Abstract
The nuclear receptor Farnesoid x receptor (FXR) is a critical regulator of multiple genes involved in bile acid homeostasis. The coactivators attracted to promoters of FXR target genes and epigenetic modifications that occur after ligand binding to FXR have not been completely defined, and it is unknown whether these processes are disrupted during cholestasis. Using a microarray, we identified decreased expression of mixed lineage leukemia 3 (MLL3), a histone H3 lysine 4 (H3K4) lysine methyl transferase at 1 and 3 days of post-common bile duct ligation (CBDL) in mice. Chromatin immunoprecipitation analysis (ChIP) analysis revealed that H3K4me3 of transporter promoters by MLL3 as part of activating signal cointegrator-2 -containing complex (ASCOM) is essential for activation of bile salt export pump (BSEP), multidrug resistance associated protein 2 (MRP2), and sodium taurocholate cotransporting polypeptide (NTCP) genes by FXR and glucocorticoid receptor (GR). Knockdown of nuclear receptor coactivator 6 (NCOA6) or MLL3/MLL4 mRNAs by small interfering RNA treatment led to a decrease in BSEP and NTCP mRNA levels in hepatoma cells. Human BSEP promoter transactivation by FXR/RXR was enhanced in a dose-dependent fashion by NCOA6 cDNA coexpression and decreased by AdsiNCOA6 infection in HepG2 cells. GST-pull down assays showed that domain 3 and 5 of NCOA6 (LXXLL motifs) interacted with FXR and that the interaction with domain 5 was enhanced by chenodeoxycholic acid. In vivo ChIP assays in HepG2 cells revealed ligand-dependent recruitment of ASCOM complex to FXR element in BSEP and GR element in NTCP promoters, respectively. ChIP analysis demonstrated significantly diminished recruitment of ASCOM complex components and H3K4me3 to Bsep and Mrp2 promoter FXR elements in mouse livers after CBDL. Taken together, these data show that the “H3K4me3” epigenetic mark is essential to activation of BSEP, NTCP, and MRP2 genes by nuclear receptors and is downregulated in cholestasis.
Keywords: hepatoma cells, microarray, epigenetics, gene regulation, activating signal cointegrator-2-containing complex, H3 lysine 4, nuclear receptors, mixed lineage leukemia 3
ligand-activated nuclear receptors (NRs), such as Farnesoid x receptor (FXR) and glucocorticoid receptor (GR), initiate transcription by binding to specific DNA regulatory elements in promoters of target genes and recruiting coactivator proteins to modify chromatin structure and induce assembly of the transcription complex containing RNA polymerase II (PolII) (2, 3, 6, 18). The primary coactivators including the p160 coactivators such as TIF2/GRIP-1/SRC-2 and related members of the p300 family bind directly to ligand-activated NRs and recruit an array of secondary coactivators such as p300/cAMP response element binding protein (CREB)-binding protein (CBP) and coactivator-associated arginine methyltransferase (CARM1), which acetylate and methylate (respectively) histones and other proteins in the transcription complex (16, 33). Secondary coactivators act synergistically in a requisite sequence to enhance transcriptional activation by binding to the p160 coactivators or other primary coactivators (25, 34).
We have been interested in how the expression of NRs and various coactivators are altered in cholestasis leading to downregulation of major bile acid transporters, bile salt export pump (BSEP), multidrug resistance associated protein 2 (MRP2), and sodium taurocholate cotransporting polypeptide (NTCP) (7, 8, 24, 26). We have found that several critical components of a steady-state complex called activating signal cointegrator-2 (ASC-2)-containing complex (ASCOM) are altered in mouse after bile duct ligation. This complex contains the transcriptional coactivator nuclear receptor coactivator 6 (NCOA6) and the histone H3 lysine 4 (H3K4) methyltransferase mixed lineage leukemia 3 (MLL3) and its paralog MLL4. Our studies in the normal mice and human liver cell lines show that these components interact directly with the bile acid receptor FXR and with GR and are involved in expression of the their target genes, BSEP, MRP2, and NTCP. Moreover, in the common bile duct ligated mouse impaired expression of critical coactivators and disruption of histone modifications in cholestasis can adversely affect FXR/GR transactivation, even if NR expression is not altered.
MATERIALS AND METHODS
Cells and cell culture.
HepG2 cells were cultured in MEM with 10% FBS and antibiotics. Huh-7 cells were cultured in RPMI with FBS and antibiotics. HEK293 cells were grown in DMEM with the same supplements. All cells were grown in 10% CO2 in a humidified incubator maintained at 37°C. When cells were treated with FXR/RXR ligands, chenodeoxycholic acid (CDCA), and 9-cis retinoic acid, respectively, the medium was changed to DMEM without phenol red and contained charcoal-adsorbed FBS.
Chemicals.
All chemicals were obtained from Sigma unless stated otherwise. Small interfering (siRNAs) for NCOA6, MLL3, and MLL4 were obtained from Dharmacon or Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to NCOA6 (400A-1) were from Bethyl Labs, while MLL3 and MLL4 (ALR) antibodies were from Santa Cruz Biotechnology. Affinity-purified antibodies to MLL4 were obtained from Dr. Kai Ge, Nuclear Receptor Laboratory (National Institute of Diabetes and Digestive and Kidney Diseases).
Common bile duct ligation.
Common bile duct ligation (CBDL) in C57Bl/6 mice were performed as described earlier using a protocol (8) approved by the Institutional Animal Care and Use Committee of Mount Sinai School of Medicine. Briefly, laparatomy was performed on the mice following which bile duct was ligated proximally and distally and severed in the middle. Serum bile acids were estimated by a kit from Trinity Biotech to ensure that successful cholestasis was achieved (see Supplemental Data; Supplemental Material for this article is available online at the Am J Physiol Gastrointest Liver Physiol website). Sham surgery was performed on control mice in which laparatomy and manipulation of the liver was performed, but bile duct was not ligated. Livers from sham-operated and bile duct-ligated mice were collected at 1, 3, and 7 days postligation. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86–23 revised 1985).
Plasmid constructs.
Human BSEP promoter-luciferase plasmid was generated as described by us previously (1). Plasmids encoding FXR and RXR were generously supplied by Dr. David Mangelsdorf (Dallas, TX) as described earlier (1). Preparation of full-length and deletion constructs representing various domains of NCOA6 has been recently described (27).
siRNA-mediated knockdown of NCOA6, MLL3, and MLL4.
siRNAs against NCOA6, MLL3, and MLL4 were obtained from Dharmacon (siGenome pool) or Santa Cruz Biotechnology (for sequences of siRNAs used, see Supplemental Table S3). For knockdown experiments, HepG2 or Huh-7 cells were plated in sixwell plates (1 × 106 cells/well) and incubated 2 days later with 50 nM siRNA using TransIT-TKO (MirusBio) at a ratio of 1:1 (μl/μl) according to manufacturer's instructions. Six hours later, medium was added to the wells, and 24 h later, spent medium was replaced with fresh DMEM. Forty-eight hours later total RNA was prepared using Trizol kit (Invitrogen, Carlsbad, CA), and real-time PCR analysis was conducted following conversion of mRNA into cDNA. Cells were harvested from the wells and lysates were prepared by resuspending in mammalian protein extraction buffer (Sigma, St. Louis, MO) after 72 h following siRNA transfection as described in Western blotting analysis.
