated to have tumour suppressor function, but its expression is downregulated in lung cancer cells (Liu et al, 2009). Our target analysis (not shown) suggests that the miRNAs in these earlier reports target regions outside of the CARE and HSR of VEGFA.
Our results indicate that at least four miRNAs, miR-297, -299, -567, and -609, target the CARE of human VEGFA 3′-UTR, and when ectopically expressed markedly inhibit VEGFA protein expression. Two of these miRNAs, miR-297 and -299, are endogenously expressed in U937 cells and in freshly isolated human PBM. These miRNAs have no effect on the amount of VEGFA mRNA, indicating that their silencing activity is at the level of mRNA translation. This conclusion was rigorously established for miR-299 that induces a shift of HSR-bearing reporter RNA from the rapidly translating polysome fractions to the non-translating mRNP pool. Although many miRNA-targeted mRNAs undergo extensive degradation (Guo et al, 2010), there are also examples of repression at the translational level, or by a combination of both processes (Eulalio et al, 2008a, 2008b). The molecular determinants and mechanisms that direct RISC-mediated degradation or translational repression have not been elucidated; however, the specific silencing pathway for a given mRNA can be tissue specific or cell specific (Lagos-Quintana et al, 2002).
The inhibitory activity of the miRNAs is prevented during hypoxia by hnRNP L, which translocates from the nucleus to the cytoplasm and binds the CARE in the VEGFA mRNA, presumably blocking the accessibility of the miRISC targeting the same element. Several recent reports have shown that RBPs can modulate repression by miRNAs in a cell state-specific manner (Bhattacharyya et al, 2006; Mishima et al, 2006; Huang et al, 2007; Kedde et al, 2007). For example, HuR binding to the 3′-UTR of CAT-1 mRNA during cellular stress prevents miR-122-mediated repression of CAT-1 expression (Bhattacharyya et al, 2006). Similarly, during germline development in zebrafish, Dnd1 binds U-rich sequences in nanos1 and TDRD7 mRNAs near the binding site of miR-430, relieving its repression (Kedde et al, 2007). In another example of an RBP-disrupting miRNA function, Lin28 selectively blocks the processing of pri-let-7 miRNAs in embryonic cells (Viswanathan et al, 2008). Finally, during synaptic development, brain-derived neurotrophic factor relieves miR-134 inhibition of Limk1 mRNA translation by an undefined mechanism (Schratt et al, 2006). In most cases, the RBP inhibits the expression or activity of an miRNA; however, other regulatory interactions between these moieties have been described. For example, several reports have revealed a cooperative interaction between an RBP and a specific miRNA where both are required for repression of gene expression (Hammell et al, 2009; Kim et al, 2009). In another atypical case, miR-328 functions as an RNA decoy by binding to the inhibitory RBP hnRNP E2 and preventing its interaction with target mRNA (Eiring et al, 2010).
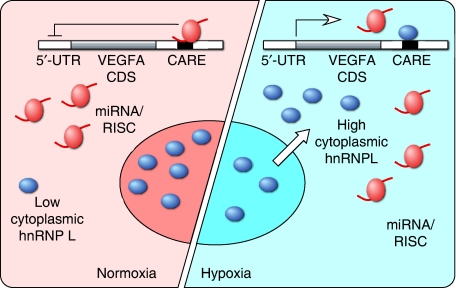
The crosstalk between hnRNP L and inhibitory miRNAs described here exhibits several unique characteristics. First, the inhibition of VEGFA expression by endogenous miRNAs occurs under basal conditions in unstressed cells, and is derepressed upon stress, that is, hypoxia. Second, hypoxia activates hnRNP L by inducing its translocation from the nucleus to the cytoplasm. The molecular mechanism regulating hnRNP L localization is not known; however, another miRNA regulator, HuR, has been shown to undergo stress-dependent translocation to the cytoplasm following dephosphorylation (Kim and Gorospe, 2008). Finally, hnRNP L and miR-297, -299, -567, and -609 share the same target site in the VEGFA 3′-UTR, namely the 20-nt CARE. To our knowledge, this is the first example of a protein-binding regulatory element identified as an miRNA target. The coincidence of the protein and miRNA target site on the VEGFA 3′-UTR, and our finding that hnRNP L binds mature VEGFA mRNA only after transport to the cytoplasm, suggests a specific mechanism. Namely, normoxic monocytic cells constitutively express miR-297 and -299; in the absence of substantial cytoplasmic hnRNP L, these miRNAs bind the 3′-UTR CARE of VEGFA mRNA and repress its expression by inhibiting translation (Figure 7). Upon switching to hypoxia, hnRNP L translocates from the nucleus to cytoplasm where it binds the same CARE region of the VEGFA mRNA, thereby