Abstract
Transcriptional cofactors are essential for proper embryonic development. One such cofactor in Drosophila, Degringolade (Dgrn), encodes a RING finger/E3 ubiquitin ligase. Dgrn and its mammalian ortholog RNF4 are SUMO-targeted ubiquitin ligases (STUbLs). STUbLs bind to SUMOylated proteins via their SUMO interaction motif (SIM) domains and facilitate substrate ubiquitylation. In this study, we show that Dgrn is a negative regulator of the repressor Hairy and its corepressor Groucho (Gro/transducin-like enhancer (TLE)) during embryonic segmentation and neurogenesis, as dgrn heterozygosity suppresses Hairy mutant phenotypes and embryonic lethality. Mechanistically Dgrn functions as a molecular selector: it targets Hairy for SUMO-independent ubiquitylation that inhibits the recruitment of its corepressor Gro, without affecting the recruitment of its other cofactors or the stability of Hairy. Concomitantly, Dgrn specifically targets SUMOylated Gro for sequestration and antagonizes Gro functions in vivo. Our findings suggest that by targeting SUMOylated Gro, Dgrn serves as a molecular switch that regulates cofactor recruitment and function during development. As Gro/TLE proteins are conserved universal corepressors, this may be a general paradigm used to regulate the Gro/TLE corepressors in other developmental processes.
Keywords: Drosophila , Hairy/Groucho, STUbL, transcriptional-repression, ubiquitin
Introduction
Transcriptional cofactors are essential for the function of sequence-specific transcription factors and are part of the machinery required to execute temporally coordinated gene expression programs. Regulation of cofactor recruitment and activity is emerging as a major level of gene expression regulation (Rosenfeld et al, 2006). For example, Hairy/Enhancer of split/Deadpan (HES) family repressors are the primary transducers of the Notch signalling pathway that has a central role in patterning, stem cell development, and is misregulated in cancers (Roy et al, 2007; Fischer and Gessler, 2007; Kopan and Ilagan, 2009). A well-studied case is the Drosophila repressor Hairy, a typical HES family member, which encodes a basic helix-loop-helix (bHLH) Orange repressor required for embryonic segmentation and adult peripheral nervous system (PNS) specification (Howard and Ingham, 1986; Carroll et al 1988; Rushlow et al, 1989; Skeath and Carroll, 1991). Hairy-mediated repression is dependent on its ability to recruit cofactors. For example, Hairy recruits the corepressor Groucho (Gro) through it C-terminal WRPW domain, an interaction that is essential for periodic repression of fushi tarazu (ftz; Paroush et al, 1994 and reviewed in Jennings and Ish-Horowicz 2008). In addition, Hairy recruits dCtBP and dSir2 through its PLSLV and basic domains, respectively (Supplementary Figure S1; Poortinga et al, 1998; Rosenberg and Parkhurst, 2002). While these cofactors are required for Hairy-mediated repression, they exhibit context-dependent recruitment and function (Bianchi-Frias et al, 2004). Interestingly, some cofactors enhance Hairy-mediated repression (e.g., Gro and dSir2), whereas others are required to refine Hairy's function (e.g., dCtBP and dTopors; Phippen et al, 2000; Secombe and Parkhurst, 2004). Consistent with this, we found that most of the genomic loci bound by Hairy in the context of Kc cells exhibit corecruitment of dSir2 and dCtBP, but are not co-bound by Gro (Bianchi-Frias et al, 2004). However, the mechanisms that regulate context-selective cofactor association with Hairy or that may regulate cofactor activities are largely unknown.
A possible mechanism is that post-translational modification of Hairy regulates its association with a given cofactor and determines its overall function. One such modification is ubiquitylation that in many cases regulates the stability of transcription factors. However, ubiquitylation can also serve as a regulatory modification that does not lead to degradation, but affects protein–protein interaction or intracellular localization (Ikeda and Dikic, 2008). Similarly, SUMOylation is a post-transcriptional modification that is involved in the regulation of gene expression and is mediated by the SUMO-specific E1-, E2-, and E3-SUMO ligase enzymes (Kerscher et al, 2006). Both ubiquitin and SUMO modifications are highly regulated (Lee et al, 2006; Carter et al, 2007; Hunter, 2007). These two modifications can also be connected through proteins collectively termed SUMO-targeted ubiquitin ligases (STUbLs; Sun et al, 2007; Geoffroy and Hay, 2009). STUbLs are RING proteins that bind non-covalently to the SUMO moiety of SUMOylated proteins via their N-terminal SUMO interaction motif (SIM) domains, and subsequently target the SUMOylated protein for ubiquitylation via their RING domain. Thus, STUbLs are able to ‘sense’ SUMOylated targets and modify them by ubiquitylation. The observation that STUbLs are associated with transcription complexes suggests that their function is directly linked to regulation of gene expression. For example, the STUbL protein RNF4 was found to be a positive regulator of steroid hormone transcription (Poukka et al, 2000). Importantly, STUbLs are structurally and functionally conserved, as the mouse and human RNF4 proteins can substitute for their yeast orthologs in functional assays (Prudden et al, 2007). STUbLs are required for the correct assembly of kinetochores, for the cell's ability to cope with genotoxic stress, and for genome stability (Kosoy et al, 2007; Prudden et al, 2007; Nagai et al, 2008; Rouse, 2009; Mukhopadhyay et al, 2010). RNF4 is highly expressed in the stem cell compartment of the developing gonads and brain, and its expression is enriched in progenitor cells, likely representing its role in ‘stemness’ (Galili et al, 2000; Ramalho-Santos et al, 2002). Recently, RNF4 was shown to regulate the SUMO- and ubiquitin-mediated degradation of PML and PML-RAR (Lallemand-Breitenbach et al, 2008; Tatham et al, 2008; Geoffroy and Hay, 2009). However, the role of STUbL proteins in transcription during development of higher eukaryotes is largely unknown.
Here, we show that Degringolade (Dgrn), the only Drosophila STUbL protein identified to date, physically and genetically interacts with Hairy and its cofactor Gro, and antagonizes Hairy/Gro-mediated repression during segmentation and neurogenesis. We find that ubiquitylation of Hairy by Dgrn affects choice of cofactor by preventing Gro, but not dCtBP, from binding to Hairy. We also find that Dgrn specifically targets SUMOylated Gro, alleviates Gro-dependent transcriptional repression, and suppresses Gro functions in vivo throughout development. DamID chromatin profiling experiments revealed that the antagonism between Dgrn and Gro is aimed at a broad array of genomic loci, suggesting that Gro-Dgrn antagonism is of general importance beyond Dgrn's interaction with Hairy.
Results
Dgrn associates with SUMOylated proteins and targets Hairy for ubiquitylation
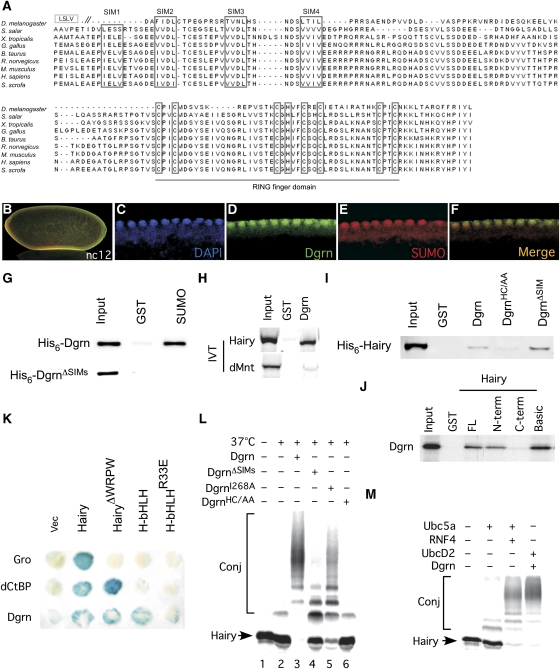
We have previously used a yeast two-hybrid assay to identify Hairy-associated proteins (Poortinga et al, 1998). One of these cDNAs encodes a 319-amino-acid protein, which we named as Dgrn (CG10981; for details regarding its genomic structure, generation of a null mutant, and function during early embryogenesis, see Barry et al, 2011). Bioinformatic analysis suggests that Dgrn is the sole Drosophila STUbL protein and an ortholog of the human RNF4 ubiquitin ligase. Sequence comparison of Dgrn with STUbL proteins from other species identified highly conserved SIM and RING domains (Figure 1A; Supplementary Figure S2). We find that Dgrn is a nuclear protein, as are SUMO or SUMOylated proteins, in developing embryos (Figure 1B–F). To test the ability of Dgrn to bind directly to SUMOylated proteins, we employed a GST pulldown assay using GST-SUMO and affinity purified histidine (His)6-Dgrn from bacteria. We find that Dgrn interacts specifically with GST-SUMO and GST-SUMO-GFP, but not with GST alone or GST-GFP, a function that requires its SIM domains (Figure 1G; data not shown). As expected, elevated levels of SUMOylated proteins are observed in Dgrn null embryos, similar to that reported for RNF4, confirming that Dgrn is a bona fide STUbL (Barry et al, 2011). As we identified Dgrn as a Hairy-associated protein, we tested the ability of Dgrn to bind to Hairy and mapped the domains within Dgrn and Hairy that mediate this interaction (Figures 1, 2 and Supplementary Figure S1). We find that GST-Dgrn interacts with 35S-Methionine in vitro translated (IVT) Hairy, but interacts very poorly with the bHLH repressor IVT-dMnt (Figure 1H).