Transient transfections and luciferase assays.
HepG2 or Huh-7 cells were plated at a concentration of 1 × 105 cells/well in 24-well plates 2 days earlier. They were transfected at day 0 with mouse or human BSEP promoter at 0.5 μg/well (in triplicate/per group) and also cotransfected with 50 ng FXR/RXR and various amouts of NCOA6 expression plasmids in OPTI-MEM (Invitrogen) where indicated. Transfections were carried out using TransIT-LT (Mirus Bio) at DNA:TransIT ratio of 1:3. On day 1, the medium was changed to DMEM without Phenol Red and with charcoal-adsorbed FBS. Ligands 9-cis retinoic (1 μM) acid and CDCA (100 μM) for RXR and FXR, respectively, were added at this time and luciferase activities were measured 24 h later using Promega kit (Promega, Madison, WI). When siRNA adenoviruses against NCOA6 were used to study their effect on promoter activity, they were infected 6 h after transfection with promoter and FXR/RXR/NCOA6 plasmids as follows: 5 × 109 viral particles/well were (multiplicity of infection of 1:1,000) added to the wells in 0.15 ml of OPTI-MEM after removal of the plasmid mixture. After incubation for 1.5 h in the incubator, 0.4 ml of serum containing DMEM were added and returned to the incubator overnight. The next day, phenol red-free DMEM and ligands were added, and luciferase activity assayed as described above. Normalization of transfection efficiencies in the different wells was achieved by cotransfection with pCMVβ galactosidase and assay of galactosidase activity. All transfection experiments were repeated twice with similar results. Representative experiments are shown in Results due to inter-experiment variations in transfection.
Glutathione S-transferase pull-down assay.
Partial recombinant proteins of NCOA6 were overexpressed as glutathione S-transferase (GST) fusion proteins in E.coli BL21 as previously described (27). Equal amounts of GST and GST fusion proteins were used for GST pull-down assays by incubating 5 μl of 35S-methionine-labeled full-length FXR obtained by in vitro translation using TNT T7 coupled transcription and translation system in a 500 μl of NETN buffer containing 1 mg/ml fatty acid free BSA in the presence and absence of CDCA (100 μM). The bound proteins were washed with three times with NETN buffer and eluted into SDS-PAGE sample buffer by heat denaturation. The proteins were separated on 4–20% gradient gels, and after the signal was fixed and amplified, the gels were dried and autoradiographed.
Mammalian two-hybrid analysis.
The mammalian two-hybrid assay was performed by modified Checkmate mammalian two-hybrid system (Promega) following manufacturer's instructions. HepG2 cells were cotransfected with the pBIND/FXR (containing ligand binding domain), pACT/NCOA6 (coding for 6 different domains), and pSG9/luc plasmids employing Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommended protocol. Briefly, cells were seeded in 24-well plates at a density of 1 × 105 cells/well and cultured to about ∼90% confluency. Plasmid constructs were combined at a ratio of 2:1:4 (promoter:NR:coactivator) to a total of 0.8 μg/well, and cells were transfected and cultured for 24 h followed by the addition of ligand CDCA (100 μM) for another 24 h. Finally, luciferase activity was determined with Lumistar Optima luminometer (LMG Biotech) with a commercially available kit (Promega). The transfection efficiency was normalized using Renilla luciferase activity, which was simultaneously expressed from the pBIND plasmids. Further details are available from the authors on request.
Chromatin immunoprecipitation analysis of cultured cell lines and mouse liver.
Chromatin immunoprecipitation analysis (ChIP) assays were conducted essentially by a combination of previously described protocols (12) and manufacturer's instructions using EZ-ChIP/MagnaChIP G kit from Upstate Biotechnology/Millipore (Millipore). Briefly, cells from 3 × 100 mm culture dishes were harvested after being fixed with 1% formaldehyde. Following lysis, genomic DNA were sonicated in Diagenode Bioruptor Sonicator for 8 × 30 s (twice, with 30-s on/30-s off cycles) resulting in DNA fragments of 200–1,000 bp. In the case of mouse liver, a suspension of nuclei prepared in 0.8 ml in an Eppendorf tube was sonicated for 8 × 10 s placing the tube in ice-cold water using a Misonix Xl Series sonicator (QSonica, Newtown, CT). The fragmented DNA was diluted in ChIP dilution buffer and preadsorbed with ProteinG-sepharose/salmon sperm DNA for 1 h at 4°C. Ten percent of the chromatin was removed and saved as “Input.” It was then incubated overnight at 4°C with 3–5 μg of the appropriate antibodies or normal mouse IgG (control). Antibody-chromatin complexes were captured by incubation with protein G-Sepharose and centrifuged. Protein G-Sepharose beads were washed with low salt, high salt, lithium chloride and finally with Tris-EDTA buffer. DNA from the beads was then eluted. Reversal of protein cross-linking and proteinase K digestion followed by purification of the DNA was then achieved. An aliquot of the DNA (5 μl) was used in a PCR reaction using specific primers flanking the FXR element (FXRE) site of human and mouse BSEP/Bsep, mouse Mrp2, and GCRE in the human NTCP. Primers flanking a site distant from the FXRE site were used as negative controls. Positive control for the ChIP consisted of immunoprecipitation with RNA PolII antibody. Quantitation of the DNA precipitated was done using quantitative (q)PCR and expressed as percent relative to the IgG control. ChIP primers used in the studies are provided in Supplemental Table S2.
Methodological details relating to 1) preparation and analysis of customized epigenetic array, 2) real-time PCR analysis, and 3) Western blot analysis can be found at the end of the Supplement Materials.
Statistical analysis.
All data are shown as means ± SD (3 animals/experiments per group). In all the experiments, comparison of data between individual groups was performed using unpaired Student's t-test. For the sake of clarity, all significant P values in Figs. 1–6 are indicated as P < 0.05, although in some cases they were even lower.