obstructing the miRISC-binding site and preventing the repression of VEGFA expression. Our results suggest that the major function of the endogenous, inhibitory miRNAs is to restrict VEGFA expression during normoxia, and the function of hnRNP L translocation is to overcome this restriction during hypoxia. Unexpectedly, VEGFA expression is not increased in hypoxia in our experiments, and we speculate that additional inhibitory mechanisms might be operative in hypoxic myeloid cells that are balanced by the suppression of miRNA activity by hnRNP L. Together, our results suggest an intricate interplay of regulatory mechanisms that might function to maintain myeloid cell VEGFA expression at a constant level in both normoxia and hypoxia. miR-297 is also expressed in non-monocytic HEK293T cells, and CARE-binding miRNAs inhibit VEGFA expression in normoxia but not hypoxia, suggesting that the mechanism might be broadly applicable to multiple cell types (Supplementary Figure S1A and B). Interestingly, hypoxia increases VEGFA expression in HEK293T cells compared with normoxic cells, even in the presence of inhibitory miRNAs thus indicating the potential for this mechanism to influence total VEGFA expression in non-myeloid cells.
Figure 7.
Schematic showing effects of CARE-binding miRNAs and hnRNP L on VEGFA expression. In normoxia, translation of VEGFA mRNA is negatively regulated by endogenous miRNA/RISC complexes binding to CARE (left). In hypoxia, hnRNP L is translocated to cytoplasm where it binds CARE, prevents miRNA/RISC activity, and increases VEGFA expression (right).
The discovery of endogenous miRNAs that target and repress VEGFA has physiological and pathophysiological implications. VEGFA is the principal agonist of angiogenesis and is essential for blood vessel formation during development and tissue repair (Ferrara, 2005). VEGFA also induces vessel permeability and leukocyte chemoattraction, events associated with chronic inflammation (Ferrara, 2005). Cancer cells ‘hijack’ VEGFA to promote tumour vascularization and growth. Interestingly, recent studies point to the critical importance of VEGFA produced by TAMs for throwing the ‘angiogenic switch’ that induces tumour angiogenesis during hypoxia (Lin et al, 2006; Qian and Pollard, 2010). Overexpression of miR-297, -299, -567, and -609 all cause robust inhibition of VEGFA expression. However, the increase in VEGFA expression observed upon inhibition of endogenous miR-297 and -299 was more modest, consistent with a ‘tuning’ function of the miRNAs rather than an all-or-none response. Recent studies in vertebrates suggest that this type of regulation by miRNAs may be the norm rather than the exception (Hobert, 2007). Experimental criteria for a tuning relationship between an miRNA and its respective target has been suggested: the miRNA and the protein product of the target mRNA must both be present in the cell, and both upregulation and downregulation of the respective target protein must be detrimental (Hobert, 2007). Certainly, the regulation of VEGFA by miR-297 and -299 fulfill these characteristics. Both of the miRNAs as well as VEGFA mRNA and protein are constitutive in monocytic cells. Accumulating evidence suggests that VEGFA expression must be regulated within a relatively narrow range. Deletion of a single VEGFA allele causes abnormal blood vessel formation and mid-gestational lethality in mice (Carmeliet et al, 1996). In contrast, modest overexpression induces aberrant vasculogenesis and heart development, and ultimately, embryonic lethality (Miquerol et al, 2000). In adult humans (and mice), elevated serum VEGFA is associated with a lethal hepatic syndrome (Wong et al, 2001). Interestingly, although reduction of VEGFA level or action by VEGFA or VEGFA receptor antagonists has been successfully applied in cancer therapy, the same repression can cause vascular disturbance, regression of blood vessels, and an array of severe side effects that hamper its clinical applicability (Kamba and McDonald, 2007). Moreover, deletion of macrophage-derived VEGFA results in accelerated tumour growth (Stockmann et al, 2008). These observations strongly indicate that VEGFA must be subject to tight bi-directional regulation. Negative regulation by miRNAs may contribute to management of VEGFA dosage within the optimal range by providing a negative regulatory effect that partially counterbalances positive transcriptional and post-transcriptional regulatory mechanisms.