Figure 1.
Dgrn is a STUBL, binds to SUMOylated proteins, and ubiquitylates Hairy. (A) Protein sequence alignment of Dgrn with its vertebrate orthologs. The conserved SIM domains and cysteine residues within the RING finger are indicated in grey boxes. (B–F) Dgrn (green) localizes with SUMO or SUMOylated proteins (red) in the nuclei of Drosophila embryos. DAPI (blue) marks nuclei. nc, embryonic nuclear cycle. (G) Bacterially purified recombinant His6-Dgrn binds to GST-SUMO, but not to GST alone. This binding requires Dgrn's SIM domains. (H) IVT labelled 35S-Met-Hairy, but not 35S-Met-dMnt, binds to GST-Dgrn. (I) Bacterially purified recombinant His6-Hairy binds to GST-Dgrn. Binding requires an intact RING domain as it is abrogated in the RING DgrnHC/AA mutant, but not the SIM domains. (J) 35S-Met-Dgrn binds to Hairy's N-terminus (N-term) or to its isolated basic region, but not to Hairy's C-terminus (C-term). (K) Dgrn and Hairy interact in vivo in a yeast two-hybrid assay. The interaction in vivo is mediated via the basic domain of Hairy. A point mutation within an isolated Hairy bHLH (bHLHR33E) abrogates binding. Binding of Dgrn to Hairy is independent of the WRPW, and neither Gro nor dCtBP bind to Hairy's bHLH. (L) Hairy ubiquitylation requires Dgrn's SIM and RING domains. (M) Both Dgrn/UbcD2 and mRNF4/Ubc5a ubiquitylate 35S-Met-Hairy in a reconstituted system. Conj denotes ubiquitin-Hairy conjugates. D. melanogaster, Drosophila melanogaster; S. salar, Salmo salar; X. tropicalis, Xenopus tropicalis; G. gallus, Gallus gallus; B. taurus, Bos taurus; R. norvegicus, Rattus norvegicus; M. musculus, Mus musculus; H. sapiens, Homo sapiens; S. scrofa, Sus scrofa.
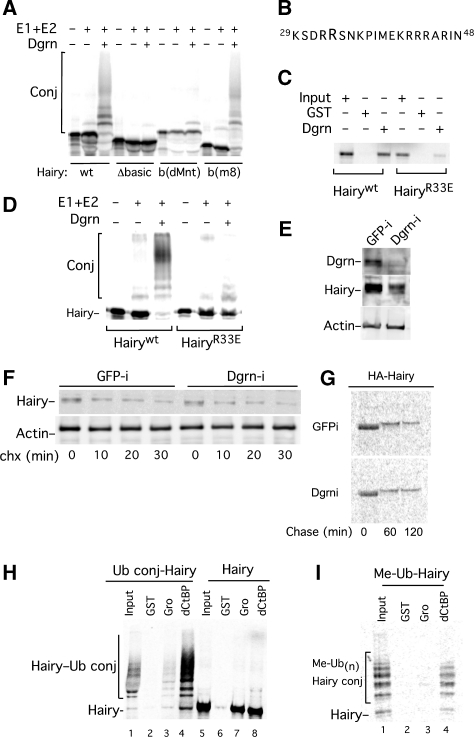
Figure 2.
Structural and mechanistic aspects of Dgrn-mediated ubiquitylation of Hairy. (A–D) Hairy's basic region and Arg33 are required for binding and ubiquitylation by Dgrn in a reconstituted system. (A) Ubiquitylation of 35S-Met-Hairy in a partially reconstituted system by Dgrn requires Hairy's basic domain. Hairy's basic domain can be substituted with the basic domain of E(spl)m8, but not by that of dMnt. (B–D) Arg33 within the basic domain is required for Dgrn recruitment to Hairy. (B) The amino acid composition of Hairy's basic region. Replacement of Arg33 by Glu33 significantly reduces Hairy-Dgrn interaction, and Hairy's ubiquitylation ((C) and (D) respectively). (E–G) Altered Dgrn levels do not affect Hairy's turnover. (E) Western blot analysis of steady state levels of endogenous Hairy protein in GFP or Dgrn (GFP-i, Dgrn-i) RNAi-treated S2R cells. (F) chx chase experiment in GFP or Dgrn RNAi-treated cells, followed by western blot analysis monitoring the protein levels of endogenous Hairy, and actin (loading control). (G) 35S-Methionine pulse-chase experiment in S2R cells using HA-Hairy as a substrate followed by αHA-IP. (H, I) Dgrn-mediated ubiquitylation of Hairy affects cofactor recruitment. (H) 35S-Met-Hairy binds to both GST-Gro and GST-dCtBP. 35S-Met Ub-Hairy conjugates fail to bind GST-Gro, but bind GST-dCtBP. (I) Poly-monoubiquitylation of Hairy by Dgrn (using Me-Ub) is sufficient to inhibit Gro recruitment, but does not interfere with dCtBP binding.
Using a yeast two-hybrid assay, we find that Dgrn binds to Hairy's bHLH domain. This binding is independent of the WRPW motif that mediates Hairy's binding to Gro (Figure 1K). Binding of Dgrn to Hairy is direct, as it can be demonstrated when both proteins are purified from bacteria, and is mediated by Dgrn's RING domain and not the SIM domain in vitro (Figure 1I; Supplementary Figure S1G). This is different from the previously reported recognition of substrates, such as GST-SUMO or PML that involves direct SUMOylation of the substrates. We also found that Hairy's basic region is required for Dgrn binding (Figure 1J and K), and that IVT-Hairy is not SUMOylated (Supplementary Figure S1E). Thus, Hairy and the other HES/bHLH proteins we examined (Barry et al, 2011) uncover a novel mode of recognition by STUbL proteins requiring the Dgrn RING domain, but not involving direct substrate SUMOylation.
We next tested the ability of Dgrn and its mouse ortholog RNF4 to ubiquitylate Hairy using an in vitro reconstituted system with IVT-Hairy or in cells using HA-Hairy as substrates. We find that both ligases efficiently ubiquitylate Hairy, an activity that requires the Drosophila E2-conjugating enzyme UbcD2 (but not UbcD1) or the mammalian Ubc5a (Figure 1M; Supplementary Figure S2E and data not shown). Dgrn's ability to ubiquitylate Hairy requires its RING domain, as replacement of its core His and cysteine residues with alanine (DgrnHC/AA; H300A+C302A) abolished the ability of Dgrn to ubiquitylate Hairy. To test whether it is Dgrn's catalytic activity that is directly required for Hairy ubiquitylation, we generated a mutant that reduces Dgrn's ligase activity without interfering with the RING structure (Ben-Saadon et al, 2006). This point mutation, DgrnI268A, has reduced ability to ubiquitylate Hairy but does not eliminate it completely, suggesting that the RING domain has a dual role; it is required not only for Dgrn catalytic activity but also to mediate Hairy recognition (Figure 1L).
While not required for binding, we find that Dgrn lacking all four of its SIM domains (DgrnΔSIMs) is compromised in its ability to ubiquitylate Hairy, similar to what has been observed for ubiquitylation of PML by RNF4 and of MATα2 by the yeast STUbL Slx5-8 (Figure 1I and L; Tatham et al, 2008; Xie et al, 2010).