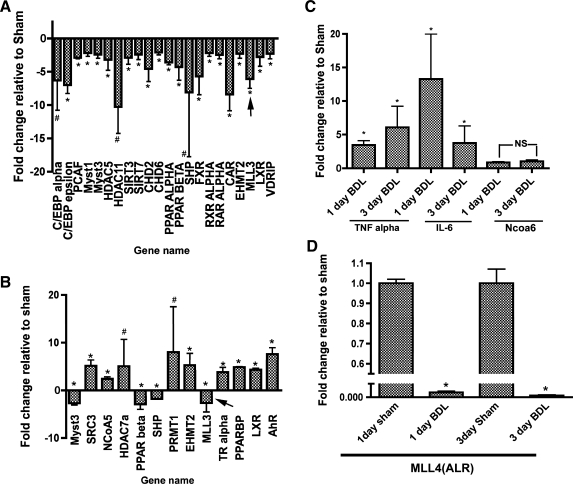
Fig. 1.
Bile duct ligation (BDL) at 1 and 3 days postsurgery leads to alterations in expression of multiple genes encoding proteins involved in epigenetic modification. Arrow indicates changes in mixed lineage leukemia 3 (MLL3) [a histone H3 lysine 4 (H3K4) methylase] expression. C and D: quantitative (q)PCR analysis of relative expression of nuclear receptor coactivator 6 (NCOA6; C) and MLL4 (D), respectively, during BDL was analyzed separately. qPCR of TNFα and IL-6 mRNA was used as positive controls in C. For the sake of clarity, the values for 1- and 3-day sham samples are not plotted and y-axis shows values relative to the sham controls (arbitrarily set as 1.0) for TNFα, IL-1β, and NCOA6 mRNAs *P value <0.05. #P value not significant.
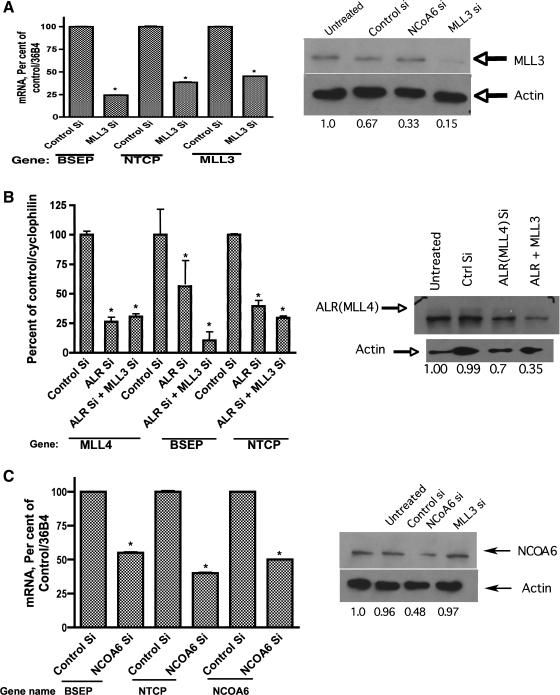
Fig. 2.
RNAi-mediated silencing of MLL3 (A), MLL4 (B), and NCOA6 (C) in HepG2 cells results in significant downregulation of bile salt export pump (BSEP) and sodium taurocholate cotransporting polypeptide (NTCP) expression. Genome siRNA pools (Dharmacon and Santa Cruz Biotechnology) against MLL3, MLL4 (ALR), and NCOA6 were obtained and transfected into HepG2 cells as described. mRNA levels for BSEP and NTCP were quantified using qPCR in addition to those for MLL3, MLL4, and NCOA6. Protein levels of cognate genes were also analyzed using specific antibodies. A representative Western blot and relative protein amounts are shown at right. *P value <0.05, compared with control small interfering (si)RNA-treatment.
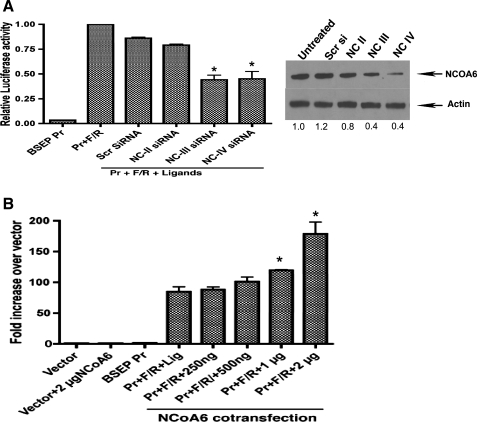
Fig. 3.
Infection of HepG2 cells with siRNA adenoviruses against NCOA6 (A) leads to a decrease in FXR-mediated transactivation of BSEP promoter while NCOA6 overexpression (B) using a plasmid encoded cDNA results in a dose-dependent increase in promoter activity. A: GFP-containing adenoviruses encoding siRNAs to fragments of NCOA6 along with a scrambled control siRNA were prepared and infection of HepG2 cells were performed as described by us earlier (10). Infection was monitored by GFP fluorescence that was 70–80%. Farnesoid x receptor (FXR) transactivation of BSEP promoter was assayed by luciferase activity. NC III and NC IV siRNA significantly (P < 0.05, compared scrambled siRNA adenovirus treatment) downregulated promoter activity while NC II did not. A representative Western blot of the protein levels is shown. B: overexpression of NCOA6 was achieved by transfection with increasing amounts of full-length cDNA plasmid and FXR transactivation of BSEP promoter was monitored by luciferase activity. Significant (P < 0.05, compared with activity in cells without NCOA6 plasmid cotransfection) stimulation of promoter activity was seen with 1 and 2 μg of NCOA6 cDNA.
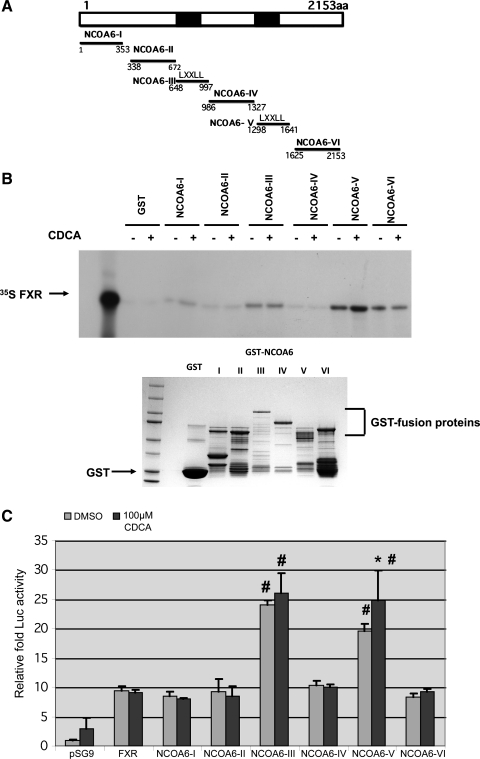
Fig. 4.