Under the conditions of our experiments, hypoxia induces hnRNP L translocation to the cytoplasm where it not only overrides miRNA-mediated inhibition of translation of VEGFA mRNA, but it also stabilizes the transcript (Claffey et al, 1998), both contributing to elevated expression of VEGFA that reverses hypoxia by inducing blood vessel formation. Although we found that hypoxia did not alter miRNA expression, there might be other stress conditions that induce CARE-binding miRNAs to a level that prevents hnRNP L-mediated induction of VEGFA, and in fact, inhibits VEGFA expression. Alternatively, the inhibitory activity of CARE-binding miRNAs against VEGFA expression might present potential therapeutic agents against tumour growth. In a recent successful application of this approach, systemic administration of miR-26a, which targets cyclins D2 and E2, inhibited hepatocellular carcinoma cell proliferation and tumour progression in mice (Kota et al, 2009). Conventional anti-VEGFA therapies target either circulating protein or cell surface receptors. In contrast, therapies using chemically stabilized miRNAs target VEGFA at the level of intracellular synthesis. This approach, combined with cell-type specific targeting strategies might permit localized inhibition or fine-tuning of VEGFA expression while minimizing adverse systemic consequences.
Materials and methods
Cells and lysates
Human monocytic cells, that is, U937 cells (CRL-1593.2, ATCC), THP1 cells (TIB-202, ATCC), and PBM were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS). Human PBM were isolated by leukapheresis, followed by countercurrent centrifugal elutriation (Czerniecki et al, 1997), under a Cleveland Clinic Institutional Review Board-approved protocol that adhered to American Association of Blood Bank guidelines. Human HEK293T cells (CRL-11268, ATCC) were cultured in DME medium supplemented with 4.5 mg/ml glucose and 10% heat-inactivated FBS. Cells were treated under either normoxic (21% ρO2) or hypoxic (1% ρO2) conditions in a humidified incubator for 24 h. Cell lysates were prepared in phosphosafe extraction buffer (Novagen) containing protease inhibitor cocktail (Thermo Scientific). Cytoplasmic and nuclear extracts were obtained using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific).
Plasmid construction, transfection, and dual luciferase activity assay
pcDNA3-FLuc-VEGFA HSR (wild type and mutant) and pcDNA3-c-Myc-hnRNP L were generated as described (Ray et al, 2009). U937 cells (2 × 106 cells) were transfected with oligomers including pre-miR miRNA precursors, miRNA-negative control, anti-miR miRNA inhibitors, anti-miR-negative control, and anti-miR mutants (200 nM, Ambion), using GenomONE-Neo EX HVJ Envelope Vector Kit (Cosmo Bio). For anti-miR mutants, nt 3 and 4 of the seed regions of miR-297 (G, U to A, A) and miR-299 (U, G to A, A) were mutated. The same kit was used to transfect U937 cells (2 × 106 cells) with plasmid DNA (40 μg). For dual luciferase assay, oligomers were cotransfected into U937 cells with FLuc reporter plasmid (40 μg) and RLuc-expressing vector pRL-SV40 (20 μg) to normalize for transfection efficiency, and relative Luc activities measured using dual luciferase assay kit (Promega). For HEK293T cells, 2 × 106 cells were transfected with pre-miR miRNA precursors (100 nM), or cotransfected with FLuc reporter plasmid (2 μg) or pcDNA3-c-Myc-hnRNP L (2 μg) and pRL-SV40 (0.5 μg) as internal transfection control using lipofectamine 2000 (Invitrogen).