Dgrn-dependent ubiquitylation of Hairy is mediated by Arg33 and does not target Hairy for degradation
We investigated the contribution of Hairy's basic region to its recognition by Dgrn in the context of the full-length Hairy protein (Figure 1K, Figure 2; Supplementary Figure S1). We find that a Hairy mutant lacking the basic region (HairyΔbasic) fails to bind or to be ubiquitylated by Dgrn. Substitution of either E(spl)-m8 or Scute (Sc) basic domains, both of which interact with Dgrn, was sufficient to promote binding and ubiquitylation. In contrast, a basic region derived from the dMnt repressor that does not bind to Dgrn failed to support binding or ubiquitylation (Figure 2A; Supplementary Figure S1). We also find that Dgrn binds and ubiquitylates Hey and all other HES members except Her (Barry et al, 2011). Comparison of the basic region of Her with that of Hairy identified a positively charged residue in Hairy, Arg33, which is replaced with the negatively charged glutamic acid (Glu) residue in Her. To determine whether Arg33 mediates Dgrn recognition, we substituted Hairy's Arg33 with Glu (HairyR33E). We find that Dgrn binds and ubiquitylates HairyR33E very poorly despite our observation that HairyR33E is still a functional repressor in reporter assays (Figure 1K, Figure 2B–D; Supplementary Figure S4C).
Next, we tested whether Dgrn targets Hairy for degradation and monitored the levels of Hairy protein as a function of Dgrn protein levels. We determined endogenous Hairy protein levels in its steady state, or in dynamic cyclohexamide (chx), as well as 35S-Methionine labelled HA-Hairy pulse-chase experiments using Drosophila S2R cells that were treated with control or Dgrn-specific RNAi (Figure 2E–G; Supplementary Figure S2F). While Hairy is a short-lived protein (5–15 min half-life), reduction in Dgrn protein levels by RNAi did not stabilize Hairy protein or attenuate its turnover. Thus, we hypothesized that Dgrn-mediated ubiquitylation of Hairy has another regulatory role. In accord, we found that Dgrn catalyses the assembly of mixed poly-ubiquitin chains, and not the lysine (Lys)48-linked poly-ubiquitin chains that are associated with targeting proteins to the 26S proteasome (Supplementary Figure S2B–D). This observation is consistent with reports that attribute specific functions to specific types of poly-ubiquitin chains depending on their intrinsic ubiquitin linkage (Ikeda and Dikic, 2008). Furthermore, using methylated ubiquitin (Me-Ub), a ubiquitin derivative that acts as a chain terminator, and partially purified Hairy substrate (lacking endogenous ubiquitin; see Materials and methods), we find that Hairy is ubiquitylated on at least five distinct residues, likely not involving N-terminal linear poly-ubiquitylation (Supplementary Figure S2D).
Dgrn-mediated ubiquitylation affects cofactor recruitment
As Dgrn does not appear to target Hairy for degradation, we hypothesized that ubiquitylation of Hairy impairs its ability to recruit its associated corepressors. We took advantage of the observation that Hairy ubiquitylation in vitro is highly efficient with ∼100% of naïve Hairy exhibiting Ub conjugation, and compared Hairy's ability to bind to two of its cofactors, Gro and dCtBP, depending on the extent of Hairy's ubiquitylation. We used naïve IVT Hairy or ubiquitylated IVT Hairy as binding substrates for Gro or dCtBP in GST pulldown assays. We find that while naïve Hairy binds both GST-Gro and GST-dCtBP, ubiquitylated Hairy fails to bind GST-Gro (Figure 2H). Using Me-Ub, we also find that poly-monoubiquitylation of Hairy is sufficient to inhibit Gro recruitment, but does not affect dCtBP recruitment (Figure 2I). Thus, our data suggest that Dgrn's ligase activity regulates protein–protein interaction, selectively inhibits Gro recruitment, affects cofactor choice, and supports the prediction that Dgrn ligase activity antagonizes Hairy/Gro-mediated repression.
Dgrn alleviates Hairy-mediated Gro-dependent repression
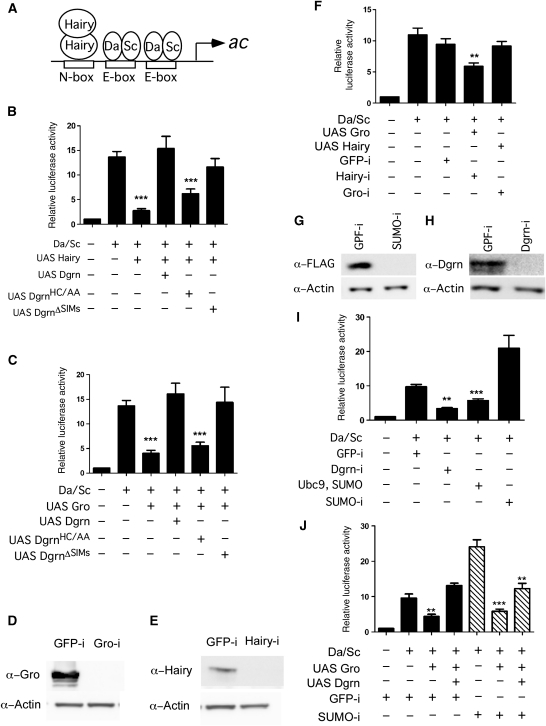
To delineate the role that Dgrn has in Hairy-mediated transcriptional repression, we tested Dgrn's ability to modulate Hairy repression of its direct target achaete (ac). ac is required for sensory bristle specification, and the core regulatory network that regulates its expression is well characterized (Skeath and Carroll, 1991). The ac promoter is activated by binding of the pro-neural bHLH proteins Sc and Daughterless (Da), and is repressed when Hairy binds in the vicinity of the Sc/Da-binding sites (Figure 3A). Using an established ac reporter system (Van Doren et al, 1994), we find that Dgrn, but not the RING mutant DgrnHC/AA, alleviates Hairy-mediated repression (Figure 3B). Dgrn's derepression activity is also attenuated in the DgrnΔSIMs mutant, albeit to a lesser degree than with wild-type Dgrn. Importantly, Dgrn binding to Hairy does not displace Hairy from DNA (Supplementary Figure S3). Dgrn's activity is highly specific and selective, as Dgrn has only minimal (statistically insignificant) effect on a reporter with mutated binding sites, and it does not affect the expression of other luciferase reporters, such as the Dorsomycin reporter that is activated by Dorsal (Supplementary Figure S4).
Figure 3.
Dgrn antagonism of Hairy and Gro repression inversely correlates with the effects of the SUMO pathway and is partially dependent on SUMOylation. (A) Schematic diagram of the ac reporter. The binding sites for sequence-specific transcription factors are indicated. ac, Sc, Da. (B) Dgrn alleviates Hairy-mediated repression of the ac luciferase reporter. Dgrn derepression activity is compromised in the Dgrn RING finger mutant (DgrnHC/AA), and is also minimally reduced in the DgrnΔSIMs mutant. (C) Dgrn alleviates Gro-mediated repression of the ac luciferase reporter. (D, E) Protein levels of Gro (D) and Hairy (E) in RNAi-treated cells used in (F) as indicated. GFP-i serves as a non-specific RNAi control. Gro-i and Hairy-i denotes Gro and Hairy RNAi, respectively. (F) Hairy-mediated transcriptional repression is dependent on Gro and is abolished in Gro-i cells. In contrast, Gro represses transcription in cells in which Hairy is inactivated using RNAi. (G, H) Protein levels of Flag-SUMO (G) and Dgrn (H) in RNAi-treated cells used in (I) and (J) as indicated. (I) Reduced Dgrn protein levels via RNAi, or expression of SUMO and Ubc9 represses transcriptional activity. Similarly, reduction in SUMO levels increases transcription from the ac reporter. (J) While Gro can mediate repression in cells with reduced SUMO levels, Dgrn's ability to alleviate repression in these cells and fully restore the activated state in SUMO-i cells is compromised. Data were collected from five independent experiments. Statistical analysis, s.e.m. and t-test comparisons were performed using the Prism5 ANOVAs software. Significance is indicated by ***P<0.001 and **P<0.01.