FXR-LBD interacts with NCOA6 nuclear receptor (NR) boxes 1 and 2 (containing LXXLL motifs) in vitro with the interaction to NR BOX 2 being enhanced by chenodeoxycholic acid (CDCA; B) and mammalian 2-hybrid analysis further support the binding of FXR to NCOA6 NR boxes 1 and 2 in vivo (C). A: schematic of the various fragments of NCOA6 used in B and C is shown. B: GST pull-down assays using various fragments of NCOA6 (schematic in C) and 35S-FXR was carried out as described. A representative autoradiogram of a short and long (Supplemental Fig. S3) exposure of the X-ray film is shown. B, bottom: Coommassie blue-stained gel. C: mammalian e-hybrid analysis between pBIND-FXR-LBD and pACT-domains I-VI of NCOA6 in the absence/presence of 100 μM CDCA and the ensuing fold increase in reporter gene activity over empty vector are shown. Note the increased reporter gene activity in the absence of ligand between domain III and V of NCOA6 and FXR-LBD (#P < 0.05, compared with domains I, II, IV, and VI). Interaction of FXR-LBD with domain V was increased significantly (*P < 0.05, compared with domain V without ligand) by addition of ligand.
Fig. 5.
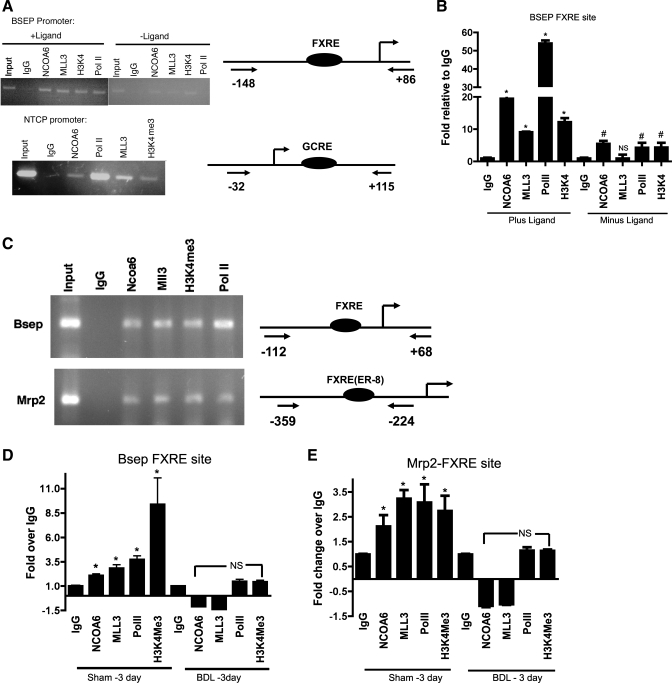
Chromatin immunoprecipitation (ChIP) analysis of hepatoma cells (HepG2) and mouse liver show recruitment of NCOA6, MLL3, and H3K4 trimethylation of histone H3 at the BSEP/Bsep and Mrp2 FXRE locus and NTCP-GCRE locus. A: representative gel showing ChIP analysis of HepG2 cells before and after the addition of ligand using antibodies to NCOA6, MLL3, H3K4Me3, and RNA polymerase II (PolII). Minimal recruitment of the proteins to the FXRE site was observed in the absence of ligand. Primers flanking the FXRE site in BSEP/Bsep, Mrp2 promoters, and GCRE site in NTCP promoter was used for PCR (see Supplemental Table S2). Schematic representation of the primers used for ChIP and the location of the sites are shown on the right. B: qPCR analysis and quantitation of ChIP signal at the FXRE locus +/− ligand. Values are plotted as fold relative to the signal obtained with IgG. *P < 0.05, compared with IgG in the presence of ligand. #P < 0.05, compared with IgG in the absence of ligand. NS, not significant. C: representative gel of ChIP analysis of the FXRE locus at the Bsep/Mrp2 promoters of sham mouse liver using antibodies to NCOA6, MLL3, and RNA PolII. Schematic of the location of the primers used for PCR relative to the FXRE locus is shown on the right. D: qPCR analysis of recruitment of the ASCOM complex proteins to the mouse liver Bsep/Mrp2 FXRE loci in 3-day sham-operated and CBDL livers plotted as fold relative to IgG. *P < 0.05, compared with IgG.
Fig. 6.
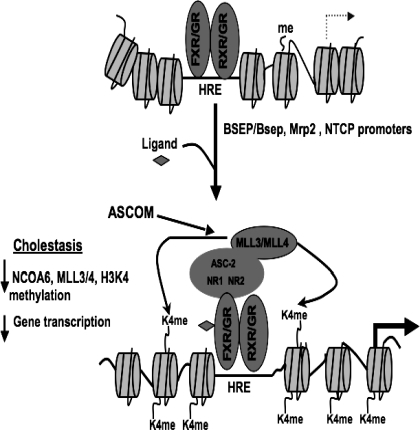
Schema of the current working model for the role of ASCOM complex in FXR/GR-transactivation of BSEP/Bsep, Mrp2, and NTCP promoters. Ligand binding to FXRE/GCRE at the cognate promoter locus leads to recruitment of ASCOM complex to the response element because of the interaction between FXR/GR and NCOA6. MLL3/MLL4, which form part of the ASCOM complex then methylates histone H3K4 to the trimethylated state, which is an activation mark. This modification of histones results in transcriptional activation of BSEP/Bsep, Mrp2, and NTCP. Conversely, downregulation of MLL3/MLL4 during cholestatic disease could lead to decreased recruitment of ASCOM complex resulting in downregulation of corresponding gene transcription of ASCOM components (such as MLL3/MLL4) resulting in decreased transporter mRNAs.
RESULTS
Expression of multiple genes encoding epigenetic modifiers, coactivators, and corepressors is altered in mouse liver during cholestasis.
As a first step towards understanding the role of epigenetic modifications in the regulation of bile acid transporter genes, a custom-made array platform using RT2 Profiler PCR Array System (SuperArray Biosciences Corp) was used to examine the expression of 84 genes encoding coactivators and histone modifying enzymes (see template with gene names in Supplemental Fig. S2). cDNA from mouse livers after 1 and 3 days post-CBDL was analyzed and compared with sham-operated controls. The results of the analysis plotted as fold change compared with sham (>2-fold) are shown in Fig. 1, A and B, for 1 and 3 days post-CBDL, respectively. There were a number of genes whose expression was downregulated significantly (P < 0.05) at 1 day ranging from >2-fold up to 11-fold. On day 3, there were many genes whose expression was upregulated that included several nuclear receptors and histone modifying enzymes, suggesting that this response might represent an adaptive mechanism by the liver. We specifically noted that MLL3, a histone H3K4 lysine methyl transferase, was downregulated at both the time points (−6.1 ± 1.4 at 1 day and −2.6 ± 1.9 at 3 days). Expression levels of these genes partially returned to control levels at 7 days after CBDL consistent with partial normalization of transporter message levels. Recent studies (14) have shown that during LXR activation of target genes, a transcription initiation complex termed ASCOM, is recruited to the promoter that contains the coactivator NCOA6 (ASC-2, PRIP) and the histone H3K4 methyl transferases MLL3 or MLL4. Catalytic activity of MLL3/MLL4 results in trimethylation of H3K4, which is an “activation mark.” Since our array did not contain NCOA6 and MLL4, their expression by qPCR analysis was assessed separately and is shown in Fig. 1, C and D, respectively. While there was no significant change in expression of NCOA6 mRNA level, there was a marked decrease in MLL4 expression at both time points. qPCR analysis of TNFα and IL-6 mRNA levels was carried out as positive controls for inflammation; both were elevated by CBDL (Fig. 1D) as reported previously (4). Since bile acid transporters (including Ntcp, mrp2, and BSEP/bsep) are downregulated early in rodent and human cholestasis (7, 8, 24, 26), these data together pointed to the potential role of MLL3/MLL4 as part of the ASCOM complex in FXR (BSEP/bsep and mrp2) and GR (NTCP) activation in liver.