Immunocytochemistry
U937 cells (about 50% confluence) on glass slides were washed three times with phosphate-buffered saline (PBS) and fixed in 1% paraformaldehyde for 30 min. Fixed cells were washed with PBS and permeabilized with 0.1% Triton X-100 for 15 min. After further washing, the cells were incubated with rabbit polyclonal anti-hnRNP L antibody (Santa Cruz) for 2 h at room temperature. After thorough washing, anti-rabbit Alexa fluor 488 (Invitrogen) was added for 60 min at room temperature. DAPI (Sigma-Aldrich) was added to stain nuclei. The slides were mounted with 90% glycerol in PBS and immunofluorescent images were acquired by confocal microscopy with LaserSharp software.
Immunoblot analysis
Cell lysates were subjected to SDS–PAGE (12%). The transferred blot was probed with rabbit anti-human VEGFA polyclonal antibody (Santa Cruz) and HRP-conjugated anti-rabbit secondary antibody (GE Healthcare), and detected with ECL Plus (Amersham). Immunoblot with mouse monoclonal anti-GAPDH-peroxidase (Sigma) antibody provided a loading control. Band intensities were measured using ImageJ software. VEGFA in conditioned media was measured following concentration with rabbit anti-human VEGFA polyclonal antibody (2 μg/ml) and GammaBind Plus Sepharose beads (GE Healthcare) followed by immunoblot with mouse monoclonal anti-VEGFA antibody (Santa Cruz) (Kim et al, 2002). Other antibodies used for immunoblotting were mouse monoclonal anti-c-Myc (Santa Cruz), rabbit polyclonal anti-hnRNP L (Santa Cruz), and mouse monoclonal anti-α-tubulin (Sigma).
Analysis of RNA by PCR
Total small RNA was extracted with miRVana miRNA Isolation kit (Ambion), and quality and quantity determined using a NanoDrop spectrophotometer. miRNA was assessed by real-time PCR using TaqMan probe and primer sets in an ABI PRISM 7000 system (Applied Biosystems), and normalized with hsa-miR-17-5p. Briefly, of total small RNA (10 ng) was reversed transcribed using Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems) and amplified using TaqMan 2x Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). To determine VEGFA and hnRNPL mRNAs, first-strand cDNAs were synthesized with Superscript III reverse transcriptase (Invitrogen) using total RNA (2 μg) extracted with Trizol (Invitrogen), and amplified with SYBR Green PCR master mix (Applied Biosystems) in an ABI PRISM 7000 system. GAPDH mRNA was used as internal normalization control. Primers for 120-nt VEGFA PCR product were TATGCGGATCAAACCTCAC (forward) and CTCGGCTTGTCACATTTTTCTTGTCTTGC (reverse); primers for 180-nt hnRNPL product were TTCTGCTTATATGGCAATGTGG (forward) and GACTGACCAGGCATGATGG (reverse); primers for 96-nt GAPDH product were TGCACCACCAACTGCTTAGC (forward) and GGCATGGACTGTGGTCATGAG (reverse).
RNA blot analysis
RNA blot analysis using locked nucleic acid probes was used to determine miRNAs (Varallyay et al, 2008). Briefly, total RNA (100 μg) was mixed with an equal volume of gel loading buffer II (Ambion), heated at 80°C for 5 min, snap-cooled, and fractionated by 15% denaturing acrylamide gel electrophoresis. RNA was transferred to Hybond N+ (GE Healthcare) by capillary blotting using 20 × SSC buffers (Invitrogen) and fixed using UV crosslinker. miRCURY LNA detection probes (10 pmol, Exiqon) complementary to the CARE-binding miRNAs were radiolabelled with 1 μl T4 polynucleotide kinase (New England Biolabs) and 1 μl [γ-32P] ATP (0.4 MBq) for 1 h at 37°C. Labelled LNA probes were heated at 95°C for 1 min, ice-cooled, diluted 1:1000 in pre-warmed (50°C) PerfectHyb Plus hybridization solution (Sigma) with denatured salmon sperm DNA (20 μg/ml, Ambion), and added to the pre-hybridized UV crosslinked membrane at 50°C for 1 h. Membranes were washed 3 × in 2 × SSC, 0.1% SDS at 50°C. Decade Marker (Ambion) was used as molecular weight markers. The membranes were stripped with boiled 0.1% SDS, 5 mM EDTA for 30 min, and reprobed with radiolabelled LNA-modified U6 as loading control.