As we find that Arg33 mediates Hairy's binding and ubiquitylation by Dgrn in vitro, it is expected that the interaction-defective HairyR33E mutant would be insensitive to Dgrn derepression activity. However, while HairyR33E efficiently represses transcription of the ac reporter, Dgrn alleviated its repression indistinguishably from that of wild-type Hairy (Supplementary Figure S4C). A possible explanation is that Dgrn's anti-repressive activity is also directed towards other factors of the repression machinery. One such factor could be Gro, a shared corepressor recruited by HES repressors during segmentation, sex-determination, and neurogenesis (Paroush et al, 1994). We used the ac reporter system to test whether Dgrn antagonizes Gro-mediated repression. Wild-type Dgrn, but not inactive Dgrn, alleviates Gro repression and restores ac reporter expression (Figure 3C). To directly test the interdependence between Hairy and Gro on ac repression, we examined their ability to repress the ac reporter in cells in which the levels of either Hairy or Gro are reduced by RNAi (Figure 3D–E, Supplementary Figure S3C). We find that Gro is essential for Hairy-mediated repression (Figure 3F). However, Gro was still able to repress transcription in the absence of detectable Hairy protein, suggesting that this activity is dependent on either residual (trace) amounts of Hairy or that factors other than Hairy are involved in Gro recruitment and ac repression, and may be the target of Dgrn action.
It is well established that SUMOylation enhances transcriptional repression, and recent reports suggest that Gro is SUMOylated (Gill, 2005; Nie et al, 2009). Thus, we tested whether SUMOylation has a role in repression of the ac promoter. Expression of SUMO together with Ubc9 (SUMO-conjugating enzyme) or reduction in Dgrn protein levels using Dgrn-specific RNAi (Figure 3H) repressed ac transcription similar to that of expressing Gro (Figure 3I; compare with Figure 3C). Consistent with this, RNAi to SUMO resulted in a marked increase in ac reporter transcription assays (Figure 3G, I and J). We examined to what extent Gro repression is dependent on SUMOylation, as well as Dgrn's ability to alleviate Gro repression in these cells. We find that Gro represses the ac reporter in cells with reduced SUMO levels (Figure 3J). However, and in contrast to control RNAi cells, the ability of Dgrn to fully restore the activated state is significantly compromised. Taken together, our transcriptional data are consistent with the observation that Dgrn ubiquitylation of Hairy specifically inhibits Gro recruitment, and indicates that Dgrn activity simultaneously targets Hairy, Gro, and the repressive chromatin environment mediated by the SUMO pathway.
Dgrn suppresses hairy phenotypes in vivo
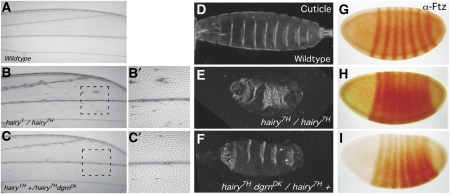
On the basis of the above results, we expected that the loss of Dgrn would suppress phenotypes associated with hypomorphic hairy alleles (Figure 4). To determine the role of Dgrn in Hairy-mediated repression in vivo during development, we examined the ability of dgrn to suppress the ectopic bristle phenotype observed in the wings of adult viable hypomorphic hairy flies, a phenotype that is associated with ac derepression. We find that numerous ectopic bristles are present on hairy1/hairy7H adult wings (Figure 4B–B’; Table IA). In this setting, halving the dose of dgrn (hairy1+/hairy7HdgrnDK) inhibited the formation of ectopic bristles predominantly within the intervein region of the wing (Figure 4C–C′; Table IA), supporting our transcriptional data (Figure 3B).
Figure 4.
dgrn antagonizes hairy-mediated repression in vivo. (A–C’) Dgrn heterozygosity suppresses the ectopic bristle phenotype associated with Hairy mutants. Wild-type wing (A). Numerous ectopic bristles are observed on hairy1/hairy7H wings (B). Wing of a hairy1/hairy7H mutant that is also heterozygous for dgrnDK (C). (B’, C’) Higher magnification of the regions outlined in B and C, respectively. (D–I) dgrn interacts genetically with hairy and suppresses Hairy transcriptional repression of ftz during embryogenesis. Larval cuticle preparations (D–F) and Ftz staining (G–I) in the wild type (D, G), homozygous hairy7H mutants (E, H), and homozygous hairy7H embryos that are also heterozygous for dgrn (h7H +/h7H dgrnDK) (F, I). Reducing the gene dose of dgrn using the dgrnDK null allele suppresses the hairy mutant phenotype.
Table 1. Genetic interactions between dgrn and hairy.
|
(A) dgrn heterozygosity partially suppresses hairy-associated bristles phenotypes (P<0.01) | ||||
|---|---|---|---|---|
| Genotype | Intervein region |
L2-vein |
||
| % Wings with extra bristles (>40) | n | % Wings with extra bristles (>40) | n | |
| w 1118 | 0 | 40 | 0 | 40 |
| hairy 1 /hairy 7H | 79 | 39 | 100 | 44 |
| hairy1+/hairy7H dgrnDK | 13 | 40 | 86 | 43 |
| (B) dgrn heterozygosity partially suppresses hairy cuticle phenotypes (P<0.01) | ||||
| Genotype | % Cuticles with >4 segments | n | ||
| w 1118 | 100 | 80 | ||
| hairy 7H /hairy 7H | 18 | 85 | ||
| hairy7H+/hairy7H dgrnDK | 50 | 101 | ||
| (C) Reduction in dgrn levels partially rescues hairy-associated embryonic lethality (P<0.01) | ||||
| Female genotype | Male genotype | % Embryonic lethality | n | |
| w 1118 | w1118 | 1 | 1509 | |
| hairy 7H /TM3 | hairy 12C /+ | 30 | 2323 | |
| hairy7H dgrnDK/TM3 | hairy 12C /+ | 19 | 2726 | |
| hairy 12C /TM3 | hairy 7H /+ | 30 | 2096 | |
| hairy 12C /TM3 | hairy7H dgrnDK/++ | 25 | 2416 | |
Hairy- and Gro-mediated repression are also required for the proper segmentation of the central portion of the embryo (Paroush et al, 1994). Embryos homozygous for a strong hypomorph (but not null) hairy allele, hairy7H, show aberrant segmentation and derepression of Hairy's downstream genetic target ftz (Figure 4D, E, G and H; see Supplementary data for the exact nature of hairy alleles used in this study). However, the severe cuticle morphology and Ftz expression are partially restored in hairy mutant embryos that are simultaneously heterozygous for dgrn (hairy7H+/hairy7HdgrnDK; Figure 4F and I; Table IB). As these hairy alleles die during embryogenesis, we tested whether reducing the dose of dgrn can rescue the embryonic lethality associated with hairy mutants. While 30% of hairy12C/hairy7H embryos die and do not hatch, only 19% embryonic lethality is observed in hairy12C+/hairy7HdgrnDK embryos (P<0.001; Table IC). Thus, we find that Dgrn limits Hairy-mediated repression during development, suppresses the phenotypes associated with hairy hypomorphic mutants during segmentation and PNS specification, and partially rescues the embryonic lethality associated with hairy mutant embryos.
Dgrn targets SUMOylated Gro protein
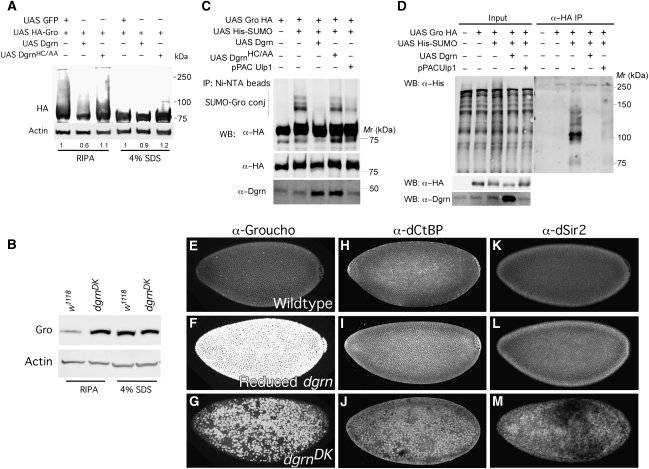
In addition to its ability to inhibit Gro recruitment to Hairy (Figures 2H and I), Dgrn may affect the Gro protein itself. To test this possibility, we expressed UAS-HA-Gro along with Dgrn, DgrnHC/AA (RING mutant), or UAS-GFP in S2R cells, and monitored the level of Gro protein (Figure 5A). While Gro is a stable protein with a half-life >6 h, we find that expression of wild-type Dgrn results in reduced HA-Gro protein level. In contrast, the level of HA-Gro in cells expressing DgrnHC/AA is not affected. Importantly, the reduction in Gro levels is only minimally reversible upon treating the cells with the proteasome inhibitor MG132 (data not shown). However, when Gro protein was extracted using 4% SDS lysis buffer, the level of Gro protein in Dgrn expressing cells was similar to that of GFP- or DgrnHC/AA-expressing cells (Figure 5A). In accord, we find that the SDS-sensitive higher forms of Gro (identified in RIPA extraction) diminish upon co-transfection of Dgrn, but not DgrnHC/AA (Figure 5A). Consistent with this, we find that endogenous Gro protein levels are elevated in dgrn mutant embryos when proteins are extracted in RIPA buffer, but not when extracted in 4% SDS buffer (Figure 5B).