siRNA-mediated silencing of MLL3, MLL4, and NCOA6 results in downregulation of BSEP and NTCP expression in hepatoma cells.
To further assess how MLL3/MLL4 and NCOA6 might regulate bile acid transporter expression, we systematically analyzed the effect of siRNA-mediated silencing of MLL3/MLL4 (individually and together) and NCOA6 (Fig. 2, A, B, and C, respectively) on BSEP and NTCP expression in HepG2 cells. siGenome pools (pools of 4 different siRNAs) representing NCOA6 and MLL3/4 were transfected into HepG2 cells. As shown in Fig. 2A, compared with control siRNA treatment, specific MLL3 siRNA reduced its own message to 45.2% of control while expression of BSEP and NTCP were reduced significantly (P < 0.05) to 24.4 and 40% of control, respectively. Although control siRNA also modestly decreased MLL3 protein, specific MLL3 siRNA reduced MLL3 protein to 16% of control. The reason for the downregulation of MLL3 protein expression by NCOA6 (Fig. 2A, right) is currently unknown. Figure 2B shows the results after treatment with MLL4 siRNA. In this experiment, an additional set of experiments were included in which both MLL3 and MLL4 (also known as ALR) siRNAs were silenced simultaneously to see if MLL3/4 silencing resulted in additive effects on expression of transporter mRNAs. There is considerable confusion in the gene nomenclature for human MLL4, and hence we refer to it as MLL4 (ALR). MLL4 and MLL4 + MLL3 siRNA treatment resulted in a decrease of MLL4 message levels to 26.3 and 30.6% of control, respectively. Under the same conditions, BSEP mRNA levels were reduced to 56.5 and 10.5% of control while NTCP levels were reduced to 39.5 and 29.5% of control, respectively. Studies by Lee et al. (14) have suggested that ASCOM complexes may contain either MLL3 or paralog MLL4, which is consistent with the fact that we observed additive effects (synergism) on bile acid transporter expression levels (with the reduction in BSEP levels being much more dramatic) with combined silencing of MLL3/MLL4. MLL4 protein levels were significantly reduced when MLL3 and MLL4 siRNAs were transfected together, and the rationale for this effect on MLL4 protein by combined treatment is not clear. Lastly, we probed the effects of NCOA6 siRNA treatment in HepG2 cells on BSEP and NTCP message levels (Fig. 2C). Silencing with NCOA6 siRNA led to a reduction of 50 ± 0.2% in cognate mRNA and 49% in its protein level compared with control. BSEP and NTCP message levels were significantly (P < 0.05) reduced to 55 ± 0.5 and 40 ± 0.6% of controls, respectively. The specificity of the siRNA effects was demonstrated from the fact that NCOA6 and MLL3 proteins levels were unaffected when siRNAs against each other had no effect on the converse protein (Fig. 2, A and C, proteins). Together these data support our observations using the array and implicated MLL3/MLL4 and NCOA6 in regulation of BSEP and NTCP expression.
NCOA6 siRNA adenovirus infection inhibits FXR activation of the BSEP promoter while overexpression of NCOA6 enhances its transactivation in HepG2 cells.
In the experiments described below, we wanted to further verify the participation of ASCOM complex in BSEP transcription by silencing via an alternate approach using adenovirus-mediated NCOA6 siRNA delivery. Therefore, we employed siRNA adenoviruses against NCOA6 generated as described earlier (27) and examined their effect on FXR-activation of BSEP promoter. NCOA6- siRNA adenoviruses (scrambled siRNA and NCOA6-specific siRNAs NC II, NC III, and NC IV corresponding to various portions of the NCOA6 mRNA sequence) were infected into HepG2 cells followed by transfection with BSEP promoter and cotransfection with FXR/RXR. Figure 3A shows luciferase activity assayed 24 h postligand addition. Infection with scrambled small interfering adenovirus and NC II did not have a significant impact on BSEP promoter activity (0.86 ± 0.01 and 0.79 ± 0.01 relative to the activity in the absence of siRNA set at 1.00). However, infection of the small interfering adenoviruses NC III and NC IV reduced the promoter activity significantly to 44–45% of that in their absence (0.44 ± 0.046 for NC III and 0.45 ± 0.073 for NC IV; P < 0.05). Analysis of protein levels show that NCOA6 protein levels were downregulated (to 67 and 41% of untreated, respectively) by NC III and NC IV. When these experiments were repeated in the absence of ligand, we did not see any change in promoter activity by adenovirus infection (data not shown), suggesting that ligand stimulation of FXR is a necessary event. These data are consistent with our earlier data wherein NC III was effective in inhibiting stimulation of CYP2C9 promoter activity stimulated by CAR and HNF4 (27). Thus these results further support the idea that NCOA6 (and a complex consisting of NCOA6/ASC-2) is a critical determinant of BSEP transcriptional activation by FXR.
Since siRNAs directed against NCOA6 inhibited promoter activity, we wanted to test, conversely, whether overexpression of NCOA6 using plasmid cotransfection will activate the promoter activity dose dependently. We transfected HepG2 cells with BSEP promoter and cotransfected with FXR/RXR in the presence of increasing amounts of NCOA6 full-length cDNA-encoding plasmid (Fig. 3B). While there was no noticeable increase using 250 ng of NCOA6 plasmid DNA compared with its absence, the promoter activity was enhanced to 120 ± 8.4 , 140 ± 1.2, and 198 ± 6.5% of control by 500 ng, 1 μg, and 2 μg of plasmid cotransfection (P < 0.05 compared with controls for 1 and 2 μg of plasmid DNAs). Taken together, these results substantiate our hypothesis that ASCOM complex is a major participant in FXR-mediated activation of BSEP promoter.
FXR interacts with NCOA6 NR boxes (LXXLL motif) 1 and 2 in GST pull-down assays and by mammalian two-hybrid analysis.