Interaction of hnRNP L with VEGFA mRNA
In vivo interaction of hnRNP L with VEGFA mRNA was determined by immunoprecipitation followed by RT–PCR as described (Shih and Claffey, 1999; Majumder et al, 2009). Briefly, nuclear and cytoplasmic extracts (500 μg) prepared in NE-PER extraction reagent (Thermo) were incubated with Halt Protease Inhibitor Cocktail (EDTA-free, Thermo), RNase inhibitor (Promega), mouse IgG (4 μg), and 50 μl of GammaBind Plus Sepharose beads for 1 h at 4°C. The supernatant was incubated overnight at 4°C with 4 μg of mouse monoclonal antibody against hnRNP L or hnRNP A1 (Santa Cruz), and then with protein G Sepharose beads (50 μl) for 4 h at 4°C. The supernatant was discarded and the beads washed four times with NE-PER cytoplasmic or nuclear extraction buffers, resuspended, and RNA extracted with an equal volume of Trizol reagent (Invitrogen) and 1/5 volume of chloroform. Total RNA was isolated from the same extracts using Trizol reagent. mRNAs and pre-mRNAs were detected by RT–PCR with Superscript reverse transcriptase III (Invitrogen) using oligo d(T) and Taq DNA polymerase (Invitrogen). For in vivo cytoplasmic interactions, PCR primers for VEGFA (Shih and Claffey, 1999) generated a 716-nt product resolved by 1.5% agarose gel electrophoresis. A 96-nt PCR product generated from GAPDH primers served as a control. For in vivo nuclear interactions, PCR primers for VEGFA exon and intron recognition generated 600- and 300-nt PCR products, respectively, and a 96-nt GAPDH PCR product served as a control. The VEGFA exon-recognizing primers giving a 600-nt PCR product were TGCGGATCCATGAACTTTCTGCTCTCTTGGG (exon 1, forward primer) and ATGCAAGCTTGCTATGGGTAGTTCTGTG (exon 8, reverse primer). The intron-recognizing primers giving a 300-nt PCR product were GTGTCATCGCCTCTCATGCAG (intron 2, forward primer) and CCACTTCCCAAAGATGCCCAC (intron 3, reverse primer).
Sucrose gradient fractionation for polysome analysis
Ribosomal fractions were prepared as described (Merrick and Hensold, 2001). Briefly, 24-h transfected HEK293T cells were lysed in TMK lysis buffer containing cycloheximide (0.1 mg/ml) and the cytosolic extract was obtained by centrifugation at 10 000 g for 20 min. The extract was overlaid on a 10–50% (w/v) sucrose gradient and centrifuged at 100 000 g for 4 h at 4 °C. Absorbance of fractions was measured at 254 nm. RNA was isolated from each fraction using Trizol reagent (Invitrogen) and used for RT–PCR.
Statistical analysis
All quantitative data are expressed as mean±s.d., n=3 independent experiments. An asterisk indicates a significant difference, P<0.05, two-tailed t-test.
Supplementary Material
Acknowledgments
We are grateful to Abul Arif, Donna Driscoll, and Fulvia Terenzi for helpful suggestions. This work was supported by NIH grants P01 HL029582, R01 GM086430, and P01 HL076491 (to PLF) and by AHA, Great Rivers Affiliate Postdoctoral Fellowships (to PY).