Figure 5.
Dgrn targets SUMOylated Gro for sequestration. (A) Western blot analysis of Gro protein levels in response to Dgrn expression in S2R cells. The levels of HA-Gro protein are reduced upon expression of a functional Dgrn in RIPA-derived extract, but not in 4% SDS-derived cell extract or by expression of DgrnHC/AA. Bottom: the relative amount of Gro compared with actin is indicated. HA: protein levels of transfected HA-Gro; actin serves as loading control. (B) Protein extracts derived from 4 h-old dgrnDK embryos generated in RIPA, but not 4% SDS buffer, show elevated levels of Gro. (C, D) Dgrn specifically targets SUMOylated Gro. S2R cells were transfected with the indicated plasmids. After 48 h, cells were lysed in denaturing buffer and SUMOylated proteins were recovered on Ni-NTA agarose (C), or in hot-lysis buffer and immunoprecipitated using anti-HA sepharose beads (D). Proteins were identified using the indicated antibodies. The input levels of Gro, Dgrn, and SUMOylated proteins are shown as indicated. (E–M) Protein expression of Hairy-associated cofactors in wild type (E, H, K), dgrnDK mutant (F, I, L), or reduced dgrn (G, J, M) embryos. dgrn mutant embryos show intense Gro protein expression compared with wild type (E–G), while the protein expression of Hairy's other cofactors, dCtBP (H–J) and dSir2 (K–M), remains the same.
Similarly, the Gro signal evident by immunostaining is stronger in dgrn mutants, whereas the protein levels of other Hairy cofactors (e.g., dSir2 or dCtBP) are relatively unchanged (Figure 5E–M). Importantly, we find that Dgrn specifically targets SUMOylated Gro, a function that requires the Dgrn RING and SIM domains (Figure 5C and D; Supplementary Figure S5F). Taken together, these findings suggest a role for Dgrn in the selective intracellular sequestration of Gro oligomers.
Dgrn antagonizes Gro function in vivo
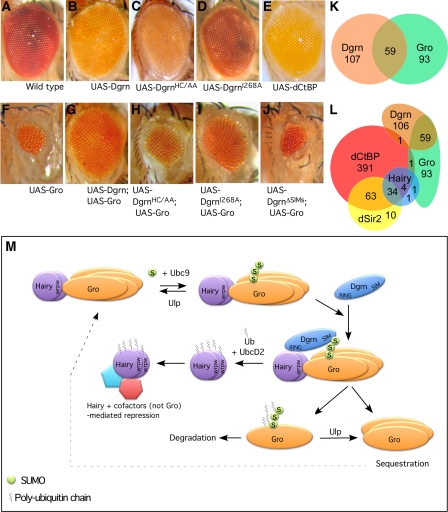
Next, we examined the structural determinant within Dgrn that mediates its antagonism to Gro in vivo. Using the eyeless-Gal4 driver, we tested the ability of Dgrn to suppress the small-eye phenotype associated with adults ectopically expressing Gro (Figure 6A–J; Supplementary Table SIA). Ectopic expression of Dgrn or its derived mutants described above (except for DgrnΔSIMs) had no significant effect on eye morphology (Figure 6A–D). However coexpression of Dgrn along with Gro efficiently suppressed the Gro phenotype, and to a large extent restored the pattern of the compound eye (compare Figure 6F and G; Supplementary Table SIA). In contrast, the DgrnHC/AA and DgrnI268A mutants failed or had limited ability to suppress the Gro eye phenotype (Figure 6H and I). Interestingly, we find that DgrnΔSIMs is unable to suppress the Gro eye phenotype (Figure 6J), indicating that the SIM domains have a functional role in Dgrn's ability to antagonize Gro-mediated repression in vivo. We noted that expression of DgrnΔSIMs alone in this setting results in lethality at the pupal stage with headless pupae (data not shown), supporting the notion that Dgrn's SIM domains are required for its function during development. Expression of the dCtBP cofactor using the eyeless Gal4 driver does not alter the eye morphology (Figure 6E).
Figure 6.
dgrn suppresses the gro eye phenotype and is co-bound with Gro to many genomic loci. (A–J) Functional Dgrn is required to suppress the small eye phenotype that results from overexpression of Gro. Expression of UAS-Gro using the eyeless-Gal4 driver results in a small-deformed eye (compare (A) to (F)). Expression of UAS-dCtBP, UAS-Dgrn, or its derived mutants alone (E, B–D) does not change the compound eye. However, coexpression of both UAS-Gro and a functional UAS-Dgrn (G), but not Dgrn that is lacking functional RING (H, I) or SIM (J) domains, suppresses the Gro eye phenotype. (K, L) Dgrn and Gro share direct targets genome-wide, as identified using DamID in Drosophila Kc cells. (K) Venn diagram depicting the genomic loci bound by Dgrn and Gro. (L) Venn diagram depicting the genomic loci bound by Hairy and its associated cofactors Dgrn, Gro, dCtBP, and dSir2. (M) A model for Dgrn function. Hairy/HES dimers are associated with Gro oligomers. A fraction of Gro proteins are SUMOylated, leading to enhanced repression and also facilitates the recruitment of Dgrn to Gro oligomers. Thus, a dual recognition event takes place: Dgrn binds to Hairy's basic region via its RING domain and simultaneously associates with SUMOylated Gro via its SIM domains. Subsequently, ubiquitylation of Hairy by Dgrn prevents association of Hairy with Gro, but not with other Hairy cofactors. Concomitantly, SUMO-Gro and its associated Gro oligomers are sequestered. SUMOylated Gro may then be degraded or alternatively de-SUMOylated. The non-SUMOylated Gro is recycled.
Another patterning/fate determination process that is regulated by Gro and governed by Notch/EGF signalling is the specification of mesothoracic sensory bristles during the development of the adult PNS (Hasson et al, 2005). In this setting, overexpression of Gro results in the loss of sensory bristles. Coexpression of wild-type Dgrn, but not Dgrn mutants, antagonizes Gro and restores bristle formation (Supplementary Table SIB). Similarly, the ectopic bristle phenotype associated with tissue-specific inactivation of Gro using the UAS-Gro RNAi transgene is suppressed by coexpression of UAS-Dgrn RNAi and UAS-Gro RNAi (Supplementary Figure S6A–D). We find that a priming phosphorylation at sites used by EGF/RTK signalling to inactivate Gro is not a prerequisite for Dgrn activity (Supplementary Figure S6E and F).
Thus, dgrn genetically interacts with gro and suppresses gro phenotypes. Dgrn function requires the SIM domains, as well as a functional RING motif to antagonize Gro in vivo. These observations fit well with the antagonism observed between Dgrn and other HES proteins during sex determination and embryonic neurogenesis, developmental processes that are regulated by Gro (Barry et al, 2011).
Dgrn and Gro co-bind to shared numerous loci genome-wide
Our observation that Dgrn antagonizes Gro-mediated repression in several in vivo developmental settings led us to analyse the genome-wide landscape co-regulated by Dgrn and Gro. We used DamID chromatin profiling to map the genomic loci co-bound by Dgrn and Gro. We have previously performed such analysis for Hairy and its cofactors Gro, dCtBP, and dSir2 (Bianchi-Frias et al, 2004). Using Dam-Dgrn chimeric protein and Drosophila Kc cells, we identified 166 genomic loci associated with Dgrn. Comparison with the loci bound by Gro and Dgrn identified 59 genomic loci co-bound by both proteins (Figure 6K; Supplementary Table SII). Interestingly, 38% of Gro direct targets are co-bound by Dgrn, suggesting that the antagonism between Dgrn and Gro takes place on a genome scale. However, this mapping also identified genomic loci exclusive to each of the factors, indicating that there must be Gro-independent regulation of genes and processes by Dgrn and vice versa. Importantly, in Kc cells, Gro and Dgrn co-bound loci do not overlap with the loci bound by Hairy and its cofactor, dSir2, and only a single gene is shared between Dgrn and dCtBP (Figure 6L). This is highly complimentary to previous reports in which Hairy binding was shown to be development and context dependent (see Bianchi-Frias et al, 2004; MacArthur et al, 2009). Specifically, in this experimental context (Kc cells), Hairy-bound loci are cooccupied by dCtBP and dSir2, but not Gro (Bianchi-Frias et al, 2004). Thus, taken together with its ability to inhibit Gro recruitment and target SUMOylated Gro for sequestration, we suggest that Dgrn may serve as a molecular selector that regulates cofactor choice, selectively inhibiting the recruitment and function of the corepressor Gro (Figure 6M).