We next reasoned that if FXR/GR recruited ASCOM complex to BSEP/bsep, mrp2, and NTCP promoters, it is likely that there was direct protein-protein interaction between these proteins. To assess such an interaction, we carried out GST pull-down assays and mammalian two-hybrid analysis employing fragments of NCOA6 and FXR and its LBD and the results are shown in Fig. 4, B and C. Figure 4B shows the results of a GST pull-down assay where 35S-FXR was incubated in vitro with fragments 1–6 (NCOA6 I to NCOA6 VI, schematically shown in Fig. 4A) of NCOA6 linked to GST. Shorter exposure of the autoradiograms shows that FXR interacts with NCOA6 III, V, and VI. The fragment V interaction was further enhanced by the addition of CDCA. Of note is the fact that both fragments III and V contain “LXXLL” motifs and hence are named NR boxes 1 and 2. Previous work (28, 32) using a variety of coactivators have shown that NRs interact with coactivators via this motif and mutation of this motif abolishes the binding. Longer exposure of the autoradiogram is shown in Supplemental Fig. S3. Figure 4B, bottom, shows a Coomassie blue-stained gel of the proteins for the various fragments of NCOA6 to indicate that all the corresponding proteins were expressed as GST-fusion proteins. Lanes 1 and 3 represent molecular weight markers and the empty GST, respectively.
To obtain additional evidence for FXR binding to NCOA6, we performed two-hybrid analysis in a mammalian cell line and the results are depicted in Fig. 4C. The slight stimulation of reporter gene activity by pSG9 vector and by FXR-LBD in the presence of CDCA might represent background activity of the system. The subsequent bars show relative luciferase activity (relative to vector alone) of FXR-LBD alone and NCOA6 I-VI (in pACT vector) when cotransfected with pBINDFXR-LBD. Similar to the pull-down data, fragments NCOA6 III and V showed significantly (P < 0.05) enhanced interaction in the absence and presence of CDCA (for NCOA6 V only) that was significant compared with FXR alone and NCOA6 fragments I, II, IV, and VI. Constructs NCOA6 I, II, IV, and VI showed no difference in their stimulation of reporter activity compared with FXR-LBD alone (with and without ligand) and ranged from 8.5-to 10.3-fold compared with vector. Taken in conjunction with the GST pull-down analysis, these results confirm that FXR does indeed interact with NCOA6 by binding to both NR boxes 1 and 2 presumably leading to recruitment of “ASCOM” complex to the BSEP promoter locus.
ChIP analysis of HepG2 cells and mouse liver reveal recruitment of MLL3, NCOA6, and H3K4 trimethylation at the BSEP/mrp2/NTCP promoter loci with no recruitment in 3-day CBDL mouse livers.
Having demonstrated that MLL3/MLL4 and NCOA6 are involved in BSEP transcriptional activation, we performed ChIP assays to show recruitment of MLL3 and NCOA6 and histone H3K4 trimethylation to BSEP/Bsep (human and mouse) and Mrp2 (in mouse) FXRE locus in hepatoma cells and in mouse liver. In addition, we also examined the recruitment of ASCOM complex to the GCRE locus in human NTCP promoter since previous studies (6, 15) have demonstrated such a recruitment to GR-target genes. Results of the ChIP data are shown in Fig. 5, A and B, for cells and Fig. 5, C, D, and E for liver tissue. Figure 5A shows a representative gel of a PCR reaction from a ChIP experiment highlighting recruitment of MLL3, NCOA6, and H3K4 trimethylation to BSEP FXRE in the presence and absence of CDCA in HepG2 cells. As would be expected, significantly diminished recruitment of the proteins were observed in the absence of ligand. Since there is endogenous synthesis of bile acids in HepG2 cells, it would be predicted that there would be some basal recruitment of these mediators to the BSEP locus even in the absence of added ligand. We also observed similar recruitment of ASCOM components to OSTα promoter FXRE, suggesting this could be a general mechanism of activation for other FXR target genes (see Supplemental Fig. S4). We were also able to demonstrate recruitment of ASCOM components to the GCRE locus in human NTCP promoter as seen in Fig. 5A, bottom, consistent with previous reports in the literature that GR target genes also recruit ASCOM complex (15). Figure 5B shows qPCR results of the ChIP assay of the BSEP FXRE locus showing a 19.5-, 9.5-, 12.3-, and 54.5-fold increase (P < 0.05, compared with the absence of ligand) in the signal relative to IgG control when antibodies to NCOA6, MLL3, H3K4, and RNA PolII (as a positive control) were used for ChIP. Fold increases (relative to IgG) of 5.6, 1.05, 4.5, and 4.4 for NCOA6, MLL3, H3K4, and PolII were obtained in the absence of ligand (P < 0.05, compared with IgG except for MLL3). ChIP assay for MLL4 could not be carried out, as the commercially available antibodies were found unsuitable for the assay.
Although these studies used hepatoma cells, we wanted to verify that the recruitment of these coactivators also occurred in vivo in mouse liver and whether changes in their recruitment could explain downregulation of Bsep and Mrp2 during cholestasis. The data in Fig 5, C–E show a representative gel and qPCR quantitation of the mouse liver ChIP assay. The data in Fig. 5C reveal that NCOA6, MLL3, and PolII are recruited to Bsep and Mrp2 FXRE locus in the mouse promoter followed by H3K4 methylation. qPCR evaluation of the precipitates obtained with the specific antibodies in sham and 3-day CBDL mouse liver are shown in Fig. 5, D and E, for Bsep and Mrp2, respectively. We chose the 3-day CBDL because previous studies (including our own: Ananthanarayanan M, Li Y, Suchy, FJ, unpublished observations) in rats and mice have shown downregulation of Bsep [at 5 days in mice by Park et al. (24)] and Mrp2 at 1–3 days post-CBDL (7, 8, 24). BSEP message has also shown to be downregulated in humans during cholestasis (26). Significantly enhanced recruitment (P < 0.05 relative to IgG) to the BSEP FXRE locus of 2.1-, 2.9-, 3.7,- and 9.4-fold over IgG for NCOA6, MLL3, PolII, and H3K4 methylation, respectively, was seen in sham-operated livers while no or “negative” enrichment was seen in the CBDL livers. For the Mrp2 FXRE locus, the enrichments were 2.1-, 3.2-, 3.2-, and 2.7-fold over IgG (P < 0.05 vs IgG) for NCOA6, MLL3, PolII, and H3K4 methylation, respectively, in the sham livers while no enrichment was observed in the livers of 3-day CBDL mice. As the FXRE site is not conserved in the human MRP2 promoter, these studies could not be carried out in HepG2 cells.