Footnotes
The authors declare that they have no conflict of interest.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA (2001) HuR and mRNA stability. Cell Mol Life Sci 58: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A (1996) Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380: 435–439 [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N (2006) Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol 71: 149–156 [DOI] [PubMed] [Google Scholar]
- Claffey KP, Shih SC, Mullen A, Dziennis S, Cusick JL, Abrams KR, Lee SW, Detmar M (1998) Identification of a human VPF/VEGF 3′ untranslated region mediating hypoxia-induced mRNA stability. Mol Biol Cell 9: 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki BJ, Carter C, Rivoltini L, Koski GK, Kim HI, Weng DE, Roros JG, Hijazi YM, Xu S, Rosenberg SA, Cohen PA (1997) Calcium ionophore-treated peripheral blood monocytes and dendritic cells rapidly display characteristics of activated dendritic cells. J Immunol 159: 3823–3837 [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA et al. (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140: 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008a) Getting to the root of miRNA-mediated gene silencing. Cell 132: 9–14 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008b) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP (2005) The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- Ferrara N (2005) The role of VEGF in the regulation of physiological and pathological angiogenesis. EXS 94: 209–231 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Goldberg-Cohen I, Furneaux H, Levy AP (2002) A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J Biol Chem 277: 13635–13640 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O (2007) miRNAs play a tune. Cell 131: 22–24 [DOI] [PubMed] [Google Scholar]
- Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y (2006) MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H (2007) Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem 282: 33632–33640 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamba T, McDonald DM (2007) Mechanisms of adverse effects of anti-VEGF therapy for cancer. Br J Cancer 96: 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H (2009) The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA 15: 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM (2007) The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131: 136–145 [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R (2007) RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286 [DOI] [PubMed] [Google Scholar]
- Kim BS, Chen J, Weinstein T, Noiri E, Goligorsky MS (2002) VEGF expression in hypoxia and hyperglycemia: reciprocal effect on branching angiogenesis in epithelial-endothelial co-cultures. J Am Soc Nephrol 13: 2027–2036 [DOI] [PubMed] [Google Scholar]
- Kim HH, Gorospe M (2008) Phosphorylated HuR shuttles in cycles. Cell Cycle 7: 3124–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11: 441–450 [DOI] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739 [DOI] [PubMed] [Google Scholar]
- Lei Z, Li B, Yang Z, Fang H, Zhang GM, Feng ZH, Huang B (2009) Regulation of HIF-1α and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One 4: e7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AP (1998) Hypoxic regulation of VEGF mRNA stability by RNA-binding proteins. Trends Cardiovasc Med 8: 246–250 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, Qian H, Xue XN, Pollard JW (2006) Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res 66: 11238–11246 [DOI] [PubMed] [Google Scholar]
- Liu B, Peng XC, Zheng XL, Wang J, Qin YW (2009) MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 66: 169–175 [DOI] [PubMed] [Google Scholar]
- Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M (2009) The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol 29: 2899–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Hensold JO (2001) Analysis of eukaryotic translation in purified and semipurified systems. Curr Protocols Cell Biol Chapter 11, Unit 11-9 [DOI] [PubMed] [Google Scholar]
- Mili S, Shu HJ, Zhao Y, Pinol-Roma S (2001) Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol Cell Biol 21: 7307–7319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquerol L, Langille BL, Nagy A (2000) Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development 127: 3941–3946 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K (2006) Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol 16: 2135–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 23: 243–249 [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S, Swanson MS, Gall JG, Dreyfuss G (1989) A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol 109: 2575–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW (2010) Macrophage diversity enhances tumor progression and metastasis. Cell 141: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM (2007) Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8: 23–36 [DOI] [PubMed] [Google Scholar]
- Ray PS, Fox PL (2007) A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J 26: 3360–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Jia J, Yao P, Majumder M, Hatzoglou M, Fox PL (2009) A stress-responsive RNA switch regulates VEGFA expression. Nature 457: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Claffey KP (1999) Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J Biol Chem 274: 1359–1365 [DOI] [PubMed] [Google Scholar]
- Stockmann C, Doedens A, Weidemann A, Zhang N, Takeda N, Greenberg JI, Cheresh DA, Johnson RS (2008) Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis. Nature 456: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varallyay E, Burgyan J, Havelda Z (2008) MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc 3: 190–196 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181 [DOI] [PubMed] [Google Scholar]
- Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP (2008) Double-stranded RNA-binding protein regulates VEGF mRNA stability, translation and breast cancer angiogenesis. Mol Cell Biol 28: 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Alfert M, Castrillon DH, Shen Q, Holash J, Yancopoulos GD, Chin L (2001) Excessive tumor-elaborated VEGF and its neutralization define a lethal paraneoplastic syndrome. Proc Natl Acad Sci USA 98: 7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.