Discussion
Transcriptional repression is, in part, governed by a dynamic equilibrium between post-transcriptional modifications, including phosphorylation, ubiquitylation, and SUMOylation. STUbL proteins are the molecular machinery that balance SUMOylation with ubiquitylation (Geoffroy and Hay, 2009). Hence, the mechanisms surrounding the recognition of substrates by STUbLs, STUbLs transcriptional activity, and the developmental context in which STUbLs operate are of great interest. Here, we characterized the role of Dgrn, the sole Drosophila STUbL protein, in Hairy-Gro-mediated transcriptional repression. We find that Dgrn limits Hairy-Gro-mediated repression during development by specifically targeting the recruitment of the corepressor Gro and affecting its localization, serving as a molecular selector regulating the cofactor recruitment.
Biochemical aspects of Dgrn function
Dgrn binds directly to Hairy and is capable of ubiquitylating Hairy in a reconstituted system and in cells (Figures 1 and 2; Supplementary Figures S1 and S2). We find that the recognition motif for Dgrn within Hairy maps to Hairy's basic region and requires a specific positive charge (Arg33). This motif is transferable and functionally conserved, not only in Hey and other HES proteins (e.g., E(spl)m8 and Dpn), but also in dMyc and other bHLH proteins including the activator Sc. Therefore, it may reflect a general property of bHLH recognition by STUbL proteins (Supplementary Figure S1; Barry et al, 2011). We find no evidence for direct SUMOylation of the HES and bHLH proteins: bacterially purified Hairy and Dgrn proteins interact, α-SUMO antibodies fail to detect SUMOylated Hairy, Hairy's mobility in SDS–PAGE is not altered upon incubation with the dUlp1 SUMO peptidase, and mutating putative SUMOylation sites within Hairy does not alter its recognition or ubiquitylation by Dgrn (Supplementary Figure S1). Accordingly, we find that Dgrn's interaction with Hairy is mediated through Dgrn's RING motif independent of the SIM domains. Similarly, the yeast STUbL Slx5–Slx8 recognizes the MATα2 repressor independent of SUMOylation (Xie et al, 2010). Hairy recognition by Dgrn/RNF4 is also different from its recognition of substrates, such as GST-SUMO or PML, that involves direct SUMOylation of the targeted protein and requires the Dgrn/RNF4 SIM domains (Sun et al, 2007; Wang and Prelich, 2009).
Importantly, SUMOylation and the SIM motifs are necessary for Dgrn to target SUMOylated Gro and for Dgrn's suppression of HES/Gro repression in vivo (Figures 5 and 6; Supplementary Figure S6). As we find that Dgrn does not bind Gro directly (Supplementary Figure S5B and C), it is likely that the SIM domains interact with the poly-SUMO chain itself (Geoffroy et al, 2010). Dgrn possessing two separate recognition modules is reminiscent of the dual recognition properties described for the RING protein UBR1 (E3α; Reiss and Hershko 1990). As the current dogma is that STUBLs recognize (via their SIM domains) poly SUMO chain(s) rather than the substrate, the dual recognition mechanism we observe with Dgrn may further substantiate substrate recognition and specificity.
The contribution of each SIM domain is additive, and a Dgrn mutant harbouring a single SIM domain is capable of binding to GST-SUMO, as well as conjugating Hairy, although to a lesser extent than wild-type Dgrn. Correspondingly, we find that elevated levels of SUMOylated proteins are detected in dgrn null embryos (Barry et al, 2011).
As an ubiquitin ligase, Dgrn catalyses the formation of mixed poly-ubiquitin chains on Hairy (Supplementary Figures S1 and S2). This ubiquitylation does not map to Hairy's basic region, its putative SUMOylation sites, or to a single Lys residue. Importantly, this poly-site ubiquitylation does not affect Hairy protein stability or integrity, but rather selectively inhibits Gro binding to Hairy (Figure 2, Supplementary Figure S1H). Furthermore, in cells in which Dgrn protein levels are reduced via RNAi, Hairy protein levels are also decreased compared with control cells, suggesting that Dgrn is likely required for Hairy expression. This is different from dTopors, a Hairy-associated PHD-RING finger protein, which catalyses Lys48-linked chains and regulates Hairy turnover (Secombe and Parkhurst, 2004). Further work will be required to determine the exact molecular events and the role that specific ubiquitin chain linkage has in Dgrn's ability to inhibit Gro from binding to Hairy in vivo.
Despite extensive efforts, we did not identify ubiquitylated Gro forms in our assays. Nonetheless, our data suggest that Dgrn specifically targets the SUMO chains on Gro, which likely serve as a signal for Gro sequestration by as yet to be identified machinery (Figure 5; Supplementary Figure S5).
Transcriptional role of Dgrn
In transcription assays, Dgrn is a potent activator of ac and Sxl transcription, a function that requires its catalytic activity. Dgrn antagonizes Hairy-, Dpn-, and Gro-mediated repression in vivo (Figures 3, 4, 5 and 6; Barry et al, 2011). We find that Dgrn specifically targets SUMOylated Gro, Dgrn function inversely correlates with SUMOylation, and that a reduction in SUMO levels impairs Dgrn's ability to fully alleviate repression. Thus, Dgrn's activity suppresses the local repressive chromatin structure generated by repressors, their associated cofactors, and the SUMO pathway. We also find that expression of DgrnHC/AA can inhibit the activation mediated by Da/Sc (Figure S4B), suggesting that Dgrn is required to alleviate repression by endogenous repressors and/or corepressors. This fits well with our observation that reduction in Dgrn protein levels via RNAi impairs Da/Sc-mediated activation (Figure 3I). While we have focused on Dgrn's effects on the repressive machinery, it is also possible that part of Dgrn ligase activity enhances the function of activators and/or coactivators. For example, Dgrn efficiently ubiquitylates the pro-neural activator Sc, and significant activation of the ac or Sxl promoters requires only Dgrn along with either Da or Sc (Supplementary Figure S1; Barry et al, 2011).
Our data suggest that part of Dgrn's activity is aimed specifically at the Gro corepressor that is shared by all HES proteins. First, Dgrn-mediated ubiquitylation of Hairy prevents Gro recruitment to Hairy. Second, Dgrn specifically targets SUMOylated Gro and its associated Gro oligomers for sequestration (Figures 2 and 5). Specifically, we find that the detected level of Gro protein is dependent on Dgrn and the method of protein extraction. For example, in embryos that lack Dgrn (dgrnDK) and when protein extracts are made in RIPA buffer, the detected levels of Dgrn in dgrnDK embryos is higher compared with that of wild type. However, if the extraction is performed in 4% SDS buffer, the detected levels of Gro protein in wild-type and dgrnDK embryo extracts is equal (Figure 5B). Likewise, in Figure 5F, G, the signal detected for Gro using immunostaining in embryos is highly complementary to the milder RIPA extraction. dgrnDK embryos show an increased signal compared with wild-type embryos (as in the absence of Dgrn, less Gro is sequestered and more Gro molecules are available for detection by the antibody). The majority of Gro appears to be sequestered. As we can recover only 90% of Gro after co-transfection of Dgrn using SDS extraction, we cannot rule out the possibility that a fraction of the SUMOylated Gro is degraded. All together, these data suggest that Dgrn is required for Gro sequestration and that loss of Dgrn ‘liberates’ sequestered Gro.
While our data support a model in which Dgrn targets SUMOylated Gro for sequestration, Dgrn may also regulate the molecular machinery that is required for Gro SUMOylation and subsequently sequestration. Furthermore, while it is established that STUbL targets SUMOylated proteins for ubiquitylation and degradation, it is also possible that Dgrn has an impact on the SUMO pathway and SUMO isopeptidases.