DISCUSSION
Ligand-induced activation of nuclear receptors for bile acids and other biliary constituents plays an essential role in regulation of genes involved in bile formation (22). This complicated process involves the cyclical assembly and disassembly of transcriptional complexes on DNA promoter elements (5). These multisubunit complexes contain a great variety and number of coregulator proteins and histone modifying enzymes that are recruited by NRs and modulate target gene activity (19). It is also clear that coactivators are differentially used by transcription factors, including NRs, in a cell-, tissue-, and promoter-specific manner (20, 21). Multiple complexes of coactivator proteins participate in local remodeling of chromatin and in recruitment and activation of RNA PolII. As the basic mechanisms and diversity of coregulators underlying ligand-dependent transcriptional activation by nuclear receptors has become better defined, it was natural to propose that their expression and function might be altered in disease. This has proven to be the case in cancer and in some genetic disorders (17, 23). However, although the expression of some nuclear receptors has been studied in humans and experimental animals with various forms of cholestasis (11, 31), there is little information on how assembly of coregulators and histone modifications are involved in perpetuating and adapting to cholestasis (9, 29, 30). Several mechanisms are likely to be operative: 1) the production of coactivators may be impaired or induced in cholestasis, 2) their recruitment to the nucleosome may be disrupted by alterations in chromatin structure, and 3) there may be active recruitment of corepressors and histone deacetylases to the promoters of these genes.
In an initial effort to understand how the expression of coregulators that regulate transporters involved in bile formation may be altered in liver disease, a custom RT2PCR Profiler array was designed to obtain an unbiased view of changes during the cholestatic process induced by common bile duct ligation. The expression profile was composed of a panel of 84 genes that include many of the known liver coactivators, corepressors, and histone modifying enzymes (see gene list in Supplement Material). We realized that although many genes will be altered during cholestasis, most may have no role in regulation of liver transporters. However, these studies could also lead to the discovery of novel coactivators whose role in regulating FXR-dependent genes had not been established. An important limitation of this approach is that it only examines coregulator gene expression that changes in cholestasis. Alternatively, recruitment of a coactivator and its translocation into the nucleus may be impaired and/or there could be formation of a repressor complex without a change in expression of the individual components.
The RT2PCR Profiler array showed that H3K4 methyltransferase MLL3 was significantly downregulated in the CBDL livers. MLL3 has recently been implicated in regulation of LXR- and FXR-mediated gene expression(13, 14). MLL3 or its paralog MLL4 belongs to a steady-state complex called ASCOM (for ASC-2 complex) containing NCOA6 (ASC-2) that serves as a coactivator of multiple nuclear receptors and transcription factors. UTX, a H3K27 demethylase, is also part of this complex. NCOA6 binds to many nuclear receptors in a ligand-dependent manner through its two LXXLL motifs or NR boxes. The first LXXLL motif of NCOA6, located near the centrally located activation domain AD2, interacts with many nuclear receptors and in a recent study (13) showed relatively weak but specific interaction with the nuclear receptor FXR. LXXLL-2 located towards the C-terminal region had been thought to be restricted in its specificity to LXR.
The down-regulation of MLL3 after 1 and 3 days of CBDL was confirmed by RT-PCR. Its paralog MLL4, which potentially could provide redundant or compensatory activity, was also decreased. NCOA6 was not represented on the RT2PCR Profiler array, but its expression at the mRNA level was not changed on analysis by qPCR. Thus a critically important function of the ASCOM complex in mediating histone H3K4 trimethylation, a mark for transcriptionally active chromatin, was disrupted by CBDL.
The importance of both NCOA6 and MLL3/4 for expression of bile acid transporters was examined in HepG2 cells. Knockdown of MLL3, MLL4, or NCOA6 expression using either siRNA or siRNA adenoviruses led to a significant decrease in mRNA and protein levels for these components of the ASCOM complex and to a marked decrease in BSEP and NTCP mRNA levels. Knockdown of either MLL3 or MLL4 led to a similar decrease in BSEP and NTCP mRNA levels, and siRNA knockdown of both methyltransferases (MLL3/4) was additive. Conversely, cotransfection of varying amounts NCOA6 into HepG2 cells along with the BSEP promoter linked to luciferase, FXR, RXR plasmids, and ligand stimulation led to a dose- dependent increase in the luciferase activity. The effects of NCOA6 and MLL3 knockdown on NTCP expression are of interest, as NCOA6 has been shown recently to interact with HNF4α (27), which we have shown previously to be critical for activation of the mouse Ntcp gene (10).
While our article was in preparation, another study (13) showed that the first LXXLL motif of NCOA6 interacted weakly, but specifically, with FXR in a GST pull-down and yeast two-hybrid assays. In contrast, we found both the LXXLL-1 and LXXLL-2 motifs bind to FXR in the GST pull-down assay, but binding to LXXLL-2 occurred more avidly and was enhanced in the presence of CDCA. These results were confirmed using the mammalian two-hybrid analysis. The reasons for this discrepancy with the work of Kim et al. (13) are not entirely clear, although it might be related to the fact that we used a mammalian two-hybrid system while Kim et al. used a yeast system. Our data thus suggest an important role for both LXXLL-1 and 2 of NCOA6 in tethering the ASCOM complex to FXR.
Next, we examined whether NCOA6 and associated methyltransferases MLL3 and MLL4 were recruited to the promoters of the FXR-target genes BSEP/Bsep, Mrp2, and OSTα and NTCP, a target gene for GR (6), using ChIP assays. In HepG2 cells NCOA6, MLL3 were recruited to FXR response elements (FXRE) of BSEP and OSTα (see Supplemental Material) in the presence of CDCA and to the GCRE in NTCP promoter confirming previous reports for other GR targets (15). H3K4 trimethylation of the histoneH3 surrounding BSEP-FXRE and NTCP-GCRE was also detected (Fig. 5). We then examined the recruitment of these ASCOM complex components to the mouse liver FXRE in Bsep and Mrp2 promoters. While we observed significant enrichment of the proteins in 3-day sham livers, no such enrichment was seen in the CBDL livers, suggesting that the downregulation of these genes observed during cholestasis in some studies in rodents and humans (7, 8, 24, 26) can be partly due to the decreased recruitment resulting in altered H3K4 methylation at these loci. Our data are consistent with preliminary studies in Mll3-null mice that exhibit (due to redundancy between Mll3 and Mll4) symptoms of mild cholestasis (Dr. Jae Woon Lee, Baylor College of Medicine, Houston, personal communication). Taking these data together, we have schematically illustrated in Fig. 6 our current working model of how ASCOM complex associates with its partners MLL3/MLL4 at the FXRE/GCRE site and activates transcription on ligand binding and the changes that occur during cholestasis.
A limitation of our work is that current approaches to the study of coactivators and epigenetic modification of histones fail to recognize the dynamic nature of changes that occur rapidly as protein complexes are recruited and others are released on ligand binding. These processes could be disrupted in disease even if the expression of these proteins is not altered and their presence on the promoter of a gene of interest is not altered in ChIP assays.
GRANTS
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-084434 (to F. J. Suchy and M. Ananthanarayanan) and by the Intramural Research Program of NIH, National Institute of Environmental Health Sciences under NIH Intramural Project Number Z01ES02124 (to S. Surapureddi and J. A. Goldstein).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the assistance of Dr. Jongsook Kim Kemper for mouse liver ChIP assays and Dr. Jae Woon Lee for MLL2 primers.
REFERENCES
- 1. Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem 276: 28857–28865, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol 69: 201–220, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Belakavadi M, Fondell JD. Role of the mediator complex in nuclear hormone receptor signaling. Rev Physiol Biochem Pharmacol 156: 23–43, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Bemelmans MH, Gouma DJ, Greve JW, Buurman WA. Cytokines tumor necrosis factor and interleukin-6 in experimental biliary obstruction in mice. Hepatology 15: 1132–1136, 1992 [DOI] [PubMed] [Google Scholar]
- 5. Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev 31: 139–170, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eloranta JJ, Jung D, Kullak-Ublick GA. The human Na+-taurocholate cotransporting polypeptide gene is activated by glucocorticoid receptor and peroxisome proliferator-activated receptor-gamma coactivator-1alpha, and suppressed by bile acids via a small heterodimer partner-dependent mechanism. Mol Endocrinol 20: 65–79, 2006 [DOI] [PubMed] [Google Scholar]
- 7. Fickert P, Zenz R, Zollner G, Pojer C, Fuchsbichler A, Zatloukal K, Denk H, Trauner M. Regulation of hepatocellular transporter expression depends on bile acid hydrophobicity in mice (Abstract). Gastroenterology 118: A934, 2000 [Google Scholar]
- 8. Gartung C, Ananthanarayanan M, Rahman MA, Schuele S, Nundy S, Soroka CJ, Stolz A, Suchy FJ, Boyer JL. Down-regulation of expression and function of the rat liver Na+/bile acid cotransporter in extrahepatic cholestasis. Gastroenterology 110: 199–209, 1996 [DOI] [PubMed] [Google Scholar]
- 9. Geier A, Fickert P, Trauner M. Mechanisms of disease: mechanisms and clinical implications of cholestasis in sepsis. Nat Clin Pract Gastroenterol Hepatol 3: 574–585, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Geier A, Martin IV, Dietrich CG, Balasubramaniyan N, Strauch S, Suchy FJ, Gartung C, Trautwein C, Ananthanarayanan M. Hepatocyte nuclear factor-4α is a central transactivator of the mouse Ntcp gene. Am J Physiol Gastrointest Liver Physiol 295: G226–G233, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geier A, Wagner M, Dietrich CG, Trauner M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta 1773: 283–308, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Kemper JK, Kim H, Miao J, Bhalla S, Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol 24: 7707–7719, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim DH, Lee J, Lee B, Lee JW. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Mol Endocrinol 23: 1556–1562, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol 22: 1312–1319, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee SK, Anzick SL, Choi JE, Bubendorf L, Guan XY, Jung YK, Kallioniemi OP, Kononen J, Trent JM, Azorsa D, Jhun BH, Cheong JH, Lee YC, Meltzer PS, Lee JW. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem 274: 34283–34293, 1999 [DOI] [PubMed] [Google Scholar]
- 16. Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci USA 102: 3611–3616, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lonard DM, Lanz RB, O'Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev 28: 575–587, 2007 [DOI] [PubMed] [Google Scholar]
- 18. Lonard DM, O'Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci 30: 126–132, 2005 [DOI] [PubMed] [Google Scholar]
- 19. McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20: 321–344, 1999 [DOI] [PubMed] [Google Scholar]
- 20. McKenna NJ, O'Malley BW. From ligand to response: generating diversity in nuclear receptor coregulator function. J Steroid Biochem Mol Biol 74: 351–356, 2000 [DOI] [PubMed] [Google Scholar]
- 21. McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O'Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol 69: 3–12, 1999 [DOI] [PubMed] [Google Scholar]
- 22. Mitro N, Gilardi F, Godio C, Scotti E, De Fabiani E, Caruso D, Crestani M. Bile acids and gene regulation: from nuclear receptors to chromatin. Front Biosci 13: 6276–6288, 2008 [DOI] [PubMed] [Google Scholar]
- 23. O'Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Res 69: 8217–8222, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park YJ, Qatanani M, Chua SS, LaRey JL, Johnson SA, Watanabe M, Moore DD, Lee YK. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology 47: 1578–1586, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Rochette-Egly C. Dynamic combinatorial networks in nuclear receptor-mediated transcription. J Biol Chem 280: 32565–32568, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Shoda J, Kano M, Oda K, Kamiya J, Nimura Y, Suzuki H, Sugiyama Y, Miyazaki H, Todoroki T, Stengelin S, Kramer W, Matsuzaki Y, Tanaka N. The expression levels of plasma membrane transporters in the cholestatic liver of patients undergoing biliary drainage and their association with the impairment of biliary secretory function. Am J Gastroenterol 96: 3368–3378, 2001 [DOI] [PubMed] [Google Scholar]
- 27. Surapureddi S, Rana R, Reddy JK, Goldstein JA. Nuclear receptor coactivator 6 mediates the synergistic activation of human cytochrome P-450 2C9 by the constitutive androstane receptor and hepatic nuclear factor-4alpha. Mol Pharmacol 74: 913–923, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teichert A, Arnold LA, Otieno S, Oda Y, Augustinaite I, Geistlinger TR, Kriwacki RW, Guy RK, Bikle DD. Quantification of the vitamin D receptor-coregulator interaction. Biochemistry 48: 1454–1461, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trauner M, Meier PJ, Boyer JL. Molecular pathogenesis of cholestasis. N Engl J Med 339: 1217–1227, 1998 [DOI] [PubMed] [Google Scholar]
- 30. Trauner M, Meier PJ, Boyer JL. Molecular regulation of hepatocellular transport systems in cholestasis. J Hepatol 31: 165–178, 1999 [DOI] [PubMed] [Google Scholar]
- 31. Trauner M, Wagner M, Fickert P, Zollner G. Molecular regulation of hepatobiliary transport systems: clinical implications for understanding and treating cholestasis. J Clin Gastroenterol 39: S111–S124, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Valadares NF, Polikarpov I, Garratt RC. Ligand induced interaction of thyroid hormone receptor beta with its coregulators. J Steroid Biochem Mol Biol 112: 205–212, 2008 [DOI] [PubMed] [Google Scholar]
- 33. Walters JR. Bile acids are physiological ligands for a nuclear receptor. Gut 46: 308–309, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu W. Nuclear receptor coactivators: the key to unlock chromatin. Biochem Cell Biol 83: 418–428, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.