Gro and its mammalian orthologs, the transducin-like enhancers of split (TLE1–4) proteins, repress transcription via several mechanisms, including oligomerization to generate local repressive chromatin structures, and are negatively regulated by phosphorylation (Nibu et al, 2001; Sekiya and Zaret 2007; Cinnamon and Paroush 2008; Jennings et al, 2008; Martinez and Arnosti 2008; Lee et al, 2009). We find that site-specific phosphorylation used by RTK signalling to inactivate Gro is not a prerequisite for Dgrn activity (Supplementary Figure S6E and F). However, the details surrounding other phosphorylations, the role of site-specific SUMOylation of Gro, and the molecular machinery mediating sequestration, as well as Dgrn's effects on specific Gro-dependent repressive mechanisms await further studies.
In vivo, we find that Dgrn antagonism of Gro is highly relevant for embryonic segmentation, PNS development, and sex determination, processes that are regulated by Gro (Barry et al, 2011). Indeed, Dgrn can suppress the gain-of-function phenotypes of Gro, as well as rescue the phenotypes associated with tissue-specific inactivation of Gro using RNAi transgenes (Figure 6; Supplementary Figure S6). We also find that the genomic targets of Gro and Dgrn are distinct from that of dCtBP or dSir2, and that 38% of Gro direct targets are shared with Dgrn (Figure 6K and L, Supplementary Table SII). Thus, we predict that Dgrn will be involved in other HES-independent, but Gro-regulated, processes as well. It is likely that both proteins have unique regulatory roles during early development. This notion stems from our observations that each of the factors has exclusive, non-overlapping, genomic binding sites (Figure 6K), and that neither of the two genes can functionally rescue the embryonic lethality associated with mutants of the other protein (i.e., Gro cannot rescue the female sterility associated with dgrn null females, and reducing the dose of Dgrn does not rescue the lethality associated with the groE48 mutant).
Finally, an open question is how can the activity of a general corepressor be temporally and spatially regulated during development. Our data to date suggest a model in which Dgrn has a regulatory role (Figure 6M). As it is suggested that SUMOylation enhances Gro-mediated repression (Ahn et al, 2009), one can imagine that ATP-dependent SUMOylation of Gro within the repressor complex will result in local augmented repression. However, concomitantly, SUMOylation will promote Dgrn recruitment, and subsequent inactivation of the repression complex on chromatin or in its vicinity, ensuring that local SUMO-augmented repression is limited in time and space. We speculate that this type of transcriptional regulation will be instrumental to define and sharpen patterning borders throughout development.
Materials and methods
Fly strains, genetic interactions, and embryo analysis
Flies were cultured on yeast–cornmeal–molasses–malt extract medium at 25°C. Alleles used in this study: h12C/TM3; h7H, rucuca/TM3; dgrnDK/TM3, Sb (dgrn null mutant that is described in Barry et al, 2011); h7HdgrnDK/TM3, Sb double mutant chromosome was generated by standard recombination. UAS-Dgrn; UAS-DgrnI268A; UAS-DgrnHC/AA and UAS-DgrnΔSIMs; and UAS-dCtBP transgenic lines were generated as described (Spradling, 1986). UAS-Gro#30 and FRT82-groE48/TM3, Sb were previously described (Orian et al, 2007). C253-Gal4, and eyeless-Gal4 were from the Bloomington Stock Center. The indicated UAS transgenic strains were expressed using the Gal4/UAS conditional system as with the following drivers: w1118; P{w[+mW.hs]=GawB}C253 (performed at 29°C), or w; eyeless-Gal4 (performed at 25°C). Scoring of the embryonic lethality was performed as described (Poortinga et al, 1998). Cuticle and wings were prepared using standard protocols. Immunofluorescence and immunostaining of embryos was performed as described (Orian et al, 2007). Statistical analysis was done using the z-test for two proportions.
Plasmids and primers
Plasmids used in yeast two-hybrid assays. Vectors for expression in yeast coding for Hairy and its mutants, dCtBP, and Gro were described in Poortinga et al, 1998.
Plasmid for bacterial expression and translation of proteins in vitro. pGEX Dgrn was generated by subcloning a PCR fragment derived from full-length dgrn ORF (BamHI–XhoI fragment) into pGEX-5X-1. pRSET-His-Dgrn (BamHI/KpnI) and its derivatives were cloned into pRSET-C. The SIM1–4 mutations correlate to deletions of 75—87 amino acids (aa), 181–184aa, 202–210aa and 239–242aa, respectively. The pRSET-Dgrn mutants C302A and I268A, and pSP65 Hairy various mutants and HairyΔBasic deletion were generated by site-directed mutagenesis. pGEX-Hairy derivatives (FL, N-Term, C-Term and Basic), pGEX-Gro and pGEX-CtBP, as well as pCITE-HairyΔPLSLV, HairyΔWRPW, pCite Hairyb−Sc and Hairyb−m8 have been previously described (Dawson et al, 1995; Poortinga et al, 1998; Phippen et al, 2000; Hasson et al, 2005). pCITE HairydMnt was generated by replacing Hairy's basic domain with that of dMnt. For in vitro translations, Dgrn-coding sequence was cloned into the pCite or pSP65 vectors (Novagen, Promega, respectively). GST, GST-SUMO, and GST-SUMO-GFP were a kind gift from A Courey.
Drosophila expression vectors. The cDNA clone containing the Dgrn-coding sequence was cloned into pUASp with KpnI–BamHI to generate UAS-Dgrn and its derivatives. UAS HA-Gro was previously described (Hasson et al, 2005). pPAC HA-dUbc9 and pPAC FLAG-dSUMO were a gift from A Courey (Bhaskar et al, 2000; Smith et al, 2004). UASp-His SUMO was generated by PCR from pPAC FLAG SUMO with the appropriate primers.
RNAi primers
Dgrn RNAi:
Forward: 5′-GAATTAATACGACTCACTATAGGGAGAGAGTCCAGTAGAAGTGATAGA-3′
Reverse: 5′-GAATTAATACGACTCACTATAGGGAGAAAGTAAATGCGAAAGAATTGAC-3′
SUMO RNAi:
Forward: 5′-CGGAATTCCGAATTAATACGACTCACTATAGGGATGTCTGACGAAAAGAAGGGAGGTG-3′
Reverse: 5′-CGGAATTCCGAATTAATACGACTCACTATAGGGTTATGGAGCGCCACCAGTCTGCTGC-3′
More information can be obtained upon request.
Protein expression, binding assays, and two-hybrid assay
Protein expression and in vitro binding experiments were performed using GST pull-down assays similar to that previously described (Orian et al, 2007). Yeast two-hybrid interaction assays were performed as described (Poortinga et al, 1998).
In vitro ubiquitylation assays
In vitro translations were carried out using the Promega in vitro TNT kit in the presence of 35S-Methionine. In vitro ubiquitylation assays with ubiquitin or its derivatives were as described (Ben-Saadon et al, 2006). For experiments involving Me-Ub, the labelled substrate was purified using a DE-52 chromatography column to remove the ubiquitin present in the IVT mixture (Orian et al, 1995).
Cell culture, transient transfections, and RNAi experiments in cells
Drosophila S2R cells were grown at 25°C and transfected using Fugene-HD® (Roche). dsRNA was prepared using the MegaScript® RNAi kit (Ambion). Proteins’ stability was determined 48 h after transfection or in dynamic experiments where 10 μM chx was added for the indicated time. Cells lysates were prepared as described (Orian et al, 2007), or where indicated using RIPA, 4% SDS, or guanidine-HCl lysis buffers. A total of 150 μg cell extract per lane was resolved via SDS–PAGE and proteins were identified using western blot analysis. 10 μg of cell extract was used to determine actin protein levels. Detection of SUMOylated Gro was done as detailed in the supplementary data and described in Herkert et al, 2010. Input material was adjusted to have equal amounts of unmodified Gro.
Luciferase assays
The ac reporter pT5 WT/luc, and the Da and Sc expression vectors have been previously described (Van Doren et al, 1994). At 48 h after transfection of S2R Drosophila cells, luciferase and renilla (control) activities were assayed using the Dual Reporter Assay (Promega).
Chromatin profiling (DamID)
A chromatin profiling experiment using Dam-Dgrn in Drosophila Kc167 cells was performed and analysed as described (Bianchi-Frias et al, 2004).
Antibodies used in these studies: mouse polyclonal anti-Dgrn (1:500, see Barry et al, 2011); α-Hairy and α-Ftz (1:50 and 1:500 from J Reinitz); α-Gro (1:100 or 1:2000 from C Delidakis); α-dSir2 (1:20, Rosenberg and Parkhurst, 2002); α-dCtBP (1:100, Phippen et al. 2000); α-Actin (1:2000, MP Biomedicals); α-FLAG M2 (1:1000, Sigma); α-SUMO-2 (1:100, Zymed); α-His (1:2000, Qiagen); and α-HA (1:2000 Covance).
Supplementary Material
Acknowledgments
We thank L Broday, D Gottschling, Z Paroush, H Rincon and members of the labs for discussions and comments on the manuscript. We are grateful to D Metzger for help at the early stages this research. We also thank H Saitoh, C Delidakis, J Reinitz, Z Paroush, R Sade, J-M Reichhart, A Courey and the Bloomington Stock Center for antibodies, DNAs, flies and other reagents used in this study. MA was supported by a Minerva grant. This work was supported by NIH Grant GM073021 (to SMP), ISF Grants (F.I.R.ST 1215/07 and 418/09) and GIF Grant 918/07 (to AO). AO is also supported by the Rappaport Research Fund.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn JW, Lee YA, Ahn JH, Choi CY (2009) Covalent conjugation of Groucho with SUMO-1 modulates its corepressor activity. Biochem Biophys Res Commun 379: 160–165 [DOI] [PubMed] [Google Scholar]
- Barry KC, Abed M, Kenyagin D, Werwie TR, Boico O, Orian A, Parkhurst SM (2011) The Drosophila STUbL protein Degringolade limits HES functions during embryogenesis. Development (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Valentine SA, Courey AJ (2000) A functional interaction between Dorsal and components of the Smt3 conjugation machinery. J Biol Chem 275: 4033–4040 [DOI] [PubMed] [Google Scholar]
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell 24: 701–711 [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow J, Vazquez J, Rosales-Nieves AE, Parkhurst SM (2004) Hairy-mediated transcriptional repression and cofactor recruitment in Drosophila. PloS Biol 2: 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Laughon A, Thalley BS (1988) Expression, function, and regulation of the Hairy segmentation protein in the Drosophila embryo. Genes Dev 7: 883–890 [DOI] [PubMed] [Google Scholar]
- Carter S, Bischof O, Dejean A, Vousden KH (2007) C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol 9: 428–435 [DOI] [PubMed] [Google Scholar]
- Cinnamon E, Paroush Z (2008) Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev 5: 435–440 [DOI] [PubMed] [Google Scholar]
- Dawson SR, Turner DL, Weintraub H, Parkhurst SM (1995) Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol 15: 6923–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Gessler M (2007) Delta-Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res 35: 4583–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Nayak S, Epstein JA, Buck CA (2000) RNF4, a RING protein expressed in the developing nervous and reproductive systems interacts with Gscl, a gene within the DiGeorge critical region. Dev Dyn 218: 102–111 [DOI] [PubMed] [Google Scholar]
- Geoffroy MC, Hay RT (2009) An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 8: 564–568 [DOI] [PubMed] [Google Scholar]
- Geoffroy MC, Jaffray EG, Walker KJ, Hay RT (2010) Arsenic induced, SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol Biol Cell 21: 4227–4239, 0.1091/mbc.E10-05-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541 [DOI] [PubMed] [Google Scholar]
- Hasson P, Egoz N, Winkler C, Volohonsky G, Jia S, Dinur T, Volk T, Courey AJ, Paroush Z (2005) EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat Genet 37: 101–105 [DOI] [PubMed] [Google Scholar]
- Herkert B, Dwertmann A, Herold S, Abed M, Naud JF, Finkernagel F, Harms GS, Orian A, Wanzel M, Eilers M (2010) The Arf tumor suppressor protein inhibits Miz1 to suppress cell adhesion and induce apoptosis. J Cell Biol 188: 905–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K, Ingham P (1986) Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell 44: 949–957 [DOI] [PubMed] [Google Scholar]
- Hunter T (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I (2008) Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Ish-Horowicz D (2008) The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol 9: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BH, Wainwright SM, Ish-Horowicz D (2008) Differential in vivo requirements for oligomerization during Groucho-mediated repression. EMBO Rep 9: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Ann Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy A, Calonge TM, Outwin EA, O′Connell MJ (2007) Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem 282: 20388–20394 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10: 547–555 [DOI] [PubMed] [Google Scholar]
- Lee MH, Lee SW, Lee EJ, Choi SJ, Chung SS, Lee JI, Cho JM, Seol JH, Baek SH, Kim KI, Chiba T, Tanaka K, Bang OS, Chung CH (2006) SUMO-specific protease SUSP4 positively regulates p53 by promoting Mdm2 self-ubiquitination. Nat Cell Biol 8: 1424–14231 [DOI] [PubMed] [Google Scholar]
- Lee W, Andrews BC, Faust M, Walldorf U, Verheyen EM (2009) Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev Biol 325: 263–272 [DOI] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keränen SV, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB (2009) Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez CA, Arnosti DN (2008) Spreading of a corepressor linked to action of long-range repressor hairy. Mol Cell Biol 28: 2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Arnaoutov A, Dasso M (2010) The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol 188: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela Em Hediger F, Gasser SM, Krogan NJ (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M (2001) Local action of long-range repressors in the Drosophila embryo. EMBO J 20: 2246–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M, Xie Y, Loo JA, Courey AJ (2009) Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One 4: e5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Delrow JJ, Rosales Nieves AE, Abed M, Metzger D, Paroush Z, Eisenman RN, Parkhurst SM (2007) A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc Natl Acad Sci USA 104: 15771–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Whiteside S, Israël A, Stancovski I, Schwartz AL, Ciechanover A (1995) Ubiquitin-mediated processing of NF-kappa B transcriptional activator precursor p105. Reconstitution of a cell-free system and identification of the ubiquitin-carrier protein, E2, and a novel ubiquitin-protein ligase, E3, involved in conjugation. J Biol Chem 270: 21707–21714 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with Hairy-related bHLH proteins. Cell 79: 805–815 [DOI] [PubMed] [Google Scholar]
- Phippen TM, Sweigart AL, Moniwa M, Krumm A, Davie JR, Parkhurst SM (2000) Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and Groucho transcriptional repression. J Biol Chem 275: 37628–37637 [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J 17: 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H, Aarnisalo P, Santti H, Janne OA, Palvimo JJ (2000) Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor-mediated transcription by different mechanisms. J Biol Chem 275: 571–579 [DOI] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA (2002) ‘Stemness’: transcriptional profiling of embryonic and adult stem cells. Science 298: 597–600 [DOI] [PubMed] [Google Scholar]
- Reiss Y, Hershko A (1990) Affinity purification of ubiquitin-protein ligase on immobilized protein substrates. Evidence for the existence of separate NH2-terminal binding sites on a single enzyme. J Biol Chem 265: 3685–3690 [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a co-activator/co-repressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428 [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Parkhurst SM (2002) Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109: 447–458 [DOI] [PubMed] [Google Scholar]
- Rouse J (2009) Control of genome stability by SLX protein complexes. Biochem Soc Trans 37: 495–510 [DOI] [PubMed] [Google Scholar]
- Roy M, Pear WS, Aster JC (2007) The multifaceted role of Notch in cancer. Curr Opin Genet Dev 17: 52–59 [DOI] [PubMed] [Google Scholar]
- Rushlow CA, Hogan A, Pinchin SM, Howe KM, Lardelli M, Ish-Horowicz D (1989) The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J 8: 3095–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Parkhurst SM (2004) Drosophila Topors is a RING finger-containing protein that functions as an ubiquitin-protein isopeptide ligase for the Hairy basic helix-loop-helix repressor protein. J Biol Chem 279: 17126–17233 [DOI] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS (2007) Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell 28: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB (1991) Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes Dev 5: 984–995 [DOI] [PubMed] [Google Scholar]
- Smith M, Bhaskar V, Fernandez J, Courey AJ (2004) Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J Biol Chem 279: 43805–43814 [DOI] [PubMed] [Google Scholar]
- Spradling AC (1986) P Element-Mediated Transformation. In Drosophila: A Practical Approach, Roberts DB (ed), pp 175–197. Oxford: IRL Press [Google Scholar]
- Sun H, Leverson JD, Hunter T (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Bailey AM, Esnayra J, Ede K, Posakony JW (1994) Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev 8: 2729–2742 [DOI] [PubMed] [Google Scholar]
- Wang Z, Prelich G (2009) Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol 7: 1694–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Rubenstein EM, Matt T, Hochstrasser M (2010) SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev 24: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.